Abstract
A unique Helicobacter species, MZ640285, was isolated from a patient with X-linked hypogammaglobulinemia suffering from recurrent abdominal abscesses and was identified by 16S rRNA gene sequence analysis. In the phylogenetic tree, the isolate fell into a cluster which included Flexispira rappini, Helicobacter bilis, and Helicobacter sp. strain Mainz. Helicobacters are being increasingly recognized as pathogens in immunocompromised hosts. These fastidious bacteria are not easily cultured in the routine diagnostic laboratory, and this is the first report of their identification by 16S rRNA gene sequencing performed directly from a clinical specimen.
Since the discovery of Helicobacter pylori in 1983, the genus Helicobacter has rapidly expanded through the isolation of other species from humans, other mammals, and birds (3). The genus now includes 28 species, as well as several other helicobacters which have not been formally named. Helicobacters have been identified in the gastrointestinal tract, liver, gallbladder, bile, peripheral blood, and joint effusions. Flexispira rappini, which is also considered a helicobacter, has been isolated from stool specimens from a diarrhea patient (3), and bacteremia in a child with pneumonia (10), in an adult patient undergoing hemodialysis (12), and in a patient with X-linked (Bruton) agammaglobulinemia (14) has been reported.
Here we describe a patient with X-linked hypogammaglobulinemia suffering from recurrent abdominal abscesses. Microscopic examination of pus revealed a number of polymorphonuclear neutrophils (PMNs) and gram-negative fusiform bacilli. 16S rRNA gene sequence analysis of DNA extracted from the pus led to the detection and identification of a unique Helicobacter species closely related to F. rappini and Helicobacter bilis.
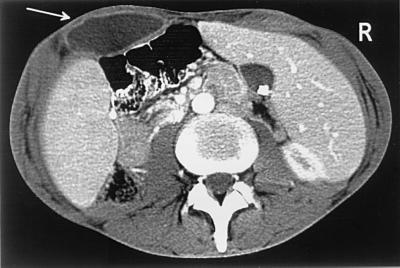
A 29-year-old man was admitted in 1997 to the department of medicine at the Mainz university hospital with persistent pain and swelling located in the left popliteal fossa and on the left side of the abdominal wall. An X-linked hypogammaglobulinemia had been diagnosed in 1986. Despite regular immunoglobulin substitution, the patient suffered from severe, recurrent respiratory infections. He had been admitted twice due to “sterile” abdominal abscesses in the last 5 years. On admission, the slim, afebrile patient complained of weight loss (6 kg) during the last 2 months. Physical examination revealed a painful and warm tumor in the patient's left popliteal fossa. Examination of the abdomen revealed scars from the previous abscesses and a warm, tender tumor of 5 by 5 cm. The patient exhibited a leukocyte count of 7.6 × 109/liter and an elevated C-reactive protein level (72 mg/dl). A high-resolution computerized tomography (CT) of the thorax showed bronchiectasis and circular areas of consolidation. A CT of the abdomen revealed an abscess in the left side of the abdominal wall (Fig. 1). Pus from the abdominal abscess was aspirated twice. Histological examination of this material revealed an acute inflammatory cellular reaction, predominantly PMNs. The patient received 400 mg of metronidazole three times a day and 500 mg of ciprofloxacin twice a day. He improved clinically, and his C-reactive protein level returned to normal. The patient was discharged after 2 weeks and was instructed to continue antibiotic therapy for an additional 2 weeks. Two months later, a control CT of the thorax showed no areas of consolidation but persistent bronchiectasis. Clinical examination revealed that the popliteal and abdominal abscesses had resolved completely.
FIG. 1.
CT of the patient's abdomen revealing the abscess in the left side of the abdominal wall.
Microscopic examination of pus from the abdominal abscess revealed numerous PMNs and faintly staining gram-negative rods with slightly curved fusiform bacteria that were best seen by acridine orange staining. Aerobic cultures were performed on blood and cysteine-lactose-electrolyte-deficient agar. Schaedler agar and thioglycolate broth were used for anaerobic cultures. Blood cultures and mycobacterial cultures were additionally inoculated. All cultures remained negative. Because bacteria could be seen by microscopy, attempts were then launched to directly extract and amplify bacterial DNA from the pus.
Genomic DNA was extracted from the abdominal pus using the QIAamp tissue kit (Qiagen, Hilden, Germany). A PCR of the 16S ribosomal DNA (rDNA) was performed with the primers pA-f (5′-AGAGTTTGATCCTGGCTCAG-3′; anneals to positions 8 to 28 of Escherichia coli 16S rRNA) and pH-b (5′-AAGGAGGTGATCCAGCCGCA-3′; positions 1542 to 1522) as described previously (2). PCR products were subcloned into the pCR 3.1 vector (TA cloning kit; Invitrogen, Groningen, The Netherlands), and two of the clones were sequenced using an Applied Biosystems model 373A DNA sequencing system and the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Weiterstadt, Germany) (5). PCR products amplified in a separate run were also directly sequenced. The sequencing primers were reported by Edwards et al. (2), and the T7 and pCR 3.1 reverse sequencing primers in the TA cloning kit (Invitrogen) are described by the manufacturer.
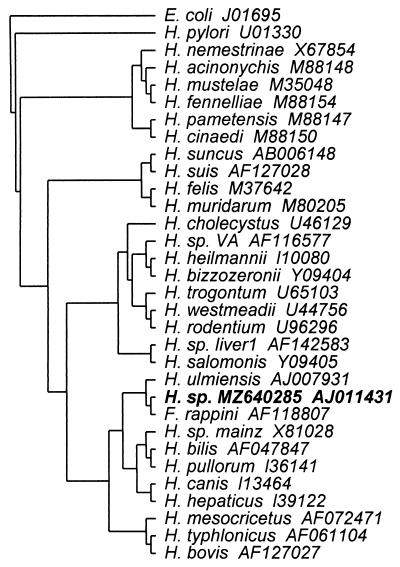
PCR amplification yielded an amplicon which was approximately 200 bp larger than typical amplicons obtained from other bacteria. Direct sequencing of PCR products as well as two different subcloned amplicons in separate PCR runs yielded identical results. DNA sequence analysis revealed the presence of an 187-bp intervening sequence (IVS) inserted at bp 210 by using E. coli numbering in the 1,668-bp fragment (2). The 16S rDNA sequence including the IVS was submitted to GenBank and a similarity search using the BLAST program indicated a close relationship to the DNA sequence of H. bilis and F. rappini (97% identity). The IVS of the Helicobacter isolate described in this report differed from those of the H. bilis and F. rappini strains by six and seven bases, respectively. A phylogenetic tree with E. coli as the outgroup was constructed by using Clustal W (12) and displayed by the TreeView program (9) (Fig. 2). The Helicobacter isolate in this report can be grouped into a cluster which includes F. rappini, H. bilis, H. sp. strain Mainz, H. pullorum, H. canis, H. hepaticus, and H. ulmiensis.
FIG. 2.
Phylogenetic tree demonstrating the relationship of Helicobacter sp. MZ640285 (in bold) to other helicobacters. The tree was rooted with E. coli as the outgroup. The GenBank accession number of each species is provided.
Helicobacters are very fastidious organisms. Isolation of some helicobacters, e.g., F. rappini, requires the use of microaerophilic conditions with H2, which is not a common feature of the microaerophilic systems used by clinical laboratories (10, 14). Even if primary cultures are obtained, subculture attempts may not turn out to be successful (11). Furthermore, only a limited range of phenotypic tests are available for the identification and differentiation of Helicobacter species. In recent years, most reports on new Helicobacter isolates relied on 16S rDNA sequencing data to identify the species. This method allows the identification of the organism without prior knowledge of the gene sequence and without the need for a culture. The Helicobacter isolate described in this report exhibited a high degree of similarity to H. bilis and F. rappini. Since studies of numerous diverse taxa demonstrated that most related species of the same genus differ in their 16S rDNA sequences by at least 1.5% (13), our Helicobacter isolate may be considered a unique Helicobacter species and is named Helicobacter sp. MZ640285. Unfortunately, we were not able to cultivate this organism.
The presence of IVSs in the 16S rRNA gene is common to several Helicobacter species. The IVS found in our Helicobacter isolate is similar to those in H. bilis and F. rappini, with a length of about 187 bp inserted at bp 210 (E. coli numbering) (4). Another type of IVS has been observed in certain strains of H. canis and H. fennelliae, which is of 235 bp inserted at nucleotide 199 (E. coli numbering) (8). Little is known on the origin and significance of IVSs in the 16S rRNA gene. In H. canis, the IVS is not retained in the mature 16S rRNA and not represented elsewhere in the genome (8).
Several species of Helicobacter, including H. cinaedi, H. fennelliae, H. westmeadii, H. sp. strain Mainz, and F. rappini, have been isolated from immunocompromised patients (1, 3, 6, 7, 10, 13–15). Recently, a case of septic shock due to H. fennelliae was reported (6). To our knowledge, the present case is the first report of a Helicobacter species identified directly in a clinical specimen. We could not identify any other bacteria in the same abscess material by conventional cultures or by DNA sequence analysis. Antibiotic therapy resulted in resolution of the abscesses. These data indicate that the Helicobacter isolate caused the abdominal abscess in this patient. The source of infection and portal of entry have not been defined.
In conclusion, helicobacters, including the unique species reported in this work, may represent an emerging group of pathogens in immunocompromised patients. 16S rRNA gene sequencing allows the rapid identification of fastidious bacteria, which cannot be cultured in the routine diagnostic laboratory.
Nucleotide sequence accession number.
The 16S rDNA sequence identified in this study was submitted to GenBank under accession no. AJ011431.
REFERENCES
- 1.Burman W J, Cohn D L, Reves R R, Wilson M L. Multifocal cellulitis and monoarticular arthritis as manifestations of Helicobacter cinaedi bacteremia. Clin Infect Dis. 1995;20:564–570. doi: 10.1093/clinids/20.3.564. [DOI] [PubMed] [Google Scholar]
- 2.Edwards U, Rogall T, Blöcker H, Emde M, Böttger E C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox J G. Helicobacters: the next generation. Bailliere's Clin Infect Dis. 1997;4:449–471. [Google Scholar]
- 4.Fox J G, Yan L L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han S-R, Schreiber H-J, Bhakdi S, Loos M, Maeurer M J. vacA genotypes and genetic diversity in clinical isolates of Helicobacter pylori. Clin Diagn Lab Immunol. 1998;5:139–145. doi: 10.1128/cdli.5.2.139-145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsueh P-R, Teng L-J, Hung C-C, Chen Y-C, Yang P-C, Ho S-W, Luh K-T. Septic shock due to Helicobacter fennelliae in a non-human immunodeficiency virus-infected heterosexual patient. J Clin Microbiol. 1999;37:2084–2086. doi: 10.1128/jcm.37.6.2084-2086.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Husmann M, Gries C, Jehnichen P, Woelfel T, Gerken G, Ludwig W, Bhakdi S. Helicobacter sp. strain Mainz isolated from an AIDS patient with septic arthritis: case report and nonradioactive analysis of 16S rRNA sequence. J Clin Microbiol. 1994;32:3037–3039. doi: 10.1128/jcm.32.12.3037-3039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linton D, Clewley J P, Burnens A, Owen R J, Stanley J. An intervening sequence (IVS) in the 16S rRNA gene of the eubacterium Helicobacter canis. Nucleic Acids Res. 1994;22:1954–1958. doi: 10.1093/nar/22.11.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 10.Sorlin P, Vandamme P, Nortier J, Hoste B, Rossi C, Pavlof S, Struelens M J. Recurrent “Flexispira rappini” bacteremia in an adult patient undergoing hemodialysis: case report. J Clin Microbiol. 1999;37:1319–1323. doi: 10.1128/jcm.37.5.1319-1323.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tee W, Leder K, Karroum E, Dyall-Smith M. “Flexispira rappini” bacteremia in a child with pneumonia. J Clin Microbiol. 1998;36:1679–1682. doi: 10.1128/jcm.36.6.1679-1682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trivett-Moore N L, Rawlinson W D, Yuen M, Gilbert G L. Helicobacter westmeadii sp. nov., a new species isolated from blood cultures of two AIDS patients. J Clin Microbiol. 1997;35:1144–1150. doi: 10.1128/jcm.35.5.1144-1150.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weir S, Cuccherini B, Whitney A M, Ray M L, MacGregor J P, Steigerwalt A, Daneshvar M I, Weyant R, Wray B, Steele J, Strober W, Gill V J. Recurrent bacteremia caused by a “Flexispira”-like organism in a patient with X-linked (Bruton's) agammaglobulinemia. J Clin Microbiol. 1999;37:2439–2445. doi: 10.1128/jcm.37.8.2439-2445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weir S C, Gibert C L, Gordin F M, Fischer S H, Gill V J. An uncommon Helicobacter isolate from blood: evidence of a group of Helicobacter spp. pathogenic in AIDS patients. J Clin Microbiol. 1999;37:2729–2733. doi: 10.1128/jcm.37.8.2729-2733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]