Abstract
We previously reported that the immunostimulatory activity of heat-killed Latilactobacillus sakei K040706 in macrophages and cyclophosphamide (CTX)-treated mice. However, identification of heat-killed L. sakei K040706 (heat-killed LS06) using a validated method is not yet reported. Further, the underlying molecular mechanisms for its immunostimulatory effects in CTX-induced immunosuppressed mice remain unknown. In this study, we developed strain-specific genetic markers to detect heat-killed L. sakei LS06. The lower detection limit of the validated primer set was 2.1 × 105 colony forming units (CFU)/mL for the heat-killed LS06 assay. Moreover, oral administration of heat-killed LS06 (108 or 109 CFU/day, p.o.) effectively improved the body loss, thymus index, natural killer cell activity, granzyme B production, and T and B cell proliferation in CTX-treated mice. In addition, heat-killed LS06 enhanced CTX-reduced immune-related cytokine (interferon-γ, interleukin (IL)-2, and IL-12) production and mRNA expression. Heat-killed LS06 also recovered CTX-altered microbiota composition, including the phylum levels of Bacteroidetes, Firmicutes, and Proteobacteria and the family levels of Muribaculaceae, Prevotellaceae, Tannerellaceae, Christensenellaceae, Gracilibacteraceae, and Hungateiclostridiaceae. In conclusion, since heat-killed L. sakei K040706 ameliorated CTX-induced immunosuppression and modulated gut microbiota composition, they have the potential to be used in functional foods for immune regulation.
Keywords: heat-killed Latilactobacillus sakei, immune improvement, cyclophosphamide, microbiota, splenocyte
1. Introduction
Immunomodulators contribute to our immunity by activating immune cells [1], and can also act as immune stimulators to protect against extracellular parasites, bacteria, allergens, and toxins [2,3], or to reduce the side effects of drug-induced immunosuppression [4]. T helper (Th) cells play a pivotal role in immunomodulation [5]. Type 1 interferon (IFN)-γ-producing Th (Th1) cells help in mounting a host defense against intracellular pathogens, including protozoa, bacteria, and viruses, and are involved in the development of certain types of autoimmune diseases [2]. In contrast, type 2 interleukin (IL)-4/IL-5/IL-13-secreting Th (Th2) cells defend against helminth infections and venom exposure and participate in different types of allergic diseases, including asthma, atopic dermatitis, allergic rhinitis, and food allergy. Increasing evidence indicates that natural immune stimulators such as probiotic strains could be an alternative to conventional therapies for maintaining immune homeostasis [6].
Lactic acid bacteria (LAB), members of the order Lactobacillales, are probiotic bacteria that distinctively produce lactic acid. LAB are commonly found in certain foods (such as yogurt) and human and animal digestive and genital systems. The specific strains of LAB can be detected using the 16S rRNA gene sequencing [7,8,9]. However, a limitation of this method is the low resolution between closely-related species. Therefore, better strain-specific detection of LAB is critical for its precise identification and quantification. In the previous study, we developed a species-specific primer set for the identification of Latilactobacillus sakei [10]. This primer set had specificity and sensitivity at a species level. For the protection and monitoring of strain, we needed a primer set at a strain level.
We previously reported the presence of Latilactobacillus sakei K040706 (formerly known as Lactobacillus sakei K040706) in the Doenjang, which is made from Meju (a brick of fermented soybeans) in brine. Heat-killed L. sakei K040706 (heat-killed LS06) enhances the phagocytic ability of recombinant IFN-γ(rIFN-γ)-primed macrophages by upregulating the production of nitric oxide, tumor necrosis factor (TNF)-α, and IL-6 via activating the nuclear factor kappa-light-chain-enhancer of activated B cells and Toll-like receptor 2. It also restores immunological parameters in cyclophosphamide (CTX)-induced immunosuppressed mice [11]. In addition, live LS06 exerts an improvement effect of colitis, involving inflammatory mediator suppressions and microbiota composition [12], and stimulates the cytokine, hematopoiesis, and immune cell production in CTX-treated mice [13]. Although we have previously demonstrated the stimulating activity of heat-killed LS06 in immune organs indexes of CTX-treated mice, the role of immunomodulation involving the molecular mechanism of heat-killed LS06 is not fully elucidated. Therefore, in this study, we developed a strain-specific real-time polymerase chain reaction (RT-PCR) method for identifying heat-killed LS06 and then examined its immunostimulatory activity in CTX-treated mice.
2. Materials and Methods
2.1. Preparation of Heat-Killed LS06 and Genomic DNA
The bacterial strains, including L. sakei K040706 (KCCM11472P) and other lactic acid bacteria, are used in this study (Table 1). Latilactobacillus strains were inoculated in fresh MRS media (De Man, Rogosa and Sharpe agar, BD Bioscience, Franklin Lakes, NJ, USA) at 30 °C for 20 h in an aerobic condition. The cultured fraction of cells (1/100) was transferred into fresh MRS media. After overnight cultivation, 1 mL of the culture was applied for DNA isolation and the pellet was collected by centrifugation at 13,000× g for 15 min, 4 °C. After washing with PBS (pH 7.0) twice, heat inactivation was carried out at 121 °C for 15 min and freeze drying was performed. DNA extraction was performed with DNeasy Blood & Tissue kits (Qiagen, Hilden, Germany) according to the manufacturer’s instruction with the following modifications: the cultivated cells were lysed with lysozyme from chicken egg white (Sigma-Aldrich, Burlington, MA, USA) for 2 h at 37 °C.
Table 1.
Lactilactobacillus sakei strains used in this study.
| Strain Number | Source |
|---|---|
| K040706 | Lab stock |
| KACC17865 | KACC (Korean Agricultural Culture Collection) |
| KACC17868 | |
| KACC17871 | |
| KACC18352 | |
| KACC17864 | |
| KACC16119 |
2.2. Whole Genome Sequencing and Comparative Genomic Analysis
Genomic DNA of L. sakei K040706 was sequenced by single-molecule, real-time sequencing technology (SMRT) on a PacBio RS II instrument (Macrogen Inc., Daejeon, Republic of Korea). Genome assembly was achieved using Hierarchical Genome Assembly Process (HGAP) version 3.0 from the SMRT portal version 2.3, with default parameters, by Macrogen, Inc. The assembled sequence was submitted, and pan-genome orthologous groups (POGs) were constructed using the EzBioCloud Comparative Genomics Database (http://cg.ezbiocloud.net, accessed on 12 March 2021). Average nucleotide identity (ANI) values and Venn diagram also were calculated and constructed by the EzBioCloud database, respectively [14]. The whole genome sequence of L. sakei K040706 was listed on NCBI database (https://www.ncbi.nlm.nih.gov/sra/SRR15647685, accessed on 26 August 2021).
2.3. Design of Strain-Specific Primer
Primers are synthesized by Macrogen (Daejeon, Republic of Korea). The strain-specific marker bands were found by analysis of POGs. After selecting the closest 4 strains by the OrthoANI algorithm (Table 2), a complementary set of L. sakei K040706 was isolated. Three primer sequences sets were designed using the sequences in these regions. The primer pairs have a similar melting temperature (Tm) and PCR product sizes (100 ~ 150 bp), which show a similar resolution by agarose gel electrophoresis. Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on 26 August 2021) was applied to design strain-specific primer.
Table 2.
The list of primer sequences for strain-specific qRT-PCR.
| Primer | Sequence (5′ to 3′) | Position * | Amplicon Size (bp) |
|---|---|---|---|
| 2017_F | AAGAGTTCGGATGGCAGCAA | 1,976,642–1,976,623 | 158 |
| 2017_R | CGCTATCCGATAAGCTCGCA | 1,976,485–1,976,504 | |
| 2018_F | ATGGGTAAAATGATTCACTCGAAATATG | 1,976,869–1,976,842 | 160 |
| 2018_R | TTATCTATTGGCCACTCTTCTATT | 1,976,711–1,976,734 | |
| 2019_F | GAAAAGGGATGCGATTGCCG | 1,977,050–1,977,031 | 170 |
| 2018_R | ATCACCCACCACTTGCCAAT | 1,976,875–1,976,894 |
* The positions are enumerated based on the complete genome sequence of L. sakei under accession number Sequence Read Archive (SRA): https://www.ncbi.nlm.nih.gov/sra/PRJNA758190, accessed on 26 August 2021.
2.4. Design of Animal Experiments
Male ICR mice (n = 50, 6 weeks old) from Central Lab. Animal Inc. (Seoul, Republic of Korea) were kept at a temperature of 20 ± 5 °C, the humidity of 40–60%, and 12 h light/dark cycle environment before use. Animal experiments were approved by the animal care committee of Kyung Hee University (KHUASP(SE)-18-158).
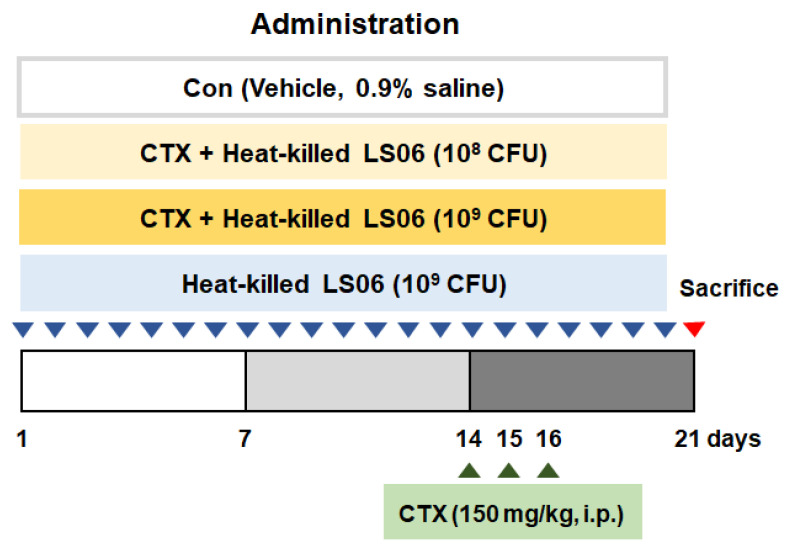
The CTX-induced immunosuppressed model was performed with some modifications [15,16]. Mice receive CTX (150 mg/kg/day, intraperitoneal (i.p.)) for immunosuppression on days 14, 15, and 16. After being acclimated for a week, mice were randomly allocated as follows (n = 10): (i) Con group (vehicle-treated control group, 0.9% saline), (ii) CTX group (vehicle + CTX 150 mg/kg), (iii) CTX + heat-killed LS06 108 colony forming units (CFU) group, (iv) CTX + heat-killed LS06 109 CFU group, (v) heat-killed LS6 108 CFU group. Vehicle and heat-killed LS06 (108 CFU or 109 CFU/day) were orally supplemented for 20 days (Figure 1).
Figure 1.
Animal study design. Establishment of cyclophosphamide (CTX)-induced immunosuppressed model and oral administration of heat-killed L. sakei K040706 (heat-killed LS06) 108 or 109 colony forming units (CFU)/day) for 20 days.
2.5. Preparation of Splenocytes and Assessment of NK Cytotoxic Activity
After sacrifice at the end of experiments, splenocytes were isolated and NK cell was activated [15]. NK cytotoxic activity was determined by measuring lactate dehydrogenase (LDH) levels, as performed in a previous report [13].
2.6. Determination of T Cell and B Cell Proliferation
To activate T cell or B cell, splenocytes were treated with concanavalin A (Con A, 5 μg/mL) or lipopolysaccharide (LPS, 1 μg/mL), respectively. The cell proliferation assay was assessed by using the Cell Titer 96® Aqueous One Solution Reagent according to the manufacturer’s instructions (Promega, Madison, WI, USA).
2.7. Determination of Cytokine and Granzyme B Production
After T and B cell activation, the productions of cytokine and granzyme B in cell culture supernatants were quantified using ELISA kits (R&D systems, Minneapolis, MN, USA).
2.8. Determination of mRNA Expression
Splenic RNA was extracted by using Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA), and the level of Th1-related cytokines and β-actin mRNA expression was determined using quantitative RT-PCR (qRT-PCR) [11]. The oligonucleotide primers are listed in Supplemental Table S1.
2.9. Microbiota Profiling
Total genomic DNA from feces was prepared by using an i-genomic stool plus kit (Intron Biotechnology, Seoul, Korea). After the preparation and amplification of the library, selection of 16S rRNAs and taxonomic assignment were performed as in a previous report [11].
2.10. Statistical Analysis
Statistical analysis was performed by GraphPad Prism 5.01 software (La Jolla, CA, USA). Data are expressed as the mean ± standard error of the mean (SEM) (n = 10). Statistical differences were analyzed using one-way analysis of variance (ANOVA) and the Dunnett’s post hoc test, and p < 0.05 was indicated statistically significant.
3. Results
3.1. Screening of Strain-Specific Genetic Markers and Primer Design
To develop strain-specific genetic markers for LAB strains, we performed a comparative genomic analysis of L. sakei K040706 with four other closely related Latilactobacillus strains. The complete genome of L. sakei K040706 contained 1,977,298 sequences. A phylogenetic tree was constructed using an up-to-date bacterial core gene set (UBGC) to compare 92 core genes [17]. Three strains belonging to L. sakei and one belonging to Latilactobacillus curvatus were selected for the pan-genome analysis (Figure 2A). For the pan-genome analysis, Heap’s law regression model was applied, which showed a robust fit model with a clear upward trend and R2 = 0.998 (Figure 2B). The core genomes had 1,394 coding sequences (CDS), from which only 81 of the L. sakei K040706 were unique. These CDSs were not part of the plasmids or phage sequences. After comparative genomics, the unique CDSs were searched in the NCBI database to compare with the CDSs of other bacteria, including Latilactobacillus sp. Subsequently, three sequences were selected, and each primer set was checked using Primer-BLAST in NCBI (Table 2).
Figure 2.
(A) Phylogenetic tree showing the relatedness of L. sakei strains based on ANI analysis. (B) Core and pan-genome curves show the trend of the core gene and pan-gene families with an increase in the number of genomes by Heap’s law model. (C) Pan-genome Venn diagram of L. sakei strains, including K040706, DSM 20017, DSM 15831, DS4, and DSM20019, showing the distribution of shared and unique pangenome orthologous groups (POGs) among the five strains.
3.2. Specificity and Efficiency of Strain-Specific Real-Time PCR Assay
Three unique genetic markers of L. sakei K040706 (K040706..2017, K040706..2018, and K040706..2019) were subjected to PCR to check their specificities against live and heat-killed L. sakei K040706, and six other L. sakei sp. The PCR was carried out at 55–57 °C. (Figure 3). The expected lengths of the PCR products of the genetic markers K040706..2017, K040706..2018, and K040706..2019 were 158, 160, and 170 bp, respectively. Amplicons appeared only in the live and heat-killed L. sakei K040706 samples, but not in the other samples. These results suggest that the designed primers bind the target sequences with high specificity.
Figure 3.
Detection of PCR product by strain-specific primers. Numbers at the top indicate the primer pairs. L, 100 bp DNA ladder; Lane 1, live LS06; Lane 2, heat-killed L. sakei LS06; Lane 3, KACC17865; Lane 4, KACC17868; Lane 5, KACC17871; Lane 6, KACC18352; Lane 7, KACC17864; Lane 8, KACC16119.
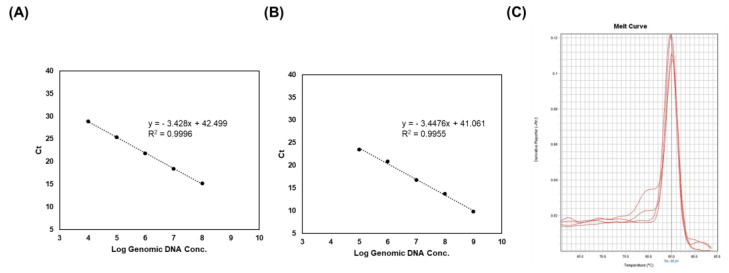
To evaluate the efficiency and limit of detection of PCR, a RT-PCR assay was conducted using the 2018 primer pair, since it showed a high density of PCR bands (Figure 4). Strain-specific primers exhibited a linear plot with a range of 2.1 × 104 to 108 CFU/mL in the live sample (Figure 4A). In addition, DNA samples from the heat-killed LS06 showed a limit of detection of 2.1 × 105 CFU/mL (in triplicate) (Figure 4B). All R2 values (correlation coefficients) were >0.99 and the efficacies of the PCR were >0.9. Melt curve analysis indicated only one melting peak varying from 85 to 86 °C (Figure 4C). These results implied that the PCR product was generated by specific primer binding.
Figure 4.
Real-time PCR analysis of genomic DNA, live, and heat-killed LS06 for a range of concentrations. (A) Standard curve of live L. sakei. (B) Standard curve of heat-killed LS06. (C) Melt curve generated by strain-specific primers in real-time PCR.
3.3. Amelioration of Thymus Index Alteration in CTX-Treated Mice by Heat-Killed LS06 Administration
To determine the immune response improvement of heat-killed LS06 in CTX-treated mice, we evaluated whether oral administration of heat-killed LS06 (108 or 109 CFU/day) recovered the CTX-reduced body weight and thymus index in mice. CTX injection (150 mg/kg, i.p.) significantly reduced body weight, while a high-dose heat-killed LS06 (109 CFU/day, p.o.) restored the bodyweight (p < 0.05, Figure 5A). In addition, high- and low-dose of heat-killed LS06 significantly improved the thymus index in CTX-treated mice (Figure 5B).
Figure 5.
Comparison of (A) body weight and (B) thymus index between CTX and heat-killed LS06 treatment in mice. Thymus index was calculated as thymus weight (g)/body weight (kg). Data are presented as the mean ± SEM (n = 10). # p < 0.05 vs. Con group; * p < 0.05, *** p < 0.001 vs. CTX group.
3.4. Heat-Killed LS06-Modulated Immune Cells in CTX-Treated Mice
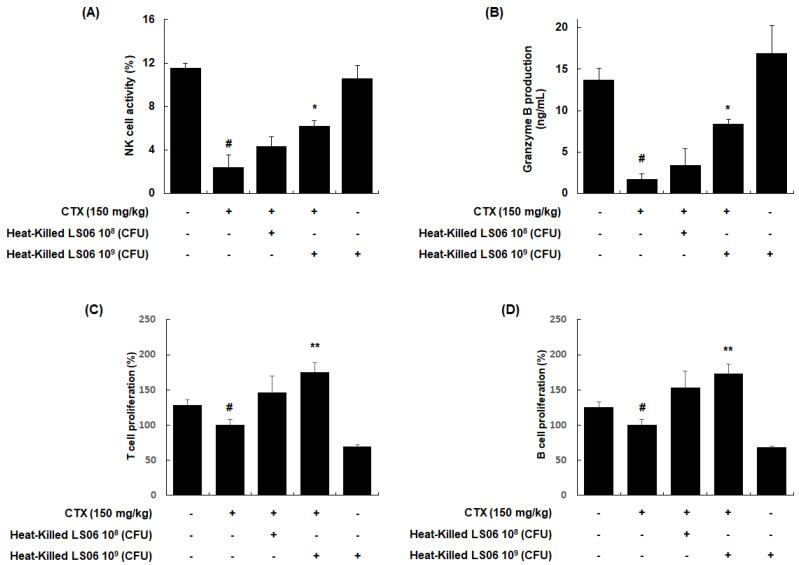
NK cells are cytotoxic and secrete granzyme B to lyse the target cancerous and virus-infected cells [18]. We investigated whether the heat-killed LS06 modulates the NK cell cytotoxicity against YAC-1 target cells. Our data showed that NK cell activity in the splenocytes was significantly reduced in CTX-treated mice compared to the vehicle-treated control group (2.40 ± 1.11% vs. 11.54 ± 0.45%, respectively, p < 0.001, Figure 6A). However, the NK cell activity in splenocytes was significantly enhanced upon high-dose heat-killed LS06 (109 CFU/day, p.o.) treatment of CTX-treated mice compared to only CTX-treatment in mice (6.23 ± 0.47% vs. 2.40 ± 1.11%, respectively, p < 0.05). Similarly, high-dose heat-killed LS06 (109 CFU/day, p.o.) treatment of CTX-treated mice significantly elevated granzyme B production in splenocytes than in only CTX-treated mice (8.42 ± 0.58% vs. 1.71 ± 0.66%, respectively, p < 0.05, Figure 6B). We further evaluated the effects of heat-killed LS06 on Con A- or LPS-induced splenocyte proliferation in CTX-immunosuppressed mice [19]. As expected, the CTX group significantly suppressed the proliferative responses of T and B cells compared to the Con group (Figure 6C,D). Meanwhile, the administration of heat-killed LS06 at 109 CFU/day significantly elevated the Con-A- or LPS-induced T cell proliferation by up to 176% or B cell proliferation by up to 173%, respectively, over the only CTX-treated group.
Figure 6.
Effects of heat-killed LS06 on the regulation of immune cells in CTX-treated mice. Heat-killed LS06 (108 or 109 CFU/day, p.o.) were provided for 20 days and the splenocytes were prepared from isolated spleen of sacrificed mice. (A) NK cell cytotoxic activity and (B) granzyme B production (C,D) T and B cell proliferation. Data are presented as the mean ± SEM (n = 10). # p < 0.05 vs. Con group; * p < 0.05, ** p < 0.01 vs. CTX group.
3.5. Heat-Killed LS06-Regulated Th1 Cytokines in Splenocytes of CTX-Treated Mice
To explore whether heat-killed LS06 control T helper (Th) cells, the effects of heat-killed LS06 on Th1-related cytokines in splenocytes were determined. CTX treatment significantly reduced the production and mRNA expression of IFN-γ, IL-2, and IL-12, whereas the administration of heat-killed LS06 significantly increased the protein and mRNA expression levels of Th1 cell-related cytokines in splenocytes isolated from CTX-immunosuppressed mice (Figure 7). Interestingly, heat-killed LS06 provision did not affect IL-10, IL-13, and IL-4 productions, indicating the Th2-related cytokines (data not shown). Our data show that the Th1-activating effect of heat-killed LS06 is mediated by increasing cytokine production and mRNA expression in CTX-treated mice.
Figure 7.
Effects of heat-killed LS06 on the expression level of Th1-related cytokines production and mRNA in splenocytes isolated from CTX-treated mice. (A–C) The production and (D–F) mRNA expression levels of the Th1-related cytokines. Data are presented as mean ± SEM (n = 10). # p < 0.05 vs. Con group; * p < 0.05, ** p < 0.01, and *** p < 0.001 vs. CTX group.
3.6. Effects of Heat-Killed LS06 on Microbiota Composition in CTX-Treated Mice
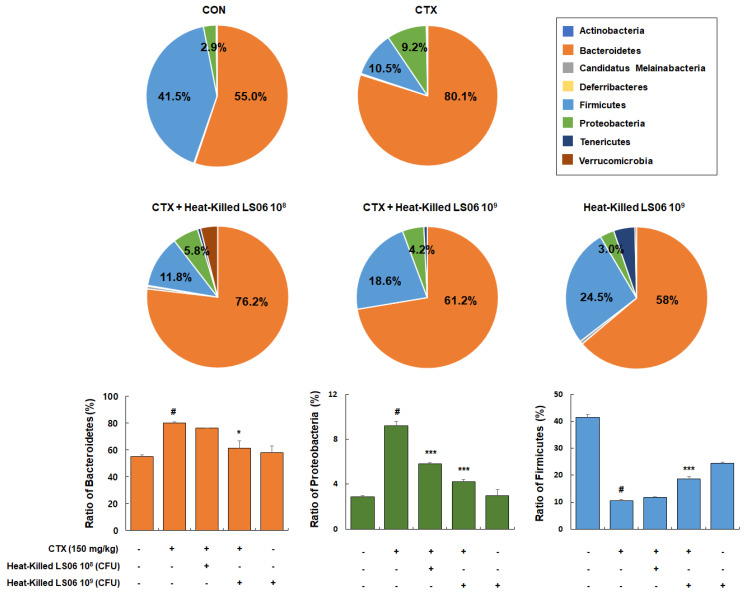
Besides being a potent immunosuppressant, CTX also reduces the intestinal tight and adherens junctions and triggers the increase in pathogenic bacteria. Hence, it is important in modulating the intestinal microbiota composition and immune function [18]. We thus determined the effects that heat-killed LS06 provide on the gut microbiota composition in CTX-treated mice. Dominant bacteria phyla mainly consist of Bacteroidetes (55.0% ± 1.3%), Firmicutes (41.5% ± 1.1%), and Proteobacteria (2.9% ± 0.1%, Figure 8). The CTX-treated group showed relative ratio of Bacteroidetes (80.1% ± 1.0%), Firmicutes (10.5% ± 0.5%), and Proteobacteria (9.2% ± 0.4%), while high-dose heat-killed LS06 supplementation to CTX-treated mice significantly altered relative abundance ratio of Bacteroidetes (61.2% ± 5.6%), Proteobacteria (4.2% ± 0.2%), and Firmicutes (18.6% ± 0.8%).
Figure 8.
Effects of heat-killed LS06 treatment on the composition of microbiota in CTX-treated mice. Genomic DNA was analyzed for bacterial composition using 16S rRNA gene sequencing. Data are presented as mean ± SEM (n = 10). # p < 0.05 vs. Con group; * p < 0.05 and *** p < 0.001 vs. CTX group.
Among families of the dominant Bacteroidetes phylum, the relative abundance ratio of Muribaculaceae, Prevotellaceae, and Tannerellaceae increased in the CTX-treated group compared to the Con group (40.4 ± 5.6%, 13.2 ± 1.7%, and 1.1 ± 0.3%, respectively), but these increases were significantly suppressed by the administration of high-dose heat-killed LS06 (31.8 ± 0.9%, 6.6 ± 0.2%, and 0.1 ± 0.02%, respectively) (Figure 9A). Furthermore, among bacterial families of the Firmicutes phylum, the relative abundance ratio of Christensenellaceae and Gracilibacteraceae significantly suppressed in the CTX-treated group compared to the Con group (0.06 ± 0.01% and 0.01 ± 0.009%, respectively), but this suppression was improved by high-dose heat-killed LS06 supplementation (0.11 ± 0.03% and 0.05 ± 0.003%, respectively) (Figure 9B). Conversely, the relative abundance rate of Hungateiclostridiaceae, which was increased by CTX treatment (0.89 ± 0.14%), was significantly reduced following a high-dose heat-killed LS06 administration (0.33 ± 0.09%). These results suggest that heat-killed LS06 administration could effectively improve the CTX-modulated gut microbiota composition and mediate immunomodulation.
Figure 9.
Effects of heat-killed LS06 treatment on family’s composition in CTX-treated mice. Relative abundance ratio of families of (A) Bacteroidetes and (B) Firmicutes. Data are presented as mean ± SEM (n = 10). # p < 0.05 vs. Con group; * p < 0.05, ** p < 0.01, and *** p < 0.001 vs. CTX group.
4. Discussion
Probiotics are microorganisms that generate a wide range of beneficial biological responses in their host, including immunostimulatory effects [20]. In our previous study, live K040706 regulated immune and inflammatory responses in vitro and in vivo experiments [11,12]. Although probiotics are primarily live microbial agents, several concerns remain about the possibility of unwanted side effects, including a higher risk of infection in sick or immunocompromised patients and very young individuals, ignition of an inflammatory response, and translocation to the locally draining tissues and blood [21]. Another concern is the safety aspects of bacteria including the acquisition of virulence genes or antimicrobial resistance [22]. These problems lead to develop alternative agents, such as probiotics killed by heat. Previous studies have demonstrated that oral rehydration therapy by Latilactobacillus acidophilus LB is effective in the treatment of children with acute diarrhea [23] and LPS-induced inflammation can be modulated by heat-killed Latilactobacillus rhamnosus GG [24]. Other studies have shown that heat-killed probiotics exhibit immunomodulatory properties under various temperature and time conditions [25,26].
There are several methods to identify LAB in probiotics and dairy products. However, these methods have only been used on living bacteria. In this study, we present a method of detecting heat-killed LAB by using novel genetic markers of L. sakei K040706 and strain-specific PCR primers based on comparative genomics. Genomic DNA can be extracted after heat inactivation and detected by real-time PCR.
Before the spread of whole-genome sequencing, random sequence (RAPD-PCR), amplification of repetitive sequences (Rep-PCR), or amplified length polymorphism analysis were used to classify bacteria. These methods are time-consuming, labor-intensive, and have low reproducibility [27]. Hence, to screen strain-specific sequences, pairwise alignment of the genome and pan-genome analysis is a more powerful tool [28,29,30]. In this study, the unique genetic markers (K040706..2017, K040706..2018, and K040706..2019) of L. sakei K040706 were selected and encoded into DNA modification methylase and two hypothetical proteins, respectively. Using BLASTN search, these proteins were identified as coding sequences with low DNA sequence homology. When the strain-specific primer was subjected to PCR with other L. sakei strains, K040706 was successfully validated.
The thymus and spleen are two vital organs in human immune regulation. The thymus is an important regulator of peripheral immune organs and immune cells, providing a place for T cell development [31]. T cells are involved in immune responses against pathogens and some cancers. In contrast, the spleen is the production site of T and B cells and can synthesize antibodies against blood-borne antigens [32]. It is well known that CTX-induced immunosuppression leads to weight loss in the thymus and spleen, reflecting the impairment of immune cells and suppressing the number of T and B cells [33,34]. In the present study, oral administration of heat-killed LS06 improved the CTX- reduced body weight, thymus index, and splenic T and B cell proliferation, indicating the recovery of immune function. Stimulating immune cells secrete cytokines, which are small-molecule soluble proteins with immune functions. After T cell differentiation into effector cells, CD4+ Th cells are divided into distinct cell types, Th1 and Th2, which secrete different types of cytokines to modulate the immune system. Th1 cells produce pro-inflammatory cytokines such as IFN-γ, IL-2, and IL-12, which stimulate phagocytosis, destroy pathogens, and are involved in organ-specific autoimmune diseases [35]. In contrast, Th2 cytokines produce the cytokines IL-4, IL-10, and IL-13, which stimulate the production of antibodies directed toward large extracellular parasites, including allergies. Hence, the balance between Th1 and Th2 cytokine production can determine the outcome of an immune response. CTX administration leads to intestinal cellular immune dysfunction, resulting in Th1/Th2 imbalance. In this study, we demonstrated that although heat-killed LS06 supplementation had no significant effect on inducing Th2-related cytokines production, the mRNA and protein expression of Th1-related cytokines recovered. This indicates that Th1 cytokines are more potently affected by heat-killed LS06 than Th2. We thus considered that heat-killed LS06-related immune improvement could modulate the Th1/Th2 balance in CTX-induced immunosuppressed mice.
The gut microbial ecosystem harbors trillions of taxonomically-diverse microorganisms that perform a range of physiological functions in the gastrointestinal tract of the host [36]. Imbalances in the gut microbial community are associated with various clinical conditions, including CTX-induced immunosuppression. CTX is a medicine for chemotherapy with evidence of efficacy and safety. However, a high-dose of CTX can lead to hepatotoxicity, mucosal barrier disruption, and an increase of pathogenic bacteria in the intestine [37]. In the present study, CTX changed the microbiota composition, including alteration in the numbers of Bacteroidetes and Firmicutes, which are the two most predominant phyla and account for almost 90% of total bacteria [37]. The CTX-induced changes in Bacteroidetes and Firmicutes were consistent with previous studies suggesting that alteration of phylum abundance may promote immunosuppression and disruption of gut homeostasis [38]. However, the heat-killed LS06 reverted the composition of phyla in the immunosuppressed mice to normal. We sequentially analyzed the abundance of Muribaculaceae, Prevotellaceae, Tannerellaceae, Christensenellaceae, Gracilibacteraceae, and Hungateiclostridiaceae. Youlong et al. demonstrated that Muribaculaceae was positively related to immune traits, whereas Prevotellaceae and Tannerellaceae were negatively correlated with immune traits [38]. In addition, members of the Tannerellaceae family are most abundant in certain gastrointestinal disorders, such as Crohn’s disease [39]. Our study showed that CTX injection significantly increased the relative abundance of Muribaculaceae, Prevotellaceae, and Tannerellaceae, but heat-killed LS06 intervention reversed these changes. CTX administration has been reported to decrease the abundance of Christensenellaceae, one of five taxa regarded as a sign of a healthy gut [40,41]. However, the abundance of Gracilibacteraceae and Hungateiclostridiaceae in the gut microbiota after CTX injection has not been elucidated. We found that CTX administration also reduced the abundance of Gracilibacteraceae and Hungateiclostridiaceae. Further, Christensenellaceae, Gracilibacteraceae, and Hungateiclostridiaceae strains in CTX-treated mice significantly increased after oral administration of heat-killed LS06. Since the compositional diversity and abundance of microbiota are fundamental to the maintenance of the immune system, further studies are needed to determine how the heat-killed LS06 performs its immunostimulatory activity to maintain immune homeostasis.
5. Conclusions
In conclusion, we demonstrated the development of a strain-specific primer and the successful quantification of heat-killed LS06 with high specificity. Further, we showed that the oral administration of heat-killed LS06 ameliorated CTX-induced suppression of immunity by recovering body weight, thymus index, immune cell proliferation, and Th1-related cytokines productions. Heat-killed LS06 improves immune responses which may be related to an alteration of the microbiota composition to normal. Therefore, heat-killed L. sakei K040706 may be utilized as an ingredient of functional food for health benefits via immune modulation and regulation of gut microbiota.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/foods10123034/s1, Table S1: The list of primer sequences for qRT-PCR.
Author Contributions
Conceptualization, J.W.C., J.-S.S., S.-Y.K. (Seo-Yeon Kim), J.-Y.K., Y.K.R. and K.-T.L.; Data curation, J.W.C., J.-S.S. and S.-Y.K. (Su-Yeon Kim); Formal analysis, K.-S.C. and J.-S.S.; Funding acquisition, J.-Y.K., Y.K.R. and K.-T.L.; Investigation, J.-S.S., S.-Y.K. (Seo-Yeon Kim) and H.-S.H.; Methodology, K.-S.C., J.W.C., J.-S.S., S.-Y.K. (Seo-Yeon Kim), H.-S.H., S.-Y.K. (Su-Yeon Kim), K.-Y.L., J.-Y.K., C.-W.C. and H.-D.H.; Project administration, J.-S.S., J.-Y.K. and Y.K.R.; Resources, J.-Y.K. and Y.K.R.; Supervision, K.-T.L.; Validation, J.-S.S.; Visualization, K.-S.C., J.W.C. and S.-Y.K. (Seo-Yeon Kim); Writing—original draft, K.-S.C., J.W.C. and J.-S.S.; Writing—review & editing, K.-S.C. and K.-T.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Medical Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2017R1A5A2014768), Main Research Program(E0201100) of the Korea Food Research Institute (KFRI), the technology transfer and commercialization Program(2020-JB-RD-125) through INNOPOLIS Foundation funded by the Ministry of Science and ICT, and Korea Industrial Complex Corp. (KICOX) funded by the Ministry of Trade, Industry, and Energy (MOTIE, RCC18005).
Institutional Review Board Statement
All animal experiments were performed under university guidelines and were approved by the ethical committee for Animal Care and Use of Kyung Hee University according to the animal protocol (KHUASP(SE)-18-158).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dhama K., Saminathan M., Jacob S.S., Singh M., Karthik K., Amarpal T.R., Malik Y.S., Singh R.K. Effect of Immunomodulation and immunomodulatory agents on health with some bioactive principles, modes of action and potent biomedical applications. Int. J. Pharmacol. 2015;11:253–290. doi: 10.3923/ijp.2015.253.290. [DOI] [Google Scholar]
- 2.Romagnani S. Th1/Th2 cells. Inflamm. Bowel. Dis. 1999;5:285–294. doi: 10.1097/00054725-199911000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Walker J.A., McKenzie A.N.J. TH2 cell development and function. Nat. Rev. Immunol. 2018;18:121–133. doi: 10.1038/nri.2017.118. [DOI] [PubMed] [Google Scholar]
- 4.Bascones-Martinez A., Mattila R., Gomez-Font R., Meurman J.H. Immunomodulatory drugs: Oral and systemic adverse effects. Med. Oral Patol. Oral Cir. Bucal. 2014;19:e24. doi: 10.4317/medoral.19087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidd P. Th1/Th2 balance: The hypothesis, its limitations, and implications for health and disease. Altern. Med. Rev. 2003;8:223–246. [PubMed] [Google Scholar]
- 6.Jantan I., Ahmad W., Bukhari S.N.A. Plant-derived immunomodulators: An insight on their preclinical evaluation and clinical trials. Front. Plant. Sci. 2015;6:655. doi: 10.3389/fpls.2015.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stackebrandt E., GOEBEL B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 1994;44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- 8.Vandamme P., Pot B., Gillis M., De Vos P., Kersters K., Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Sullivan D.J. Methods for analysis of the intestinal microflora. Curr. Issues Intest. Microbiol. 2000;1:39–50. [PubMed] [Google Scholar]
- 10.Choi J.W., Rhee Y.K., Hong H.D., Cho C.W., Lee Y.C., Lim T.G., Kim J.S. A primer set for Specifically Detecting Lactobacillus sakei and uses thereof. No. 1021454080000. K.R. Patent. 2020 August 11;
- 11.Jung J.Y., Shin J.S., Lee S.G., Rhee Y.K., Cho C.W., Hong H.D., Lee K.T. Lactobacillus sakei K040706 evokes immunostimulatory effects on macrophages through TLR 2-mediated activation. Int. Immunopharmacol. 2015;28:88–96. doi: 10.1016/j.intimp.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 12.Seo S.H., Shin J.S., Lee W.S., Rhee Y.K., Cho C.W., Hong H.D., Lee K.T. Anti-colitis effect of Lactobacillus sakei K040706 via suppression of inflammatory responses in the dextran sulfate sodium-induced colitis mice model. J. Funct. Foods. 2017;29:256–268. doi: 10.1016/j.jff.2016.12.045. [DOI] [Google Scholar]
- 13.Kim S.Y., Shin J.S., Chung K.S., Han H.S., Lee H.H., Lee J.H., Kim S.Y., Ji Y.W., Ha Y., Kang J. Immunostimulatory effects of live Lactobacillus sakei K040706 on the CYP-induced immunosuppression mouse model. Nutrients. 2020;12:3573. doi: 10.3390/nu12113573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon S.H., Ha S.M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin J.S., Chung S.H., Lee W.S., Lee J.Y., Kim J.L., Lee K.T. Immunostimulatory effects of cordycepin-enriched WIB-801CE from Cordyceps militaris in splenocytes and cyclophosphamide-induced immunosuppressed mice. Phytother. Res. 2018;32:132–139. doi: 10.1002/ptr.5960. [DOI] [PubMed] [Google Scholar]
- 16.Han H.S., Shin J.S., Song Y.R., Rhee Y.K., Cho C.W., Ryu J.H., Inn K.S., Hong H.D., Lee K.T. Immunostimulatory effects of polysaccharides isolated from young barley leaves (Hordeum vulgare L.) with dual activation of Th1 and Th2 in splenic T cells and cyclophosphamide-induced immunosuppressed mice. Int. J. Biol. Macromol. 2020;147:954–964. doi: 10.1016/j.ijbiomac.2019.10.062. [DOI] [PubMed] [Google Scholar]
- 17.Na S.-I., Kim Y.O., Yoon S.-H., Sung-min H., Baek I., Chun J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018;56:281–285. doi: 10.1007/s12275-018-8014-6. [DOI] [PubMed] [Google Scholar]
- 18.Belkaid Y., Hand T.W. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerqueira F., Cordeiro-Da-Silva A., Gaspar-Marques C., Simoes F., Pinto M.M., Nascimento M.S. Effect of abietane diterpenes from Plectranthus grandidentatus on T- and B-lymphocyte proliferation. Bioorg. Med. Chem. 2004;12:217–223. doi: 10.1016/j.bmc.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Gill H.S., Guarner F. Probiotics and human health: A clinical perspective. Postgrad. Med. J. 2004;80:516–526. doi: 10.1136/pgmj.2003.008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kataria J., Li N., Wynn J.L., Neu J. Probiotic microbes: Do they need to be alive to be beneficial? Nutr. Rev. 2009;67:546–550. doi: 10.1111/j.1753-4887.2009.00226.x. [DOI] [PubMed] [Google Scholar]
- 22.Wassenaar T.M., Klein G. Safety aspects and implications of regulation of probiotic bacteria in food and food supplements. J. Food Prot. 2008;71:1734–1741. doi: 10.4315/0362-028X-71.8.1734. [DOI] [PubMed] [Google Scholar]
- 23.Lievin-Le Moal V., Sarrazin-Davila L.E., Servin A.L. An experimental study and a randomized, double-blind, placebo-controlled clinical trial to evaluate the antisecretory activity of Lactobacillus acidophilus strain LB against nonrotavirus diarrhea. Pediatrics. 2007;120:e795–e803. doi: 10.1542/peds.2006-2930. [DOI] [PubMed] [Google Scholar]
- 24.Li N., Russell W.M., Douglas-escobar M., Hauser N., Lopez M., Neu J. Live and heat-killed Lactobacillus rhamnosus GG: Effects on proinflammatory and anti-inflammatory cytokines/chemokines in gastrostomy-fed infant rats. Pediatr. Res. 2009;66:203–207. doi: 10.1203/PDR.0b013e3181aabd4f. [DOI] [PubMed] [Google Scholar]
- 25.Matsuzaki T., Yamazaki R., Hashimoto S., Yokokura T. The effect of oral feeding of Lactobacillus casei strain Shirota on immunoglobulin E production in mice. J. Dairy Sci. 1998;81:48–53. doi: 10.3168/jds.S0022-0302(98)75549-3. [DOI] [PubMed] [Google Scholar]
- 26.Ou C.C., Lin S.L., Tsai J.J., Lin M.Y. Heat-killed lactic acid bacteria enhance immunomodulatory potential by skewing the immune response toward Th1 polarization. J. Food Sci. 2011;76:M260–M267. doi: 10.1111/j.1750-3841.2011.02161.x. [DOI] [PubMed] [Google Scholar]
- 27.Treven P. Strategies to develop strain-specific PCR based assays for probiotics. Benef. Microbes. 2015;6:887–898. doi: 10.3920/BM2015.0009. [DOI] [PubMed] [Google Scholar]
- 28.Shen-Gunther J., Xia Q., Stacey W., Asusta H.B. Molecular pap smear: Validation of HPV genotype and host methylation profiles of ADCY8, CDH8, and ZNF582 as a predictor of cervical cytopathology. Front. Microbiol. 2020;11:595902. doi: 10.3389/fmicb.2020.595902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim E., Yang S.M., Cho E.J., Kim H.Y. Novel real-time PCR assay for Lactobacillus casei group species using comparative genomics. Food Microbiol. 2020;90:103485. doi: 10.1016/j.fm.2020.103485. [DOI] [PubMed] [Google Scholar]
- 30.Chenchen Z., Xingtong Y., Dai W., Ya G., Chunlei W., Qiming L., Jiaqi W., Boxing Y., Zhiming P., Ruixia G. Rapid strain-specific identification of two Lactobacillus rhamnosus strains using PCR based on gene family analysis. LWT. 2021;146:111395. [Google Scholar]
- 31.Thapa P., Farber D.L. The role of the thymus in the immune response. Thorac. Surg. Clin. 2019;29:123–131. doi: 10.1016/j.thorsurg.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie Z., Bai Y., Chen G., Rui Y., Chen D., Sun Y., Zeng X., Liu Z. Modulation of gut homeostasis by exopolysaccharides from Aspergillus cristatus (MK346334), a strain of fungus isolated from Fuzhuan brick tea, contributes to immunomodulatory activity in cyclophosphamide-treated mice. Food Funct. 2020;11:10397–10412. doi: 10.1039/D0FO02272A. [DOI] [PubMed] [Google Scholar]
- 33.Thomson A.W., Mathie I.H., Sewell H.F. Cyclophosphamide-induced eosinophilia in the rat: Concomitant changes in T-cell subsets, B cells and large granular lymphocytes within lymphoid tissues. Immunology. 1987;60:383–388. [PMC free article] [PubMed] [Google Scholar]
- 34.Misra R.R., Bloom S.E. Roles of dosage, pharmacokinetics, and cellular sensitivity to damage in the selective toxicity of cyclophosphamide towards B and T cells in development. Toxicology. 1991;66:239–256. doi: 10.1016/0300-483X(91)90196-8. [DOI] [PubMed] [Google Scholar]
- 35.Delcenserie V., Martel D., Lamoureux M., Amiot J., Boutin Y., Roy D. Immunomodulatory effects of probiotics in the intestinal tract. Curr. Issues Mol. Biol. 2008;10:37–54. [PubMed] [Google Scholar]
- 36.Guo H., Chou W.-C., Lai Y., Liang K., Tam J.W., Brickey W.J., Chen L., Montgomery N.D., Li X., Bohannon L.M. Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science. 2020;370:9097. doi: 10.1126/science.aay9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Routy B., Letendre C., Enot D., Chénard-Poirier M., Mehraj V., Séguin N.C., Guenda K., Gagnon K., Woerther P.-L., Ghez D. The influence of gut-decontamination prophylactic antibiotics on acute graft-versus-host disease and survival following allogeneic hematopoietic stem cell transplantation. Oncoimmunology. 2017;6:e1258506. doi: 10.1080/2162402X.2016.1258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y., Yan Y., Zhou W., Chen D., Huang K., Yu S., Mi J., Lu L., Zeng X., Cao Y. Effects of polysaccharides from bee collected pollen of Chinese wolfberry on immune response and gut microbiota composition in cyclophosphamide-treated mice. J. Funct. Foods. 2020;72:104057. doi: 10.1016/j.jff.2020.104057. [DOI] [Google Scholar]
- 39.Hernández M., de Frutos M., Rodríguez-Lázaro D., López-Urrutia L., Quijada N.M., Eiros J.M. Fecal microbiota of toxigenic Clostridioides difficile-associated diarrhea. Front. Microbiol. 2019;9:3331. doi: 10.3389/fmicb.2018.03331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Sun M., Jin H., Yang J., Kang S., Liu Y., Yang S., Ma S., Ni J. Effects of Lycium barbarum polysaccharides on immunity and the gut microbiota in cyclophosphamide-induced immunosuppressed mice. Front. Microbiol. 2021;12:701566. doi: 10.3389/fmicb.2021.701566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mancabelli L., Milani C., Lugli G.A., Turroni F., Cocconi D., van Sinderen D., Ventura M. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol. Ecol. 2017;93:fix153. doi: 10.1093/femsec/fix153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.