Abstract
Our systematic review investigated the additional effect of artificial intelligence-based devices on human observers when diagnosing and/or detecting thoracic pathologies using different diagnostic imaging modalities, such as chest X-ray and CT. Peer-reviewed, original research articles from EMBASE, PubMed, Cochrane library, SCOPUS, and Web of Science were retrieved. Included articles were published within the last 20 years and used a device based on artificial intelligence (AI) technology to detect or diagnose pulmonary findings. The AI-based device had to be used in an observer test where the performance of human observers with and without addition of the device was measured as sensitivity, specificity, accuracy, AUC, or time spent on image reading. A total of 38 studies were included for final assessment. The quality assessment tool for diagnostic accuracy studies (QUADAS-2) was used for bias assessment. The average sensitivity increased from 67.8% to 74.6%; specificity from 82.2% to 85.4%; accuracy from 75.4% to 81.7%; and Area Under the ROC Curve (AUC) from 0.75 to 0.80. Generally, a faster reading time was reported when radiologists were aided by AI-based devices. Our systematic review showed that performance generally improved for the physicians when assisted by AI-based devices compared to unaided interpretation.
Keywords: artificial intelligence, deep learning, computer-based devices, radiology, thoracic diagnostic imaging, chest X-ray, CT, observer tests, performance
1. Introduction
Artificial intelligence (AI)-based devices have made significant progress in diagnostic imaging segmentation, detection, and disease differentiation, as well as prioritization. AI has emerged as the cutting-edge technology to bring diagnostic imaging into the future [1]. AI may be used as a decision support system, where radiologists reject or accept the algorithm’s diagnostic suggestions, which was investigated in this review, but there is no AI-based device that fully autonomously diagnose or classify findings in radiology yet. Some products have been developed for the purpose of radiological triage [2]. Triage and notification of a certain finding have been a task that has had some autonomy since there is no clinician assigned to re-prioritize the algorithm’s suggestions. Other uses of AI algorithms could be suggestion of treatment options based on disease specific predictive factors [3] and automatic monitoring and overall survival prognostication to aid the physician in deciding the patient’s future treatment plan [4].
The broad application of plain radiography in thoracic imaging and the use of other modalities, such as computed tomography (CT), to delineate abnormalities adds to the number of imaging cases that can provide information to successfully train an AI-algorithm [5]. In addition to providing large quantities of data, chest X-ray is one of the most used imaging modalities. Thoracic imaging has, therefore, not only a potential to provide a large amount of data for developing AI-algorithms successfully, but there is also potential for AI-based devices to be useful in a great number of cases. Because of this, several algorithms in thoracic imaging have been developed—most recently in the diagnosis of COVID-19 [6].
AI has attracted increasing attention in diagnostic imaging research. Most studies demonstrate their AI-algorithm’s diagnostic superiority by separately comparing the algorithm’s diagnostic accuracy to the accuracy achieved by manual reading [7,8]. Nevertheless, several factors seem to prevent AI-based devices from diagnosing pathologies in radiology without human involvement [9], and only few studies conduct observer tests where the algorithm is being used as a second or concurrent reader to radiologists: a scenario closer to a clinical setting [10,11]. Even though diagnostic accuracy of an AI-based device can be evaluated by testing it independently, this may not reflect the true clinical effect of adding AI-based devices, since such testing eliminates the factor of human-machine interaction and final human decision making.
Our systematic review investigated the additional effect AI-based devices had on physicians’ abilities when diagnosing and/or detecting thoracic pathologies using different diagnostic imaging modalities, such as chest X-ray and CT.
2. Materials and Methods
2.1. Literature Search Strategy
The literature search was completed on 24 March 2021, from 5 databases: EMBASE, PubMed, Cochrane library, SCOPUS, and Web of Science. The search was restricted to peer-reviewed publications of original research written in English from 2001–2021, both years included.
The following specific MESH terms were used in PubMed: “thorax”, “radiography, thoracic”, “lung”, “artificial intelligence”, “deep Learning”, “machine Learning”, “neural networks, computer”, “physicians”, “radiologists”, “workflow”, “physicians”. MESH terms were combined with the following all-fields specific search words and their bended forms: “thorax”, “chest”, “lung”, “AI”, “artificial intelligence”, “deep learning”, “machine learning”, “neural networks”, “computer”, “computer neural networks”, “clinician”, “physician”, “radiologist”, “workflow”.
To perform the EMBASE search, the following combination of text word search and EMTREE terms were used: (“thorax” (EMTREE term) OR “lung” (EMTREE term) OR “chest” OR “lung” OR “thorax”) AND (“artificial intelligence (EMTREE term) OR “machine learning” (EMTREE term) OR “deep learning” (EMTREE term) OR “convolutional neural network” (EMTREE term) OR “artificial neural network” (EMTREE term) OR “ai” OR “artificial intelligence” OR “neural network” OR “deep learning” OR “machine learning”) AND (“radiologist (EMTREE term) OR “ physician” (EMTREE term) OR “clinician” (EMTREE term) OR “workflow” (EMTREE term) OR “radiologist” OR “clinician” OR “physician” OR “workflow”).
We followed the PRISMA guidelines for literature search and study selection. After removal of duplicates, all titles and abstracts retrieved from the search were independently screened by two authors (D.L. and L.M.P.). In case of unresolved disagreements, that could not be determined by consensus vote between D.L. and L.M.P., a third author (J.F.C.) was appointed to assess and resolve the disagreement. Data were extracted by D.L. and L.M.P. using pre-piloted forms. To describe the performance of the radiologists without and with assistance of AI-based devices, we used a combination of narrative synthesis and compared measures of accuracy, area under the ROC curve (AUC), sensitivity, specificity, and time measurements.
For evaluating the risk of bias and assess quality of research, we used the QUADAS-2 tool [12].
2.2. Study Inclusion Criteria
Peer-reviewed original research articles published in English, between 2001 and 2021, were reviewed for inclusion. Inclusion criteria were set at follows:
-
AI-based devices, either independent or incorporated into a workflow, used for imaging diagnosis and/or detection of findings in lung tissue, regardless of thoracic imaging modality;
and
-
an observer test where radiologists or other types of physicians used the AI-algorithm as either a concurrent or a second reader;
and
-
within the observer test, the specific observer that diagnosed/detected the findings without AI-assistance must also participate as the observer with AI-assistance;
and
outcome measurements of observer tests included either sensitivity, specificity, AUC, accuracy, or some form of time measurement recording observers’ reading time without and with AI-assistance.
Studies where one set of physicians, with the aid of AI, retrospectively re-evaluate another set of physicians’ diagnoses without AI were excluded. AI-based devices that did not detect specific pulmonary tissue findings/pathology, e.g., rib fracture, aneurisms, thyroid enlargements etc. were also excluded.
3. Results
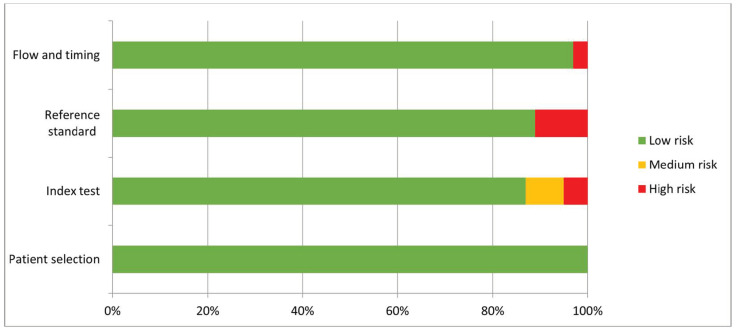
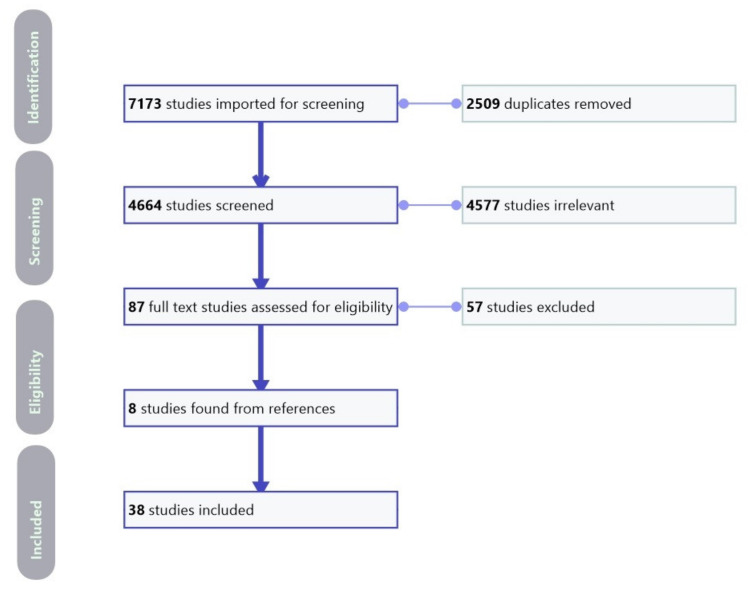
We included a total of 38 studies [13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50] in our systematic review. The QUADAS-2 tool is presented in Figure 1, and a PRISMA flowchart of the literature search is presented in Figure 2.
Figure 1.
The QUADAS-2 tool for evaluating risk of bias and assess quality of research.
Figure 2.
Preferred reporting items for systematic reviews and meta-analyses (PRISMA) flowchart of the literature search and study selection.
We divided the studies into two groups: The first group, consisting of 19 studies [13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31], used an AI-based device as a concurrent reader in an observer test, where the observers were tasked with diagnosing images with assistance from an AI-based device, while not being allowed (blinded) to see their initial diagnosis made without assistance from AI (Table 1a). The second group, consisting of 20 studies [19,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50] used the AI-based device as a second reader in an un-blinded sequential observer test, thus allowing observers to see and change their original un-assisted diagnosis (Table 1b).
Table 1.
(a) Included studies with artificial intelligence-based devices as concurrent readers in the observer test. (b) Included studies with artificial intelligence-based devices in an observer test with a sequential test design.
| Author | Year | Standard of Reference | Type of Artificial Intelligence-Based CAD | Pathology | No. of Cases | Test Observers | Image Modality |
| a | |||||||
| Bai et al. [13] | 2021 | RT-PCR | EfficientNet-B3 Convolutional Neural Network | COVID-19 pneumonia | 119 | 6 radiologists (10–20 years of chest CT experience) | CT |
| Beyer et al. [19] | 2007 | Radiologist identified and consensus vote | Commercially available (LungCAD prototype version, Siemens Corporate Research, Malvern, PA, USA) |
Pulmonary nodules | 50 | 4 radiologists (2–11 years experience) | CT |
| de Hoop et al. [20] | 2010 | Histologically confirmed | Commercially available (OnGuard 5.0; Riverain Medical, Miamisburg, OH, USA) | Pulmonary nodules | 111 | 1 general radiologist, 1 chest radiologist, and 4 residents | Chest X-ray |
| Dorr et al. [14] | 2020 | RT-PCR | DenseNet 121 architecture | COVID-19 pneumonia | 60 | 23 radiologists and 31 emergency care physicians | Chest X-ray |
| Kim et al. [15] | 2020 | Bacterial culture and RT-PCR for viruses | Commercially available (Lunit INSIGHT for chest radiography, version 4.7.2; Lunit, Seoul, South Korea) | Pneumonia | 387 | 3 emergency department physicians (6–7 years experience) | Chest X-ray |
| Koo et al. [21] | 2020 | Pathologically confirmed | Commercially available (Lunit Insight CXR, ver. 1.00; Lunit, Seoul, South Korea) | Pulmonary nodules | 434 | 2 thoracic radiologists and 2 residents | Chest X-ray |
| Kozuka et al. [22] | 2020 | Radiologist identified and majority vote | Faster Region-Convolutional Neural Network | Pulmonary nodules | 120 | 2 radiologists (1–4 years experience) | CT |
| Lee et al. [23] | 2012 | Pathologically confirmed | Commercially available (IQQA-Chest, EDDA Technology, Princeton Junction, NJ, USA) | Pulmonary nodules malignant/benign | 200 | 5 chest radiologists and 5 residents | Chest X-ray |
| Li et al. [24] | 2011 | CT | Commercially available (SoftView, version 2.0; Riverrain Medical, Miamisburg, OH, USA-Image normalization, feature extraction and regression networks) | Pulmonary nodules | 151 | 3 radiologists (10–25 years experience) | Chest X-ray |
| Li et al. [25] | 2011 | Pathologically confirmed and radiology assessed | Commercially available (SoftView, version 2.0; Riverain Medical) | Pulmonary nodules | 80 | 2 chest radiologists, 4 general radiologists, and 4 residents | Chest X-ray |
| Liu et al. [16] | 2020 | - | Segmentation model with class attention map including a residual convolutional block | COVID-19 pneumonia | 643 | - | Chest X-ray |
| Liu et al. [26] | 2019 | Radiologist identified and majority vote | DenseNet and Faster Region-Convolutional Neural Network | Pulmonary nodule | 271 | 2 radiologists (10 years experience) | CT |
| Martini et al. [27] | 2021 | Radiologist consensus | Commercially available (ClearRead-CT, Riverrain Technologies, Miamisburg, OH, USA) | Pulmonary consolidations/nodules | 100 | 2 senior radiologists, 2 final-year residents, and 2 inexperienced residents | MDCT |
| Nam et al. [29] | 2021 | RT-PCR and CT | Deep learning-based algorithm (Deep convolutional neural network) | Pneumonia, pulmonary edema, active tuberculosis, interstitial lung disease, nodule/mass, pleural effusion, acute aortic syndrome, pneumoperitoneum, rib fracture, pneumothorax, mediastinal mass. | 202 | 2 thoracic radiologists, 2 board-certified radiologists, and 2 residents | Chest X-ray |
| Rajpurkar et al. [31] | 2020 | Positive culture or Xpert MTB/RIF test | Convolutional Neural Network | Tuberculosis | 114 | 13 physicians (6 months–25 years of experience) | Chest X-ray |
| Singh et al. [28] | 2021 | Radiologically reviewed | Commercially available (ClearRead CT Vessel Suppression and Detect, Riverain Technologies TM) | Subsolid nodules (Incl ground-glass and/or part-solid) | 123 | 2 radiologists (5–10 years experience) | CT |
| Sung et al. [30] | 2021 | CT and clinical information | Commercially available (Med-Chest X-ray system (version 1.0.0, VUNO, Seoul, South Korea) | Nodules, consolidation, interstitial opacity, pleural effusion, pneumothorax | 128 | 2 thoracic radiologists, 2 board-certified radiologists, 1 radiology resident, and 1 non-radiology resident | Chest X-ray |
| Yang et al. [17] | 2021 | RT-PCR | Deep Neural Network | COVID-19 pneumonia | 60 | 3 radiologists (5–20 years experience) | CT |
| Zhang et al. [18] | 2021 | RT-PCR | Deep Neural Network using the blur processing method to improve the image enhancement algorithm | COVID-19 pneumonia | 15 | 2 physicians (13–15 years experience) | CT |
| Author | Year | Standard of Reference | Type of Artificial Intelligence-Based CAD | Pathology | No. of Cases | Test Observers | Image Modality |
| b | |||||||
| Abe et al. [47] | 2004 | Radiological review and clinical correlation | Single three-layer, feed-forward Artificial Neural Network with a back-propagation algorithm | Sarcoidosis, miliary tuberculosis, lymphangitic carcinomatosis, interstitial pulmonary edema, silicosis, scleroderma, P. Carinii pneumonia, Langerhals cell histiocytosis, idiopathic pulmonary fibrosis, viral pneumonia, pulmonary drug toxicity | 30 | 5 radiologists (6–18 years experience) | Chest X-ray |
| Abe et al. [48] | 2003 | Radiology consensus | Fourier transformation and Artificial Neural Network | Detection of interstitial lung disease | 20 | 8 chest radiologists, 13 other radiologists, and 7 residents | Chest X-ray |
| Clinical correlation and bacteriological | Artificial Neural Network | Differential diagnosis of 11 types of interstitial lung disease | 28 | 16 chest radiologists, 25 other radiologists, and 12 residents | Chest X-ray | ||
| Pathology | Artificial Neural Network | Distinction between malignant and benign pulmonary nodules | 40 | 7 chest radiologists, 14 other radiologists, and 7 residents | Chest X-ray | ||
| Awai et al. [33] | 2004 | Radiological review | Artificial Neural Network | Pulmonary nodules | 50 | 5 board-certified radiologists and 5 residents | CT |
| Awai et al. [32] | 2006 | Histology | Neural Network | Pulmonary nodules malignant/benign | 33 | 10 board-certified radiologists and 9 radiology residents | CT |
| Beyer et al. [19] | 2007 | Radiologist identified and consensus vote | Commercially available (LungCAD prototype version, Siemens Corporate Research, Malvern, PA, USA) | Pulmonary nodules | 50 | 4 radiologists (2–11 years experience) | CT |
| Bogoni et al. [34] | 2012 | Majority of agreement | Commercially available (Lung CAD VC20A, Siemens Healthcare, Malvern, PA, USA) | Pulmonary nodules | 43 | 5 fellowship-trained chest radiologists (1–10 years experience) | CT |
| Chae et al. [35] | 2020 | Pathologically confirmed and radiologically reviewed | CT-lungNET (Deep Convolutional Neural Network) | Pulmonary nodules | 60 | 2 medical students, 2 residents, 2 non-radiology physicians, and 2 thoracic radiologists | CT |
| Chen et al. [36] | 2007 | Surgery or biopsy | Deep Neural Network | Pulmonary nodules malignant/benign | 60 | 3 junior radiologists, 3 secondary radiologists, and 3 senior radiologists | CT |
| Fukushima et al. [49] | 2004 | Pathological, bacteriological and clinical correlation | Single three-layer, feed-forward Artificial Neural Network with a back-propagation algorithm | Sarcoidose, diffuse panbronchioloitis, nonspecific interstitial pneumonia, lymphangitic carcinomatosis, usual interstitial pneumonia, silicosis, BOOP or chronic eopsinophilic pneumonia, pulmonary alveolar proteinosis, miliary tuberculosis, lymphangiomyomatosis, P, carinii pneumonia or cytomegalovirus pneumonia | 130 | 4 chest radiologists and 4 general radiologists | High Resolution CT |
| Hwang et al. [50] | 2019 | Pathology, clinical or radiological | Deep Convolutional Neural Network with dense blocks | 4 different target diseases (pulmonary malignant neoplasms, tuberculosis, pneumonia, pneumothorax) classified in to binary classification of normal/abnormal | 200 | 5 thoracic radiologists, board-certified radiologists, and 5 non-radiology physicians | Chest X-ray |
| Kakeda et al. [41] | 2004 | CT | Commercially available (Trueda, Mitsubishi Space Software, Tokyo, Japan) | Pulmonary nodules | 90 | 4 board-certified radiologists and 4 residents | Chest X-ray |
| Kasai et al. [40] | 2008 | CT | Three Artificial Neural Networks | Pulmonary nodules | 41 | 6 chest radiologists and 12 general radiologists | Lateral chest X-ray only |
| Kligerman et al. [42] | 2013 | Histology and CT | Commercially available (OnGuard 5.1; Riverain Medical, Miamisburg, OH, USA) | Lung cancer | 81 | 11 board-certified general radiologists (1–24 years experience) | Chest X-ray |
| Liu et al. [37] | 2021 | Histology, CT, and biopsy/surgical removal | Convolutional Neural Networks | Pulmonary nodules malignant/benign | 879 | 2 senior chest radiologists, 2 secondary chest radiologists, and 2 junior radiologists | CT |
| Matsuki et al. [38] | 2001 | Pathology and radiology | Three-layer, feed-forward Artificial Neural Network with a back-propagation algorithm | Pulmonary nodules | 50 | 4 attending radiologists, 4 radiology fellows, 4 residents | High Resolution CT |
| Nam et al. [43] | 2019 | Pathologically confirmed and radiologically reviewed | Deep Convolutional Neural Networks with 25 layers and 8 residual connections | Pulmonary nodules malignant/benign | 181 | 4 thoracic radiologists, 5 board-certified radiologists, 6 residents, and 3 non-radiology physicians | Chest X-ray |
| Oda et al. [44] | 2009 | Histology, cytology, and CT | Massive training Artificial Neural Network | Pulmonary nodules | 60 | 7 board-certified radiologists and 5 residents | Chest X-ray |
| Rao et al. [39] | 2007 | Consensus and majority vote | LungCAD | Pulmonary nodules | 196 | 17 board-certified radiologists | MDCT |
| Schalekamp et al. [45] | 2014 | Radiologically reviewed, pathology and clinical correlation | Commercially available (ClearRead +Detect 5.2; Riverain Technologies and ClearRead Bone Suppression 2.4; Riverain Technologies) | Pulmonary nodules | 300 | 5 radiologists and 3 residents | Chest X-ray |
| Sim et al. [46] | 2020 | Biopsy, surgery, CT, and pathology | Commercially available (ALND, version 1.00; Samsung Electronics, Suwon, South Korea) | Cancer nodules | 200 | 5 senior chest radiologists, 4 chest radiologists, and 3 residents | Chest X-ray |
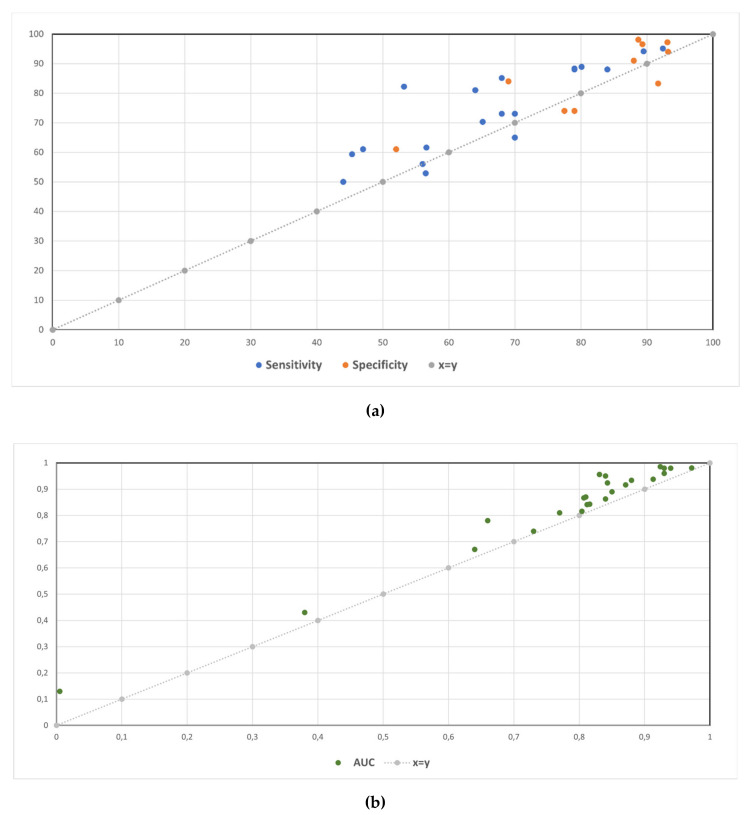
Visual summaries of the performance change in sensitivity, specificity, and AUC for all studies are shown in Figure 3a,b.
Figure 3.
Sensitivity and specificity (a) and AUC (b) without and with the aid of an AI-based device.
3.1. Studies Where Human Observers Used AI-Based Devices as Concurrent Readers
In 19 studies observers were first tasked to diagnose the image without an AI-based device. After a washout period, the same observers were then tasked to diagnose the images again. They were not allowed to see and change their original un-aided radiological diagnosis before making their diagnosis aided by and AI-based device (Table 1a). The results of the observer tests are listed in Table 2a–c for concurrent reader studies.
Table 2.
Sensitivity and specificity (a); accuracy and AUC (b); and time measurement results (c) for observer tests without and with AI-based devices as a concurrent reader.
| Author | Without AI-Based CAD | With AI-Based CAD | Change | Statistical Significance between Difference | ||
| Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | |||
| a | ||||||
| Bai et al. [13] | 79 | 88 | 88 | 91 | ↑ | p < 0.001 |
| Beyer et al. [19] | 56.5 | - | 61.6 | - | ↑ | p < 0.001 |
| de Hoop et al. [20] | 56 * | - | 56 * | - | ↑ | - |
| Dorr et al. [14] | 47 | 79 | 61 | 75 | ↑ | p < 0.007 |
| Kim et al. [15] | 73.9 | 88.7 | 82.2 | 98.1 | ↑ | p < 0.014 |
| Koo et al. [21] | 92.4 | 93.1 | 95.1 | 97.2 | ↑ | - |
| Kozuka et al. [22] | 68 | 91.7 | 85.1 | 83.3 | ↑ | p < 0.01 ** |
| Lee et al. [23] | 84 | - | 88 | - | ↑ | - |
| Rajpurkar et al. [31] | 70 | 52 | 73 | 61 | ↑ | - |
| Singh et al. [28] | 68 * | 77.5 * | 73 * | 74 * | ↑ | - |
| Sung et al. [30] | 80.1 | 89.3 | 88.9 | 96.6 | ↑ | p < 0.01 |
| Yang et al. [17] | 89.5 | - | 94.2 | - | ↑ | p < 0.05 |
| Author | Without AI-Based CAD | With AI-Based CAD | Change | Statistical Significance between Difference | ||
| Accuracy (%) | AUC | Accuracy (%) | AUC | |||
| b | ||||||
| Bai et al. [13] | 85 | - | 90 | - | ↑ | p < 0.001 |
| Kim et al. [15] | - | 0.871 | - | 0.916 | ↑ | p = 0.002 |
| Koo et al. [21] | - | 0.93 | - | 0.96 | ↑ | p < 0.0001 |
| Li et al. [24] | - | 0.840 | - | 0.863 | ↑ | p = 0.01 |
| Li et al. [25] | - | 0.807 | - | 0.867 | ↑ | p < 0.001 |
| Liu et al. [26] | - | 0.66 * | - | 0.78 * | ↑ | - |
| Nam et al. [29] | 66.3 * | - | 82.4 * | - | ↑ | p < 0.05 |
| Rajpurkar et al. [31] | 60 | - | 65 | - | ↑ | p = 0.002 |
| Singh et al. [28] | - | 0.73 * | - | 0.74 * | ↑ | Not statistically significant |
| Sung et al. [30] | - | 0.93 | - | 0.98 | ↑ | p = 0.003 |
| Yang et al. [17] | 94.1 | - | 95.1 | - | ↑ | p = 0.01 |
| Author | Without AI-Based CAD | With AI-Based CAD | Change | Statistical Significance between Difference | ||
| Time | Time | |||||
| c | ||||||
| Beyer et al. [19] | 294 s (1) | 337 s (1) | ↓ | p = 0.04 | ||
| Kim et al. [15] | 165 min (2) | 101 min (2) | ↑ | - | ||
| Kozuka et al. [22] | 373 min(2) | 331 min (2) | ↑ | - | ||
| Liu et al. [16] | 100.5 min (3) | 34 min (3) | ↑ | p < 0.01 | ||
| Liu et al. [26] | 15 min (1) | 5–10 min (1) | ↑ | - | ||
| Martini et al. [27] | 194 s (1) | 154 s (1) | ↑ | p < 0.001 | ||
| Nam et al. [29] | 2771.2 s * (1) | 1916 s * (1) | ↑ | p < 0.002 | ||
| Sung et al. [30] | 24 s (1) | 12 s (1) | ↑ | p < 0.001 | ||
| Zhang et al. [18] | 3.623 min (2) | 0.744 min (2) | ↑ | - | ||
a: * our calculated average; ** for sensitivity only; - not applicable; ↑ positive change. b: * our calculated average; - not applicable; ↑ positive change. c: (1) per image/case reading time; (2) total reading time for multiple cases; (3) station survey time; * our calculated average; - not applicable; ↑ positive change; ↓ negative change.
3.1.1. Detection of Pneumonia
Bai et al. [13], Dorr et al. [14], Kim et al. [15] Liu et al. [16], Yang et al. [17], and Zhang et al. [18] had AI-based algorithms to detect pneumonia findings of different kinds, e.g., Covid-19 pneumonia from either non-Covid-19 pneumonia or non-pneumonia. Bai et al. [13], Yang et al. [17], Dorr et al. [14], and Zhang et al. [18] investigated detection of Covid-19 pneumonia. Bai et al. [13], Dorr et al. [14], and Yang et al. [17] all had significant improvement in performance measured in sensitivity after being aided by their AI-based devices (Table 2a), and Zhang et al. [18] reported shorter reading time per image but there was not any mention of statistical significance (Table 2c). Liu et al. [16] incorporated an AI-algorithm into a novel emergency department workflow for Covid-19 evaluations: a clinical quarantine station, where some clinical quarantine stations were equipped with AI-assisted image interpretation, and some did not. They compared the overall median survey time at the clinical quarantine stations in each condition and reported statistically significant shortened time (153 min versus 35 min, p < 0.001) when AI-assistance was available. Median survey time specific to the image interpretation part of the clinical quarantine station was also significantly shortened (Table 2c), but they did not report if the shortened reading time were accompanied by the same level of diagnostic accuracy. While the previously mentioned studies specifically investigated Covid-19 pneumonia, Kim et al. [15] used AI-assistance to distinguish pneumonia from non-pneumonia and reported significant improvement in performance measured in sensitivity and specificity after AI-assistance (Table 2a).
Detection of Pulmonary Nodules
Beyer et al. [19], de Hoop et al. [20], Koo et al. [21], Kozuka et al. [22], Lee et al. [23], Li et al. [24], Li et al. [25], Liu et al. [26], Martini et al. [27], and Singh et al. [28] used AI-based devices to assist with detection of pulmonary nodules. Even though de Hoop et al. [20] found a slight increase in sensitivity in residents (49% to 51%) and change in radiologists (63% to 61%) for nodule detection, both changes were not statistically significant (Table 2a). In contrast, Koo et al. [21], Li et al. [24], and Li et al. [25] reported improvement of AUC for every individual participating radiologist when using AI-assistance, regardless of experience level (Table 2b). Lee et al. [23] reported improved sensitivity (84% to 88%) when using AI as assistance (Table 2a) but did not mention if the change in sensitivity was significant. However, their reported increase in mean figure of merit (FOM) was statistically significant. Beyer et al. [19] had performed both blinded and un-blinded observer tests; in the blinded, concurrent reader test, radiologists had significant improved sensitivity (56.6% to 61.6%, p < 0.001) (Table 2a) but also significantly increased time for reading when assisted by AI (increase of 43 s per image, p = 0.04) (Table 2c). Martini et al. [27] reported improved interrater agreement (17–34%) in addition to improved mean reading time (Table 2c), when assisted by AI. Results for the effects of AI assistance on radiologists by Kozuka et al. [22], Liu et al. [26], and Singh et al. [28] are also shown in Table 2a,b, but only Kozuka et al. [22] reported significant improvement (sensitivity from 68% to 85.1%, p < 0.01). In addition to change in accuracy, Liu et al. [26] reported a reduction of reading time per patient from 15 min to 5–10 min without mentioning statistical significance.
Detection of Several Different Findings and Tuberculosis
Nam et al. [29] tested an AI-based device in detecting 10 different abnormalities and measured the accuracy by dividing them into groups of urgent, critical, and normal findings. Radiologists significantly improved their detection of critical (accuracy from 29.2% to 70.8%, p = 0.006), urgent (accuracy from 78.2% to 82.7%, p = 0.04), and normal findings (accuracy from 91.4% to 93.8%, p = 0.03). Reading times per reading session were only significantly improved for critical (from 3371.0 s to 640.5 s, p < 0.001) and urgent findings (from 2127.1 to 1840.3, p < 0.001) but significantly prolonged for normal findings (from 2815.4 s to 3267.1 s, p < 0.001). Even though Sung et al. [30] showed overall improvement in detection (Table 2a–c), per-lesion sensitivity only improved in residents (79.7% to 86.7%, p = 0.006) and board-certified radiologists (83.0% to 91.2%, p < 0.001) but not in thoracic radiologists (86.4% to 89.4%, p = 0.31). Results from a study by Rajpurkar et al. [31] for the effects of AI-assistance on radiologists detecting tuberculosis show that there were significant improvement in both sensitivity, specificity, and accuracy when aided by AI (Table 2a,b).
3.2. Studies Where Human Observers Used AI-Based Devices as a Second Reader in a Sequential Observer Test Design
In 20 studies, observers were first tasked to diagnose the image without an AI-based device. Immediately afterwards, they were tasked to diagnose the images aided by an AI-based device and were also allowed to see and change their initial diagnosis (Table 1b). The results of the observer tests are listed in Table 3a–c for sequential observer test design studies.
Table 3.
Sensitivity and specificity (a); accuracy and AUC (b); and time measurement results (c) for sequential observer tests without and with AI-based devices as a second reader.
| Author | Without AI-Based CAD | With AI-Based CAD | Change | Statistical Significance between Difference | ||
| Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | |||
| a | ||||||
| Abe et al. [48] | 64 | - | 81 | - | ↑ | p < 0.001 |
| Beyer et al. [19] | 56.5 | - | 52.9 | - | ↓ | p < 0.001 |
| Bogoni et al. [34] | 45.34 * | - | 59.34 * | - | ↑ | p < 0.03 |
| Chae et al. [35] | 70 * | 69 * | 65 * | 84 * | ↓ | Not statistically significant |
| Hwang et al. [50] | 79 * | 93.2 * | 88.4 * | 94 * | ↑ | p = 0.006–0.99 |
| Kligerman et al. [42] | 44 | - | 50 | - | ↑ | p < 0.001 |
| Sim et al. [46] | 65.1 | - | 70.3 | - | ↑ | p < 0.001 |
| Author | Without AI-Based CAD | With AI-Based CAD | Change | Statistical Significance between Difference | ||
| Accuracy (%) | AUC | Accuracy (%) | AUC | |||
| b | ||||||
| Abe et al. [47] | - | 0.81 | - | 0.87 | ↑ | p = 0.031 |
| Abe et al. [48] | - | 0.94 | - | 0.98 | ↑ | p < 0.01 |
| Abe et al. [48] | - | 0.77 | - | 0.81 | ↑ | p < 0.001 |
| Awai et al. [33] | - | 0.64 | - | 0.67 | ↑ | p < 0.01 |
| Awai et al. [32] | - | 0.843 | - | 0.924 | ↑ | p = 0.021 |
| Chae et al. [35] | 69 * | 0.005 * | 75 * | 0.13 * | ↑ | Not statistically significant |
| Chen et al. [36] | - | 0.84 * | - | 0.95 * | ↑ | p < 0.221 |
| Fukushima et al. [49] | - | 0.972 * | - | 0.982 * | ↑ | p < 0.071 |
| Hwang et al. [50] | - | 0.880 * | - | 0.934 * | ↑ | p <0.002 |
| Kakeda et al. [41] | - | 0.924 | - | 0.986 | ↑ | p < 0.001 |
| Kasai et al. [40] | - | 0.804 | - | 0.816 | ↑ | Not statistically significant |
| Kligerman et al. [42] | - | 0.38 | - | 0.43 | ↑ | p = 0.007 |
| Liu et al. [37] | - | 0.913 | - | 0.938 | ↑ | p = 0.0266 |
| Matsuki et al. [38] | - | 0.831 | - | 0.956 | ↑ | p < 0.001 |
| Nam et al. [43] | - | 0.85 * | - | 0.89 * | ↑ | p < 0.001-0.87 |
| Oda et al. [44] | - | 0.816 | - | 0.843 | ↑ | p = 0.011–0.310 |
| Rao et al. [39] | 78 | - | 82.8 | - | ↑ | p < 0.001 |
| Schalekamp et al. [45] | - | 0.812 | - | 0.841 | ↑ | p = 0.0001 |
| Author | Without AI-Based CAD | With AI-Based CAD | Change | Statistical Significance between Difference | ||
| Time | Time | |||||
| c | ||||||
| Beyer et al. [19] | 294 s (1) | 274 s (1) | ↑ | p = 0.04 | ||
| Bogoni et al. [34] | 143 s (1) | 225 s (1) | ↓ | - | ||
a:* our calculated average; - not applicable; ↑ positive change; ↓ negative change. b: * our calculated average; - not applicable; ↑ positive change. c: (1) per image/case reading time; - not applicable; ↑ positive change; ↓ negative change.
3.2.1. Detection of Pulmonary Nodules Using CT
A total of 16 studies investigated the added value of AI on observers in the detection of pulmonary nodules; nine studies [19,32,33,34,35,36,37,38,39] used CT scans, and seven studies [40,41,42,43,44,45,46] used chest X-rays (Table 1b). Although Awai et al. [33], Liu et al. [37], and Matsuki et al. [38] showed statistically significant improvement across all radiologists (Table 3b) when using AI, other studies reported only significant increase in a sub-group of their test observers. Awai et al. [32] and Chen et al. [36] reported only significant improvement in the groups with the more junior radiologists; Awai et al. [32] reported an AUC from 0.768 to 0.901 (p = 0.009) in residents but no significant improvement in the board-certified radiologists (AUC 0.768 to 0.901, p = 0.19), and Chen et al. [36] reported an AUC from 0.76 to 0.96 (p = 0.0005) in the junior radiologists and 0.85 to 0.94 (p = 0.014) in the secondary radiologists but no significant improvement in the senior radiologists (AUC 0.91 to 0.96, p = 0.221). In concordance, Chae et al. [35] only reported significant improvement in the non-radiologists (AUC from 0.03 to 0.19, p < 0.05) but not for the radiologists (AUC from −0.02 to 0.07). While the results from Bogoni et al. [34] confirm the results from Beyer et al.’s [19] concurrent observer test, Beyer et al. [19] showed in the sequential observer test the opposite: decreased sensitivity (56.5 to 52.9, p < 0.001) with shortened reading time (294 s to 274 s per image, p = 0.04) (Table 3a,c). In addition to overall increase in accuracy (Table 3b), Rao et al. [39] also reported that using AI resulted in greater number of positive actionable management (averaged 24.8 patients), i.e., recommendations for additional images and/or biopsy, that were missed without AI.
3.2.2. Detection of Pulmonary Nodules Using Chest X-ray
As with detection of pulmonary nodules using CT, there were also contrasting results regarding radiologist experience level when using chest X-rays as the test set. Kakeda et al. [41] (AUC 0.924 to 0.986, p < 0.001), Kligerman et al. [42] (AUC 0.38 to 0.43, p = 0.007), Schalekamp et al. [45] (AUC 0.812 to 0.841, p = 0.0001), and Sim et al. [46] (sensitivity 65.1 to 70.3, p < 0.001) showed significant improvement across all experience levels when using AI (Table 3a,b). Nam et al. [43] showed significant increase in average among every radiologist experience level (AUC 0.85 to 0.89, p < 0.001–0.87), but, individually, there were more observers with significant increase among non-radiologists, residents, and board-certified radiologists than thoracic radiologists. Only one out of four thoracic radiologists had a significant increase. On the other hand, Oda et al. [44] only showed significant improvement for the board-certified radiologists (AUC 0.848 to 0.883, p = 0.011) but not for the residents (AUC 0.770 to 0.788, p = 0.310). Kasai et al. [40] did not show any statistically significant improvement(Table 3b), but they reported that sensitivity improved when there were only lateral images available (67.9% to 71.6%, p = 0.01).
3.2.3. Detection of Several Different Findings
Abe et al. [47], Abe et al. [48], Fukushima et al. [49], and Hwang et al. [50] explored the diagnostic accuracy in detection of several different findings besides pulmonary nodules with their AI-algorithm (Table 1b). While Abe et al. [47] found significant improvement in all radiologists (Table 3b), Fukushima et al. [49] only found significant improvement in the group of radiologists that had more radiological task experience (AUC 0.958 to 0.971, p < 0.001). In contrast, Abe et al. [48] found no significant improvement in the more senior radiologists for detection of interstitial disease (p > 0.089), and Hwang et al. [50] found no significant improvement in specificity for the detection of different major thoracic diseases in the more senior radiologists (p > 0.62). However, there were significant improvements in average among all observers for both studies (Table 3a,b).
4. Discussion
The main finding of our systematic review is that human observers assisted by AI-based devices had generally better detection or diagnostic performance using CT and chest X-ray, measured as sensitivity, specificity, accuracy, AUC, or time spent on image reading compared to human observers without AI-assistance.
Some studies suggest that physicians with less radiological task experience benefit more from AI-assistance [30,32,35,36,48,50], while others showed that physicians with greater radiological task experience benefitted the most from AI-assistance [44,49]. Gaube et al. [51] suggested that physicians with less experience were more likely to accept and deploy the suggested advice given to them by AI. They also reported that observers were generally not averse to following advice from AI compared to advice from humans. This suggests that the lack of improvement in the radiologists’ performance with AI-assistance, was not caused by lack of trust in the AI-algorithm but more by the presence of confidence in own abilities. Oda et al. [44] did not find that the group of physicians with less task experience improved from assistance by AI-based device and had two possible explanations. Firstly, the less experienced radiologists had a larger interrater variation of diagnostic performance, leading to insufficient statistical power to show statistical significance. This was also an argument used by Fukushima et al. [49]. Secondly, they argued that the use of AI-assistance lowers false-negative more than false-positive findings, and radiologists with less task experienced generally had more false-positive findings. However, Nam et al. [43] found that physicians with less task experience were more inclined to change their false-negative diagnosis’ and not their false-positive findings; therefore, they benefitted more from AI-assistance. Nam et al. [43], confirmed Oda et al.’s [44] finding in that there was a higher acceptance rate for false-negative findings. Brice [52] also confirmed this and suggested that correcting false-negative findings could have the most impact on reducing errors in radiological diagnosis. Although Oda et al. [44], Nam et al. [43], and Gaube et al. [51] had different reports on which level of physicians could improve their performance the most from the assistance of AI-based devices, they all confirm that AI-assistance lowers false-negative findings, which warrants advancing development and implementation of AI-based devices in to the clinics.
A limitation of our review is the heterogeneity of our included studies, e.g., the different methods for observer testing; some of our studies used a blinded observer test where AI-based devices was used as a concurrent reader (Table 1a), some studies used an un-blinded, sequential observer test (Table 1b), and some used both [19]. To the best of our knowledge, Kobayashi et al. [53] was one of the first to use and discuss both test types. Even though they concluded that there was no statistical significance in the difference of the results obtained from the two methods, they argue that an un-blinded, sequential test type would be less time consuming and practically easier to perform. Since then, others have adopted this method of testing [54] not only in thoracic diagnostic imaging and accepted it as a method for comparing effect of diagnostic tests [55]. Beyer et al. [19] also performed both methods of testing, but they did not come to the same conclusions about the results as Kobayashi et al. [53]. Their results of the two test methods were not the same; In the blinded concurrent reader test, they used more reading time per image (294 s to 337 s, p = 0.04) but achieved higher sensitivity (56.5 to 61.6, p < 0.001), and, in the un-blinded sequential reader test, they were quicker to interpret each image (294 s to 274 s, p = 0.04) but had worse sensitivity (56.5 to 52.9, p < 0.001) when assisted by AI. The test observers in the study by Kobayashi et al. [53] did not experience prolonged reading time, even though Bogoni et al. [34] confirmed the results by Beyer et al. [19] and also argued that correcting false-positives would prolong the time spent on an image. Roos et al. [56] also reported prolonged time spent on rejecting false positive cases when testing their computer-aided device and explained that false-positive cases may be harder to distinguish from true-positive cases. This suggests that the sequential observer test design could result in prolonged time spent on reading an image when assisted by a device since they are forced to decide on previous findings. Future observer test studies must, therefore, be aware of this bias, and more studies are needed to investigate this aspect of observer tests.
A pre-requisite for AI-based devices to have a warranted place in diagnostic imaging is that it has higher accuracy than the intended user, since human observers with less experience may have a higher risk of also being influenced by inaccurate advice due to availability bias [57] and premature closure [58]. To be able to include a larger number of studies, we allowed the possibility of some inter-study variability in the performance of the AI-based devices because of different AI-algorithms being used. We recognize this as a limitation adding to the heterogeneity of our systematic review. In addition, we did not review the diagnostic performance of the AI-algorithm by itself, and we did not review the training or test dataset that was used to construct the AI-algorithm. Because of the different AI-algorithms, the included studies may also have been subjected to publication bias since there may be a tendency to only publish well-performing AI-algorithms.
Improved performance in users is a must before implementation can be successful. Our systematic review focused on observer tests performed in highly controlled environments where they were able to adjust their study settings to eliminate biases and variables. However, few prospective clinical trials have been published where AI-based devices have been used, in a more dynamic and clinically realistic environment [59,60]. No clinical trials have been published using AI-based devices on thoracic CT or chest X-rays, whether it be as a stand-alone diagnostic tool or as an additional reader to humans [61]. Our systematic review has, therefore, been a step towards the integration of AI in the clinics by showing that it generally has a positive influence on physicians when used as an additional reader. Further studies are warranted not only on how AI-based devices influence human decision making but also on their performance and integration into a more dynamic, realistic clinical setting.
5. Conclusions
Our systematic review showed that sensitivity, specificity, accuracy, AUC, and/or time spent on reading diagnostic images generally improved when using AI-based devices compared to not using them. Disagreements still exist, and more studies are needed to uncover factors that may inhibit an added value by AI-based devices on human decision-making.
Author Contributions
Conceptualization, D.L., J.F.C., S.D., H.D.Z., L.T., D.E., M.F. and M.B.N.; methodology, D.L., L.M.P., C.A.L. and J.F.C.; formal analysis, D.L., L.M.P. and J.F.C.; investigation, D.L., L.M.P., J.F.C. and M.B.N.; writing—original draft preparation, D.L.; writing—review and editing, D.L., L.M.P., C.A.L., H.D.Z., D.E., L.T., M.F., S.D., J.F.C. and M.B.N.; supervision, J.F.C., S.D. and M.B.N.; project administration, D.L.; funding acquisition, S.D. and M.B.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Innovation Fund Denmark (IFD) with grant no. 0176-00013B for the AI4Xray project.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sharma P., Suehling M., Flohr T., Comaniciu D. Artificial Intelligence in Diagnostic Imaging: Status Quo, Challenges, and Future Opportunities. J. Thorac. Imaging. 2020;35:S11–S16. doi: 10.1097/RTI.0000000000000499. [DOI] [PubMed] [Google Scholar]
- 2.Aidoc. [(accessed on 11 November 2021)]. Available online: https://www.aidoc.com/
- 3.Mu W., Jiang L., Zhang J., Shi Y., Gray J.E., Tunali I., Gao C., Sun Y., Tian J., Zhao X., et al. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat. Commun. 2020;11:5228. doi: 10.1038/s41467-020-19116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trebeschi S., Bodalal Z., Boellaard T.N., Bucho T.M.T., Drago S.G., Kurilova I., Calin-Vainak A.M., Pizzi A.D., Muller M., Hummelink K., et al. Prognostic Value of Deep Learning-Mediated Treatment Monitoring in Lung Cancer Patients Receiving Immunotherapy. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.609054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willemink M.J., Koszek W.A., Hardell C., Wu J., Fleischmann D., Harvey H., Folio L.R., Summers R.M., Rubin D.L., Lungren M.P. Preparing Medical Imaging Data for Machine Learning. Radiology. 2020;295:4–15. doi: 10.1148/radiol.2020192224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laino M.E., Ammirabile A., Posa A., Cancian P., Shalaby S., Savevski V., Neri E. The Applications of Artificial Intelligence in Chest Imaging of COVID-19 Patients: A Literature Review. Diagnostics. 2021;11:1317. doi: 10.3390/diagnostics11081317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pehrson L.M., Nielsen M.B., Lauridsen C. Automatic Pulmonary Nodule Detection Applying Deep Learning or Machine Learning Algorithms to the LIDC-IDRI Database: A Systematic Review. Diagnostics. 2019;9:29. doi: 10.3390/diagnostics9010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D., Vilmun B.M., Carlsen J.F., Albrecht-Beste E., Lauridsen C., Nielsen M.B., Hansen K.L. The Performance of Deep Learning Algorithms on Automatic Pulmonary Nodule Detection and Classification Tested on Different Datasets That Are Not Derived from LIDC-IDRI: A Systematic Review. Diagnostics. 2019;9:207. doi: 10.3390/diagnostics9040207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strohm L., Hehakaya C., Ranschaert E.R., Boon W.P.C., Moors E.H.M. Implementation of artificial intelligence (AI) applications in radiology: Hindering and facilitating factors. Eur. Radiol. 2020;30:5525–5532. doi: 10.1007/s00330-020-06946-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner R.F., Metz C.E., Campbell G. Assessment of Medical Imaging Systems and Computer Aids: A Tutorial Review. Acad. Radiol. 2007;14:723–748. doi: 10.1016/j.acra.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Gur D. Objectively Measuring and Comparing Performance Levels of Diagnostic Imaging Systems and Practices. Acad. Radiol. 2007;14:641–642. doi: 10.1016/j.acra.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Whiting P.F., Rutjes A.W.S., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., Leeflang M.M., Sterne J.A., Bossuyt P.M., QUADAS-2 Group QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 13.Bai H.X., Wang R., Xiong Z., Hsieh B., Chang K., Halsey K., Tran T.M.L., Choi J.W., Wang D.-C., Shi L.-B., et al. Artificial Intelligence Augmentation of Radiologist Performance in Distinguishing COVID-19 from Pneumonia of Other Origin at Chest CT. Radiology. 2021;299:E225. doi: 10.1148/radiol.2021219004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorr F., Chaves H., Serra M.M., Ramirez A., Costa M.E., Seia J., Cejas C., Castro M., Eyheremendy E., Slezak D.F., et al. COVID-19 pneumonia accurately detected on chest radiographs with artificial intelligence. Intell. Med. 2020;3-4:100014. doi: 10.1016/j.ibmed.2020.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J.H., Kim J.Y., Kim G.H., Kang D., Kim I.J., Seo J., Andrews J.R., Park C.M. Clinical Validation of a Deep Learning Algorithm for Detection of Pneumonia on Chest Radiographs in Emergency Department Patients with Acute Febrile Respiratory Illness. J. Clin. Med. 2020;9:1981. doi: 10.3390/jcm9061981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P.-Y., Tsai Y.-S., Chen P.-L., Tsai H.-P., Hsu L.-W., Wang C.-S., Lee N.-Y., Huang M.-S., Wu Y.-C., Ko W.-C., et al. Application of an Artificial Intelligence Trilogy to Accelerate Processing of Suspected Patients With SARS-CoV-2 at a Smart Quarantine Station: Observational Study. J. Med. Internet Res. 2020;22:e19878. doi: 10.2196/19878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Lure F.Y., Miao H., Zhang Z., Jaeger S., Liu J., Guo L. Using artificial intelligence to assist radiologists in distinguishing COVID-19 from other pulmonary infections. J. X-ray Sci. Technol. 2021;29:1–17. doi: 10.3233/XST-200735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D., Liu X., Shao M., Sun Y., Lian Q., Zhang H. The value of artificial intelligence and imaging diagnosis in the fight against COVID-19. Pers. Ubiquitous Comput. 2021:1–10. doi: 10.1007/s00779-021-01522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beyer F., Zierott L., Fallenberg E.M., Juergens K.U., Stoeckel J., Heindel W., Wormanns D. Comparison of sensitivity and reading time for the use of computer-aided detection (CAD) of pulmonary nodules at MDCT as concurrent or second reader. Eur. Radiol. 2007;17:2941–2947. doi: 10.1007/s00330-007-0667-1. [DOI] [PubMed] [Google Scholar]
- 20.De Hoop B., de Boo D.W., Gietema H.A., van Hoorn F., Mearadji B., Schijf L., van Ginneken B., Prokop M., Schaefer-Prokop C. Computer-aided Detection of Lung Cancer on Chest Radiographs: Effect on Observer Performance. Radiology. 2010;257:532–540. doi: 10.1148/radiol.10092437. [DOI] [PubMed] [Google Scholar]
- 21.Koo Y.H., Shin K.E., Park J.S., Lee J.W., Byun S., Lee H. Extravalidation and reproducibility results of a commercial deep learning-based automatic detection algorithm for pulmonary nodules on chest radiographs at tertiary hospital. J. Med. Imaging Radiat. Oncol. 2020;65:15–22. doi: 10.1111/1754-9485.13105. [DOI] [PubMed] [Google Scholar]
- 22.Kozuka T., Matsukubo Y., Kadoba T., Oda T., Suzuki A., Hyodo T., Im S., Kaida H., Yagyu Y., Tsurusaki M., et al. Efficiency of a computer-aided diagnosis (CAD) system with deep learning in detection of pulmonary nodules on 1-mm-thick images of computed tomography. Jpn. J. Radiol. 2020;38:1052–1061. doi: 10.1007/s11604-020-01009-0. [DOI] [PubMed] [Google Scholar]
- 23.Lee K.H., Goo J.M., Park C.M., Lee H.J., Jin K.N. Computer-Aided Detection of Malignant Lung Nodules on Chest Radiographs: Effect on Observers’ Performance. Korean J. Radiol. 2012;13:564–571. doi: 10.3348/kjr.2012.13.5.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F., Hara T., Shiraishi J., Engelmann R., MacMahon H., Doi K. Improved Detection of Subtle Lung Nodules by Use of Chest Radiographs with Bone Suppression Imaging: Receiver Operating Characteristic Analysis With and Without Localization. Am. J. Roentgenol. 2011;196:W535–W541. doi: 10.2214/AJR.10.4816. [DOI] [PubMed] [Google Scholar]
- 25.Li F., Engelmann R., Pesce L.L., Doi K., Metz C.E., MacMahon H. Small lung cancers: Improved detection by use of bone suppression imaging-comparison with dual-energy subtraction chest radiography. Radiology. 2011;261:937–949. doi: 10.1148/radiol.11110192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu K., Li Q., Ma J., Zhou Z., Sun M., Deng Y., Tu W., Wang Y., Fan L., Xia C., et al. Evaluating a Fully Automated Pulmonary Nodule Detection Approach and Its Impact on Radiologist Performance. Radiol. Artif. Intell. 2019;1:e180084. doi: 10.1148/ryai.2019180084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martini K., Blüthgen C., Eberhard M., Schönenberger A.L.N., De Martini I., Huber F.A., Barth B.K., Euler A., Frauenfelder T. Impact of Vessel Suppressed-CT on Diagnostic Accuracy in Detection of Pulmonary Metastasis and Reading Time. Acad. Radiol. 2020;28:988–994. doi: 10.1016/j.acra.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Singh R., Kalra M.K., Homayounieh F., Nitiwarangkul C., McDermott S., Little B.P., Lennes I.T., Shepard J.-A.O., Digumarthy S.R. Artificial intelligence-based vessel suppression for detection of sub-solid nodules in lung cancer screening computed tomography. Quant. Imaging Med. Surg. 2021;11:1134–1143. doi: 10.21037/qims-20-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nam J.G., Kim M., Park J., Hwang E.J., Lee J.H., Hong J.H., Goo J.M., Park C.M. Development and validation of a deep learning algorithm detecting 10 common abnormalities on chest radiographs. Eur. Respir. J. 2020;57:2003061. doi: 10.1183/13993003.03061-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung J., Park S., Lee S.M., Bae W., Park B., Jung E., Seo J.B., Jung K.-H. Added Value of Deep Learning-based Detection System for Multiple Major Findings on Chest Radiographs: A Randomized Crossover Study. Radiology. 2021;299:450–459. doi: 10.1148/radiol.2021202818. [DOI] [PubMed] [Google Scholar]
- 31.Rajpurkar P., O’Connell C., Schechter A., Asnani N., Li J., Kiani A., Ball R.L., Mendelson M., Maartens G., Van Hoving D.J., et al. CheXaid: Deep learning assistance for physician diagnosis of tuberculosis using chest x-rays in patients with HIV. NPJ Digit. Med. 2020;3:1–8. doi: 10.1038/s41746-020-00322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Awai K., Murao K., Ozawa A., Nakayama Y., Nakaura T., Liu D., Kawanaka K., Funama Y., Morishita S., Yamashita Y. Pulmonary Nodules: Estimation of Malignancy at Thin-Section Helical CT—Effect of Computer-aided Diagnosis on Performance of Radiologists. Radiology. 2006;239:276–284. doi: 10.1148/radiol.2383050167. [DOI] [PubMed] [Google Scholar]
- 33.Awai K., Murao K., Ozawa A., Komi M., Hayakawa H., Hori S., Nishimura Y. Pulmonary Nodules at Chest CT: Effect of Computer-aided Diagnosis on Radiologists’ Detection Performance. Radiology. 2004;230:347–352. doi: 10.1148/radiol.2302030049. [DOI] [PubMed] [Google Scholar]
- 34.Bogoni L., Ko J.P., Alpert J., Anand V., Fantauzzi J., Florin C.H., Koo C.W., Mason D., Rom W., Shiau M., et al. Impact of a Computer-Aided Detection (CAD) System Integrated into a Picture Archiving and Communication System (PACS) on Reader Sensitivity and Efficiency for the Detection of Lung Nodules in Thoracic CT Exams. J. Digit. Imaging. 2012;25:771–781. doi: 10.1007/s10278-012-9496-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chae K.J., Jin G.Y., Ko S.B., Wang Y., Zhang H., Choi E.J., Choi H. Deep Learning for the Classification of Small (≤2 cm) Pulmonary Nodules on CT Imaging: A Preliminary Study. Acad. Radiol. 2020;27:e55–e63. doi: 10.1016/j.acra.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 36.Chen H., Wang X.-H., Ma D.-Q., Ma B.-R. Neural network-based computer-aided diagnosis in distinguishing malignant from benign solitary pulmonary nodules by computed tomography. Chin. Med. J. 2007;120:1211–1215. doi: 10.1097/00029330-200707020-00001. [DOI] [PubMed] [Google Scholar]
- 37.Liu J., Zhao L., Han X., Ji H., Liu L., He W. Estimation of malignancy of pulmonary nodules at CT scans: Effect of computer-aided diagnosis on diagnostic performance of radiologists. Asia-Pacific J. Clin. Oncol. 2020;17:216–221. doi: 10.1111/ajco.13362. [DOI] [PubMed] [Google Scholar]
- 38.Matsuki Y., Nakamura K., Watanabe H., Aoki T., Nakata H., Katsuragawa S., Doi K. Usefulness of an Artificial Neural Network for Differentiating Benign from Malignant Pulmonary Nodules on High-Resolution CT. Am. J. Roentgenol. 2002;178:657–663. doi: 10.2214/ajr.178.3.1780657. [DOI] [PubMed] [Google Scholar]
- 39.Rao R.B., Bi J., Fung G., Salganicoff M., Obuchowski N., Naidich D. LungCAD: A clinically approved, machine learning system for lung cancer detection; Proceedings of the 13th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; San Jose, CA, USA. 12–15 August 2007; pp. 1033–1037. [Google Scholar]
- 40.Kasai S., Li F., Shiraishi J., Doi K. Usefulness of Computer-Aided Diagnosis Schemes for Vertebral Fractures and Lung Nodules on Chest Radiographs. Am. J. Roentgenol. 2008;191:260–265. doi: 10.2214/AJR.07.3091. [DOI] [PubMed] [Google Scholar]
- 41.Kakeda S., Moriya J., Sato H., Aoki T., Watanabe H., Nakata H., Oda N., Katsuragawa S., Yamamoto K., Doi K. Improved Detection of Lung Nodules on Chest Radiographs Using a Commercial Computer-Aided Diagnosis System. Am. J. Roentgenol. 2004;182:505–510. doi: 10.2214/ajr.182.2.1820505. [DOI] [PubMed] [Google Scholar]
- 42.Kligerman S., Cai L., White C.S. The Effect of Computer-aided Detection on Radiologist Performance in the Detection of Lung Cancers Previously Missed on a Chest Radiograph. J. Thorac. Imaging. 2013;28:244–252. doi: 10.1097/RTI.0b013e31826c29ec. [DOI] [PubMed] [Google Scholar]
- 43.Nam J.G., Park S., Hwang E.J., Lee J.H., Jin K.-N., Lim K.Y., Vu T.H., Sohn J.H., Hwang S., Goo J.M., et al. Development and Validation of Deep Learning-based Automatic Detection Algorithm for Malignant Pulmonary Nodules on Chest Radiographs. Radiology. 2019;290:218–228. doi: 10.1148/radiol.2018180237. [DOI] [PubMed] [Google Scholar]
- 44.Oda S., Awai K., Suzuki K., Yanaga Y., Funama Y., MacMahon H., Yamashita Y. Performance of Radiologists in Detection of Small Pulmonary Nodules on Chest Radiographs: Effect of Rib Suppression With a Massive-Training Artificial Neural Network. Am. J. Roentgenol. 2009;193 doi: 10.2214/AJR.09.2431. [DOI] [PubMed] [Google Scholar]
- 45.Schalekamp S., van Ginneken B., Koedam E., Snoeren M.M., Tiehuis A.M., Wittenberg R., Karssemeijer N., Schaefer-Prokop C.M. Computer-aided Detection Improves Detection of Pulmonary Nodules in Chest Radiographs beyond the Support by Bone-suppressed Images. Radiology. 2014;272:252–261. doi: 10.1148/radiol.14131315. [DOI] [PubMed] [Google Scholar]
- 46.Sim Y., Chung M.J., Kotter E., Yune S., Kim M., Do S., Han K., Kim H., Yang S., Lee D.-J., et al. Deep Convolutional Neural Network-based Software Improves Radiologist Detection of Malignant Lung Nodules on Chest Radiographs. Radiology. 2020;294:199–209. doi: 10.1148/radiol.2019182465. [DOI] [PubMed] [Google Scholar]
- 47.Abe H., Ashizawa K., Li F., Matsuyama N., Fukushima A., Shiraishi J., MacMahon H., Doi K. Artificial neural networks (ANNs) for differential diagnosis of interstitial lung disease: Results of a simulation test with actual clinical cases. Acad. Radiol. 2004;11:29–37. doi: 10.1016/S1076-6332(03)00572-5. [DOI] [PubMed] [Google Scholar]
- 48.Abe H., MacMahon H., Engelmann R., Li Q., Shiraishi J., Katsuragawa S., Aoyama M., Ishida T., Ashizawa K., Metz C.E., et al. Computer-aided Diagnosis in Chest Radiography: Results of Large-Scale Observer Tests at the 1996–2001 RSNA Scientific Assemblies. RadioGraphics. 2003;23:255–265. doi: 10.1148/rg.231025129. [DOI] [PubMed] [Google Scholar]
- 49.Fukushima A., Ashizawa K., Yamaguchi T., Matsuyama N., Hayashi H., Kida I., Imafuku Y., Egawa A., Kimura S., Nagaoki K., et al. Application of an Artificial Neural Network to High-Resolution CT: Usefulness in Differential Diagnosis of Diffuse Lung Disease. Am. J. Roentgenol. 2004;183:297–305. doi: 10.2214/ajr.183.2.1830297. [DOI] [PubMed] [Google Scholar]
- 50.Hwang E.J., Park S., Jin K.-N., Kim J.I., Choi S.Y., Lee J.H., Goo J.M., Aum J., Yim J.-J., Cohen J.G., et al. Development and Validation of a Deep Learning-Based Automated Detection Algorithm for Major Thoracic Diseases on Chest Radiographs. JAMA Netw. Open. 2019;2:e191095. doi: 10.1001/jamanetworkopen.2019.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaube S., Suresh H., Raue M., Merritt A., Berkowitz S.J., Lermer E., Coughlin J.F., Guttag J.V., Colak E., Ghassemi M. Do as AI say: Susceptibility in deployment of clinical decision-aids. Npj Digit. Med. 2021;4:1–8. doi: 10.1038/s41746-021-00385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brice J. To Err is Human; Analysis Finds Radiologists Very Human. [(accessed on 1 October 2021)]. Available online: https://www.diagnosticimaging.com/view/err-human-analysis-finds-radiologists-very-human.
- 53.Kobayashi T., Xu X.W., MacMahon H., E Metz C., Doi K. Effect of a computer-aided diagnosis scheme on radiologists’ performance in detection of lung nodules on radiographs. Radiology. 1996;199:843–848. doi: 10.1148/radiology.199.3.8638015. [DOI] [PubMed] [Google Scholar]
- 54.Petrick N., Haider M., Summers R.M., Yeshwant S.C., Brown L., Iuliano E.M., Louie A., Choi J.R., Pickhardt P.J. CT Colonography with Computer-aided Detection as a Second Reader: Observer Performance Study. Radiology. 2008;246:148–156. doi: 10.1148/radiol.2453062161. [DOI] [PubMed] [Google Scholar]
- 55.Mazumdar M., Liu A. Group sequential design for comparative diagnostic accuracy studies. Stat. Med. 2003;22:727–739. doi: 10.1002/sim.1386. [DOI] [PubMed] [Google Scholar]
- 56.Roos J.E., Paik D., Olsen D., Liu E.G., Chow L.C., Leung A.N., Mindelzun R., Choudhury K.R., Naidich D., Napel S., et al. Computer-aided detection (CAD) of lung nodules in CT scans: Radiologist performance and reading time with incremental CAD assistance. Eur. Radiol. 2009;20:549–557. doi: 10.1007/s00330-009-1596-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gunderman R.B. Biases in Radiologic Reasoning. Am. J. Roentgenol. 2009;192:561–564. doi: 10.2214/AJR.08.1220. [DOI] [PubMed] [Google Scholar]
- 58.Busby L.P., Courtier J.L., Glastonbury C.M. Bias in Radiology: The How and Why of Misses and Misinterpretations. RadioGraphics. 2018;38:236–247. doi: 10.1148/rg.2018170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang P., Liu X., Berzin T.M., Brown J.R.G., Liu P., Zhou C., Lei L., Li L., Guo Z., Lei S., et al. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): A double-blind randomised study. Lancet Gastroenterol. Hepatol. 2020;5:343–351. doi: 10.1016/S2468-1253(19)30411-X. [DOI] [PubMed] [Google Scholar]
- 60.Lin H., Li R., Liu Z., Chen J., Yang Y., Chen H., Lin Z., Lai W., Long E., Wu X., et al. Diagnostic Efficacy and Therapeutic Decision-making Capacity of an Artificial Intelligence Platform for Childhood Cataracts in Eye Clinics: A Multicentre Randomized Controlled Trial. EClinicalMedicine. 2019;9:52–59. doi: 10.1016/j.eclinm.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagendran M., Chen Y., Lovejoy C.A., Gordon A., Komorowski M., Harvey H., Topol E.J., A Ioannidis J.P., Collins G., Maruthappu M. Artificial intelligence versus clinicians: Systematic review of design, reporting standards, and claims of deep learning studies. BMJ. 2020;368:m689. doi: 10.1136/bmj.m689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.