Abstract
The thermogenic peroxisome proliferator-activated receptor γ (PPAR-γ) coactivator 1 (PGC-1) has previously been shown to activate mitochondrial biogenesis in part through a direct interaction with nuclear respiratory factor 1 (NRF-1). In order to identify related coactivators that act through NRF-1, we searched the databases for sequences with similarities to PGC-1. Here, we describe the first characterization of a 177-kDa transcriptional coactivator, designated PGC-1-related coactivator (PRC). PRC is ubiquitously expressed in murine and human tissues and cell lines; but unlike PGC-1, PRC was not dramatically up-regulated during thermogenesis in brown fat. However, its expression was down-regulated in quiescent BALB/3T3 cells and was rapidly induced by reintroduction of serum, conditions where PGC-1 was not detected. PRC activated NRF-1-dependent promoters in a manner similar to that observed for PGC-1. Moreover, NRF-1 was immunoprecipitated from cell extracts by antibodies directed against PRC, and both proteins were colocalized to the nucleoplasm by confocal laser scanning microscopy. PRC interacts in vitro with the NRF-1 DNA binding domain through two distinct recognition motifs that are separated by an unstructured proline-rich region. PRC also contains a potent transcriptional activation domain in its amino terminus adjacent to an LXXLL motif. The spatial arrangement of these functional domains coincides with those found in PGC-1, supporting the conclusion that PRC and PGC-1 are structurally and functionally related. We conclude that PRC is a functional relative of PGC-1 that operates through NRF-1 and possibly other activators in response to proliferative signals.
Nuclear respiratory factor 1 (NRF-1) was originally identified as a nuclear transcription factor that trans-activates the promoters of a number of mitochondrion-related genes in vitro (5, 9,10, 31). These include respiratory subunits and the rate-limiting heme biosynthetic enzyme, as well as factors involved in the replication and transcription of mitochondrial DNA (reviewed in reference 25). In addition to its proposed role in respiratory chain expression, NRF-1 has also been implicated in other cellular functions. Most recently, genes encoding two rate-limiting enzymes in purine nucleotide biosynthesis (6), a receptor involved in chemokine signal transduction (32), a subunit of a neural receptor (20), and the human polio virus receptor CD155 (27) were all shown to have functional NRF-1 binding sites in their promoters. Moreover, we recently established that targeted disruption of the NRF-1 gene in mice results in early embryonic lethality associated with a deficiency in mitochondrial DNA (15). These observations are consistent with a broad role for NRF-1 in growth and development.
NRF-1 has recently been implicated in the transcriptional control of mitochondrial biogenesis during adaptive thermogenesis through its interaction with the cold-inducible coactivator, PGC-1 (for peroxisome proliferator-activated receptor γ [PPAR-γ] coactivator 1) (33). This protein was originally cloned as an interacting partner of the nuclear hormone receptor PPAR-γ by two-hybrid screening (22). It was also shown to have a broad specificity for interaction with several nuclear hormone receptors (22) and more recently was found to interact with PPAR-α in the transcription of nuclear genes encoding mitochondrial fatty acid oxidation enzymes (30). Interestingly, PGC-1 is predominantly expressed in heart, brown adipose tissue (BAT), skeletal muscle (SKM), kidney, and to some extent liver (16, 22), tissues with abundant mitochondria. Furthermore, its expression is rapidly induced in cold-exposed animals (22), consistent with a role in mitochondrial biogenesis.
Ectopic overexpression of PGC-1 in both NIH 3T3 cells and the myogenic cell line C2C12 resulted in increased expression of both nuclear and mitochondrial genes encoding mitochondrial proteins (33). Many of these genes are either directly or indirectly controlled by NRF-1 and/or NRF-2 (reviewed in reference 25). Overexpression of PGC-1 resulted in increased expression of both NRF-1 and -2 mRNA, and PGC-1 interacted physically with NRF-1 to augment transcriptional activation of NRF-1-dependent promoters. Furthermore, expression of a dominant negative NRF-1 inhibited the PGC-1-mediated increase in mitochondrial biogenesis (33). These findings have recently been extended to cultured cardiomyocytes and to cardiac tissue in vivo (17). Most notably, heart-specific overexpression of PGC-1 in transgenic mice led to excessive mitochondrial proliferation, resulting in cardiac pathology. Taken together, the results support an important role for the interplay between NRF-1 and PGC-1 in the physiological control of respiratory chain expression.
The expression of PGC-1 is limited to certain tissues and physiological conditions. Thus, it was of interest to determine whether there are other regulated coactivators that function through NRF-1 and display different physiological and/or tissue specificities. Here, we describe the characterization of a novel PGC-1-related coactivator (PRC) that is expressed in a cell cycle-dependent fashion. PRC is functionally related to PGC-1 in that it interacts directly with NRF-1, has a potent amino-terminal transcriptional activation domain, and requires NRF-1 to activate NRF-1 target genes.
MATERIALS AND METHODS
Plasmids.
A 4.8-kb KIAA0595 cDNA containing a large open reading frame and cloned into SalI/NotI of pBluescript II SK(+) (KIAA0595/pBSII) was obtained from the Kazusa DNA Research Institute. No function was assigned to this cDNA, and we refer to it here as the PRC cDNA because of its structural and functional relatedness to PGC-1. To obtain the 5′ end of PRC, Marathon-ready cDNAs from human brain and testis were amplified using Marathon adapter primers AP1 and AP2 (Clontech). Antisense gene-specific primers, PRCrev1 (5′-GAG GTG GGG ATT GGC CCC AGC GTG G-3′) and the nested primer PRCrev2 (5′-CTC CAG CTA GGA AGC TTG GGG GAA-3′), were designed from the 5′ end of the truncated PRC cDNA. The nested PCR products from both tissues were subsequently ligated into pGEM-T (Promega), sequenced, and found to be identical. This plasmid was digested with NotI (a site present in the Marathon adapter), blunted with Klenow enzyme, and digested with a unique HindIII site in PRC. This released a 0.7-kb fragment corresponding to the 5′ end of PRC. This fragment was ligated into KIAA0595/pBSII, which had been cut with XhoI, blunted, and then cut with HindIII, thus creating plasmid FL-PRC/pBSII. Ligation of the blunt NotI site in the 5′ end cDNA with the blunted XhoI site in KIAA0595/pBSII recreated the XhoI site in the 5′ end of the plasmid FL-PRC/pBSII.
A 5.3-kb XhoI/NotI fragment from FL-PRC/pBSII, containing the full-length cDNA of PRC, was ligated into SalI/NotI of pSV-SPORT1 to create the mammalian expression vector FL-PRC/pSV-SPORT. Bacterial expression of recombinant PRC was accomplished using the pET32 series (Novagen), which expresses proteins as S-tagged thioredoxin fusions. PRC(N221)/pET32b was made by inserting a 670-bp XhoI/HindIII fragment of FL-PRC/pBSII into the SalI/HindIII sites of pET32b. PRC(95–533)/pET32a was made by first introducing a BamHI site in front of amino acid residue 95 by means of PCR, using sense primer BamPRC95s (5′-TGG GAT CCA TGC AGA GCT ACA TGG ATG-3′) and antisense primer PRCrev1with FL-PRC/pBSII as template. The PCR product was digested with BamHI/HindIII, combined with a 1-kb HindIII/EcoRI fragment from FL-PRC/pBSII, and ligated into the BamHI/EcoRI site of pET32a. PRC(529–1022)/pET32c was made by ligating a 1.5-kb PvuII fragment from FL-PRC/pSV-SPORT into the EcoRV site of pET32c. PRC(739–1047)/pET32c and PRC(1047–1379)/pET32c were made by first isolating a 1.9-kb Acc65I/StuI fragment from FL-PRC/pBSII and then digesting this fragment with SacI. The resulting 997-bp Acc65I/SacI and 925-bp SacI/StuI fragments were ligated into Acc65I/SacI- and SacI/XhoI-digested pET32c, respectively (the XhoI site was blunted with Klenow enzyme prior to EcoRI digestion). PRC(1379–1664)/pET32c was made by ligating a 1.2-kb StuI/NotI fragment from FL-PRC/pBSII into the EcoRV/NotI sites of pET32c.
To map the activation domain of PRC, a series of amino-terminal Gal4 DNA binding domain fusions was constructed using the vector pSG424 (24). For carboxy-terminal deletions, a suitable restriction site for in-frame cloning into pSG424 was introduced. To this end, a 2.4-kb fragment from FL-PRC/pBSII was amplified using sense primer 5′-TCT CGA GGA TCC AGA TGG CGG CGC GCC GGG GA-3′ and antisense primer 5′-TGC CTG GGG CTG GTG GGA TGA CAA G-3′, thereby creating both an XhoI and a BamHI site in frame with the Gal4 DNA binding domain (Gal4DBD) just upstream from the start codon of PRC (shown in boldface). This fragment was then cloned back into FL-PRC/pBSII by digesting the product with XhoI and HindIII, purifying the 0.7-kb fragment, and ligating it into similarly digested FL-PRC/pBSII, thus creating FL-Bam-PRC/pBSII. This plasmid was then digested to create a series of carboxy-terminal deletions in PRC/pSG424 as follows (designations for the deletions are in parentheses): BamHI/DraI (N-C), BamHI/StuI (N-1379), BamHI/Acc65I (N-739), BamHI/XbaI (N-550), BamHI/HindIII (N-221), and BamHI/BglII (the BglII site was blunted with Klenow enzyme prior to BamHI digestion) (N-133). Two amino-terminal deletions of PRC, 133–550 and 221–550 in pSG424, were constructed by first linearizing FL-PRC/pBSII with BglII (at amino acid [aa] 133) or HindIII (at aa 221). The overhangs were blunted with Klenow enzyme and then digested with XbaI (at aa 550). The released fragments were then ligated in frame with SmaI/XbaI-digested pSG424. Two additional amino-terminal deletions were created by inserting an EcoRI site in frame with Gal4DBD by PCR with sense primers 5′-GCT GAA TTC ACG ATG TCT AGC CCT AAG AAC-3′ and 5′-TGC GAA TTC ACC ATG AAC ACT AGG ACT CCC-3′ and, as antisense primer, the T3 promoter primer (in pBluescript II SK). The products were ligated into pGEM-T, thus creating PRC(535-C)/pGEM5Zf and PRC(1387-C)/pGEM5Zf. A 3.5-kb EcoRI/DraI (DraI beyond the PRC stop codon) fragment of PRC(535-C)/pGEM5Zf was isolated and ligated into an EcoRI/XbaI-digested pSG424 (where the XbaI site was blunted with Klenow enzyme prior to EcoRI digestion), thus creating PRC(535-C)/pSG424. A 1-kb EcoRI/SacI fragment of PRC(1387-C)/pGEM5Zf (where the SacI site came from the multiple cloning site of pBSII) was ligated into an EcoRI/SacI-digested pSG424, thus creating PRC(1387-C)/pSG424.
The rat δ-aminolevulinate synthase (δ-ALAS) promoter was constructed by PCR using primers 5′-AAC TGC AGC CCC TTA GCA TCT-3′ and 5′-GAA TGG GCA TCT GCG AAC GAC-3′ based on the published sequence (4). The resulting PCR product was subcloned into pGEM-T, and the sequence was verified. A PstI (in the forward primer) and SmaI (in the 5′ untranslated region of the 5-ALAS gene) fragment from this plasmid was further subcloned into pBluescript that had been digested with PstI/EcoRV. An XbaI/HindIII fragment from this plasmid was inserted into pGL3 basic (Promega) that had been digested with NheI/HindIII, yielding the pGL3/δ-ALAS(-479) plasmid. The two NRF-1 sites in pGL3/δ-ALAS(-479) were mutated with mutagenesis primers 5′-GGC CGA CTC CGG TGC ATG TAT GCG CGG CAG GCC GC-3′ and 5′-GCC GCA CCC ACA GCA TAT ATG CAG CGG TCA CCC CCG-3′ (the mutated nucleotides are shown in boldface letters) with the QuikChange Site-Directed Mutagenesis kit (Stratagene), thus generating pGL3/ALAS(m1) and pGL3/ALAS(m2), respectively. The double mutant pGL3/ALAS(m1m2) was constructed in the same manner.
For RNase protection assays of murine cells, a 1,552-bp mouse PRC cDNA, homologous to positions 3325 to 4877 of the human sequence, was amplified from mouse liver cDNA with sense primer 5′-TGA GAC CCA GGA GAA CAG ACC AAA GGA GA-3′ and antisense primer 5′-TTC GGC CCC CAA AGC AGA GAT-3′, designed from a compilation of expressed sequence tag clones displaying more than 80% nucleic acid identity with human PRC. The PCR product was cloned into pGEM5Zf (Promega), thus creating plasmid mPRC/pGEM5Zf. Sequencing revealed >90% identity to PRC at the nucleic acid level and >95% identity at the amino acid level and the construct could thus safely be considered to be the mouse homologue to human PRC. mPRC/pGEM5Zf was linearized with Tth111I for riboprobe synthesis. PGC-1(0.6)/pBluescript was made by inserting an EcoRI fragment from PGC-1/pSV-SPORT (22) into pBluescript SK(+). This plasmid was linearized with NheI for riboprobe synthesis. Other plasmids used in the RNase protection assay (mouse cytochrome c, rat cytochrome oxidase subunit IV [COXIV], and mouse NRF-1) have been described elsewhere (14, 15).
The plasmids 4xNRF1/Luc, pSG5/NRF-1 and its deletion mutants used in the S-tag pull-down assay, pSG5/NRF-1–3xHA, 5xGal/Luc, and the RC4/-326 promoter series have all been described previously (11, 31, 33).
Cell culture and transfections.
BALB/3T3, COS, and C6 glioma cells were maintained in Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with 10% calf serum (HyClone) and 1% penicillin–streptomycin solution (Sigma). C2C12 and HepG2 cells were maintained in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS) (HyClone) and 1% penicillin–streptomycin solution (Sigma). For transient transfections, cells were plated at appropriate density 24 h prior to transfection and transfected by a standard calcium phosphate precipitation protocol (2). After 6 to 8 h, the cells were washed, fed, and, following an additional 40-h incubation, harvested. Cell extracts were prepared, and luciferase assays were performed with reagents purchased from PharMingen. Spectrophotometric β-galactosidase assays were performed with the β-galactosidase enzyme assay system (Promega). For immunostaining, the cells were trypsinized directly after transfection and reinoculated on chamber slides at the desired cell number as described below.
Serum starvation experiments with BALB/3T3 cells were performed as follows. A total of 875,000 cells were inoculated in 150-mm petri dishes, and the cells were grown for 48 h, except proliferating cells, which were harvested 24 h postinoculation. The medium was removed and replaced with starvation medium (DMEM supplemented with 0.5% FBS). Starved cells were harvested after 72 h in starvation medium. For serum induction, the medium was replaced with DMEM supplemented with 20% FBS, and the cells were harvested at various time points. The medium was changed 48 h after induction, and confluent cells were harvested 24 h later.
Cold induction.
Twenty-eight-day-old BALB/c mice were maintained at 23°C with water and food ad libitum. The mice were then placed at 4°C for the indicated times. BAT and the soleus muscle were dissected, minced with scissors, and homogenized in TRIzol with a VirTishear tissue homogenizer. Total RNA was isolated, and the RNase protection assay was performed with 10 μg of total RNA.
RNA and protein analysis.
For preparation of whole-cell extracts, cells were washed once in ice-cold phosphate-buffered saline (PBS) supplemented with 0.2 mM phenylmethylsulfonyl fluoride (PMSF) and 20 M N-acetyl-leu-leu-norleucinal (LLnL), scraped in 1 ml of the same buffer, and pelleted by a brief centrifugation. The cell pellet was lysed in NP-40 lysis buffer (150 mM NaCl; 50 mM Tris [pH 8]; 1% NP-40; 2.5 mM Na2V3O4; 5 mM NaF; 1 mM each PMSF, EDTA, benzamidine, and dithiothreitol; 1 mg each of pepstatin A, leupeptin, and aprotinin per ml; and 20 μM LLnL). The lysate was incubated for 20 min on ice, sonicated three times for 3 s at the lowest setting on a Branson Sonifier 450, and spun for 5 min at 20,000 × g to remove cell debris. Protein concentrations were measured by a Bio-Rad protein assay. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were performed according to established methods (2). Total RNA was isolated using TRIzol (Gibco), and 5 to 10 μg of total RNA was analyzed by an RNase protection assay performed as described previously (1).
Antibodies.
Anti-PRC antiserum was produced (Bethyl Laboratories, Montgomery, Tex.) with recombinant PRC expressed from PRC(95–533)/pET32 and purified using S-protein agarose (Novagen) as described below. Crude serum was affinity purified as described (12). Briefly, serum was diluted 1/10 in 10 mM Tris and passed through a CNBr-activated Sepharose 4B (Pharmacia) affinity column to which purified PRC(95–533)/thioredoxin had been coupled. The column was washed with 20 bed volumes of 10 mM Tris and 0.5 M NaCl, and antibodies were eluted with 100 mM glycine (pH 2) into 0.1 volume of 2.5 M Tris (pH 8). The eluate was dialyzed three times against 100 volumes of PBS with 0.1% sodium azide and concentrated. Because PRC was expressed as an S-tagged fusion protein to bacterial thioredoxin, the antibodies directed against thioredoxin were absorbed out by incubating the serum with thioredoxin immobilized on protein S-agarose in PBS overnight at 4°C. The resulting antibodies primarily recognized a 150-kDa protein in whole-cell extracts (see Fig. 2B). The antibodies were also reactive to overexpressed PRC in mammalian and bacterial cells (data not shown). Goat anti-NRF-1 antiserum (31) that was used to detect NRF-1 in coimmunoprecipitation experiments was affinity purified with recombinant NRF-1 as described above.
FIG. 2.
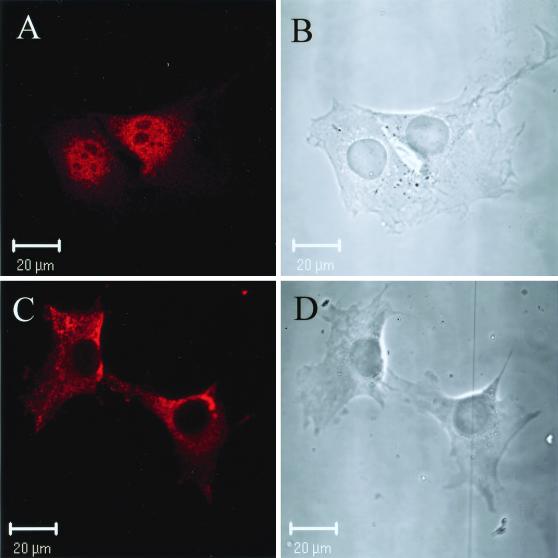
Nuclear localization of PRC by confocal immunofluorescence microscopy. (A) BALB/3T3 cells were stained with anti-PRC serum as the primary antibody. (B) The phase-contrast image of the results shown in panel A. (C) BALB/3T3 cells were stained with anti-PRC serum that had been preadsorbed with recombinant PRC [PRC(95–533)/thioredoxin]. (D) The phase-contrast image of results shown in panel C. Rhodamine-conjugated goat anti-rabbit IgG was used as the secondary antibody in the results shown in panels A and C.
S-tag pull-down assay.
The different PRC/pET32 plasmids were transformed into BL21 CodonPlus(DE3)-RIL cells (Stratagene), and a 1/100 dilution of overnight cultures was grown for 3 h. Isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mM was then added, and the cells were grown for an additional 2.5 h at room temperature. Cells were resuspended in 1/50 of the original culture volume in BLB (50 mM NaPO4 [pH 8.0], 300 mM NaCl, 10% glycerol, 0.1 mM EDTA, 0.1% Triton X-100, 10 mM β-mercaptoethanol, 1 mM each benzamidine and PMSF, and 1 μg each of pepstatin A, aprotinin, and leupeptin/ml) and treated for 30 min on ice with 1 mg of lysozyme/ml. The cells were then lysed by one freeze-thaw cycle and then made less viscous by six 10-s bursts on a Branson Sonifier 450 at 20% output level. Cell debris was removed by centrifugation at 20,000 × g for 20 min, and the integrity of expressed PRC protein fragments in the supernatant was analyzed by Western blotting with horseradish peroxidase-conjugated protein S (Novagen). The PRC fragments were then purified by combining 1 ml of the cleared lysate with 25 μl of protein S-agarose beads and incubated for 60 min at room temperature on a rocking table. The protein S-agarose was then washed four times in 1 ml of BLB with 1.3 M NaCl and then twice with BWB (20 mM Tris [pH 7.5], 150 mM NaCl, and 0.1% Triton X-100) and finally resuspended in 100 μl of BWB to make a 25% slurry. Approximately 1 μg of protein S-immobilized protein was then combined with nonbound protein S-agarose to a final volume of 25 μl, washed twice with BWB, and then resuspended in 45 μl of BWB. Five microliters of 35S-labeled in vitro-translated NRF-1 (TnT Reticulocyte Lysate system; Promega) was then added and incubated for 90 min at room temperature. The agarose beads were washed five times with 1 ml of BWB and finally eluted in 20 μl of SDS-PAGE sample buffer, separated on an SDS–10% PAGE gel, dried, and analyzed by autoradiography.
Coimmunoprecipitation.
Six 150-mm dishes of 50% confluent C2C12 cells were lysed in NP-40 lysis buffer as described above. Affinity-purified anti-PRC antibody (1 μg) or preimmune serum (1 μl) was added to 750 μg of cleared lysate in a total volume of 500 μl. Following 1 h of incubation on ice, 25 μl of protein A-agarose (Roche) was added, and the incubation was continued for another hour at 4°C on a rocking table. The immunoprecipitate was centrifuged at 4°C 150 × g for 1 min and washed four times with 1 ml of NP-40 lysis buffer. The anti-PRC precipitate was then eluted in 20 μl of SDS-PAGE loading buffer and run on an SDS–7.5% PAGE gel. After immunoblotting was done, the filter was probed with affinity-purified goat anti-NRF-1 serum (0.5 μg/ml).
Immunofluorescence microscopy.
Approximately 20,000 BALB/3T3 cells that in some cases had been transfected with pSG5/NRF1–3xHA as described above were plated on four-well glass chamber slides (Nalge Nunc). The cells were fixed 24 h later in ice-cold 4% paraformaldehyde in PBS for 16 h at 4°C. Fixed cells were subsequently blocked and permeabilized for 30 min at room temperature in PBS with 1% bovine serum albumin and 0.02% Triton X-100, which was then used in all following steps. Rabbit anti-PRC antibody (2 μg/ml) and mouse monoclonal antihemagglutinin antibody (1:600 dilution; Babco) were incubated for 60 min at room temperature. Cells were washed three times for 5 min each, and secondary antibodies were used at a 1:200 dilution for tetramethyl rhodamine isocyanate-conjugated goat anti-rabbit immunoglobulin G (IgG), at a 1:50 dilution for Alexa Fluor 488-conjugated goat anti-rabbit IgG (Molecular Probes; all other secondary antibodies from Jackson ImmunoResearch), and at a 1:100 dilution for rhodamine red X-conjugated goat anti-mouse IgG (the latter two were used for colocalization staining) and incubated for 60 min at room temperature. The cells were washed as above, rinsed in H2O, and mounted in Mount-Quik (Electron Microscopy Sciences). Confocal images were generated with an LSM 510 confocal microscope (Zeiss).
Nucleotide sequence accession number.
The 5.3-kb cDNA that resulted from the full-length cloning of KIAA0595 has been submitted to GenBank with accession number AF325193.
RESULTS
Structural similarities between PRC and PGC-1.
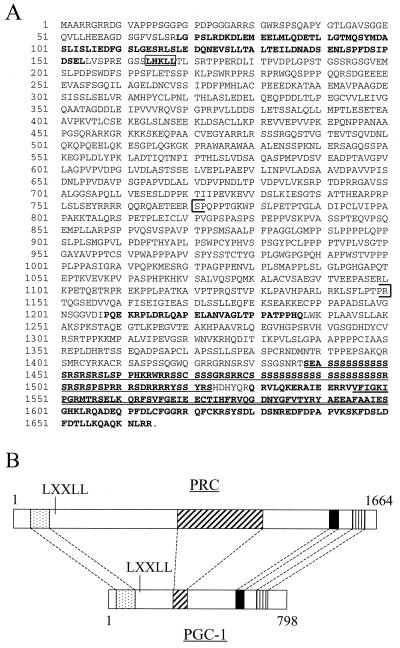
In order to identify novel proteins that interact with NRF-1, we searched the databases for sequences with similarities to PGC-1. The closest related sequence had been deposited as the hypothetical protein KIAA0595 in the HUGE database (Kazusa DNA Research Institute) and had not been assigned any function, nor was it submitted as a full-length coding sequence. Its closest sequence similarity was with PGC-1 and a high-molecular-weight nuclear antigen from chicken (26). The most striking sequence similarity between these proteins was mainly located in the carboxy-terminal region. This region has recently been implicated in efficient splicing of target genes, an activity that apparently is linked to the coactivation function of PGC-1 (19). Full-length cloning of KIAA0595 revealed a 5.3-kb cDNA with an open reading frame of 1,664 aa residues. The amino-terminal region included an additional segment of similarity with PGC-1 (Fig. 1A) along with an LXXLL motif. These similarities in primary structure suggested that KIAA0595 might be a PGC-1-related coactivator, which has been here designated PRC.
FIG. 1.
Primary structure of PRC. (A) Predicted amino acid sequence of PRC. Regions of significant sequence similarity with PGC-1 are in boldface letters. The coactivator signature LXXLL is boxed; the proline-rich region is within brackets. The underlined sequence represents the RS domain, and the double underlined sequence represents the RNA recognition motif. (B) Alignment of similar domains in PRC and PGC-1. Regions of similarity between the two proteins are as follows: activation domain (stippled box), proline-rich region (cross-hatched box), RS domain (solid box), and RNA recognition motif (vertically hatched box). Amino acid coordinates are indicated above and below.
Although the overall sequence of PRC is not significantly related to PGC-1, the spatial pattern of specific regions of significant sequence similarity is conserved (Fig. 1B). These include an acidic region in the amino-terminal domain, followed by an LXXLL motif that is believed to be a nuclear receptor coactivator signature (13). In addition, a proline-rich region (approximately 20% proline) between residues 770 and 1150 is more extensive than that found in PGC-1 and is consistent with an unstructured conformation in the middle of the molecule. Finally, like PGC-1, PRC has an RS-rich domain, followed by the RNA recognition motif in its carboxy terminus. Taken together, these features are suggestive of related function.
If PRC is a transcriptional coactivator, one might expect it to be localized to the nucleus. To examine the subcellular distribution of PRC, rabbit anti-PRC serum was prepared. Confocal imaging of BALB/3T3 cells revealed that PRC is predominantly a nuclear protein (Fig. 2). Following adsorption of the antibody with the same fragment of PRC that was used as an antigen to raise the antibody, the nuclear staining observed in the results shown in Fig. 2A was eliminated (Fig. 2C). The cytoplasmic staining that remained most likely resulted from cross-reactivity, although the possibility that some fraction of PRC is cytoplasmic has not been eliminated. Nevertheless, the clear nuclear localization of PRC is consistent with coactivator function.
Regulated expression of PRC.
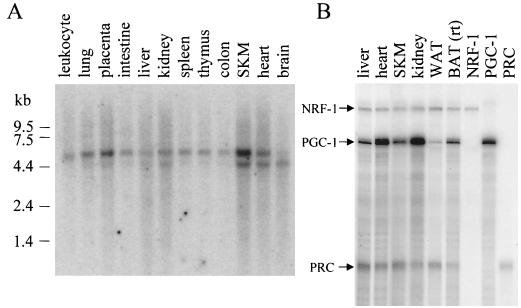
To investigate whether, in analogy with PGC-1 (22), PRC also displays tissue-specific expression, a 1.6-kb amino-terminal cDNA fragment of PRC was hybridized to a Northern blot of poly(A) RNA from multiple human tissues. The results shown in Fig. 3A demonstrate that two different human PRC mRNAs of approximately 5 and 6 kb are present in all tissues tested with somewhat higher levels, especially of the smaller transcript, in skeletal muscle and heart. Both human transcripts are long enough to encode the entire coding sequence, and murine cells appear to have only the 5-kb transcript (results not shown). PRC mRNA levels were compared to those of PGC-1 and NRF-1 in several mouse tissues by a sensitive RNase protection assay. As shown in Fig. 3B, PRC resembles NRF-1 in that it is present in all tissues but does not exhibit large fluctuations in expression between tissues. This contrasts with PGC-1, which displays enhanced expression in heart and kidney. The most notable difference between PRC and PGC-1 is that PGC-1 mRNA is elevated in brown fat compared to white fat, whereas PRC mRNA is somewhat lower in brown fat than in white fat. This suggests that PRC may not contribute to the thermogenic properties of brown fat.
FIG. 3.
Expression profile of PRC mRNA. (A) A human multiple-tissue Northern blot (Clontech) probed with a 32P-labeled 1.6-kb PRC cDNA fragment comprising the 5′ end of the cDNA. Positions of RNA standards of known length in kilobases are indicated at the left. (B) Comparison of NRF-1, PGC-1, and PRC mRNA expression by RNase protection in various mouse tissues. Protected fragments (365, 297, and 192 bp for NRF-1, PGC-1, and PRC, respectively) from individual probes using liver RNA are shown in the last three lanes. WAT, white adipose tissue, rt, room temperature.
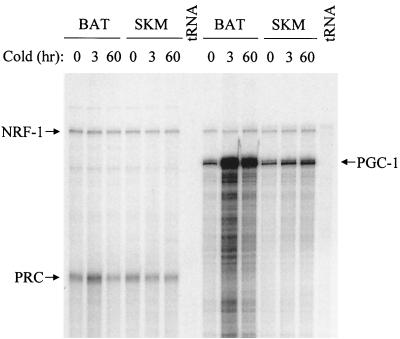
Since PGC-1 mRNA is markedly up-regulated in brown fat in response to cold exposure, it was of interest to determine whether PRC was regulated similarly. Figure 4 shows a comparison of NRF-1, PRC, and PGC-1 transcripts in both brown fat and skeletal muscle during a time course of cold exposure. NRF-1 mRNA expression was essentially constant in both tissues under all conditions and thus serves as an ideal negative control. As expected, PGC-1 mRNA is dramatically induced in brown fat upon cold exposure but much less so in skeletal muscle. In comparison, PRC mRNA shows a weak transient induction in brown fat and is unchanged in skeletal muscle. These results establish that PRC is only modestly cold regulated compared to PGC-1 and suggest that PRC is not functionally equivalent to PGC-1 in cold adaptation.
FIG. 4.
Response of NRF-1, PGC-1, and PRC transcripts to cold exposure. Transcripts were detected by RNase protection in BAT and SKM obtained from mice exposed to 4°C for the indicated times. Probes were the same as those used in the results shown in Fig. 3. tRNA serves as a negative control.
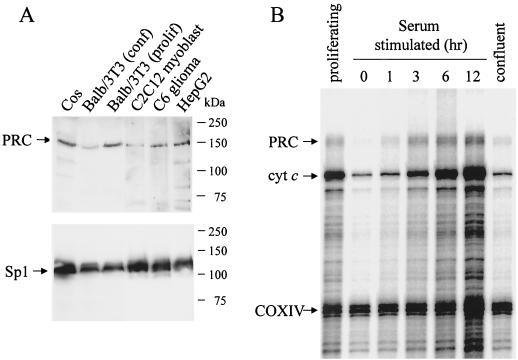
Immunoblotting with the anti-PRC serum revealed a protein of approximately 150 kDa in extracts from both murine and human cell lines (Fig. 5A). This is in reasonable agreement with the 177-kDa predicted molecular mass of PRC. Interestingly, inclusion of the calpain-proteasome inhibitor, LLnL, was required to consistently prepare whole-cell extracts in which the 150-kDa protein was undegraded. Moreover, even in the presence of LLnL, the 150-kDa protein was degraded when stored overnight at 4°C (not shown). It is also apparent that significantly less PRC was expressed in BALB/3T3 cells that had reached confluence than in proliferating cells.
FIG. 5.
Cell cycle regulation of PRC expression. (A) Immunoblot of whole-cell extracts (30 μg per lane) from murine cell lines (BALB/3T3 fibroblasts, C2C12 myoblasts, and C6 glioma cells) and primate cell lines (Cos and HepG2) with rabbit anti-PRC antibodies or rabbit anti-Sp1 serum as indicated. Molecular mass standards in kilodaltons are indicated at the right. (B) BALB/3T3 cells were cultured, and total RNA was isolated from proliferating, serum-starved, serum-stimulated, or confluent cells as described in Materials and Methods. mRNA levels for PRC, cytochrome c (cyt c), and COXIV were analyzed by RNase protection assay. Protected fragments are 192, 175, and 130 bp for PRC, cyt c, and COXIV, respectively. (C) Comparison of NRF-1, PGC-1, and PRC transcripts upon serum stimulation of quiescent BALB/3T3 cells. Protected fragments (365, 297, and 192 bp for NRF-1, PGC-1, and PRC, respectively) from individual probes with mouse liver RNA are shown in the first three lanes with tRNA serving as a negative control. RNA samples were isolated at the indicated times following serum addition.
The notably lower level of PRC protein in nonproliferating BALB/3T3 cells suggested that PRC expression might be enhanced during cell proliferation. To test this possibility, cells were serum starved to induce G0 arrest, and the PRC mRNA level was measured during a time course of serum stimulation as well as in proliferating and confluent cells. Cytochrome c mRNA was measured simultaneously as a positive control, while COXIV mRNA served as a negative control (14). As shown in Fig. 5B, the PRC mRNA level was much lower in serum-starved cells than in proliferating BALB/3T3 cells. Following reintroduction of serum, PRC expression was largely recovered within 3 h and was maintained for at least 12 h. When the cells were allowed to grow until confluency, PRC mRNA dropped again to levels similar to those of serum-starved cells, even though the confluent cells were maintained in the serum induction medium (20% FBS). The expression of the CREB–NRF-1-responsive gene, cytochrome c, was also rapidly induced by serum, as demonstrated previously (14). However, the dramatic reduction in cytochrome c expression in confluent cells had not previously been noted. The expression of COXIV mRNA remained largely unaffected during the course of the experiment.
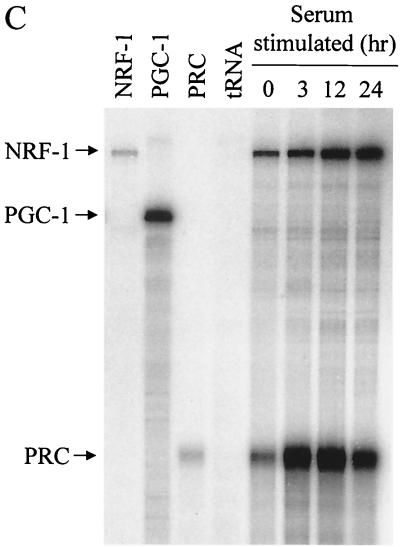
An RNase protection assay was also used to compare PRC to NRF-1 and PGC-1 mRNAs during a time course of serum induction. No PGC-1 mRNA was detectable under conditions where PRC mRNA was substantially induced (Fig. 5C). This suggests that PRC provides a proliferative function under conditions where PGC-1 is not required. The rapid and robust induction of PRC transcript in response to serum was in contrast to the more modest and temporally delayed induction of NRF-1. The results demonstrate that PRC is cell cycle regulated and that its induction is a relatively early event in the G0-to-G1 transition.
PRC transcriptional coactivation through NRF-1.
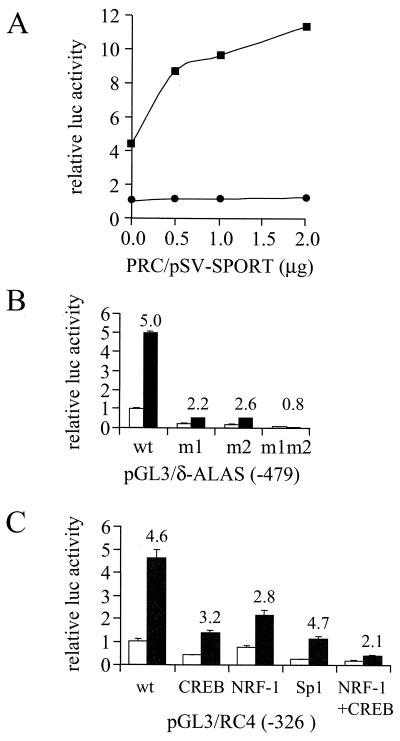
It has previously been demonstrated that PGC-1 is a potent activator of NRF-1-dependent promoters and that this activation was most likely due to physical interaction between the two proteins (33). To determine whether PRC could function as an NRF-1-dependent coactivator, a luciferase reporter plasmid driven by four tandem NRF-1 sites (4xNRF-1/Luc) was cotransfected with a PRC expression vector. As shown in Fig. 6A, PRC alone was unable to activate this promoter. However, when NRF-1 was expressed simultaneously, a substantial activation of expression was observed (Fig. 6A) reminiscent of that previously obtained with PGC-1 (33).
FIG. 6.
NRF-1-dependent coactivation by PRC. (A) Increasing amounts of the mammalian expression vector PRC/pSV-SPORT were cotransfected into BALB/3T3 cells with CMV/β-gal (Clontech) and reporter plasmid 4xNRF-1/luc. Graphs indicate the stimulation of luciferase expression in the presence (squares) or absence (circles) of NRF-1 expression vector pSG5/NRF1. Data are means ± standard deviations (SD) of two independent experiments and are expressed as the ratio of the luciferase and β-galactosidase activities. Control activity was normalized to a value of 1. (B) The δ-ALAS(-479)/pGL3 wild-type (wt) promoter plasmid or mutated derivatives having site-directed point mutations in either one (m1 or m2) or both (m1m2) NRF-1 recognition sites were cotransfected with either PRC/pSV-SPORT (black bars) or the pSV-SPORT control (gray bars). Data represent the means ± SD of the ratio of the luciferase and β-galactosidase activities for a representative experiment. Numbers above the bars refer to the fold induction achieved in the presence of PRC expression vector. (C) The pGL3/RC4 (-326) wild-type (wt) cytochrome c promoter plasmid or derivatives with mutations in CREB, NRF-1, Sp1, or CREB plus NRF-1 recognition sites were cotransfected with PRC/pSV-SPORT (black bars) or empty pSV-SPORT (gray bars). Data represent the means ± SD of the ratio of the luciferase and β-galactosidase activities for two independent experiments. Numbers above the bars refer to the fold induction achieved in the presence of PRC expression vector.
This result prompted the question of whether PRC could also activate a natural promoter that depended on NRF-1 for full activity. The δ-ALAS promoter has two functional NRF-1 recognition sites that are essential for basal promoter activity (4). In agreement with these findings, we found that when either of the two NRF-1 elements was mutated, about 70 to 80% of promoter activity was lost; when both were mutated, full activity was diminished by approximately 95% (Fig. 6B). When the wild-type δ-ALAS promoter was cotransfected with the PRC expression vector, a fivefold activation of the basal activity was achieved. Mutation of one of the two NRF-1 sites (m1 and m2) diminished PRC activation to two- to threefold; in the double mutant, a complete ablation of the coactivating properties of PRC was observed. These data are consistent with the ability of PRC to coactivate through NRF-1 in the context of a natural promoter.
The cytochrome c promoter is also dependent on NRF-1, although to a lesser extent, because of the presence of other proximal promoter elements (9). As shown in Fig. 6C, PRC also activated the cytochrome c promoter. Although mutation of the single NRF-1 site diminished activation by PRC, mutation of both NRF-1 and CREB sites was necessary for substantial loss of PRC-dependent activation. This suggested that PRC was able to utilize both NRF-1 and CREB in mediating full promoter activity. It is of interest to note that both NRF-1 and CREB participate in the serum induction of cytochrome c expression (14). In contrast, mutation of the Sp1 sites markedly reduced promoter activity but had no effect on promoter activation by PRC. In addition, PRC had no effect on β-actin promoter expression under conditions where NRF-1-dependent promoters were markedly activated (data not shown). These observations are consistent with selectivity on the part of PRC in its ability to utilize transcriptional activators.
A direct interaction between PRC and NRF-1.
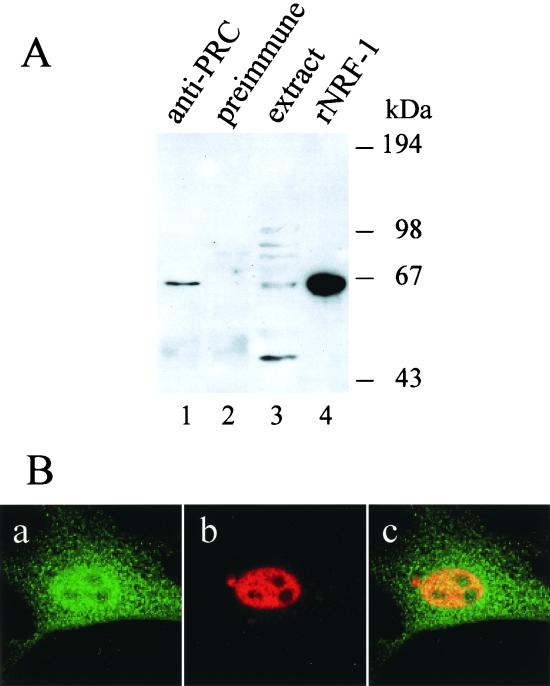
The above experiments suggested that PRC may interact directly with NRF-1 to augment transcription from NRF-1-dependent promoters. To test whether NRF-1 and PRC were indeed associated in vivo, PRC was immunoprecipitated from a whole-cell lysate of C2C12 cells, and the immunoprecipitate was probed with goat anti-NRF-1 antibodies in a Western blot analysis. As seen in Fig. 7A, NRF-1, which migrates at approximately 68 kDa, was efficiently precipitated using anti-PRC antibodies (lane 1), whereas no detectable NRF-1 was precipitated with preimmune serum (lane 2). Identical results were obtained with BALB/3T3 cells and C6 glioma cell extracts (results not shown). These results strongly suggest that NRF-1 is associated with PRC in vivo. In addition, confocal laser scanning microscopy was used to investigate the in vivo colocalization of NRF-1 and PRC. As shown in Fig. 7B, both PRC (panel a) and NRF-1 (panel b) exhibit intense nuclear staining. Overlay of the PRC (green) and NRF-1 (red) images reveals yellow staining within the nucleoplasm. This is consistent with the in vivo colocalization of the two molecules.
FIG. 7.
Interaction between PRC and NRF-1 in vivo. (A) Coimmunoprecipitation of PRC and NRF-1 from cell extracts. C2C12 myoblast whole-cell extracts (750 μg) were subjected to immunoprecipitation using either anti-PRC serum (lane 1) or preimmune serum (lane 2). Immune complexes were brought down with protein A-agarose, washed, and run on an SDS–7.5% PAGE gel. As positive controls, 50 μg of the extract and 3 ng of the recombinant NRF-1 were run in lanes 3 and 4, respectively. After transfer, the immunoblot was probed with affinity-purified goat anti-NRF1 antibody. Molecular mass standards in kilodaltons are indicated on the right. (B) Colocalization of PRC and NRF-1 by confocal laser scanning fluorescence microscopy. BALB/3T3 cells transfected with NRF-1–3xHA were stained with anti-PRC serum (green) (a) or anti-hemagglutinin antibody (red) (b). Green and red images were merged (panel c) to visualize nuclear colocalization. Panel dimensions are 66.5 by 66.5 μm. Confocal images were generated with an LSM 510 confocal microscope (Zeiss).
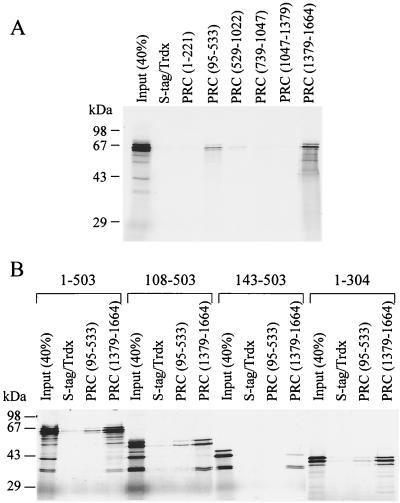
An in vitro binding assay was developed to further test NRF-1 binding to PRC and to map the domains of interaction for both proteins. Protein S-tagged thioredoxin-PRC fusion proteins were expressed and utilized to perform a pull-down assay with protein S-agarose serving as the insoluble matrix (see Materials and Methods). Full-length PRC was not efficiently expressed in Escherichia coli, making it necessary to use subfragments of the protein to assay binding to NRF-1. Two of the PRC fusion proteins displayed a specific interaction with 35S-labeled NRF-1 (Fig. 8A). A fragment encompassing the carboxy-terminal domain of PRC (1379–1664) bound strongly to NRF-1 with up to 40% of the input material bound. This domain corresponds to the region most highly conserved between PRC and PGC-1 (Fig. 1). A second fusion protein encompassing PRC aa 95 to 533 showed weaker but reproducible binding to NRF-1. This second NRF-1 binding domain, although not conserved in sequence, is in a similar position (between the activation domain and the RS domain) as the NRF-1 binding domain in PGC-1 (33).
FIG. 8.
Molecular determinants required for interaction between PRC and NRF-1. (A) Mapping of the NRF-1 binding domains in PRC. Six different fragments of PRC were fused to S-tagged thioredoxin, purified, immobilized on protein S-agarose, and tested for binding with 35S-labeled NRF-1. After binding was complete, the complexes were washed five times, eluted in SDS-PAGE sample buffer, and separated on SDS–10% PAGE gels. Gels were dried, and bound proteins were visualized by autoradiography. As a negative control, S-tagged thioredoxin was used. Molecular mass standards are indicated on the left. (B) Mapping of the PRC binding domain of NRF-1. The two major NRF-1-interacting domains of PRC fused to S-tagged thioredoxin and thioredoxin alone were purified and immobilized on protein S-agarose and incubated with different 35S-labeled NRF-1 fragments as indicated. The bound proteins were analyzed as described in the legend to panel A.
Since PGC-1 interacts with NRF-1 via the NRF-1 DNA binding domain (33), it was important to determine whether PRC also utilized the DNA binding domain for docking with NRF-1. Binding of thioredoxin alone and of the two PRC fragments that bound NRF-1 was tested for binding against full-length NRF-1 and three deletion mutants. The results shown in Fig. 8B demonstrate that removal of the carboxy-terminal intrinsic activation domain of NRF-1 (1–304) or the amino-terminal domain containing multiple sites of phosphorylation (108–503) had no effect on binding either the amino- or carboxy-terminal PRC fragment. However, an amino-terminal deletion to aa 143 (143–503) that is unable to bind DNA (31) failed to interact efficiently with either PRC subfragment. These data are consistent with the results obtained for PGC-1 binding to NRF-1 and establish that interaction with both proteins requires an intact NRF-1 DNA binding domain.
An amino-terminal domain required for transcriptional activation by PRC.
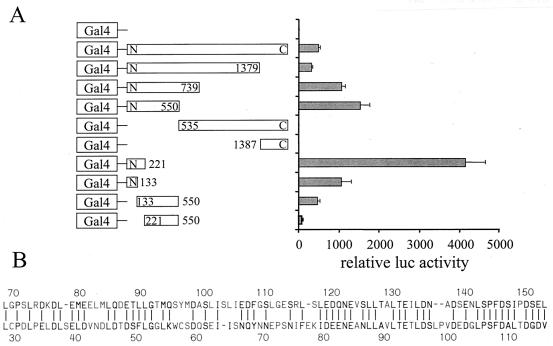
One means by which PRC could potentiate gene expression is through an intrinsic transcriptional activation domain. The activation domain may be targeted to the transcription machinery through the interaction of PRC with DNA binding transcription factors such as NRF-1. To elucidate such a domain, a series of fusions between the Gal4DBD and PRC were expressed in mammalian cells and tested for their ability to activate the 5xGal4 luciferase reporter plasmid in transiently transfected cells. As shown in Fig. 9A, the full-length PRC fusion stimulated reporter activity 500-fold over the Gal4DBD alone. Progressive carboxy-terminal truncation of PRC to residue 221 increased transcriptional activation more than 4,000-fold over control. A carboxy-terminal truncation to residue 133 diminished activity about fourfold compared to the deletion to residue 221. Amino-terminal deletions to residues 539 and 1387 were indistinguishable from the Gal4 control, supporting the conclusion that a potent activation domain resided in the amino-terminal region. This was further confirmed by amino-terminal deletions to residues 133 and 221 in the context of the N-550 fragment. These deletions progressively diminished activation function. All fusion proteins were expressed at similar levels except the full-length N-C, which was expressed at a somewhat lower level, as determined by either immunoblotting or gel shift assays (not shown). We conclude that sequences essential for full activity lie on either side of residue 133.
FIG. 9.
Mapping of the PRC activation domain. (A) BALB/3T3 cells were transfected with a 5xGal4 luciferase reporter plasmid, CMV/β-gal, and either pSG424 control vector (Gal4) or the various PRC/pSG424 fusions, as indicated. Results are expressed as the ratio of the luciferase to β-galactosidase activities and are the means ± standard errors of the mean for at least three independent experiments, each performed in duplicate. (B) Comparison of the putative activation domains in PRC and PGC-1. A Lipman-Pearson protein alignment of aa 69 to 154 of PRC and aa 29 to 117 of PGC-1 is shown. Identical or homologous (by 1 distance unit) amino acids are indicated by a vertical line (|). (C) Comparison of wild-type PRC to the amino-terminal deletion PRC/222C in the coactivation of NRF-1-dependent transcription from a 4xNRF-1/luc reporter plasmid. Results are expressed as in shown in panel A.
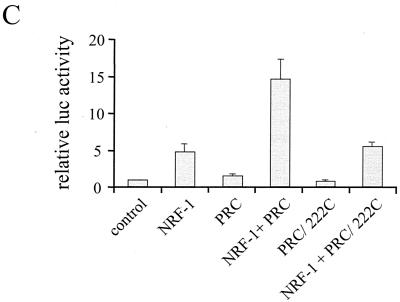
A Lipman-Pearson alignment of PRC and PGC-1 within the amino-terminal domain revealed an 80-aa stretch of considerable sequence similarity that includes PRC residues 69 to 154 (Fig. 9B). Interestingly, this region of sequence alignment has also recently been mapped to contain the PGC-1 activation domain (16, 30). To test whether this domain is required for NRF-1-dependent activation by PRC, the wild-type and the 222-C deletion of PRC were compared for their ability to trans-activate through NRF-1. The results show that deletion of the first 222 residues of PRC completely eliminates the coactivation of NRF-1-dependent transcription by PRC (Fig. 9C). This confirms that the activation domain is essential for the coactivator function of PRC.
DISCUSSION
This work describes the identification and characterization of a novel transcriptional coactivator designated PRC. The evidence supports the conclusion that PRC is both structurally and functionally related to PGC-1. Like PGC-1, PRC is a nuclear protein with an RNA recognition motif near its carboxy terminus and a potent transcriptional activation domain near its amino terminus. Furthermore, PRC augments NRF-1-dependent transcription of several promoters presumably through its direct interaction with the DNA binding domain of NRF-1. Both in vivo and in vitro experiments support the specific interaction of PRC with NRF-1. Two distinct domains in PRC participate in NRF-1 recognition. One is a carboxy-terminal domain that is conserved between PRC and PGC-1 and has recently been found to mediate a ligand-independent interaction between PGC-1 and the estrogen receptor (29). This region has also been implicated in the association of PGC-1 with RNA processing factors but was not required for transcriptional activation (19). Likewise, we find that a deletion of this same region in PRC does not affect transcriptional activation of NRF-1 target genes (not shown). As in PGC-1, the carboxy-terminal domain of PRC may participate in RNA processing. The second NRF-1 recognition motif is in the amino-terminal one-third of PRC. Although this motif is in a location similar to the NRF-1 and PPAR-γ recognition domains in PGC-1 (33), it shares no obvious sequence similarity with PGC-1.
By contrast, the transcriptional activation domain in PRC shares significant sequence similarity with that in PGC-1. This region lies between residues 69 and 154 and is highly acidic (24% acidic amino acids) and hydrophobic (40% hydrophobic amino acids). The hydrophobic residues are interspersed with acidic residues in a manner reminiscent of the herpes simplex virus VP-16 activation domain (7). This contrasts with the NRF-1 activation domain, which has essential hydrophobic residues interspersed with glutamines and relatively few acidic amino acids (11). It is likely that activation function spans this conserved region in PRC because either amino- or carboxy-terminal deletions to residue 133 impair, but do not eliminate, transcriptional activation.
As might be expected of a master regulator of mitochondrial biogenesis, PGC-1 mRNA expression in mouse tissues is reflective of their mitochondrial contents and capacities for aerobic energy production. PGC-1 levels are high in heart and kidney and dramatically elevated in brown fat during adaptive thermogenesis. In contrast, PRC mRNA expression is similar in all mouse tissues tested, and its cold induction in brown fat is modest compared to that of PGC-1. The small PRC induction observed may result from the proliferation of a subpopulation of brown preadipocytes during cold adaptation (23). Thus, it seems unlikely that PRC and PGC-1 function identically in regulating mitochondrial content in postmitotic tissues. However, PRC is rapidly up-regulated when quiescent fibroblasts are induced to reenter the cell cycle in response to serum growth factors and down-regulated when they are contact inhibited to cease growth. These changes occur concomitantly with the expression of the cytochrome c gene, which has recently been shown to have a central role in the induction of respiration that occurs upon entry to the cell cycle (14). Here, PRC is shown to require both NRF-1 and CREB recognition sites for maximal stimulation of the cytochrome c promoter. Both of these elements are also necessary for maximal serum induction of the cytochrome c promoter (14), suggesting that PRC may activate transcription through CREB or CREB-related transcription factors. It is noteworthy that PGC-1 is not expressed in quiescent or proliferating BALB/3T3 cells. In addition, PGC-1 is not expressed in proliferating C2C12 (33) and HepG2 cells where PRC is highly expressed (not shown). Thus, PGC-1 appears not to be essential for mitochondrial maintenance and respiratory function in growing cells.
PRC and PGC-1 are indistinguishable in their interaction with NRF-1 and the coactivation of NRF-1 target genes. Thus, it is likely that they provide complementary functions in governing mitochondrial biogenesis. PGC-1 clearly responds to sympathetic enervation in mediating the thermogenic response, whereas PRC is most responsive to proliferative signals and is regulated according to the cell cycle. It is also possible that the two molecules have overlapping rather than identical specificities for transcription factor interactions, and thus both are required to implement distinct programs of gene expression. The structural and functional similarities between PGC-1 and PRC suggest that PRC may also be involved in mediating transcriptional responses from nuclear hormone receptors. The presence of the LXXLL motif, which is believed to be a mediator of nuclear hormone coactivator interaction (13), also supports this notion. Preliminary pull-down assays have demonstrated that estrogen receptor and thyroid hormone receptor β do interact with both the amino- and carboxy-terminal domains of PRC in a ligand-independent manner (data not shown). This contrasts with the ligand-dependent interaction of these receptors with PGC-1. Interestingly, unlike NRF-1, both the nuclear hormone receptors tested interacted with the PRC fragment N221, which contains the LXXLL motif. Further studies are required to determine the functional significance of these in vitro interactions.
Enigmatically, attempts to overexpress PRC, either by establishing a stable cell line or by retrovirus infection, have not resulted in the induction of NRF-1 target genes (unpublished observations). This contrasts with PGC-1, where ectopic expression stimulates the expression of NRF-1 target genes related to mitochondrial respiratory function (33). Although we have overexpressed PRC mRNA to high levels, this was not accompanied by increased expression of PRC protein. Using the same conditions, we do observe overexpression of PGC-1 and the concomitant biological effect on NRF-1 expression. This could be interpreted to mean that PRC is under posttranscriptional regulation or that the protein is actively degraded to prevent its accumulation beyond physiological levels. It is of interest in this context that PRC is highly unstable and that LLnL, an inhibitor of the proteosome, calpain, and cathepsins, stabilizes the protein (not shown).
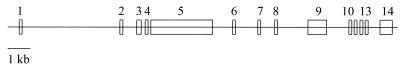
The PRC locus (originally designated KIAA0595) was initially assigned to human chromosome 10 (21). Two cosmid clones (AC006179 and AL160011) deposited in GenBank contained regions of identity with PRC. We assembled these into a continuous sequence of approximately 25 kb containing all coding information of the PRC cDNA. This was distributed among 14 exons, each with exon-intron boundaries conforming to the consensus for donor and acceptor splice junctions (Fig. 10). Clone AC006179 maps to 10q24.2-q24.3, which places the human PRC gene more precisely at this genomic location.
FIG. 10.
Organization of the human PRC gene. A linear representation of the human PRC locus shows the relative positions and approximate sizes of 14 exons (open boxes) spanning 25 kb of chromosome 10. The PRC cDNA coordinates for the exons are as follows: 1, 1–192; 2, 193–381; 3, 382–528; 4, 529–630; 5, 631-3535; 6, 3536–3589; 7, 3590–3647; 8, 3648–3718; 9, 3719–4439; 10, 4440–4589; 11, 4590–4656; 12, 4657–4778; 13, 4779–4930; and 14, 4931–5332. The 5′ end of the cDNA (accession number AF325193) determined by rapid amplification of cDNA ends and PCR was designated 1.
Interestingly, Finnish and Pakistani pedigree studies have allowed the assignment of an autosomal dominant progressive external ophthalmoplegia, type 1, to 10q23.3-24.3 and 10q23.31-q25.1, respectively (18, 28). Another disease mapped to the vicinity of the PRC locus is Thiel-Behnke corneal dystrophy, which has been linked to 10q23-24 (34). While these ocular diseases, at a first glance, do not appear to be linked to NRF-1-dependent gene expression, it has been shown that a loss of function of NRF-1 in zebra fish led to extensive degeneration of photoreceptors and their precursors (3). Moreover, certain forms of progressive external ophthalmoplegia have been associated with mutations in mitochondrial DNA that result in reduced respiratory function (reviewed in reference 8). It is intriguing to speculate that mutations in the PRC locus reduce NRF-1 target gene expression, which in turn compromises mitochondrial respiratory function. Such a pathway may account for the autosomally inherited forms of progressive external ophthalmoplegia and other neurological defects that resemble those associated with mitochondrial DNA mutations.
ACKNOWLEDGMENTS
This work was supported by United States Public Health Service grant GM32525-18.
We thank Kristel Vercauteren and Raymond A. Pasko for excellent technical assistance and Lei Huo for critical comments.
REFERENCES
- 1.Andersson U, Houstek J, Cannon B. ATP synthase subunit c expression: physiological regulation of the P1 and P2 genes. Biochem J. 1997;323:379–385. doi: 10.1042/bj3230379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1990. [Google Scholar]
- 3.Becker T S, Burgess S M, Amsterdam A H, Allende M L, Hopkins N. not really finished is crucial for development of the zebrafish outer retina and encodes a transcription factor highly homologous to human nuclear respiratory factor 1 and avian initiation binding repressor. Development. 1998;124:4369–4378. doi: 10.1242/dev.125.22.4369. [DOI] [PubMed] [Google Scholar]
- 4.Braidotti G, Borthwick I A, May B K. Identification of regulatory sequences in the gene for 5-aminolevulinate synthase from rat. J Biol Chem. 1993;268:1109–1117. [PubMed] [Google Scholar]
- 5.Chau C A, Evans M J, Scarpulla R C. Nuclear respiratory factor 1 activation sites in genes encoding the gamma-subunit of ATP synthase, eukaryotic initiation factor 2α, and tyrosine aminotransferase. Specific interaction of purified NRF-1 with multiple target genes. J Biol Chem. 1992;267:6999–7006. [PubMed] [Google Scholar]
- 6.Chen S H, Nagy P L, Zalkin H. Role of NRF-1 in bidirectional transcription of the human GPAT-AIRC purine biosynthesis locus. Nucleic Acids Res. 1997;25:1809–1816. doi: 10.1093/nar/25.9.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cress W D, Triezenberg S J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 8.DiMauro S, Moraes C T. Mitochondrial encephalomyopathies. Arch Neurol. 1993;50:1197–1208. doi: 10.1001/archneur.1993.00540110075008. [DOI] [PubMed] [Google Scholar]
- 9.Evans M J, Scarpulla R C. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF and intron Sp1 recognition sites. J Biol Chem. 1989;264:14361–14368. [PubMed] [Google Scholar]
- 10.Evans M J, Scarpulla R C. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990;4:1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- 11.Gugneja S, Virbasius C A, Scarpulla R C. Nuclear respiratory factors 1 and 2 utilize similar glutamine-containing clusters of hydrophobic residues to activate transcription. Mol Cell Biol. 1996;16:5708–5716. doi: 10.1128/mcb.16.10.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 13.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 14.Herzig R P, Scacco S, Scarpulla R C. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J Biol Chem. 2000;275:13134–13141. doi: 10.1074/jbc.275.17.13134. [DOI] [PubMed] [Google Scholar]
- 15.Huo L, Scarpulla R C. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knutti D, Kaul A, Kralli A. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol Cell Biol. 2000;20:2411–2422. doi: 10.1128/mcb.20.7.2411-2422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehman J J, Barger P M, Kovacs A, Saffitz J E, Medeiros D M, Kelly D P. Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Investig. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F Y, Tariq M, Croxen R, Morten K, Squier W, Newsom-Davis J, Beeson D, Larsson C. Mapping of autosomal dominant progressive external ophthalmoplegia to a 7-cM critical region on 10q24. Neurology. 1999;53:1265–1271. doi: 10.1212/wnl.53.6.1265. [DOI] [PubMed] [Google Scholar]
- 19.Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman B M. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 20.Myers S J, Peters J, Huang Y, Comer M B, Barthel F, Dingledine R. Transcriptional regulation of the GluR2 gene: neural-specific expression, multiple promoters, and regulatory elements. J Neurosci. 1998;18:6723–6739. doi: 10.1523/JNEUROSCI.18-17-06723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding region sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- 22.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 23.Rehnmark S, Nedergaard J. DNA synthesis in brown adipose tissue is under β-adrenergic control. Exp Cell Res. 1989;180:574–579. doi: 10.1016/0014-4827(89)90086-4. [DOI] [PubMed] [Google Scholar]
- 24.Sadowski I, Ptashne M. A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scarpulla R C. Nuclear control of respiratory chain expression in mammalian cells. J Bioenerg Biomembr. 1997;29:109–119. doi: 10.1023/a:1022681828846. [DOI] [PubMed] [Google Scholar]
- 26.Shimada K, Masahiko H, Mizuno S. A nuclear matrix-associated high molecular mass nuclear antigen, HMNA, of chicken and marked decrease of its immunoreactivity during the progression of S phase. J Cell Sci. 1997;110:3031–3041. doi: 10.1242/jcs.110.24.3031. [DOI] [PubMed] [Google Scholar]
- 27.Solecki D, Bernhardt G, Lipp M, Wimmer E. Identification of a nuclear respiratory factor-1 binding site within the core promoter of the human polio virus receptor/CD155 gene. J Biol Chem. 2000;275:12453–12462. doi: 10.1074/jbc.275.17.12453. [DOI] [PubMed] [Google Scholar]
- 28.Suomalainen A, Kaukonen J, Amati P, Timonen R, Haltia M, Weissenbach J, Zeviani M, Somer H, Peltonen L. An autosomal locus predisposing to deletions of mitochondrial DNA. Nat Genet. 1995;9:146–151. doi: 10.1038/ng0295-146. [DOI] [PubMed] [Google Scholar]
- 29.Tcherepanova I, Puigserver P, Norris J D, Spiegelman B M, McDonnell D P. Modulation of estrogen receptor-α transcriptional activity by the coactivator PGC-1. J Biol Chem. 2000;275:16302–16308. doi: 10.1074/jbc.M001364200. [DOI] [PubMed] [Google Scholar]
- 30.Vega R B, Huss J M, Kelly D P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Virbasius C A, Virbasius J V, Scarpulla R C. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993;7:2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- 32.Wegner S A, Ehrenberg P K, Chang G, Dayhoff D E, Sleeker A L, Michael N M. Genomic organization and functional characterization of the chemokine receptor CXCR4, a major entry co-receptor for human immunodeficiency virus type 1. J Biol Chem. 1998;273:4754–4760. doi: 10.1074/jbc.273.8.4754. [DOI] [PubMed] [Google Scholar]
- 33.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla R C, Spiegelman B M. Mechanisms controlling mitochondrial biogenesis and function through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 34.Yee R W, Sullivan L S, Lai H T, Stock E L, Lu Y, Khan M N, Blanton S H, Daiger S P. Linkage mapping of Thiel-Behnke corneal dystrophy (CDB2) to chromosome 10q23–q24. Genomics. 1997;46:152–154. doi: 10.1006/geno.1997.5028. [DOI] [PubMed] [Google Scholar]