Abstract
SARS, a new type of respiratory disease caused by SARS-CoV, was identified in 2003 with significant levels of morbidity and mortality. The recent pandemic of COVID-19, caused by SARS-CoV-2, has generated even greater extents of morbidity and mortality across the entire world. Both SARS-CoV and SARS-CoV-2 spreads through the air in the form of droplets and potentially smaller droplets (aerosols) via exhaling, coughing, and sneezing. Direct detection from such airborne droplets would be ideal for protecting general public from potential exposure before they infect individuals. However, the number of viruses in such droplets and aerosols is too low to be detected directly. A separate air sampler and enough collection time (several hours) are necessary to capture a sufficient number of viruses. In this work, we have demonstrated the direct capture of the airborne droplets on the paper microfluidic chip without the need for any other equipment. 10% human saliva samples were spiked with the known concentration of SARS-CoV-2 and sprayed to generate liquid droplets and aerosols into the air. Antibody-conjugated submicron particle suspension is then added to the paper channel, and a smartphone-based fluorescence microscope isolated and counted the immunoagglutinated particles on the paper chip. The total capture-to-assay time was <30 min, compared to several hours with the other methods. In this manner, SARS-CoV-2 could be detected directly from the air in a handheld and low-cost manner, contributing to slowing the spread of SARS-CoV-2. We can presumably adapt this technology to a wide range of other respiratory viruses.
Keywords: Bioaerosol, Respiratory virus, Airborne pathogens, COVID-19, Smartphone microscope, Paper microfluidics
1. Introduction
We have recently witnessed the worldwide pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing the coronavirus disease 2019 (COVID-19). While morbidity and mortality have occurred mostly with individuals with pre-existing conditions, they can also occur in healthy individuals (Fozouni et al., 2020). The current number of COVID-19 deaths amounts to 5,373,552 worldwide at the time of writing, clearly indicating the seriousness of this pandemic (https://www.worldometers.info/coronavirus/). COVID-19 diagnostics have typically been carried out on the nasopharyngeal swabs and recently on the nasal swabs from the infected patients (Zenhausern et al., 2021). However, since SARS-CoV-2 spreads through the air, it may be too late to contain its spread by testing the infected individuals. The best approach would be direct detection from the air.
Bioaerosol refers to particulate matter with life characteristics suspended in the air (Hinds, 1982). Bioaerosol sizes range from 0.02 to 30 μm. In SARS-CoV-2, water droplets are generated from the human mouth through exhaling, coughing, and sneezing. SARS-CoV-2, with a diameter of 0.1 μm, has been believed to spread through the droplets >5 μm (thus “droplets” but not “aerosols”). Therefore, 6-feet or 2-m physical distancing has emerged as a safety precaution (WHO, 2021). However, several reports have been made that SARS-CoV-2 can spread with droplets smaller than 5 μm (i.e., aerosols) (Vuorinen et al., 2020; Chia et al., 2020; Hwang et al., 2021; Chen et al., 2020).
While the best practice to prevent SARS-CoV-2 infection is a face mask (Zhang et al., 2020), it is not perfect in capturing all bioaerosols, especially when SARS-CoV-2 spreads through smaller droplets and aerosols. In addition, some people cannot wear face masks due to medical conditions, and some others are not willing to wear face masks. The best practice might be the direct detection from the air. However, the number of viruses in such air samples tends to be very low, which cannot easily be detected conventionally. Typically, air samplers are necessary to increase the number of captured viruses. There are three widely adopted methods for collecting viruses (or bacteria) from the air: impaction, impinging, and filtration (Terzieva et al., 1996; Fronczek and Yoon, 2015). The impaction method is the most common, where aerosols are aerodynamically separated by size according to their diameter. In the impinging method, the air is sucked in and passed through a device containing a liquid buffer to collect viruses or bacteria. The filtration method is economical and straightforward, where bioaerosols are collected using a fibrous or membrane filter. For all three methods, an additional incubation for more than 24 h may be required due to the small number of viruses and bacteria in the air. Very few papers have been published to detect the COVID-19 aerosols from the air directly. In the small number of such published works, droplets/aerosols were collected using the traditional air samplers for a long time, and conventional laboratory analyses were used, such as reverse transcription polymerase chain reaction (RT-PCR) and enzyme-linked immunosorbent assay (ELISA) (Piri et al., 2021; Su et al., 2020). These methods are not easy to be implemented in the field and cannot rapidly provide results.
In this study, we propose a handheld, rapid, low-cost, smartphone-based paper microfluidic assay capable of directly detecting SARS-CoV-2 in the droplets/aerosols from the air, without the need for an air sampler and the long collection time. We designed a system that simulates a human cough using a simple sprayer in a controlled chamber. Droplets/aerosols are passively collected directly on the paper microfluidic chips without using any collector, pump, fan, or filter. Antibody-conjugated submicron fluorescent particle suspension is then added to the paper microfluidic chip, inducing antibody-antigen binding and subsequent particle aggregation. A low-cost smartphone-based fluorescence microscope was fabricated, used to quantify the extent of this particle aggregation from the microscopic images, and confirmed the presence of SARS-CoV-2 from the air. To the best of our knowledge, this work is the first demonstration of direct bioaerosol capture and subsequent immunoassay on a single platform. The device and method can slow the spread of SARS-CoV-2 and other emerging respiratory viruses.
2. Materials and methods
2.1. SARS-CoV-2 samples
SARS-CoV-2 Isolate USA-WA1/2020, was deposited by Dr. Natalie J. Thornburg at the U.S. Centers for Disease Control Prevention (CDC) and obtained from the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA). SARS-CoV-2 was passaged on mycoplasma negative Vero cells (ATCC #CCL-81) at a MOI of 0.005 for 48 h. Supernatant and cell lysate were combined, subjected to a single freeze-thaw, and then centrifuged at 3000 RPM for 10 min to remove cell debris. Concentration ranged from 106 PFU/mL to 107 PFU/mL, corresponding to 2–6 ng/mL, respectively. Virus stock was poured into a 15-cm petri dish and irradiated in a Bio-Rad GS Gene Linker UV Chamber on the ‘sterilize’ setting twice for 90 s with a brief swirl in between. Complete inactivation of virus was confirmed by standard plaque assay or 50% tissue culture infectious dose (TCID50). All live virus manipulations were performed in a biosafety level 3 laboratory and procedures were approved by the University of Arizona's Institutional Biosafety Committee.
2.2. Antibody conjugation to fluorescent polystyrene particles
Yellow-green fluorescent carboxylated polystyrene particles (CAYF500NM; Magsphere, Inc., Pasadena, CA, USA) were used for assaying SARS-CoV-2. According to the manufacturer, their diameter was 500 nm, the excitation peak was 488 nm (blue), and the emission peak was 509 nm. Rabbit polyclonal antibody to SARS-CoV-2 (40588-T30; SinoBiological, Inc., Wayne, PA, USA) was covalently conjugated to the particles via carbodiimide reaction. Detailed information and optimizations can be found in the previous works (Chung et al., 2019, 2021; Kim et al., 2021; Park and Yoon, 2015). The final particle concentration was 0.06 μg/μL, optimized for showing the maximum extent of particle aggregation (Chung et al., 2021; Kim et al., 2021). Particle stocks were stored in a refrigerator.
2.3. Paper microfluidic chip
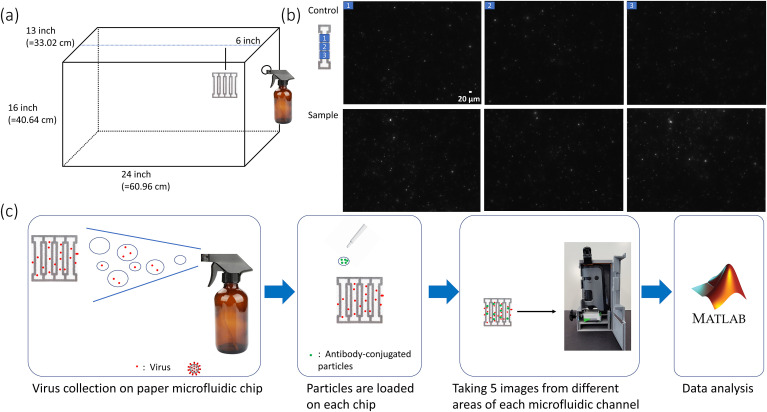
The paper microfluidic chip was designed, optimized, and wax-printed in the same way used in the previous work (Kim et al., 2021; Chung et al., 2021). There were four channels in each chip. Each channel was 21 mm long and 2.4 wide, with dumbbell-shaped squares at both ends (Fig. 1 ). Nitrocellulose paper was used (CN95; Sartorius, Goettingen, Germany). Capillary flow rate is 65–115 s over 40 mm as reported by the manufacturer. Thickness is 240–270 μm.
Fig. 1.
Assay procedure. (a) Chamber design with dimensions and the placements of a paper microfluidic chip and a sprayer. (b) Fluorescence images of a paper microfluidic channel for 0 ng/mL (control) and 600 pg/mL (sample) UV-inactivated SARS-CoV-2 in 10% saliva. Three different images from a single channel were taken with a benchtop fluorescence microscope. (c) The assay procedure, starting from the air collection, antibody-particles, smartphone-based fluorescence microscopic imaging, and image processing with the MATLAB script.
2.4. Air chamber
A fish tank was purchased and used as an air chamber (Fish Tanks Direct, North Venice, FL, USA). A hole was drilled on one side, where a sprayer (Amber Spray Bottles, Maredash, Shenzhen, China) was inserted and sealed. Fig. 1A shows the schematics and dimensions of this air chamber. A photograph of this chamber is shown in Supplementary Fig. S1. The chamber dimension (e.g., 24-inch length) roughly corresponds to the one-tenth scale of a typical classroom, as demonstrated in the previous work (Kwon et al., 2014). Following the requirements from Research Laboratory and Safety Services at the University of Arizona, the chamber was inspected, approved for this research, placed within a chemical hood, sprayed with ethanol, thoroughly wiped before and after all experiments. Air collection experiments were performed at the biosafety level 2 laboratory.
2.5. Direct air sampling on paper microfluidic chips and assay procedure
UV-inactivated SARS-CoV-2 was spiked into the 10% v/v human saliva solutions (from human donors, confirmed negative for SARS-CoV-2) with varying concentrations and loaded to the sprayer. The final concentration was 600 pg/mL, comparable to that in human saliva from active COVID-19 patients (Zhu et al., 2020). Concentrations of 0, 200, 400, and 800 pg/mL were additionally used to demonstrate the quantification capability. A paper microfluidic chip was placed 6 in (15.2 cm; one-quarter point of the chamber length), 12 in (30.5 cm; one-half point), or 24 in (61 cm; full length) away from the sprayer nozzle. We sprayed two times manually (simulating typical human coughs), consuming 3 mL, as well as five times (simulating repetitive human coughs) consuming 7.5 mL. Spraying made the paper chips wet. We waited for 15 min and collected the paper microfluidic chips from the chamber. The chips were incubated at room temperature for additional 10 min, allowing liquid to be evaporated. 4 μL of antibody conjugated particle suspension was then pipetted to the center of each channel and incubated for another 5 min allowing the particles to interact with the viruses.
2.6. Smartphone-based fluorescence microscopic imaging of paper microfluidic chips
A smartphone-based fluorescence microscope was designed and built following the method described in Chung et al. (2021), with the modifications described below. A commercial microscope attachment to a smartphone was purchased and used (MicroFlip 100-250x High Power Pocket Microscope; Carson Optical, NY, USA). A 460 nm LED (WP7113QBC/G; Digi-Key Electronics, MN, USA) was used as a light source for the excitation of fluorescent particles. The excitation wavelength is slightly shorter than the peak excitation of the particles (488 nm) to avoid the overlap with the emission signal while providing sufficient excitation to the particles, as confirmed with the fluorescence images. A 9-V battery powered this LED. Acrylic films (#382 and #15; Color Filter Booklet; Edmund Optics, AZ, USA) were used as the low-cost excitation and emission filters, placed between the microscope attachment and smartphone camera. A smartphone camera (Samsung Galaxy S20 FE 5G; Samsung Electronics America, Inc., NJ, USA) was used to image each paper channel and isolate only the aggregated particles, which will be discussed in the following section. All components (a microscope attachment, an LED, a 9-V battery, acrylic filter, and a smartphone) are mounted on a foldable stand and stage designed in SolidWorks and 3D-printed using Creality Ender-3 (Shenzhen Creality 3D Technology Co. Ltd.; Shenzhen, Guangdong, China) with PETG filament (Overture; Wilmington, DE, USA). For the comparison purpose, a benchtop fluorescence microscope was also used to image the paper chips, whose procedure can be found in the previous work (Kim et al., 2021).
2.7. Image processing
The MATLAB (The MathWorks, Inc.; Natick, MA, USA) script used in this study is based on the previous work (Chung et al., 2019; Kim et al., 2021). Fluorescence images were acquired from five different locations of each channel since the field-of-view (FOV) was small and could not represent the overall particle aggregation behavior over the length of the channel. Each image was split into three color channels (red, green, and blue). The green channel images were used to capture the green fluorescence (peaked at 509 nm) of the particles. The color histogram of the fluorescence signal was investigated to optimize the intensity threshold value. The pixels with the intensities lower than 60 (out of 255) were eliminated to remove background noise, which was optimized through a series of experiments using only the fluorescent particles on the paper. (For the benchtop fluorescence microscope analysis, the threshold value was 200). The images were then binarized (black and white). Next, size thresholding was applied to isolate only the aggregated particles. Only the spheres with 10–400 pixels were considered (the same size thresholding was used for the benchtop fluorescence microscope analysis); all other spheres are considered noise or artifacts (especially dust).
2.8. Air sampling on paper microfluidic chips under forced fan circulation
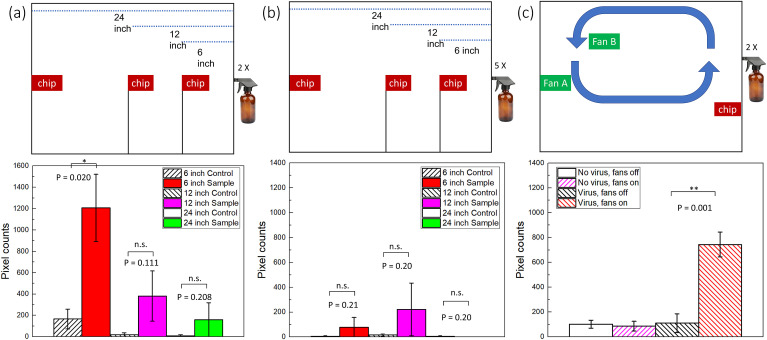
Two computer case cooling fans (NFD1293259B–2F; Y.S. Tech., Garden Grove, CA, USA, and FD08025, Masscool, Walnut, CA, USA) were installed in the chamber, as shown in Fig. 3. A simple voltage divider circuit controlled the speed of fans and subsequently circulation rate. Two fans are installed to circulate the air in one circular direction. All other conditions were the same as those described in section 2.5.
Fig. 3.
Assay results with a smartphone-based fluorescence microscope. Pixel counts from five different images of a single channel were summed and used as a single data point. Experiments were repeated four times (each time with five images), each using a different paper microfluidic chip. Average pixel counts (n = 4) are plotted with the error bars representing standard errors. (a) With two-times spraying, (b) with five-times spraying, and (c) two-times spraying and fans installed. n = 4, each time with different spraying/capture and with different paper microfluidic chips; * denotes p ≤ 0.05; ** denotes p ≤ 0.01; n.s. denotes not significant.
3. Results and discussion
Two types of antibodies were used to assay SARS-CoV-2 from the air samples. One was rabbit monoclonal antibody, and the other was rabbit polyclonal antibody, both to SARS-CoV-2. Experiments were initially conducted by sequentially pipette-adding the SARS-CoV-2 solution (0 and 600 pg/mL in 10% saliva) and the antibody-particle suspension on each paper microfluidic channel to demonstrate the initial feasibility. A benchtop fluorescence microscope was used for this initial investigation, and the results are shown in Fig. 1B. A substantial number of particle aggregations were found from the fluorescence microscopic images of the paper microfluidic channels, demonstrating the capability of identifying SARS-CoV-2 using this method. (We have conducted the same experiments using monoclonal antibody conjugated particles, using rabbit monoclonal anti-SARS-CoV-2, catalog number 40588-R0004 from SinoBiological, Inc., and found that the extent of particle aggregation was much inferior to that with a polyclonal antibody. Refer to Supplementary Fig. S2.)
The overall procedure is shown in Fig. 1C. Droplets and aerosols containing SARS-CoV-2 are generated from a sprayer and passively collided with the paper microfluidic chips. The SARS-CoV-2 concentration of 600 pg/mL (or 600 fg/μL) was determined from the typical SARS-CoV-2 concentration found among COVID-19 infected patients (Zhu et al., 2020). The amount of SARS-CoV-2 colliding to the paper microfluidic chip should be much lower than that inside the sprayer, and the fraction settling within a single channel would be even lower than that. The control was the 10% saliva without any SARS-CoV-2. Only the pixels representing aggregated particles were summed using the MATLAB script from the fluorescence microscope images.
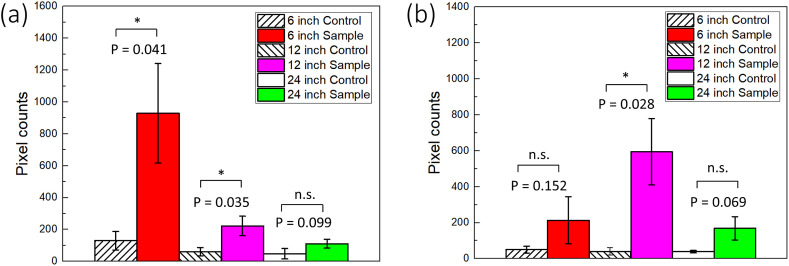
We first investigated whether our sensor system works with a benchtop fluorescence microscope. Fig. 2 A shows the pixel counts of aggregated particles on the paper microfluidic chip, where the SARS-CoV-2 samples were sprayed at the distances of 6, 12, and 24 inches (15.2, 30.5, and 60 cm, respectively), corresponding to the quarter, half, and full length of the air chamber (Fig. 1A). Polyclonal antibody conjugated particles were used in all further experiments. Total pixel counts decreased as the distance increased from 6 to 24 inches, indicating that the number of captured droplets/aerosols in each channel decreased with the distance. Means and standard errors were collected from four different experiments, each time using different paper microfluidic chips. The results with 6 and 12 inches were significantly different from those of the negative controls (0 pg/mL 10% saliva) (p < 0.05). Fig. 2B shows the result with five-times spraying. Higher pixel counts were recorded at 12 and 24 inches than those with two-times spraying, improving the results. However, the pixel counts at 6 inches were significantly lower than those of 12 inches. They were also lower than those with two-times spraying at 6 inches. We have noticed that liquids were falling off from the paper microfluidic chip, as shown in Supplementary Fig. S3, leading to the loss of liquid and subsequently inefficient capturing. All raw images are summarized in Supplementary Fig. S4.
Fig. 2.
Assay results with a benchtop fluorescence microscope. Pixel counts from three different images of a single channel were summed and used as a single data point. Experiments were repeated four times (each time with three images), each using a different paper microfluidic chip. Average pixel counts (n = 4) are plotted with the error bars representing standard errors, each time with different spraying/capture and with different paper microfluidic chips. (a) With two-times spraying, and (b) with five-times spraying. * denotes p ≤ 0.05, **p ≤ 0.01, and n.s. = not significant.
Experiments were repeated using a smartphone-based fluorescence microscope. The diagram of the internal light path and the photographs of the smartphone-based fluorescence microscope are shown in Supplementary Fig. S5. Five images were collected from a single channel since the FOV of a smartphone microscope was smaller than that of a benchtop fluorescence microscope. Fig. 3A and B shows the results. Overall, the pixel counts are lower than those with a benchtop fluorescence microscope. However, the trend is identical: the pixel counts decreased as the distance increased, and the five-times spraying at 6-inch distance did not work. The sample data was significantly different from that of the control group at a 6-inch distance. All raw images used in Fig. 3A and B are summarized in Supplementary Fig. S6.
To demonstrate the quantification capability, we have conducted additional experiments with varying concentrations (0, 200, 400, 600, and 800 pg/mL) with two-times spraying at a 6-inch distance. The result was shown in Supplementary Fig. S7, showing satisfactory linearity (R2 = 0.956). Most data points are significantly different (p < 0.05) from that of 0 pg/mL except for 800 pg/mL, although its p-value of 0.053 is still close to 0.05. Using this linear regression equation, we can convert the sample assay results shown in Fig. 3A to the concentration: 570, 40, and 0 pg/mL, respectively. Considering 600 pg/mL concentration, the paper microfluidic chips at 12-inch and 24-inch distances captured 7% and 0% of the same at 6-inch distance, respectively.
Additional experiments were performed to see if this method could be applied to the environment that can better represent the air-conditioned rooms. As shown in Fig. 3C and Supplementary Fig. S8, two fans were installed inside the chamber, designed to circulate the air. A paper chip was installed on the same side of the sprayer. Larger droplets can be visually identified using a bright light bulb, as shown in Supplementary Fig. S8. Most larger droplets traveled from the right-side sprayer to the other direction at least 24 inches, indicating that larger droplets must travel for a minimum of 48 inches to reach the paper chip. Smaller droplets and aerosols can travel farther than 48 inches, although they cannot be identified from the photographs.
The assay results are summarized in Fig. 3C. With viruses and fans off (third column), the pixel counts were not significantly different from those with no virus (i.e., deionized water) and fans off (first column) and fans on (second column). Comparison of third and fourth columns (fans off vs. on, both with viruses) indicates that viruses could not be captured on the paper chip with no air circulation. Comparison of second and fourth columns (with vs. without viruses, both with fans on) shows that there were no false positives. The average pixel counts with viruses and fans (fourth column) are 740 ± 100, showing a significant difference from the other three (p ≤ 0.01). This number is smaller than the 6-inch sample data shown in Fig. 3A but higher than the 12-inch and 24-inch sample data, indicating the role of air circulation towards effective virus capture on paper chips. All raw images used in Fig. 3C are summarized in Supplementary Fig. S9.
4. Conclusion
We collected the SARS-CoV-2 from the sprayed droplets/aerosols directly on a paper microfluidic chip without using any sampler device. Neither pumps nor fans were necessary (besides the fans used to simulate air conditioning) to collect these droplets, and the collection was entirely passive. Assays were conducted directly on the paper microfluidic chip, without the need for sample collection, transfer, dilution, and purification. Antibody conjugated particle suspension was pipette-added to the channel, allowing particle aggregation induced by antibody-virus binding. A smartphone-based fluorescence microscope captured fluorescent images, and a MATLAB script isolated and quantified particle aggregation. Despite the small number of viruses captured on each channel, we could detect them with only two sprays (presumably equivalent to two coughs) at a 6-inch (15.2 cm) distance. Successful capture and detection were also demonstrated with the air circulated through the chamber. The method requires a paper microfluidic chip (neither pre-loading nor immobilization was necessary), antibody-particle suspension, and a smartphone-based fluorescence microscope (requiring low-cost and off-the-shelf components, such as an LED, a 9-V battery, acrylic film, and a microscope attachment. The total cost of parts and supplies for a smartphone-based fluorescence microscope was US$46.60 (excluding a smartphone), as shown in Supplementary Table S1. It is also foldable and handheld, whose dimension is 10 cm × 5 cm x 10 cm, as shown in Supplementary Fig. S10. The method and device can be adapted for detecting other respiratory viruses and bacteria by changing the antibody and optimizing the particle concentration, etc.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the University of Arizona's Test All Test Smart Program as well as Tech Launch Arizona's Asset Development Program. We thank Alanna V. Zubler and Babak Safavinia for their help in the chamber design and construction. K.S. acknowledges the Computational and Mathematical Modeling of Biomedical Systems Training Grant from the National Institute of General Medical Sciences (NIGMS), U.S. National Institutes of Health, grant number GM132008.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113912.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Chen W., Zhang N., Wei J., Yen H.L., Li Y. Build. Environ. 2020;176:106859. doi: 10.1016/j.buildenv.2020.106859. [DOI] [Google Scholar]
- Chia P.Y., Coleman K.K., Tan Y.K., Ong S.W.X., Gum M., Lau S.K., Lim X.F., Lim A.S., Sutjipto S., Lee P.H., Son T.T., Young B.E., Milton D.K., Gray G.C., Schuster S., Barkham T., De P.P., Vasoo S., Chan M., Ang B.S.P., Tan B.H., Leo Y.S., Ng O.T., Wong M.S.Y., Marimuthu K. Nat. Commun. 2020;11:2800. doi: 10.1038/s41467-020-16670-2. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Breshears L.E., Perea S., Morrison C.M., Betancourt W.Q., Reynolds K.A., Yoon J.Y. ACS Omega. 2019;4:11180–11188. doi: 10.1021/acsomega.9b00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Breshears L.E., Gonzales A., Jennings C.M., Morrison C.M., Betancourt W.Q., Reynolds K.A., Yoon J.Y. Nat. Protoc. 2021;16:1452–1475. doi: 10.1038/s41596-020-00460-7. [DOI] [PubMed] [Google Scholar]
- Fozouni P., Son S., Díaz de León Derby M., Knott G.J., Gray C.N., D'Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I., Boehm D., Tsou C.L., Shu J., Bhuiya A., Armstrong M., Harris A.R., Chen P.Y., Osterloh J.M., Meyer-Franke A., Joehnk B., Walcott K., Sil A., Langelier C., Pollard K.S., Crawford E.D., Puschnik A.S., Phelps M., Kistler A., DeRisi J.L., Doudna J.A., Fletcher D.A., Ott M. Cell 184: 1–11. 2020. [DOI]
- Fronczek C.F., Yoon J.Y. J. Lab. Autom. 2015;20:390–410. doi: 10.1177/2211068215580935. [DOI] [PubMed] [Google Scholar]
- Hinds W.C. John Wiley & Sons; Hoboken: 1982. Aerosol Technology: Properties, Behaviour, and Measurement of Airborne Particles. [Google Scholar]
- Hwang S.E., Chang J.H., Oh B., Heo J. Int. J. Infect. Dis. 2021;104:73–76. doi: 10.1016/j.ijid.2020.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Romero-Lozano A., Hwang D.S., Yoon J.Y. J. Hazard Mater. 2021;413:125338. doi: 10.1016/j.jhazmat.2021.125338. [DOI] [PubMed] [Google Scholar]
- Kwon H.J., Fronczek C.F., Angus S.V., Nicolini A.M., Yoon J.Y. J. Lab. Autom. 2014;19:322–331. doi: 10.1177/2211068213504205. [DOI] [PubMed] [Google Scholar]
- Park T.S., Yoon J.Y. IEEE Sensor. J. 2015;15:1902–1907. doi: 10.1109/JSEN.2014.2367039. [DOI] [Google Scholar]
- Piri A., Kim H.R., Park D.H., Hwang J. J. Hazard Mater. 2021;413:125417. doi: 10.1016/j.jhazmat.2021.125417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Sutarlie L., Loh X.J. Chem. Asian J. 2020;15:4241–4255. doi: 10.1002/asia.202001051. [DOI] [PubMed] [Google Scholar]
- Terzieva S., Donnelly J., Ulevicius V., Grinshpun S.A., Willeke K., Stelma G.N., Brenner K.P. Appl. Environ. Microbiol. 1996;62:2264–2272. doi: 10.1128/aem.62.7.2264-2272.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorinen V., Aarnio M., Alava M., Alopaeus V., Atanasova N., Auvinen M., Balasubramanian N., Bordbar H., Erästö P., Grande R., Hayward N., Hellsten A., Hostikka S., Hokkanen J., Kaario O., Karvinen A., Kivistö I., Korhonen M., Kosonen R., Kuusela J., Lestinen S., Laurila E., Nieminen H.J., Peltonen P., Pokki J., Puisto A., Råback P., Salmenjoki H., Sironen T., Österberg M. Saf. Sci. 2020;130:104866. doi: 10.1016/j.ssci.2020.104866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations. https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations
- Zenhausern R., Chen C.H., Yoon J.Y. Biomicrofluidics 15. 2021 doi: 10.1063/5.0041089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Li Y., Zhang A.L., Wang Y., Molina M.J. Proc. Natl. Acad. Sci. U.S.A. 2020;117:14857–14863. doi: 10.1073/pnas.2009637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Guo J., Xu Y., Chen X. J. Infect. 2020;81:e48–e50. doi: 10.1016/j.jinf.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.