Abstract
Mycobacterium celatum type 1 was found to cross-react in the AccuProbe Mycobacterium tuberculosis complex assay. Subsequently, we found a statistically significant increase in the relative light units with lower temperatures, suggesting that it is necessary to perform this AccuProbe assay at between 60 and 61°C. We also recommend the inclusion of M. celatum type 1 as a negative control.
The introduction and routine application of nucleic acid probes, such as the AccuProbe Mycobacterium tuberculosis complex assay (TB AccuProbe; Gen-Probe Incorporated, San Diego, Calif.), has considerably shortened the time required for identification of the M. tuberculosis complex while providing high sensitivity and specificity (15). However, in a study by Butler et al. (2) cross-reactivity was observed between M. celatum type 1, but not type 2, in the TB AccuProbe. DNA sequencing showed that M. celatum type 1 differs by a single nucleotide from the probe used in the assay (6), while type 2 differs by four nucleotides. Recently, Bull and coworkers identified a novel subtype of the pathogen M. celatum, type 3, that also showed cross-reactivity in the TB AccuProbe (1); however, the DNA sequence for the probe region was not reported.
M. celatum is a newly recognized, slow-growing, nonpigmented species whose biochemical characteristics and colony morphology are similar to those of M. avium complex, M. malmoense, M. shimoidei, and M. xenopi (1, 3, 11). The three types of M. celatum can be distinguished from one another and from other mycobacteria by DNA sequencing or restriction fragment length polymorphism analysis of selected genes and by high-performance liquid chromatography (3, 10, 11, 17; J. Baldus-Patel, D. G. Leonard, X. Pan, J. M. Musser, R. E. Berman, and I. Nachamkin, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. U-18, p. 637, 1999). In recent years, a considerable amount of clinical evidence has indicated that M. celatum can lead to fatal disease in both immunocompetent and immunocompromised patients (4, 8, 14, 18). Therefore, the impact of misidentification in the TB AccuProbe can be significant (5).
Thus, M. celatum type 1 (ATCC 51131) was submitted for identification to 137 laboratories participating in New York State's proficiency testing program. Ninety of the 137 laboratories used the TB AccuProbe, and one-third of those reported false-positive (>30,000) relative light units (RLU). These results indicate that in spite of Gen-Probe's previous modification of the selection time from 5 to 10 min (16; 1993 original and 1994 revised package inserts for the AccuProbe cultural identification test, Gen-Probe Incorporated), a considerable number of laboratories still obtained false-positive TB AccuProbe results with M. celatum.
To identify possible reasons for the false-positive results, we requested procedural details from the 90 laboratories. Unfortunately, this information did not reveal any significant differences in methodology between those with positive and those with negative RLU values. However, in a previous report on cross-reactivity of M. terrae with the TB AccuProbe (7), it was found that changes in temperature influenced the specificity of the test results. Thus, we sought to determine the effects of both selection step time and temperature by testing M. celatum type 1 (ATCC 51131) in the TB AccuProbe in one of our laboratories. First, the M. celatum isolate was tested to ensure that M. tuberculosis was not also present in the suspension. This test was negative. We then performed experiments using M. celatum previously grown on either Löwenstein-Jensen medium or in Middlebrook 7H9 broth. When broth was used, 1.5 ml of broth was centrifuged, the supernatant was removed, and the pellet was resuspended in 500 μl of broth. One-hundred-microliter aliquots of loopfuls of growth from solid media or resuspended broth were tested according to the TB AccuProbe protocol (AccuProbe culture identification test revised package insert). It was noted that the suspensions from the solid media contained considerably more bacteria than those prepared from broth-grown cultures. Experiments were carried out in triplicate with selection times of 9, 10, 11, and 12 min at 60°C (standard temperature) and with temperatures of 58, 59, 60, and 61°C at 10 min (standard selection time). Suspensions of M. tuberculosis and M. avium previously grown on solid media were included as positive and negative controls. The RLU values are expressed as means + standard errors of the means (SEM). Comparisons were performed by the Mann-Whitney U test.
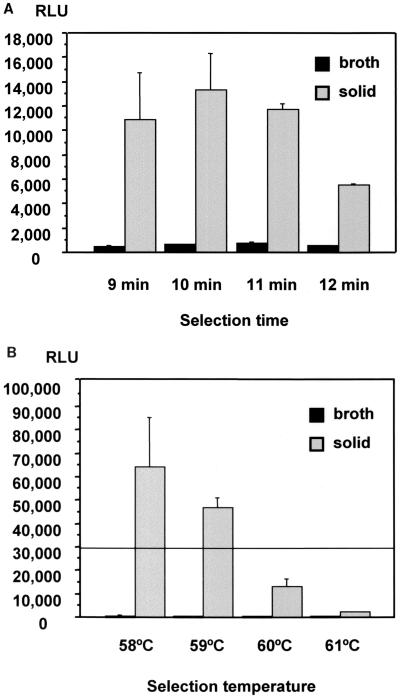
The results on the effect of the selection time are shown in Fig. 1A. There were no statistically significant differences between the mean RLU values at the different selection times for suspensions from the same medium. However, the RLU values from the solid medium suspension were significantly higher than those from the broth-grown suspension (P < 0.05). As stated before, the solid medium suspension contained a greater cell density than the broth-grown suspension. Thus, these results suggest that the M. celatum cell density could influence the RLU value. However, the highest RLU values obtained were still under the cutoff of 30,000 (Fig. 1A).
FIG. 1.
(A) Effect of selection time on RLU value in the AccuProbe M. tuberculosis complex assay with M. celatum. There were no statistically significant differences between the mean RLU values with solid or liquid medium for the different selection times. The RLU values from the solid medium suspension were significantly higher than those from the broth-grown suspension at all selection times (P < 0.05). Data are expressed as means plus SEM. The RLU cutoff is 30,000. (B) Effect of temperature on RLU value in the AccuProbe M. tuberculosis complex assay with M. celatum. The Mann-Whitney U test revealed significant differences between the mean RLU values with solid medium for 58°C versus 60 and 61°C, between 59°C versus 60 and 61°C, and 60°C versus 61°C (P < 0.05). The RLU values from the solid medium suspension were significantly higher than those from the broth-grown suspension at all selection temperatures (P < 0.05). Data are expressed as means plus SEM. The RLU cutoff is indicated by a horizontal line.
As to the effect of the temperature, the RLU value obtained with a suspension from solid media was found to decrease as the temperature was increased, as shown in Fig. 1B. Statistical analysis revealed a significant difference between the RLU values from the solid medium suspension at 58°C versus 60 and 61°C, at 59°C versus 60 and 61°C, and at 60 versus 61° (P < 0.05). There were no significant differences between the RLU values from the broth-grown suspension at the different temperatures. But once again, the RLU values from the solid medium suspension were significantly higher than those from the broth-grown suspension at all temperatures (P < 0.05).
In summary, our experiments revealed that the number of organisms present could strongly influence the RLU value, as there was a statistically significant difference between the RLU values of suspensions derived from broth versus solid medium. These results suggest that the presence of a larger biomass of M. celatum from the solid medium resulted in increased signal. Furthermore, in agreement with the previous findings of Ford et al. (7) for cross-reaction of M. terrae in the TB AccuProbe, we also found that changes in temperature can significantly affect the RLU value when a heavy inoculum of M. celatum is present. We found a statistically significant increase in RLU with lower temperatures; however, varying the selection time did not appear to have an effect on the signal with M. celatum. These results suggest that in order to eliminate cross-reactivity with M. celatum, it is imperative to perform the test with a selection temperature between 60 and 61°C rather than the presently recommended 60 ± 1°C.
Of the 90 participating laboratories that used the TB AccuProbe, only one reported a selection temperature (59.5°C) lower than the 60°C recommended by Gen-Probe. However, only 9 (11%) participants checked the temperature of their heating instruments before each testing, while the majority (51%) checked the temperatures only annually, and some did not check at all (3%). The controlled experimental results of this study strongly suggest that frequent temperature checks on heating instruments are warranted. The guidelines of the National Committee for Clinical Laboratory Standards for molecular diagnostic methods for infectious diseases state the following (13): “Quality control charts that indicate acceptable ranges should be posted on all water baths, incubators, and heating blocks. Temperatures should be checked and recorded on these charts daily.”
In order to monitor the performance of the test, we recommend that M. celatum type 1 (ATCC 51131) be used as a negative control either instead of or along with the presently suggested M. avium (ATCC 25291). Furthermore, when identifying members of the M. tuberculosis complex, cellular and colony morphology should be taken into consideration (9, 12). Finally, since M. celatum may be difficult to identify with conventional biochemical tests, we recommend the use of molecular diagnostic techniques and/or high-performance liquid chromatography for the most accurate identification of this organism (3, 10, 11, 13a, 17).
Acknowledgments
We thank Olivia Montaño for excellent technical assistance.
Á. Somoskövi was supported in part by grant no. 1D43TW00915 from the Fogarty International Center, National Institutes of Health.
REFERENCES
- 1.Bull T J, Shanson D C, Archard L C, Yates M D, Hamid M E, Minnikin D E. A new group (type 3) of Mycobacterium celatum isolated from AIDS patients in the London area. Int J Syst Bacteriol. 1995;45:861–862. doi: 10.1099/00207713-45-4-861. [DOI] [PubMed] [Google Scholar]
- 2.Butler W R, O'Connor S P, Yakrus M A, Gross W M. Cross-reactivity of genetic probe for detection of Mycobacterium tuberculosis with newly described species Mycobacterium celatum. J Clin Microbiol. 1994;32:536–538. doi: 10.1128/jcm.32.2.536-538.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler W R, O'Connor S P, Yakrus M A, Smithwick R W, Plikaytis B B, Moss C W, Floyd M M, Woodley C L, Kilburn J O, Vadney F S, Gross W M. Mycobacterium celatum sp. nov. Int J Syst Bacteriol. 1993;43:539–548. doi: 10.1099/00207713-43-3-539. [DOI] [PubMed] [Google Scholar]
- 4.Bux-Gewehr I, Hagen H P, Rüsch-Gerdes S, Feurle G E. Fatal pulmonary infection with Mycobacterium celatum in an apparently immunocompetent patient. J Clin Microbiol. 1998;36:587–588. doi: 10.1128/jcm.36.2.587-588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahl D M, Klein D, Morgentaler A. Penile mass caused by the newly described organism Mycobacterium celatum. Urology. 1996;47:266–268. doi: 10.1016/S0090-4295(99)80433-6. [DOI] [PubMed] [Google Scholar]
- 6.Emler S, Ninet B, Rohner P, Auckenthaler R, Jäger D, Hirschel B. Molecular basis for cross-reactivity between a strain of Mycobacterium terrae and DNA probes for Mycobacterium tuberculosis complex. Eur J Clin Microbiol Infect Dis. 1995;14:627–629. doi: 10.1007/BF01690741. [DOI] [PubMed] [Google Scholar]
- 7.Ford E G, Snead S J, Todd J, Warren N G. Strains of Mycobacterium terrae complex which react with DNA probes for M. tuberculosis complex. J Clin Microbiol. 1993;31:2805–2806. doi: 10.1128/jcm.31.10.2805-2806.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haase G, Skopnik H, Batge S, Bottger E C. Cervical lymphadenitis caused by Mycobacterium celatum. Lancet. 1994;344:1020–1021. doi: 10.1016/s0140-6736(94)91680-2. [DOI] [PubMed] [Google Scholar]
- 9.Kaminski D A, Hardy D J. Selective utilization of DNA probes for identification of Mycobacterium species on the basis of cord formation in primary BACTEC 12B cultures. J Clin Microbiol. 1995;33:1548–1550. doi: 10.1128/jcm.33.6.1548-1550.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim B-J, Lee S-H, Lyu M-A, Kim S-J, Bai G-H, Kim S-J, Chae G-T, Kim E-C, Cha C-Y, Kook Y-H. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB) J Clin Microbiol. 1999;37:1714–1720. doi: 10.1128/jcm.37.6.1714-1720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metchock B G, Nolte F S, Wallace R J., Jr . Mycobacterium. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 399–437. [Google Scholar]
- 12.Morris A J, Reller L B. Reliability of cord formation in BACTEC media for presumptive identification of mycobacteria. J Clin Microbiol. 1993;31:2533–2534. doi: 10.1128/jcm.31.9.2533-2534.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Molecular diagnostic methods for infectious diseases. Approved guideline. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1995. [Google Scholar]
- 13a.Patel J B, Leonard D G B, Pan X, Musser J M, Berman R E, Nackamkin I. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J Clin Microbiol. 2000;38:246–251. doi: 10.1128/jcm.38.1.246-251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piersimoni C, Tortoli E, De Sio G. Disseminated infection due to Mycobacterium celatum in patients with AIDS. Lancet. 1994;344:332. doi: 10.1016/s0140-6736(94)91369-2. [DOI] [PubMed] [Google Scholar]
- 15.Salfinger M, Pfyffer G E. The new diagnostic mycobacteriology laboratory. Eur J Clin Microbiol Infect Dis. 1994;11:961–979. doi: 10.1007/BF02111498. [DOI] [PubMed] [Google Scholar]
- 16.Stockman L, Springer B, Bottger E C, Roberts G D. Mycobacterium tuberculosis nucleic acid probes for rapid diagnosis. Lancet. 1993;341:1486. doi: 10.1016/0140-6736(93)90935-a. [DOI] [PubMed] [Google Scholar]
- 17.Tortoli E, Bartoloni A, Burrini C, Mantella A, Simonetti M T. Utility of high-performance liquid chromatography for identification of mycobacterial species rarely encountered in clinical laboratories. Eur J Clin Microbiol Infect Dis. 1995;14:240–243. doi: 10.1007/BF02310365. [DOI] [PubMed] [Google Scholar]
- 18.Tortoli E, Piersimoni C, Bacosi D, Bartoloni A, Betti F, Bono L, Burrini C, De Sio G, Lacchini C, Mantella A, Orsi P G, Penati V, Simonetti M T, Böttger E C. Isolation of the newly described species Mycobacterium celatum from AIDS patients. J Clin Microbiol. 1995;33:137–140. doi: 10.1128/jcm.33.1.137-140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]