Abstract
The correlation between response to antiviral therapy and pretreatment viral load in patients with chronic hepatitis C has prompted the development of quantitative assays to measure viral load. The aim of our study was to assess the clinical relevance of the newly developed semiautomated PCR system COBAS HCV MONITOR version 2.0 in comparison with (i) the AMPLICOR HCV MONITOR version 1.0 assay, which underestimates RNA concentration of hepatitis C virus (HCV) genotypes 2 to 6, and (ii) the QUANTIPLEX HCV RNA version 2.0 assay, which achieves equivalent quantification for each HCV genotype, with samples from 174 patients diagnosed with chronic hepatitis C before therapy. The level and range of quantification measured with AMPLICOR HCV MONITOR version 1.0 were 1 log lower than when measured with the COBAS HCV MONITOR version 2.0, at 0.261 × 106 RNA copies/ml (range, 0.001 × 106 to 2.50 × 106 RNA copies/ml) and 4.032 × 106 RNA copies/ml (range, 0.026 × 106 to 72.6 × 106 RNA copies/ml), respectively. The two assays showed a poor correlation (r2 = 0.175). The level and range of quantification were similar when measured with the COBAS HCV MONITOR version 2.0 and QUANTIPLEX HCV RNA version 2.0 assays, at 3.03 × 106 RNA copies/ml (range, 0.023 × 106 to 72.6 × 106 RNA copies/ml) and 4.91 Meq/ml (range, 0.200 to 49.5 Meq/ml), respectively. The two assays showed a strong correlation (r2 = 0.686) for each HCV genotype. The duration of treatment (6 or 12 months) is modulated according to HCV genotype and viral load. Our results indicate that COBAS HCV MONITOR version 2.0 and QUANTIPLEX HCV RNA version 2.0 assays showing an equal dynamic range for each HCV genotype are suitable tools to assess patients before therapy.
Quantification of hepatitis C virus (HCV) RNA has become an important issue in the evaluation of patients with chronic hepatitis C. It is well established that, in patients with chronic hepatitis C, the response to alpha interferon therapy is correlated to serum HCV RNA levels before therapy (2, 13, 14). A low viral load (less than 2 × 106 RNA copies/ml) is a strong predictor of a sustained response to therapy. Furthermore, recent studies have shown that the duration of combination therapy might be modulated according to pretreatment viral load and HCV genotype (3, 6, 16, 20), recommending a longer duration of therapy (12 months) in patients with a high viral load (>2 × 106 RNA copies/ml) who are infected with HCV genotype 1. Thus, the assessment of viral load is helpful for monitoring antiviral drug therapy. Consequently, quantification of serum HCV RNA levels needs to be specific, accurate, reproducible, and standardized in order to provide an accurate prediction of treatment response and comparisons between the clinical studies. Commercial standardized assays have been developed for the evaluation of viral load using either competitive reverse transcription (RT)-PCR (21) or direct quantification with the branched DNA (bDNA) assay (9). The first-generation assays showed limited accuracy with marked genotypic variability (7, 10, 12, 18). Recently, significant progress has been made with the introduction of the QUANTIPLEX HCV bDNA second-generation (bDNA v2.0) assay (4) and the AMPLICOR HCV MONITOR version 2.0 assay (17), a well-calibrated, more-sensitive, and non-genotype-dependent (4, 17, 18) assay which is now available in a semiautomated quantitative RT-PCR version, the COBAS HCV MONITOR version 2.0 (COBAS v2.0) assay (1a, 5, 21). The aim of our study was to evaluate, with pretreatment serum samples, the clinical relevance of the new semiautomated system COBAS v2.0 assay modified to quantify all HCV genotypes equally, in comparison with the AMPLICOR HCV MONITOR version 1.0 (AMPLICOR v1.0) and the bDNA v2.0 assays routinely used in our center.
MATERIALS AND METHODS
We analyzed 174 serum samples. Ninety-four samples, tested retrospectively, were from patients enrolled in a controlled trial for the treatment of chronic hepatitis C; 80 samples, tested consecutively, were from patients diagnosed with chronic hepatitis C, prior to beginning therapy.
Direct RNA quantification.
The bDNA v2.0 assay was used according to the manufacturer's instructions (QUANTIPLEX HCV RNA v2.0; Bayer, Puteaux, France). The concentration of HCV RNA in each clinical specimen was calculated from the standard curve created with the four standards (RNA transcripts) included in each assay run. Results are expressed as HCV RNA genome megaequivalents per milliliter. The linear dynamic quantification range of the assay was between 0.2 and 120 Meq/ml (4).
RT-PCR quantification.
The AMPLICOR v1.0 assay was used according to the manufacturer's instructions (Roche Diagnostics Systems). The results are expressed as the number of RNA copies per milliliter. The linearity of the assay range was between 103 and 5 × 105 RNA copies/ml (21).
Semiautomated RT-PCR quantification.
The COBAS v2.0 assay was used according to the manufacturer's instructions (Roche Diagnostics Systems). The principle of the assay is similar to that of the AMPLICOR v1.0 assay (17). The AMPLICOR v1.0 assay provides manual serum HCV RNA extraction, automated RT-PCR, and detection and calculation of the number of viral RNA copies. The linearity of the assay range was between 103 and 106 RNA copies/ml (1).
Genotyping of HCV.
HCV genotyping was performed using reverse hybridization with the line probe assay (InGeN, Rungis, France) (22).
Statistical analysis.
For the analysis, logarithm transformation (log10) was used for all data. The bDNA v2.0 assay was used as a comparison assay. The results are shown as geometric means and ranges. Correlations between the assay values were calculated by regression analysis. The Spearman rank correlation coefficient test was used to evaluate the correlations between the quantitative values. The level of statistical significance was set at P < 0.05.
RESULTS
A total 100, 100, and 98% of the samples had detectable serum HCV RNA levels with the AMPLICOR v1.0, COBAS v2.0, and bDNA v2.0 assays, respectively. The geometric means were 0.261 × 106 RNA copies/ml (range, 0.001 × 106 to 2.5 × 106 RNA copies/ml), 3.03 × 106 RNA copies/ml (range, 0.023 × 106 to 72.6 × 106 RNA copies/ml), and 4.91 Meq/ml (range, <0.200 to 49.5 Meq/ml) for the AMPLICOR v1.0, COBAS v2.0, and bDNA v2.0 assays, respectively.
Quantification of HCV RNA with the COBAS v2.0 assay.
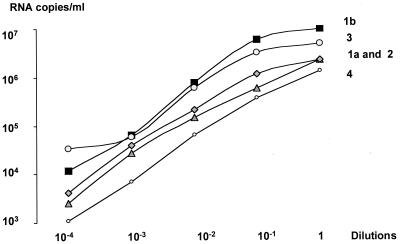
The linearity of the assay was measured with serial dilutions of serum samples for HCV genotypes 1a, 1b, 2, 3, and 4. The results showed linearity ranging between 103 and 106 RNA copies/ml for each HCV genotype (Fig. 1). In order to evaluate the proportion of serum samples with levels above 106 RNA copies/ml, 40 samples were quantified undiluted and with a 1/100 dilution. A total of 33 of 40 (82%) of the samples had a titer above 106 RNA copies/ml when measured before dilution. After dilution, none of the samples had a titer above 106 RNA copies/ml, and 2 of 40 (5%) had titers below 103 RNA copies/ml and had to be retested in undiluted form. All the samples showed a higher titer when measured after dilution, with 2.20 × 106 and 4.8 × 106 RNA copies/ml in undiluted and diluted samples, respectively (P = 0.001). Consequently, in order to have titers measured within the linear range of the assay for the study, all the samples were measured after a 1/100 dilution.
FIG. 1.
Measurement of COBAS v2.0 assay linearity range, with serial dilutions, for each HCV genotype.
Comparison of AMPLICOR v1.0 and COBAS v2.0 assays.
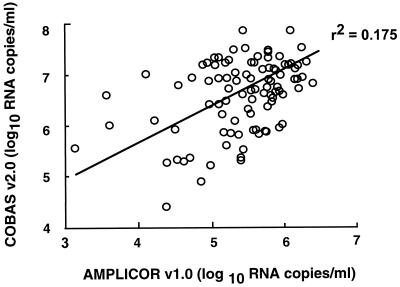
A comparison between the two assays was performed retrospectively with pretreatment serum samples of 94 patients enrolled in a controlled trial. The geometric means of the serum HCV RNA levels were 0.261 × 106 RNA copies/ml (range, 0.001 × 106 to 2.50 × 106 RNA copies/ml) and 4.032 × 106 RNA copies/ml (range, 0.026 × 106 to 72.6 × 106 RNA copies/ml) with the AMPLICOR v1.0 and COBAS v2.0 assays, respectively. The viral load measured with the two assays showed a poor correlation (r2 = 0.175) (Fig. 2). The correlation between the two assays for each HCV genotype is shown in Table 1. For each HCV genotype, the geometric mean serum HCV RNA level measured with the AMPLICOR v1.0 assay was more than 1 log unit lower than when measured with the COBAS v2.0 assay (Table 1). The difference was more marked for HCV genotypes 2 and 3, with a mean factor of 21 and 30, respectively, than for genotypes 1a and 1b, each with a mean factor of 8.
FIG. 2.
Correlation between HCV RNA titers (log10) measured with AMPLICOR v1.0 assay and COBAS v2.0 assay in serum samples from 94 patients with chronic hepatitis C.
TABLE 1.
Comparison of the results obtained with the two RT-PCR quantitative assays, according to HCV genotype, of serum samples from 94 patients with chronic hepatitis C
| Assay and value | HCV genotype (no. of patients)
|
||||
|---|---|---|---|---|---|
| 1a (9) | 1b (32) | 2 (17) | 3 (29) | Other (7) | |
| AMPLICOR v1.0 | |||||
| Geometric meana | 5.41 | 5.53 | 5.18 | 5.21 | 5.75 |
| Range | 4.62–5.94 | 4.49–6.40 | 4.11–6.21 | 3–6.29 | 5.10–6.01 |
| COBAS v2.0 | |||||
| Geometric meana | 6.33 | 6.47 | 6.49 | 6.75 | 6.81 |
| Range | 5.30–7.23 | 4.92–7.86 | 5.21–7.47 | 4.41–7.86 | 6.03–7.35 |
| r2b | 0.273 | 0.299 | 0.211 | 0.211 | 0.222 |
Values are log10 RNA copies per milliliter.
Spearman rank correlation coefficient between the two assays.
Comparison of COBAS HCV MONITOR v2.0 and bDNA v2.0 assays.
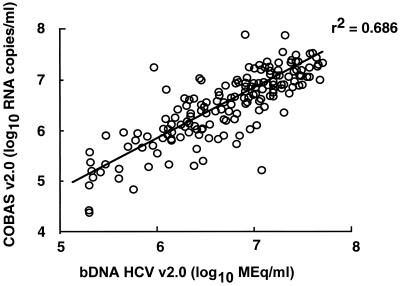
The comparison between the two assays was performed with 174 serum samples (94 pretreatment serum samples measured retrospectively and 80 serum samples measured consecutively before the initiation of therapy). The 174 serum samples were diluted (1/100) for quantification with the COBAS v2.0 assay. After dilution, none of the samples was measured above the high end of the assay (106 RNA copies/ml); 132 of 174 (76%) samples showed a viral titer of <106 RNA copies/ml, and 4 of 174 (2%) samples were measured below the low end of the assay (103 RNA copies/ml) and had to be retested undiluted. The geometric means of serum HCV RNA levels were 3.03 × 106 RNA copies/ml (range, 0.023 × 106 to 72.6 × 106 RNA copies/ml) and 4.91 Meq/ml (range, 0.200 to 49.5 Meq/ml) with the COBAS v2.0 assay and the bDNA v2.0 assay, respectively. The two assays are highly correlated (r2 = 0.686) (Fig. 3). The relationship between values obtained by both assays generated the following equation: log10 bDNA = 0.314 + 0.918 log10 COBAS. For each HCV genotype, a good correlation was observed between the two assays, and the geometric means of serum HCV RNA levels were similar (Table 2).
FIG. 3.
Correlation between hepatitis C virus RNA titers (log10) measured with COBAS v2.0 assay and bDNA v2.0 assay in serum samples from 174 patients with chronic hepatitis C.
TABLE 2.
Comparison of the results obtained with COBAS v2.0 and bDNA v2.0 assays, according to HCV genotype, of serum samples from 174 patients with chronic hepatitis C
| Assay and value | HCV genotype (no. of patients)
|
|||||
|---|---|---|---|---|---|---|
| 1a (20) | 1b (55) | 2 (24) | 3 (37) | 4 (16) | Other (22) | |
| COBAS v2.0 | ||||||
| Geometric meana | 6.47 | 6.41 | 6.52 | 6.67 | 5.77 | 6.57 |
| Range | 5.21–7.23 | 4.92–7.86 | 5.08–7.47 | 4.41–7.86 | 4.36–6.41 | 5.69–7.35 |
| bDNA v2.0 | ||||||
| Geometric meana | 6.75 | 6.53 | 6.66 | 6.82 | 6.47 | 6.78 |
| Range | 5.32–7.67 | 5.30–7.65 | 5.32–7.46 | 5.3–7.69 | 5.30–7.30 | 5.70–7.48 |
| rb | 0.773 | 0.749 | 0.480 | 0.804 | 0.700 | 0.893 |
Values are log10 RNA copies per milliliter.
Spearman rank correlation coefficient between the two assays.
DISCUSSION
Access to reliable assays to measure viral load are useful to clinicians, as it appears that pretreatment serum HCV RNA levels either alone or in combination with HCV genotype can be used to predict therapeutic response to interferon or combination therapy in patients with chronic hepatitis C (2, 3, 6, 13, 14, 16, 20). Likewise, HCV genotype has been implicated as a factor predictive of response to interferon therapy (2, 3, 6, 13, 14, 15, 16, 20). As HCV genotypic variability may influence the efficiency of HCV RNA quantification, one obstacle to understanding the clinical relevance of HCV genotype and viremia levels has been, until recently, the lack of a reliable method for HCV RNA quantification. Indeed, the first-generation quantification assays were HCV genotype dependent, since most had primers and probes based on the originating HCV prototype strain (i.e., HCV genotype 1) until the genetic diversity of the HCV genome became clear (12, 14, 20).
The bDNA assay has been recently refined with target probes designed to overcome the consequence of HCV genotypic variability and to achieve equal quantification of all HCV genotypes, as compared to the first-generation assay, which underestimated HCV genotypes 2 and 3 (4, 10, 12, 19).
Reduced amplification efficiency in the AMPLICOR v1.0 assay (8) results from secondary structures that could form at relatively low annealing and extension temperatures employed in the AMPLICOR v1.0 assay. Adding dimethyl sulfoxide to the reaction mixture and modifying the thermal cycling conditions in the AMPLICOR HCV MONITOR VERSION 2.0 assay most likely promotes unfolding of these secondary structures, resulting in an improved amplification efficiency. These modifications result in equal quantification of each HCV genotype (17) and in higher viral titers (approximately 10-fold) when the automated system (the COBAS v2.0 assay) is used (1). Our results, showing an equal quantification for each HCV genotype with the COBAS v2.0 assay with clinical samples, are in accordance with those of Mellor et al. (17) with RNA transcripts. Furthermore, in our study the titers obtained with the COBAS v2.0 and the bDNA v2.0 assays with clinical samples are similar, suggesting that the two assays are equivalently calibrated. Overcoming the consequences of HCV genotypic variability was a real challenge, since HCV genotypes other than 1a and 1b are found in roughly 30 to 40% of patients with chronic hepatitis C in the United States and in western Europe (11, 15), and the EASL International Consensus Conference on Hepatitis C has recommended the duration of therapy be modulated according to HCV genotype and level of viremia (6).
Despite the difference in low-end quantification limits defined for the bDNA v2.0 assay (0.2 Meq/ml) and the COBAS v2.0 assay (3 × 103 RNA copies/ml), the abilities of the two assays to identify viremic patients were quite similar (98 and 100%). The high-end limit for the COBAS v2.0 assay (106 RNA copies/ml) requires a dilution step for a large number of samples (82% in our study) in order to obtain an accurate quantification, while the bDNA v2.0 assay is able to quantitate up to this limit. Since the majority of the patients eligible for therapy have a viral load of >106 RNA copies/ml and since a cutoff value of 2 × 106 RNA copies/ml (representing 60 to 76% of the patients) is recommended for determining the duration of therapy (3, 6, 20), the addition of a standardized dilution step in the COBAS v2.0 assay by the manufacturer would avoid the risk of interlaboratory discrepancies. Finally, our study suggests that in clinical practice, the assessment of patients with chronic hepatitis C before antiviral therapy makes a high-amplitude range for a quantitative assay more relevant than one with a low sensitivity.
Both the COBAS v2.0 and bDNA v2.0 assays are suitable for assessing patients with chronic hepatitis C before therapy and for evaluating treatment regimens. This new approach of serum HCV RNA quantification will lead to a better standardization and comparison of controlled trials of chronic hepatitis C treatments.
ACKNOWLEDGMENTS
We thank C. Hézode (Centre Hospitalier Henri-Mondor, Créteil), K. Barange (Clinique Dieulafoy, Toulouse), P. Couzigou (Hôpital Haut-Levêque, Pessac), D. Larrey (Hôpital Saint-Eloi, Montpellier), O. Rosmorduc (Hôpital Saint-Antoine, Paris), A. Tran (Hôpital de l'Archet 2, Nice), J. P. Zarsky (Centre Hospitalier, Grenoble), C. Trépo (Hôtel-Dieu, Lyon), C. Bréchot (Hôpital Neker, Paris), M. A. Bigard (CHU Nancy Brabois, Vandoeuvre Les Nancy), J. D. Grange (Hôpital Tenon, Paris), H. Bismuth (Hôpital Paul-Brousse, Villejuif), C. Eugène (CHI, Poissy), J. P. Miguet (Hôpital Jean-Minjoz, Besançon), C. Buffet (Hôpital Bicêtre, Le Kremlin Bicêtre), P. Blanc (Hôpital Saint-Eloi, Montpellier), and the French Multicenter Study Group, Produit Roche, Neuilly-sur-Seine, France.
REFERENCES
- 1.Afonso A M, Didier J, Plouvier E, Falissard B, Ferey M P, Bogard M, Dussaix E. Performance of an automated system for quantification of hepatitis C virus RNA. J Virol Methods. 2000;86:55–60. doi: 10.1016/s0166-0934(99)00179-2. [DOI] [PubMed] [Google Scholar]
- 1a.Albadalejo J, Alonzo R, Antinozzi R, Bogard M, Bourgault A M, Colucci G, Fenner T, Petersen H, Sala E, Vincelette J, Young C. Multicenter evaluation of the COBAS AMPLICOR HCV assay, an integrated PCR system for detection of hepatitis C virus RNA in the diagnostic laboratory. J Clin Microbiol. 1998;36:862–865. doi: 10.1128/jcm.36.4.862-865.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis G, Lau J. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology. 1997;20(Suppl.):S123–S127. doi: 10.1002/hep.510260721. [DOI] [PubMed] [Google Scholar]
- 3.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. New Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 4.Detmer J, Lagier R, Flynn J, Zayat C, Kolberg J, Collins M, Urdea M, Sanchez-Pescador R. Accurate quantification of hepatitis C virus (HCV) RNA from all genotypes by using branched-DNA technology. J Clin Microbiol. 1996;34:901–907. doi: 10.1128/jcm.34.4.901-907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doglio A, Laffont C, Caroli-Bosc F X, Rochet P, Lefebvre J C. Second generation of the automated Cobas Amplicor HCV assay improves sensitivity of hepatitis C virus RNA detection and yields results that are more clinically relevant. J Clin Microbiol. 1999;37:1567–1569. doi: 10.1128/jcm.37.5.1567-1569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EASL International Consensus Conference on Hepatitis C. Consensus statement. J Hepatol. 1999;30:956–961. [PubMed] [Google Scholar]
- 7.Gretch D R, De la Rosa C, Carithers R L, Jr, Willson R A, Williams B, Corey L. Assessment of hepatitis C viremia using molecular amplification technologies: correlations and clinical implications. Ann Intern Med. 1995;123:321–329. doi: 10.7326/0003-4819-123-5-199509010-00001. [DOI] [PubMed] [Google Scholar]
- 8.Hawkins A, Davidson F, Simmonds P. Comparison of plasma virus load among individuals infected with hepatitis C virus (HCV) genotypes 1, 2, and 3 by Quantiplex HCV RNA assay version 1 and 2, Roche Monitor assay, and an in-house limiting dilution method. J Clin Microbiol. 1997;35:187–192. doi: 10.1128/jcm.35.1.187-192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolberg J A, Sanchez-Pescador R, Detmer J, Collins M, Sheridan P, Neuwald P, Wilber J, et al. Groupe Français d'Études Moléculaires des hépatites virales (GEMHEP) (ed.), Hepatitis C virus: new diagnostic tools. Paris, France: John Libbey Eurotext; 1994. Branched DNA quantitation of hepatitis C viral RNA in patient sera; pp. 57–70. [Google Scholar]
- 10.Lau J Y N, Simmonds P, Urdea M. Implication of variations of “conserved” regions of hepatitis C virus genome. Lancet. 1995;346:425–426. doi: 10.1016/s0140-6736(95)92786-7. [DOI] [PubMed] [Google Scholar]
- 11.Lau J Y N, Davis G L, Prescott L E, Maertens G, Lindsay K L, Quian K P, Mizokami M, Simmonds P the Hepatitis Interventional Therapy Group. Distribution of hepatitis C virus genotypes determined by line probe assay in patients with chronic hepatitis C seen at tertiary referral centers in United States. Ann Intern Med. 1996;124:868–876. doi: 10.7326/0003-4819-124-10-199605150-00002. [DOI] [PubMed] [Google Scholar]
- 12.Lunel F, Cresta P, Vitour D, Payan C, Dumont B, Frangeul L, Reboul D, Brault C, Piette J C, Huraux J M. Comparative evaluation of hepatitis C virus RNA quantitation by bDNA, NASBA, and Monitor assays. Hepatology. 1999;29:528–535. doi: 10.1002/hep.510290237. [DOI] [PubMed] [Google Scholar]
- 13.Martinot-Peignoux M, Marcellin P, Pouteau M, Castelnau C, Boyer N, Poliquen M, Degott C, Descombes V, Lebreton V, Milotova V, Benhamou J D, Erlinger S. Pretreatment serum HCV RNA levels and HCV genotype are the main and independent prognostic factors of sustained response to alpha interferon therapy in chronic hepatitis C. Hepatology. 1995;22:1050–1056. [PubMed] [Google Scholar]
- 14.Martinot-Peignoux M, Boyer N, Pouteau M, Castelnau C, Giuily N, Duchatelle V, Aupérin A, Degott C, Benhamou J P, Erlinger S, Marcellin P. Predictors of sustained response to alpha interferon therapy in chronic hepatitis C. J Hepatol. 1998;29:214–223. doi: 10.1016/s0168-8278(98)80006-8. [DOI] [PubMed] [Google Scholar]
- 15.Martinot-Peignoux M, Roudot-Thoraval F, Mendel I, Coste J, Izopet J, Duverlie G, Payan C, Pawlotsky J M, Defer C, Bogard M, Gerolami V, Halfon P, Buisson Y, Fouqueray B, Loiseau P, Lamoril J, Lefrère J J, Marcellin P the GEMHEP. Hepatitis C virus genotypes in France: relationship with epidemiology, pathogenicity and response to interferon therapy. J Viral Hepatol. 1999;6:435–443. doi: 10.1046/j.1365-2893.1999.00187.x. [DOI] [PubMed] [Google Scholar]
- 16.McHutchinson J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z, Ling M H, Cort S, Albrecht J. Interferon alpha-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. New Engl J Med. 1999;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 17.Mellor J, Hawkins A, Simmonds P. Genotype dependence of hepatitis C virus load measurement in commercially available quantitative assays. J Clin Microbiol. 1999;37:2525–2532. doi: 10.1128/jcm.37.8.2525-2532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pawlotsky J M, Lonjon I, Hézode C, Raynard B, Darthuy F, Rémiré J, Soussy C J, Dhumeaux D. What strategy should be used for diagnosis of hepatitis C virus infection in clinical laboratories? Hepatology. 1998;27:1700–1702. doi: 10.1002/hep.510270632. [DOI] [PubMed] [Google Scholar]
- 19.Pawlotsky J M, Martinot-Peignoux M, Poveda J D, Bastie A, Le Breton V, Darthuy F, Remeri J, Erlinger S, Dhumeaux D, Marcellin P. Quantification of hepatitis C virus RNA in serum by branched DNA-based signal amplification assays. J Virol Methods. 1999;79:227–235. doi: 10.1016/s0166-0934(99)00024-5. [DOI] [PubMed] [Google Scholar]
- 20.Poynard T, Marcellin P, Lee S S, Niederau C, Minuk G S, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C, Albrecht J. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet. 1998;352:1426–1432. doi: 10.1016/s0140-6736(98)07124-4. [DOI] [PubMed] [Google Scholar]
- 21.Roth W K, Lee J H, Ruster B, Zeuzem S. Comparison of two quantitative hepatitis C virus reverse transcriptase PCR assays. J Clin Microbiol. 1996;34:261–264. doi: 10.1128/jcm.34.2.261-264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuyver L, Rossau R, Wyseur A, Duhamel M, Vanderborght B, Van Heuverswyn H, Maertens G. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]