Abstract
Spread of the carbapenemase-encoding and mobilized colistin resistance (mcr) genes among Enterobacteriales poses a great threat to global public health, especially when the both genes are transferred by a single plasmid. Here, we identified a blaNDM–1- and mcr-9-co-encoding plasmid harbored by a clinical isolate of Klebsiella pneumoniae (KPN710429). KPN710429 was recovered from a blood sample from an inpatient in a tertiary hospital in China, and antimicrobial susceptibility testing showed that it was multidrug-resistant and only susceptible to aztreonam, colistin, and tigecycline. KPN710429 belongs to sequence type (ST) 1308 and capsular serotype KL144. The string test of KPN710429 was negative, and this strain didn’t exhibit a hypervirulent phenotype according to serum-killing and Galleria mellonella lethality assessments. Whole-genome sequencing revealed the KPN710429 genome comprises a single chromosome and three plasmids. All virulence associated genes were harbored by chromosome. Most of its antimicrobial resistance genes, including blaNDM–1 and mcr-9 were carried by plasmid pK701429_2, belonging to the incompatibility (Inc) HI2/HI2A group and ST1. Comparative genomics assays indicates that pK710429_2 could be a hybrid plasmid, formed by a Tn6696-like blaNDM–1 region inserting into a mcr-9-positive-IncHI2/HI2A plasmid. pK710429_2 contained the conjugative transfer gene regions, Tra1 and Tra2, with some structural variations, and conjugation assays revealed that pK710429_2 was transferable. Although pK710429_2 lacked the qseB-qseC regulatory genes, mcr-9 expression was upregulated after pretreatment with colistin for 6 h, leading to colistin resistance in KPN710429. To our knowledge, this is the first report of a blaNDM–1- and mcr-9-co-encoding transferable plasmid harbored by a bloodstream-infection-causing K. pneumoniae strain in China. Effective surveillance should be implemented to assess the prevalence of the plasmid co-harboring carbapenemase-encoding gene and mcr-9.
Keywords: Klebsiella pneumoniae, carbapenem-resistant, bla NDM–1 , mcr-9, IncHI2, plasmid
Introduction
Klebsiella pneumoniae is one of the most notorious members of Enterobacteriales, contributing to both community and nosocomial infections. Rapid spread of carbapenem-resistant K. pneumoniae (CRKP) poses a great threat to global public health (Effah et al., 2020). Most CRKP isolates produce carbapenemases, which are grouped into molecular classes A, B, and D. New Delhi metallo-beta-lactamase (NDM)-1, one of the most common class B carbapenemases, hydrolyze carbapenems and other β-lactams except aztreonam (Wu et al., 2019). The blaNDM–1 was first identified in Enterobacteriales from a Swedish patient hospitalized in India in 2008 (Yong et al., 2009). Subsequently, widespread distribution of blaNDM–1-positive Enterobacteriales has been reported worldwide. The blaNDM–1 gene is typically carried by plasmids, which are mostly transferable and co-exist with many other resistance determinants, further complicating clinical anti-infective treatment (Wu et al., 2019). Colistin, a cationic cyclic-peptide, is concern as the last-line therapeutic options for treating severe infections caused by carbapenem-resistant Enterobacteriales, especially blaNDM–1-harboring CRKP (Poirel et al., 2017). However, concerns were further raised regarding the emergence and spread of mobilized colistin-resistance (mcr) genes. Since mcr-1 was initially discovered in China in 2015, a growing number of mcr-like genes (from mcr-2 to mcr-10) have been identified worldwide (Liu et al., 2016; Li et al., 2020). Among them, mcr-9 is a newly emerging variant first identified from a clinical Salmonella enterica isolate in the United States in 2019 (Carroll et al., 2019). Notably, co-transfer of mcr-9 and carbapenemase-encoding genes (e.g., blaNDM, blaIMP, and blaVIM) by a single plasmid has been detected in clinical isolates of Enterobacter spp., but not yet reported in Klebsiella species (Chavda et al., 2019; Li et al., 2020; Ai et al., 2021).
In this study, we characterized the genomic features of a blaNDM–1- and mcr-9-co-encoding plasmid harbored by a clinical CRKP isolate. These findings will supplement existing data on the molecular epidemiological characteristics of plasmids co-harboring carbapenemase-encoding gene and mcr-9.
Materials and Methods
Strain Identification
The K. pneumoniae strain KPN710429 was isolated from a blood sample from a 69-year-old woman with duodenal cancer at the Second Hospital of Anhui Medical University in Anhui, China, in June 2020. This bloodstream infection was hospital-acquired, and the patient was not treated with polymyxin B or colistin before this strain was isolated. The species was identified using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) (Bruker, Bremen, Germany).
Antimicrobial Susceptibility Testing
In vitro antimicrobial susceptibility tests (ASTs) were performed for amikacin, aztreonam, cefepime, cefotaxime, cefoxitin, ceftazidime, ceftazidime-avibactam, ceftriaxone, cefuroxime, ciprofloxacin, colistin, ertapenem, gentamicin, imipenem, levofloxacin, meropenem, piperacillin-tazobactam, ticarcillin-clavulanic acid, tigecycline, and tobramycin using the broth microdilution method. The susceptibility breakpoints of tigecycline and colistin were interpreted in according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoint (EUCAST, 2020); all other antimicrobial agents were interpreted using Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2020). The standard Escherichia coli strain ATCC25922 was used for quality control.
Hypermucoviscous Phenotype Detection and Virulence Assessments
The hypermucoviscous phenotype was detected using the string test as described previously (Mazloum et al., 2016). The virulence of KPN710429 were assessed by serum-killing and Galleria mellonella lethality assays according to previously described (Liu et al., 2021). For references, a hypervirulent K. pneumoniae (designated as KPN54798H–ctrl) and classic K. pneumoniae (designated as KPN49L–ctrl) were employed as hypervirulence and low-virulence control, respectively (Liu Z. et al., 2019b).
Whole-Genome Sequencing, Assembly, and Comparative Analysis
The total genomic DNA of strain KPN710429 was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Whole-genome sequencing (WGS) and assembly were performed as previously described (Liu et al., 2021). Virulence associated genes were predicted using VFDB1 (Liu B. et al., 2019a). Antimicrobial resistance genes (ARGs) were identified by using Resfinder2 (Bortolaia et al., 2020). The multilocus sequence type (MLST) was identified by submitting the genome sequence to MLST 2.0 (Larsen et al., 2012). Capsular serotyping was achieved by submitting the genome sequence to Kaptive Web3 (Wyres et al., 2016). Each plasmid identified in this study was analyzed using PlasmidFinder4 and pMLST5 to investigate the replicons (Carattoli et al., 2014), and sequences were compared using BLAST analysis and the BLAST Ring Image Generator (Alikhan et al., 2011). Sequence alignments for the genetic environments of blaNDM–1 and mcr-9 were performed using BLAST and visualized with Easyfig v 2.2.3 (Sullivan et al., 2011).
Conjugation Experiments
The transferability of the blaNDM–1 and mcr-9 genes was assessed using conjugation as previously described (Ai et al., 2021). Approximately 1 × 108 colony-forming units (CFU)/mL of the donor strain (KPN710429) and the recipient strain (sodium azide-resistant E. coli J53) were mixed at 1:1 ratio and spotted onto filter membrane, which were placed on Muller-Hinton (MH) agar plates and incubated for 18–24 h at 37°C. Transconjugants were selected on MH agar containing both sodium azide (300 μg/mL) and ceftriaxone (32 μg/mL). Presence of blaNDM–1 and mcr-9 in the transconjugants (designated as J53-KPN710429) was confirmed by PCR and sequencing analysis, Supplementary Table 1 lists the primers sequences. The minimum inhibitory concentrations (MICs) of antimicrobial agents for the transconjugants were assessed using the broth microdilution method.
Colistin Induction Assays
Colistin induction assays were performed as previously described with some modifications (Kieffer et al., 2019). KPN710429 were inoculated in MH broth supplemented with colistin (0.5, 1.0, and 2.0 μg/mL) or without colistin at final bacterial suspensions of 1.0 MCF, respectively. After shaking at 37°C for 6 h, the strains were enriched by centrifugation and washed three times with sterile saline, then 0.5 MCF bacterial suspensions were prepared for colistin susceptibility testing and mRNA extraction. The relative expression level of mcr-9 and IncHI2-repA were determined by quantitative real-time (qRT)-PCR as described previously (Pfaffl, 2001). Expression of the K. pneumoniae rpoB housekeeping gene was used to normalize the transcript levels.
Additionally, the mgrB gene of induced strains was amplified and sequenced as previously described (Haeili et al., 2017), then the above sequences were aligned with that of the original strain to determine whether the mgrB was mutated during the colistin induction assay. Supplementary Table 1 lists the primers sequences.
Statistics
The Shapiro–Wilk method was used to test normality. Normally data are presented as means ± standard deviations. Bliss method were used to calculated the 50% lethal dose (LD50) of G. mellonella larvae (72 h after infection), and the LD50 values were expressed as log10(lg) transformed values. Statistical analysis was performed with GraphPad Prism (San Diego, CA, United States) using unpaired Student’s t-tests. P-values of <0.05 was considered statistically significant.
Nucleotide Sequence Accession Numbers
The K. pneumoniae KPN710429 genome sequence was submitted to GenBank under accession numbers CP073657 (plasmid pK710429_1), CP073658 (plasmid pK710429_2), CP073659 (plasmid pK710429_3), and CP073660 (chromosome of KPN710429).
Ethics Statement
The K. pneumoniae strain KPN710429 was isolated from a clinical specimen, generated as part of routine clinical laboratory procedure. The Ethics Committee of The Second Hospital of Anhui Medical University exempted this study from review because it focused only on bacteria.
Results
Antimicrobial Susceptibility and Virulence Assessments
Strain KPN710429 was identified as K. pneumoniae using MALDI-TOF-MS, and its string test was negative. In vitro AST results showed that strain KPN710429 was resistant to all cephalosporins, carbapenems, β-lactam/β-lactamase inhibitors, aminoglycosides, and quinolones, but susceptible to aztreonam, tigecycline and colistin (Table 1). The serum-killing assays demonstrated that KPN710429 was serum intermediate sensitive (grade 3) (Supplementary Figure 1). The lgLD50 of G. mellonella larvae due to KPN710429 (5.54 ± 0.16) was higher than that due to KPN54798H–ctrl (4.93 ± 0.08) (P = 0.0043), and there was no significant different from that due to KPN49L–ctrl (5.86 ± 0.19) (P = 0.0907).
TABLE 1.
Antimicrobial susceptibility of strain KPN710429, its transconjugants and E. coli J53.
| Antimicrobial agents | KPN710429 | J53-KPN710429 | E. coli J53 | |
| MICa | MICa | MICa | ||
| Aminoglycosides | AMK | 64(R) | 32(I) | 2(S) |
| GEN | 64(R) | 8(I) | 1(S) | |
| TOB | 64(R) | 8(I) | 1(S) | |
| Monocyclic β-lactam | ATM | 1(S) | 1(S) | 1(S) |
| Cephalosporin | FEP | 64(R) | 16(R) | 0.125(S) |
| FOX | 128(R) | 64(R) | 2(S) | |
| CAZ | 128(R) | 64(R) | 0.125(S) | |
| CRO | 128(R) | 64(R) | 0.25(S) | |
| CXM | 128(R) | 64(R) | 0.25(S) | |
| CTX | 128(R) | 64(R) | 0.25(S) | |
| β-lactam/β-lactamase inhibitor | TIC | 128(R) | 64(R) | 1(S) |
| TZP | 128(R) | 128(R) | 1(S) | |
| CZA | 64(R) | 32(R) | 1(S) | |
| Carbapenems | ETP | 32(R) | 16(R) | 0.125(S) |
| IPM | 32(R) | 8(R) | 1(S) | |
| MEM | 32(R) | 8(R) | 0.25(S) | |
| Quinolones | CIP | 4(R) | 0.5(I) | 0.0625(S) |
| LEV | 8(R) | 1(I) | 0.125(S) | |
| Others | TGC | 0.5(S) | 0.5(S) | 0.5(S) |
| COL | 1(S) | 1(S) | 0.5(S) | |
aThe unit for MIC is μg/mL. MIC, minimum inhibitory concentration; S, susceptible; I, intermediate; R, resistant; AMK, amikacin; ATM, aztreonam; FEP, cefepime; FOX, cefoxitin; CAZ, ceftazidime; CZA, ceftazidime-avibactam; CRO, ceftriaxone; CTX, cefotaxime; CXM, cefuroxime; CIP, ciprofloxacin; COL, colistin; ETP, ertapenem; GEN, gentamicin; IPM, imipenem; LEV, levofloxacin; MEM, meropenem; TIC, ticarcillin-clavulanic acid; TZP, piperacillin-tazobactam; TGC, tigecycline; and TOB, tobramycin.
Genome Feature of K. pneumoniae Strain KPN710429
Whole-genome sequencing revealed that the genome of K. pneumoniae strain KPN710429 comprised a single chromosome (5,124,764 bp) and three plasmids ranging in size from 70,448 bp to 288,244 bp (Table 2). In silico analysis assigned this isolate to sequence type (ST) 1308 and serotype KL144. ResFinder analysis was performed to identify a number of ARGs. Among them, blaOKP, fosA, oqxA, and oqxB were located on the chromosome, while all other ARGs, aac (6′)-Ib-cr, aadA2b, ant (2″)-Ia, blaNDM–1, mcr-9, qacE, qnrA1, sul1, and tet(A), were harbored by the largest plasmid pK710429_2. Virulence gene analysis showed that the aerobactin receptor gene (iutA), enterobactin gene cluster (entABCDEFS/fepABCDG), salmochelin gene cluster (iroEN), type 1 fimbriae gene cluster (fimABCDEFHIK), type 3 fimbriae gene cluster (mrkABCDFHIJ), were present on the chromosome, while no virulence associated genes were detected in the three plasmids.
TABLE 2.
Genomic features of the strain KPN710429.
| Parameter | Chromosome | pK710429_1 | pK710429_2 | pK710429_3 |
| Genome size (bp) | 5,124,764 | 170,592 | 288,244 | 70,448 |
| GC content (%) | 58.05 | 51.67 | 46.91 | 52.44 |
| Plasmid Inc. type | NA | IncFIB | IncHI2/HI2A | IncR |
| MLST/pMLST | ST1308 | None | ST1 | None |
| Antimicrobial resistance gene | blaOKP, fosA, oqxA, oqxB | None | aac (6′)-Ib-cr, aadA2b, ant (2″)-Ia, blaNDM–1, mcr-9, qacE, qnrA1, sul1, tet(A) | None |
| Virulence gene | entABCDEFS, fepABCDG, iutA, iroEN, mrkABCDFHIJ, fimABCDEFGHIK | None | None | None |
Inc, incompatibility; NA, not applicable.
Comparative Genomic Analysis of pK710429_2 and Conjugation Experiment
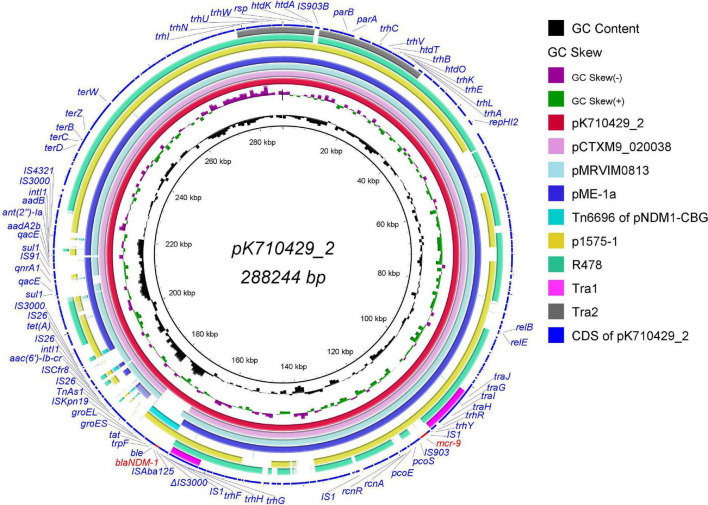
Sequence analysis showed that pK710429_2 was a large multidrug resistant (MDR) plasmid with a length of 288,244 bp and containing 302 coding genes. This plasmid belongs to the IncHI2/HI2A group with an average G + C content of 46.91%. With 81% query coverage and 99.97% identity in BLASTn, the sequence of pK55602_2 was partially consistent with that of plasmid p1575-1 (accession no. CP068288), a recently reported blaNDM–1-mcr-9-co-positive-IncHI2 plasmid harbored by a clinical isolate of Enterobacter hormaechei (Ai et al., 2021; Figure 1). Further analyses revealed that the pK710429_2 sequence can be roughly divided into two distinct regions based on its similarity to different plasmids. The first mcr-9-harboring-region (accounted for 97% of the plasmid sequences) was highly similar to the sequences of several previously reported mcr-9-harboring-plasmids, including pCTXM9_020038 (99% query coverage and 99.99% identity, accession no. CP031724), pMRVIM0813 (98% query coverage and 99.99% identity, accession no. KP975077), and pME-1a (96% query coverage and 99.99% identity, accession no. CP041734). Seven ARGs [aac(6′)-Ib-cr, aadA2b, ant(2″)-Ia, qacE, qnrA1, sul1, and tet(A)] and two class 1 integrons were found clustered within a 39,671-bp-long array of IS26/IS4321-bounded gene cluster (positions 168, 917-176, 764 bp) in this region. The second blaNDM–1-harboring-region, accounted for 3% of the plasmid sequences (positions 168, 917-176, 764 bp), was highly similar to the sequences of the Tn6696 region in plasmid pNDM1-CBG (100% query coverage and 99.99% identity, accession no. CP046118) (Figure 1).
FIGURE 1.
Genomic sequence of plasmid pK710429_2 compared with other IncHI2 plasmids, the Tn6696 region and the Tra1/Tra2 region. The complete plasmid pK710429_2 sequence was used as the reference, and the white and colored regions of the circles indicate absence and presence, respectively. The circles from inside to outside indicate the GC content of plasmid pK710429_2, GC skew of plasmid pK710429_2, plasmid pK710429_2, plasmid pCTXM_020038 (accession no. CP031724), plasmid pMRVIM0813 (accession no. KP975077), plasmid pME-1a (accession no. CP041734), the Tn6696 region of plasmid pNDM1-CBG (accession no. CP046118), plasmid p1575-1 (accession no. CP068288), plasmid R478 (accession no. BX664015), the Tra1/Tra2 region in R478 and coding sequence (CDS) of plasmid pK710429_2. blaNDM–1 and mcr-9 are indicated in red; the other genes are in blue. Δ indicates the truncated gene.
Additionally, with 76% query coverage and 99.96% identity in BLASTn, the sequence of pK55602_2 was partially consistent with that of plasmid R478 (accession no. BX664015), the first prototype IncHI2 plasmid in which all the conjugative transfer genes were present in the Tra1 and Tra2 regions (Gilmour et al., 2004). Different from plasmid R478, the Tra1 region in pK710429_2 was divided into two parts at 101,547–111,838 bp and 161,470–167,880 bp, and the Tra2 region was split by an IS903B-like element (Figure 1). In the conjugation experiments, pK710429_2 was transferred to E. coli J53 at the frequency of 1.2 × 10–6. PCR detection confirmed that the transconjugant (J53-KPN710429) was positive for blaNDM–1 and mcr-9. Phenotypic testing of the transconjugant showed that it was resistance to all β-lactams except aztreonam (Table 1).
Genetic Environment of blaNDM–1 and mcr-9
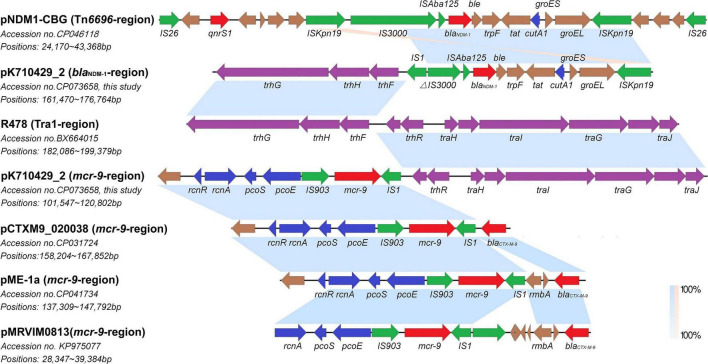
In plasmid pK710429_2, blaNDM–1 gene was located on a region with the cassette structure of ΔIS3000-ISAba125-blaNDM–1-ble-trpF-tat-cutA1-groES-groEL-ISKpn19, which was partly similar to that of Tn6696 in pNDM1-CBG. Parts of the Tar1 region were located upstream of this cassette, with an IS1 between them. Interestingly, the remaining parts of the Tar1 region were located downstream of mcr-9, also with an IS1 between them. The rcnR-rcnA-pcoE-pcoS-IS903 structure, locating upstream of mcr-9 in pK710429_2, was consistent with that of plasmid pCTXM9_020038, pME-1a, and pMRVIM0813 (Figure 2). However, qseB-qseC regulatory genes were not found in plasmid pK710429_2.
FIGURE 2.
Co-linear analyses for the genetic environment of blaNDM–1 and mcr-9 in plasmid pK710429_2. The blaNDM–1-harboring-region in plasmid pK710429_2 was compared with TN6696-region in plasmid pNDM1-CBG and Tra1-region in plasmid R478; the mcr-9-harboring-region in plasmid pK710429_2 was compared with Tra1-region in plasmid R478, mcr-9-harboring-region in plasmid pCTXM9_020038, pME-1a, and pMRVIM0813. Blue or orange shading denotes regions of shared homology or inversion among different plasmids, respectively. Colored arrows indicate open reading frames, with red, green, blue, purple, and brown arrows representing antimicrobial resistance genes, mobile elements, heavy metal resistance genes, conjugative transfer genes, and plasmid backbone genes, respectively. Δ indicates the truncated gene.
Colistin Induction Assays
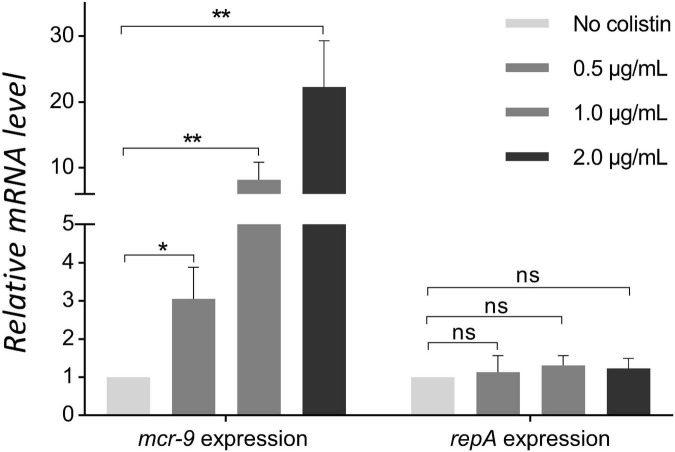
The inducibility of mcr-9 expression was detected using colistin induction assays. After pretreatment with 0.5, 1, and 2 μg/mL of colistin, the KPN710429 exhibited colistin MICs of 4, 16, and 32 μg/mL, respectively. The mcr-9 gene expressions were upregulated accordingly, while IncHI2 plasmid replicase gene repA expression remained unchanged (Figure 3). The mgrB gene was not mutated during the colistin induction assay (Supplementary Figure 2).
FIGURE 3.
Comparison of mcr-9 and repA expression levels in strain KPN710429 with and without colistin pretreatment. Error bars indicate the standard deviation for three triplicate samples. Asterisks indicate statistical significance at different levels by Student’s t-tests. *P < 0.05; **P < 0.01; ns, not significant.
Discussion
Few clinical anti-infective therapeutic options exist to combat the emergence and spread of MDR gram-negative pathogens co-harboring carbapenemase-encoding and mcr-family genes (Mmatli et al., 2020). Although mcr-9 was first identified in 2019, recent research indicates that it has existed for a long time and has been spreading among CRKP isolates of human origin in Europe since 2013 (Wang et al., 2020). In China, clinical isolates of Enterobacteriales co-harboring blaNDM–1 and mcr-9 have been reported, all of which belong to Enterobacter spp., and the blaNDM–1 and mcr-9 genes were harbored by different plasmids in most strains (Yuan et al., 2019; Lin et al., 2020; Ai et al., 2021). Here, we report a blaNDM–1 and mcr-9 co-harboring clinical isolate of K. pneumoniae, with both blaNDM–1 and mcr-9 located on the same plasmid. To our knowledge, this is the first report of a blaNDM–1 and mcr-9 co-encoding plasmid harbored by clinical CRKP isolate in China.
Carbapenem-resistant K. pneumoniae strain KPN710429 belongs to ST1308, a rarely sequence type which has been reported as a singleton clone and recovered from inanimate surfaces in hospital environments (Zenati et al., 2017). The string test of KPN710429 was negative, and this strain didn’t exhibit a hypervirulent phenotype according to in vitro and in vivo virulence assessments. AST results and the international classification scheme (Magiorakos et al., 2012) classify strain KPN710429 as an MDR bacterium, with an MDR phenotype closely related to the ARGs it harbored. WGS data revealed that most ARGs, including blaNDM–1 and mcr-9, were harbored by the IncHI2/HI2A-ST1 plasmid, pK710429_2. Notably, IncHI2-ST1 plasmids were found to be the predominant replicon type carrying mcr-9, and the rcnR-rcnA-pcoE-pcoS-IS903-mcr-9-wbuC structure was consistent in most mcr-9 cassettes (Li et al., 2020). pK710429_2 also harbored this core structure but without wbuC. Additionally, no qseC-qseB, a two-component system involved in mcr-9 expression (Kieffer et al., 2019), was found in pK710429_2, which might explain the colistin susceptibility phenotype of strain KPN710429. Further analysis indicated that pK710429_2 was a conjugative plasmid, and pK710429_2 contained the conjugative transfer gene regions, Tra1 and Tra2, with some structural variations.
In addition to the conjugative plasmid, the accumulation and dissemination of resistance genes was largely due to the actions of mobile genetic elements, including insertion sequences, transposons, gene cassettes, and integrons. In this study, the cassette structure harboring blaNDM–1 was flanked by several mobile elements. Notably, the blaNDM–1 cassette was similar to that of Tn6696, a recently reported transposon (Chen et al., 2020), suggesting that the plasmid pK710429_2 could be a hybrid plasmid, formed by a Tn6696-like blaNDM–1 region inserting into a mcr-9-positive-IncHI2/HI2A plasmid.
Unlike other alleles of mcr genes which consistently display colistin resistance, most of the mcr-9-positive isolates were found to susceptible to colistin (Carroll et al., 2019; Chavda et al., 2019; Tyson et al., 2020). The mcr-9 is an inducible gene encoding phosphoethanolamine transferase, and mcr-9 expression was induced in the presence of colistin (Kieffer et al., 2019). In this study, the colistin resistance phenotype remained absent in KPN710429, but the induction assays showed that after pretreatment with different concentrations of colistin for 6 h, KPN710429 showed the colistin resistance phenotype, and the mcr-9 gene expressions were upregulated accordingly. Because mutation of mgrB gene is one of the mechanisms that contributes to colistin resistance in K. pneumoniae (Haeili et al., 2017), we further confirmed that no mgrB mutation occurred in KPN710429 during the colistin induction assay. A previous study revealed that the lacking of two-component regulatory genes qseC-qseB was commonly observed among the mcr-9-positive plasmid (Li et al., 2020). In this study, plasmid pK710429_2 also lacked the qseB-qseC regulatory genes. More research is needed to confirm the essential role of the qseC-qseB module or the involvement of other genes in mcr-9 induction.
Additionally, the novel antibiotic ceftazidime-avibactam was ineffectively against blaNDM–1-positive bacteria (Zasowski et al., 2015), thus, clinical CRKP isolates co-harboring blaNDM–1 and mcr-9 usually exhibit a colistin sensitive but ceftazidime-avibactam resistant phenotype, which may mislead colistin to become a very possible anti-infection option. Therefore, if the blaNDM–1-and mcr-9-co-harboring plasmid becomes widely spread among Enterobacteriales, it will likely bring more serious adverse consequences. Therefore, it is imperative to establish a molecular screening method for mcr-9 to facilitate its rapid and accurate detection.
Conclusion
Here, we reported the blaNDM–1- and mcr-9-co-encoding transferable IncHI2/HI2A plasmid, pK710429_2, obtained from the ST1308 MDR clinical K. pneumoniae strain, KPN710429, recovered in China. WGS and comparative genomics assays revealed that plasmid pK710429_2 could be a hybrid plasmid, formed by a Tn6696-like blaNDM–1 region inserting into a mcr-9-positive-IncHI2/HI2A plasmid, indicating that horizontal gene transfer events play a key role in the plasmid evolution. Although pK710429_2 lacked qseC-qseB, the mcr-9 showed increased expression when induced with colistin, leading to colistin resistance in KPN710429. This finding indicated that the co-transfer of carbapenemase-encoding gene and mcr-9 in clinically important pathogens poses a high risk to the clinical treatment. Effective surveillance is needed to assess the prevalence of the plasmid co-harboring carbapenemase-encoding gene and mcr-9.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XiL, QZ, and JL conceived and designed the experiments. ZL, XH, and XX performed the experiments. WC, XinL, and YL analyzed the data. ZL, XX, and XiL wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 81973983 and 82172306), Anhui Natural Science Foundation (No.1908085QH366), and the Project Supported by Public Technology Research Projects of Zhejiang Province, China (No. LGD21H190001).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.756201/full#supplementary-material
Serum-killing assays of the tested and references strains. Data are presented as the mean ± standard error, and lg-transformed values were utilized to normalize the data (n = 3 for each isolate). KPN54798H–ctrl, KPN710429, and KPN49L–ctrl were serum resistant (grade 6), serum intermediate sensitive (grade 3), and serum sensitive (grade 1), respectively.
Alignment analysis of the mgrB gene sequences between the original KPN710429 strain and the three induced strains. All four mgrB gene sequences were identical, and no mismatch was detected.
References
- Ai W., Zhou Y., Wang B., Zhan Q., Hu L., Xu Y., et al. (2021). First Report of Coexistence of blaSFO–1 and blaNDM–1 β-Lactamase Genes as Well as Colistin Resistance Gene mcr-9 in a Transferrable Plasmid of a Clinical Isolate of Enterobacter hormaechei. Front. Microb. 12:676113. 10.3389/fmicb.2021.676113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genom. 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolaia V., Kaas R. S., Ruppe E., Roberts M. C., Schwarz S., Cattoir V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75 3491–3500. 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58 3895–3903. 10.1128/aac.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll L. M., Gaballa A., Guldimann C., Sullivan G., Henderson L. O., Wiedmann M. (2019). Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. MBio 10 e853–e819. 10.1128/mBio.00853-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavda K. D., Westblade L. F., Satlin M. J., Hemmert A. C., Castanheira M., Jenkins S. G., et al. (2019). First Report of blaVIM–4- and mcr-9-Coharbouring Enterobacter Species Isolated from a Pediatric Patient. mSphere 4 e629–e619. 10.1128/mSphere.00629-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Lin Y., Li Z., Lu L., Li P., Wang K., et al. (2020). Characterization of a New Transposon, Tn6696, on a blaNDM–1-Carrying Plasmid From Multidrug-Resistant Enterobacter cloacae ssp. dissolvens in China. Front. Microbiol. 11:525479. 10.3389/fmicb.2020.525479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2020). Performance Standards for Antimicrobial Susceptibility Testing, 30th Edn. Wayne, PA: CLSI supplement M100. [Google Scholar]
- Effah C. Y., Sun T., Liu S., Wu Y. (2020). Klebsiella pneumoniae: an increasing threat to public health. Ann. Clin. Microbiol. Antimicrob. 19:1. 10.1186/s12941-019-0343-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST (2020). Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0. [Google Scholar]
- Gilmour M. W., Thomson N. R., Sanders M., Parkhill J., Taylor D. E. (2004). The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52 182–202. 10.1016/j.plasmid.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Haeili M., Javani A., Moradi J., Jafari Z., Feizabadi M. M., Babaei E. (2017). MgrB Alterations Mediate Colistin Resistance in Klebsiella pneumoniae Isolates from Iran. Front. Microbiol. 8:2470. 10.3389/fmicb.2017.02470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer N., Royer G., Decousser J. W., Bourrel A. S., Palmieri M., Ortiz, et al. (2019). mcr-9, an Inducible Gene Encoding an Acquired Phosphoethanolamine Transferase in Escherichia coli, and Its Origin. Antimicrob. Agents Chemother. 63 e965–e919. 10.1128/AAC.00965-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50 1355–1361. 10.1128/jcm.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Dai X., Zeng J., Gao Y., Zhang Z., Zhang L. (2020). Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci. Rep. 10:8113. 10.1038/s41598-020-65106-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Yang Y., Yang Y., Chen G., He R., Wu Y., et al. (2020). Co-Occurrence of mcr-9 and blaNDM–1 in Enterobacter cloacae Isolated from a Patient with Bloodstream Infection. Infect. Drug Resist. 13 1397–1402. 10.2147/idr.s248342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zheng D., Jin Q., Chen L., Yang J. (2019a). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47 D687–D692. 10.1093/nar/gky1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Liu Z., Chu W., Li X., Tang W., Ye J., Zhou Q., et al. (2021). Genomic Features and Virulence Characteristics of a Community-Acquired Bloodstream Infection-Causing Hypervirulent Klebsiella pneumoniae ST86 Strain Harbouring KPC-2-Encoding IncX6 Plasmid. Microb. Drug Resist. 27 360–368. 10.1089/mdr.2019.0394 [DOI] [PubMed] [Google Scholar]
- Liu Z., Gu Y., Li X., Liu Y., Ye Y., Guan S., et al. (2019b). Identification and Characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China. Ann. Lab. Med. 39 167–175. 10.3343/alm.2019.39.2.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Mazloum M., Le Meur M., Barnaud G., Messika J. (2016). Hypermucoviscous Klebsiella pneumoniae pneumonia: follow the string! Intensive Care Med. 42 2092–2093. 10.1007/s00134-016-4363-y [DOI] [PubMed] [Google Scholar]
- Mmatli M., Mbelle N. M., Maningi N. E., Osei Sekyere J. (2020). Emerging Transcriptional and Genomic Mechanisms Mediating Carbapenem and Polymyxin Resistance in Enterobacteriaceae: a Systematic Review of Current Reports. mSystems 5 e783–e720. 10.1128/mSystems.00783-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Jayol A., Nordmann P. (2017). Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 30 557–596. 10.1128/cmr.00064-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson G. H., Li C., Hsu C. H., Ayers S., Borenstein S., Mukherjee S., et al. (2020). The mcr-9 Gene of Salmonella and Escherichia coli Is Not Associated with Colistin Resistance in the United States. Antimicrob. Agents Chemother. 64 e573–e520. 10.1128/AAC.00573-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu F., Hu Y., Zhang G., Zhu B., Gao G. F. (2020). Detection of mobile colistin resistance gene mcr-9 in carbapenem-resistant Klebsiella pneumoniae strains of human origin in Europe. J. Infect. 80 578–606. 10.1016/j.jinf.2019.12.016 [DOI] [PubMed] [Google Scholar]
- Wu W., Feng Y., Tang G., Qiao F., McNally A., Zong Z. (2019). NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 32 e00115–e00118. 10.1128/cmr.00115-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyres K. L., Wick R. R., Gorrie C., Jenney A., Follador R., Thomson N. R., et al. (2016). Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2:e000102. 10.1099/mgen.0.000102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D., Toleman M. A., Giske C. G., Cho H. S., Sundman K., Lee K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, blaNDM–1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53 5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Li Y., Wang G., Li C., Xiang L., She J., et al. (2019). Coproduction Of MCR-9 And NDM-1 By Colistin-Resistant Enterobacter hormaechei Isolated From Bloodstream Infection. Infect. Drug Resist. 12 2979–2985. 10.2147/idr.s217168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasowski E. J., Rybak J. M., Rybak M. J. (2015). The β-Lactams Strike Back: Ceftazidime-Avibactam. Pharmacotherapy 35 755–770. 10.1002/phar.1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenati K., Sahli F., Garcia V., Bakour S., Belhadi D., Rolain J. M., et al. (2017). Occurrence and clonal diversity of multidrug-resistant Klebsiella pneumoniae recovered from inanimate surfaces in Algerian hospital environment: First report of armA, qnrB and aac(6’)-Ib-cr genes. J. Glob. Antimicrob. Resist. 10 148–153. 10.1016/j.jgar.2017.05.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Serum-killing assays of the tested and references strains. Data are presented as the mean ± standard error, and lg-transformed values were utilized to normalize the data (n = 3 for each isolate). KPN54798H–ctrl, KPN710429, and KPN49L–ctrl were serum resistant (grade 6), serum intermediate sensitive (grade 3), and serum sensitive (grade 1), respectively.
Alignment analysis of the mgrB gene sequences between the original KPN710429 strain and the three induced strains. All four mgrB gene sequences were identical, and no mismatch was detected.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.