Abstract
Background:
ArfGAP with GTPase domain, Ankyrin repeat and PH domain 2 Antisense 1 (AGAP2-AS1) is a promising long noncoding RNA that may possess prognostic value for different types of tumors. The objective of this meta-analysis is to evaluate the prognostic value of long noncoding RNA AGAP2-AS1 in cancer patients.
Methods:
A systematic literature search of the PubMed, Cochrane Library, EMBASE, Medline, Web of Science, CNKI, Weipu, and Wanfang electronic databases were carried out in this meta-analysis. Synthetic hazard ratios (HRs) or odd ratios (ORs) with 95% confidence intervals (CIs) were obtained to determine the prognostic and clinicopathological significance of AGAP2-AS1 expression in tumors.
Results:
The final meta-analysis included 10 studies that contained 948 patients. The pooled results provided evidence that AGAP2-AS1 overexpression predicted reduced overall survival (OS) (HR = 1.77, 95% CI: 1.49–2.09, P < .00001), disease-free survival (HR = 1.84, 95% CI: 1.40–2.41, P < .0001), and progression-free survival (HR = 1.84, 95% CI: 1.01–3.33, P = .04) and for various cancers. Additionally, the AGAP2-AS1 overexpression was concerned with lymph node metastasis (positive vs negative, OR = 2.95, 95% CI: 1.96–4.45, P < .00001), advanced tumor node metastasis stage (III/IV vs I/II, OR = 3.73, 95% CI: 2.71–5.13, P < .00001), and tumor size (larger vs smaller, OR = 2.28, 95% CI: 1.24–4.18, P = .008). Besides, data from gene expression profiling interactive analysis dataset verified the results in our meta-analysis. The results showed that the expression level of AGAP2-AS1 was higher in most tumor tissues than in the corresponding normal tissues and was linked to poor OS and disease-free survival.
Conclusions:
Our results indicated that AGAP2-AS1 overexpression was closely correlated with shorter OS in multiple cancer types, suggesting that AGAP2-AS1 might function as a promising predictor for clinical outcomes in cancer.
Keywords: AGAP2-AS1, cancer, long noncoding RNA, overall survival, prognosis
1. Introduction
Cancer from various systems and organs is one of the diseases that pose a great threat to human health globally.[1] A substantial majority of cancers have the characteristics of occult onset, difficult diagnosis, and rapid progression, which are the main causes of the high rate of mortality. Meanwhile, tumors of different origins are not the same thing in terms of biological features, lesion involvement, clinical manifestations, efficacy, and prognosis.[2] Recently, multi-disciplinary treatment mode, a fixed expert group composed of multi-disciplinary experts, having been proposing appropriate treatment schemes for cancer patients.[3] Despite proper management of their disease, the prognosis for many cancer patients continues dismal, partly due to the lack of prognostic and diagnostic markers. Thus, it is necessary in order to identify effective prognostic markers that can provide urgently needed treatment strategies.
Non-coding RNA refers to RNA that is not translated into polypeptides.[4] These RNA can be divided into 2 categories based on length: small noncoding RNAs that are shorter than 200 nucleotides and long non-coding RNAs that are longer than 200 nucleotides.[5] Long noncoding RNA (lncRNAs) have recently gained more attention in the medical community for their potential prognostic value in cancer. Additionally, the relationship between lncRNAs, signal pathways in cancer, and cancer phenotypes has become a topical issues.[6] Previous studies identified the pivotal role of lncRNAs in biological processes, such as genomic imprinting, histone modification, chromatin remodeling, and post-transcriptional regulation.[7] In recent years, lncRNAs have also been shown to be involved in tumor occurrence and progression. Moreover, it was reported that dysregulation of lncRNAs was significantly correlated with clinical characteristics and cancer prognosis. These data suggested that lncRNAs are novel biomarkers and therapeutic targets in cancer.[8]
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 antisense 1 (AGAP2-AS1), a component of lncRNAs, has recently been investigated for its involvement in promoting cancer deterioration and progression, and the dysregulation of AGAP2-AS1 has been detected in different types of cancer.[9,10] It has been reported that upregulated AGAP2-AS1 expression can induce specific biological phenotypes and poor prognosis.[11] Subsequently, another study demonstrated that increased AGAP2-AS1 expression played a vital role in promoting tumor cell proliferation and invasion, which was indicative of a poor prognosis for cancer patients.[12] To date, there is no meta-analysis that provides an assessment of the effect of SNHG3 on the prognosis of cancer patients. Therefore, our aim was to evaluate the prognostic value of lncRNA AGAP2-AS1 expression in tumors.
2. Material and methods
2.1. Literature search
Two independent reviewers searched the PubMed, Cochrane Library, EMBASE, Medline, Web of Science, CNKI, Weipu, and Wanfang from February 02, 2016 to February 02, 2021. The search was conducted irrespective of the region or language. The following keywords and Medical Subject Headings were included: “AGAP2-AS1”, “ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 antisense 1”, “lncRNA”, “long noncoding RNA”, “cancer”, “carcinoma”, “neoplasm”, “prognosis”, and “survival”.
The following criteria for inclusion in our meta-analysis to select eligible studies: a definite diagnosis or histopathological diagnosis of cancer patients; information about survival and clinical prognostic parameters of lncRNA AGAP2-AS1 in patients with cancer was reported; and enough information were available for calculating the pooled hazard risk (HR) and 95% confidence interval (CI). Exclusion criteria for the studies were as follows: studies with absent information of prognostic outcomes; duplicate publications; and nonhuman studies, letters, case reports, review articles, and other studies without original data.
2.2. Data extraction and quality assessment
Data were drawn from each study by 3 authors independently and a consensus was reached. The following information was extracted: author, country, publication year, tumor type, cancer size, follow-up time, detection method, and cutoff value. Patient number for each group was divided based on the positive or negative lymph node metastasis (LNM), distant metastasis (DM), tumor size, tumor node metastasis stage, and patient number for high or low AGAP2-AS1 expression in each group.
When only Kaplan–Meier curves were available, HRs and 95% CIs were extracted from graphical survival plots by using Engauge Digitizer V4.1 (https://sourceforge.net/projects/digitizer/).[13] If reported directly in univariate or multivariate analyses, HRs with corresponding 95% CIs were extracted from multivariate analyses.
A quality assessment for all of the included studies depended on the Newcastle–Ottawa quality assessment scale, which is composed of the following 3 dimensions: selection, comparability, and exposure. Each study was scored from 0 to 9 according to these dimensions. A study with a Newcastle–Ottawa quality assessment scale score ≥6 was considered to be of high quality.[14]
2.3. Statistical analyses
All statistical analyses of the data were calculated using Review Manager (RevMan) 5.3 software and Stata/SE 14.1 software. Sensitivity analysis was performed by omitting literature one by one to determine whether the results were stable and the publication bias of this meta-analysis was evaluated by using the Begg test according to Stata/SE 14.1 software. The Q test and I2 statistics were applied to estimate the heterogeneity of results. A fixed-effects model was selected when I2 < 50% was observed. The synthetic estimate was calculated depending on the random-effects model when the heterogeneity was obvious (I2 > 50%). A two-tailed P-value < .05 was considered as statistically significant.
3. Results
3.1. Literature search and selection
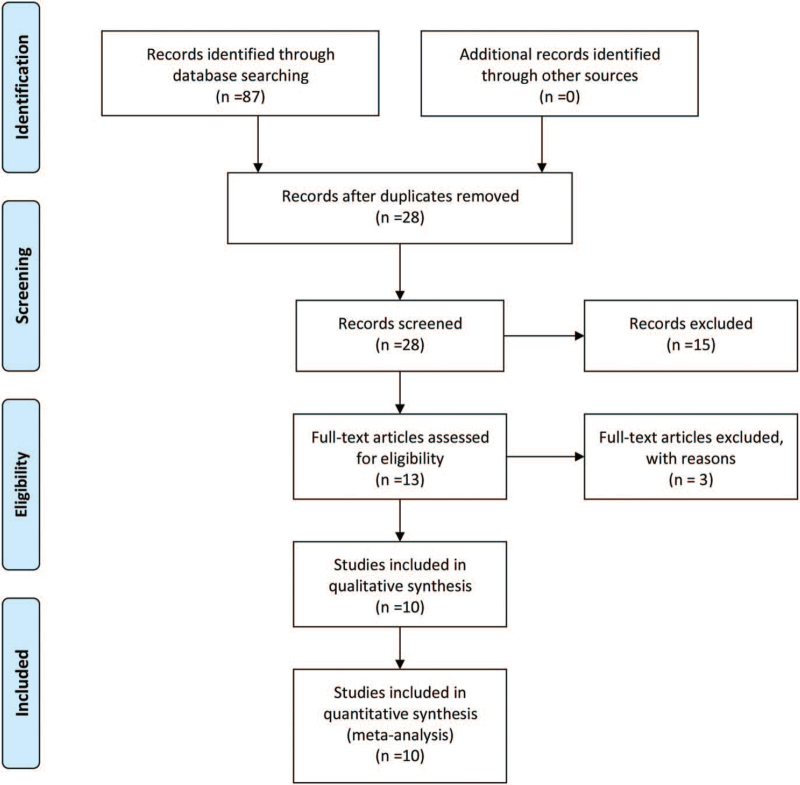
The literature selection process is shown in Figure 1. Preliminarily, 87 relevant studies in total were yielded from the search of the PubMed, Cochrane Library, EMBASE, CNKI, Weipu, and Wanfang electronic databases. Among these, 59 studies were excluded as duplicating articles. Then we further excluded 15 studies by reviewing the title and abstract. Subsequently, 3 more studies were not able to be included because of insufficient data and being unrelated to our study. Finally, 10 studies containing 948 patients were eligible for this meta-analysis and were highly consistent with the inclusion criteria. All of the included studies were published between 2016 and 2020 and came from China. Multiple forms of cancers were analyzed in the present meta-analysis, including pancreatic cancer,[11] hepatocellular carcinoma,[15] glioblastoma,[16] gastric cancer,[12] nonsmall cell lung cancer (NSCLC),[9,17] colorectal cancer (CRC),[18] epithelial ovarian cancer,[19] papillary thyroid cancer,[20] glioma.[21] The detailed information obtained from the studies is summarized in Table 1.
Figure 1.
Flow diagram of the study selection procedure in this meta-analysis.
Table 1.
The main characteristics of the included studies in the meta-analysis.
| AGAP2-AS1 expression | ||||||||||
| Study | Region | Tumor type | Sample size | TNM stage | High | Low | Cutoff value | Detection method | Outcome measure | NOS |
| Hui et al 2019[11] | China | PC | 46 | I–IV | 23 | 23 | Median | qRT-PCR | OS | 6 |
| Liu et al 2019[15] | China | HCC | 137 | I–IV | 69 | 68 | Median | qRT-PCR | OS, DFS | 7 |
| Tian et al 2018[16] | China | GBM | 40 | NA | 20 | 20 | Median | qRT-PCR | OS | 7 |
| Qi et al 2017[12] | China | GC | 50 | I–IV | 25 | 25 | Median | qRT-PCR | OS, PFS | 8 |
| Li et al 2016[17] | China | NSCLC | 80 | NA | 40 | 40 | Median | qRT-PCR | OS, DFS | 6 |
| Hong et al 2020[18] | China | CRC | 116 | I–IV | 58 | 58 | Median | qRT-PCR | OS, DFS | 7 |
| Fan et al 2017[9] | China | NSCLC | 198 | I–IV | 99 | 99 | Median | qRT-PCR | OS | 8 |
| Tingting et al 2020[19] | China | EOC | 80 | NA | 40 | 40 | Median | qRT-PCR | NA | 8 |
| Shao et al 2020[20] | China | PTC | 110 | I–IV | 55 | 55 | Median | qRT-PCR | NA | 8 |
| Zheng et al 2019[21] | China | Glioma | 91 | I–IV | 49 | 42 | Median | qRT-PCR | OS | 7 |
AGAP2-AS1 = ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 antisense 1, CRC = colorectal cancer, DFS = disease-free survival, EOC = epithelial ovarian cancer, GBM = glioblastoma, GC = gastric cancer, HCC = hepatocellular carcinoma, NA = not available, NOS = Newcastle–Ottawa quality assessment scale, NSCLC = nonsmall cell lung cancer, OS = overall survival, PC = pancreatic cancer, PFS = progression-free survival, PTC = papillary thyroid cancer.
3.2. AGAP2-AS1 expression is highly correlated with OS, DFS, and PFS
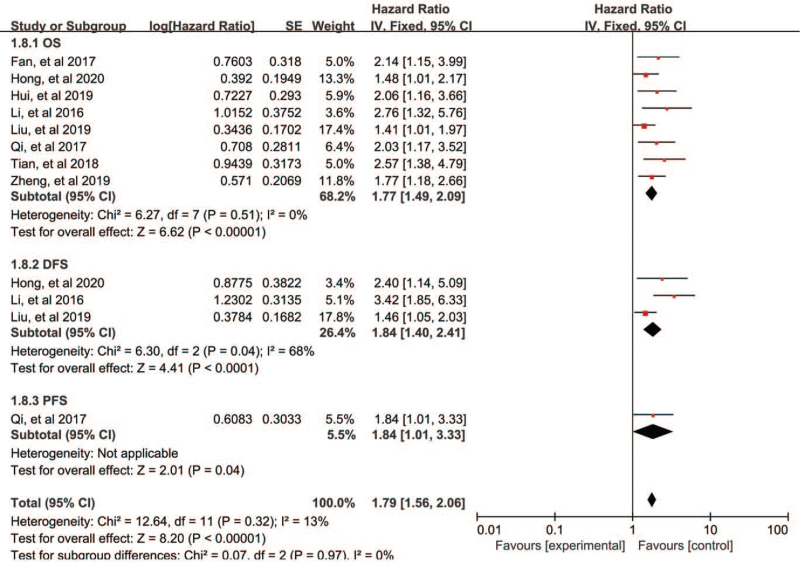
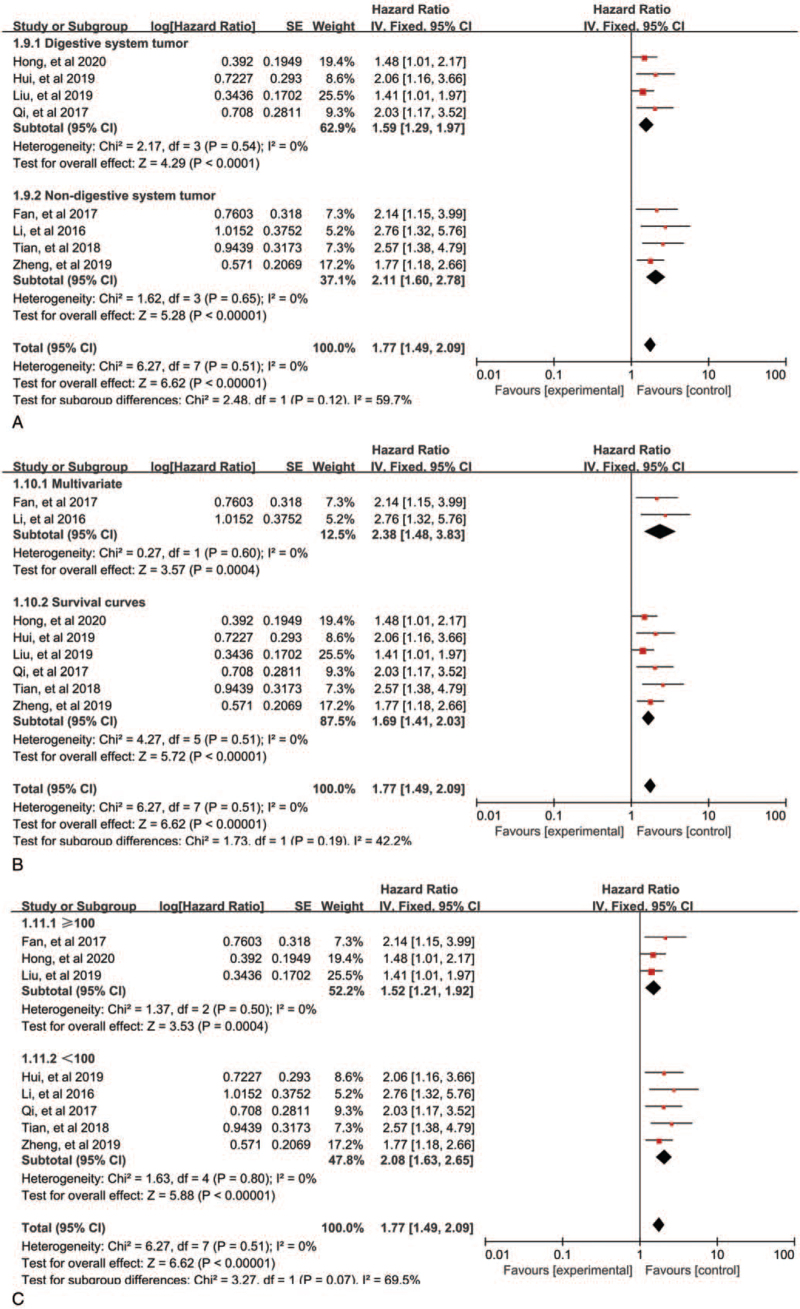
Overall, 8 of the 10 studies investigated cancer prognosis. A total of 758 patients were assessed for the HR and 95% CI of overall survival (OS). The fixed-effects model was performed to analyze the pooled HR and its 95% CI depended on no obvious heterogeneity (P = .51, I2 = 0%). We further elucidated the relationship between AGAP2-AS1 expression and the OS, as illustrated in Figure 2. The pooled results revealed that the high expression of AGAP2-AS1 was related to poor prognosis of cancers (HR = 1.77, 95% CI: 1.49–2.09, P < .00001, Fig. 2). In the subgroup analysis stratified by tumor type, analysis method, and sample, as exhibited in Figure 3A–C, we found that elevated AGAP2-AS1 could act as a prognostic predictor for patients with digestive system tumors (HR = 1.59, 95% CI: 1.29–1.97, P < .0001) or patients with non-digestive system tumors (HR = 2.11, 95% CI: 1.60–2.78, P < .00001) Fig. 3A). Thus, the prognosis of cancer patients with AGAP2-AS1 overexpression was worse than those with low expression of AGAP2-AS1. In terms of disease-free survival (DFS), only 3 studies were included, and the pooled results indicated that patients with high expression of AGAP2-AS1 had poor DFS (HR = 1.84, 95% CI: 1.40–2.41, P < .0001). Only one focused on the relationship between AGAP2-AS1 and progression-free survival (PFS) (HR = 1.84, 95% CI: 1.01–3.33, P = .04) (Fig. 2).
Figure 2.
Forest plots for the association between AGAP2-AS1 expression with OS (A), DFS (B), and PFS (C). AGAP2-AS1 = ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 antisense 1, DFS = disease-free survival, OS = overall survival, PFS = progression-free survival.
Figure 3.
Forest plots of subgroup analysis for OS: subgroup analysis by tumor type (A), subgroup analysis by analysis method (B), and subgroup analysis by sample size (C).
3.3. Relationship between AGAP2-AS1 expression and clinicopathological characteristics
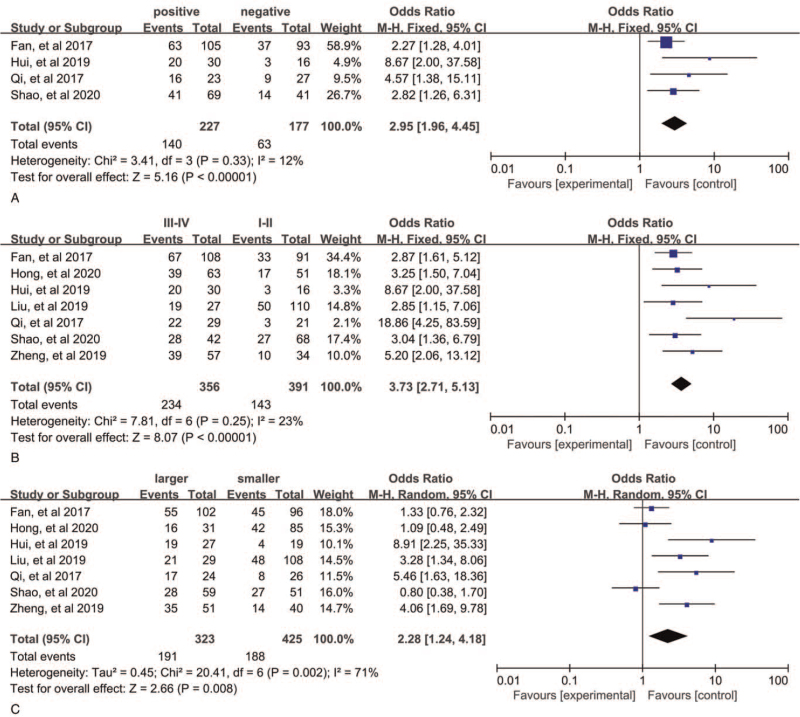
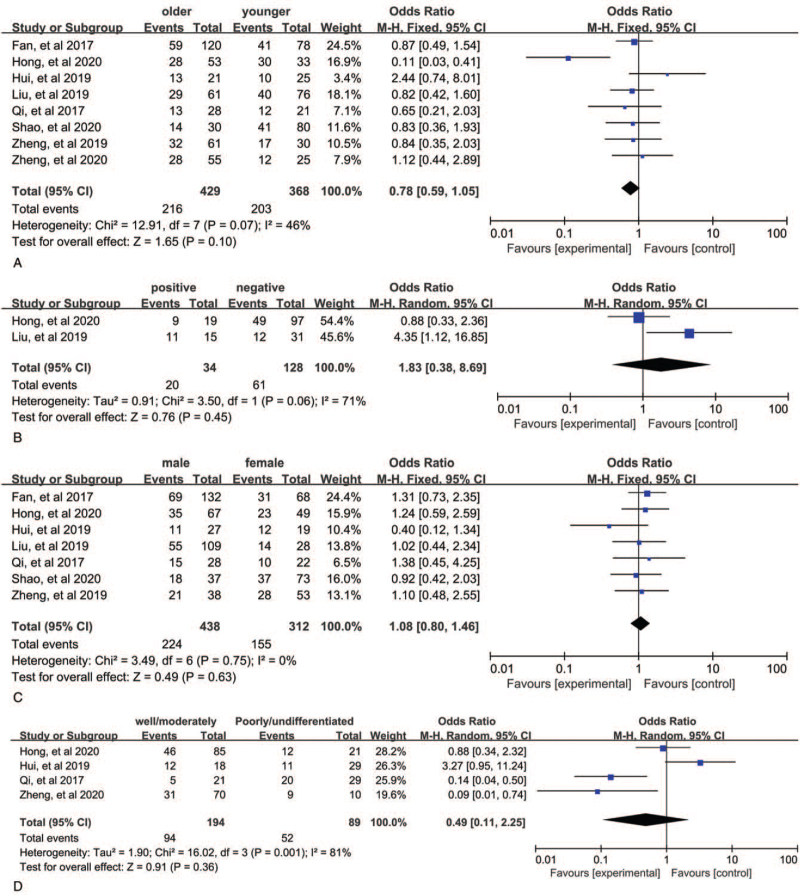
The merged results from 7 studies with 747 patients demonstrated that patients with AGAP2-AS1 overexpression have a more advanced stage (III/IV) cancer (III/IV vs I/II, OR = 3.73, 95% CI: 2.71–5.13, P < .00001, Fig. 4B). Here we used a fixed-effects model because of no obvious heterogeneity (P = .23, I2 = 23%). In addition, these 4 studies contained 404 individuals showed correlation between AGAP2-AS1 and LNM in various cancers. A fix-effects model was utilized again because of obvious heterogeneity (P = .33, I2 = 12%), and the pooled results showed that LNM was more susceptible to occur in the upregulated AGAP2-AS1 expression group than the downregulated AGAP2-AS1 expression group (OR = 2.95, 95% CI: 1.96–4.45, P < .00001, Fig. 4A). These 7 studies provided information for tumor size. The pooled results indicated that patients with high AGAP2-AS1 expression have larger tumor size (OR = 2.28, 95% CI: 1.24–4.18, P = .008, Fig. 4C), and a random-effects model was used (P = .002, I2 = 71%). Furthermore, we did an investigation on the relationship between SNHG3 expression and age, gender, DM, and histological grade. However, the pooled results suggested that AGAP2-AS1 expression was not positively associated with these characteristics (Fig. 5A–D). The details are shown in Table 2.
Figure 4.
Forest plots for the correlation between AGAP2-AS1 expression and clinicopathological characteristics: (A) lymph node metastasis, (B) TNM stage, and (C) tumor size. AGAP2-AS1 = ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 antisense 1.
Figure 5.
Forest plots for the correlation between AGAP2-AS1 expression and clinicopathological characteristics: (A) age, (B) distant metastasis, (C) gender, and (D) histological grade. AGAP2-AS1 = ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 antisense 1.
Table 2.
Summary of the relationship between AGAP2-AS1 over-expressed and clinicopathological parameters.
| Heterogeneity | |||||||
| Clinicopathological parameters | Studies | Patients | OR (95% CI) | P-value | I 2 | P-value | Model |
| Age (older vs younger) | 8 | 797 | 0.78 (0.59, 1.05) | .10 | 46% | .07 | Fixed |
| Gender (male vs female) | 7 | 750 | 1.08 (0.80, 1.46) | .63 | 0% | .75 | Fixed |
| Tumor size (larger vs smaller) | 7 | 748 | 2.28 (1.24, 4.18) | .008 | 71% | .002 | Random |
| TNM stage (III + IV vs I + II) | 7 | 747 | 3.73 (2.71, 5.13) | <.00001 | 23% | .25 | Fixed |
| LNM (positive vs negative) | 4 | 404 | 2.95 (1.96, 4.45) | <.00001 | 12% | .33 | Fixed |
| DM (positive vs negative) | 2 | 162 | 1.83 (0.38, 8.69) | .45 | 71% | .06 | Random |
| Histological grade (well/moderately vs poorly) | 4 | 283 | 0.49 (0.11, 2.25) | .36 | 81% | .001 | Random |
AGAP2-AS1 = ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 antisense 1, CI = confidence interval, DM = distant metastasis, LNM = lymph node metastasis, OR = odds ratio.
Significance of bold values P <0.05.
3.4. Validation of the role of lncRNA AGAP2-AS1 in human tumors
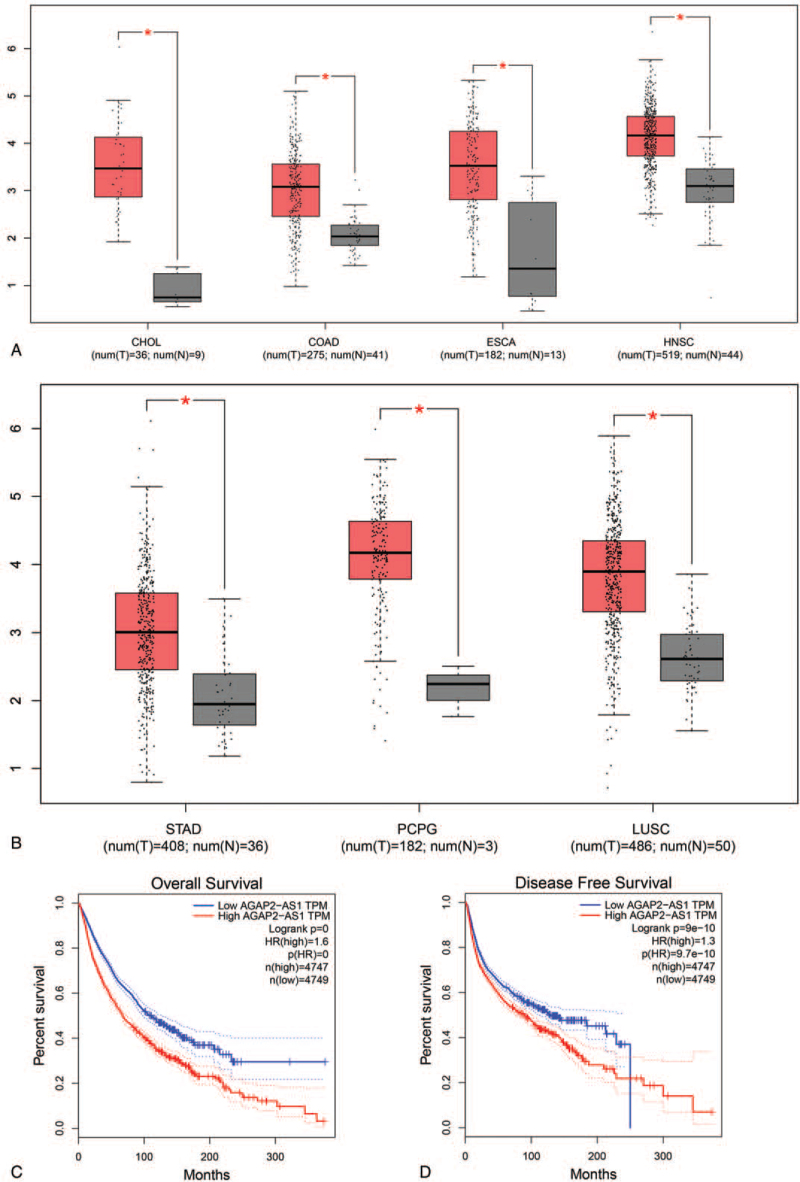
Based on gene expression profiling interactive analysis (GEPIA), the expression level of lncRNA AGAP2-AS1 in multiple cancers was shown in Figure 6A and B, and the results demonstrated that AGAP2-AS1 was obviously higher overexpressed in tumor tissue than in the corresponding normal tissues, including cholangiocarcinoma, colon adenocarcinoma, esophageal carcinoma, head and neck squamous cell carcinoma, kidney renal papillary cell carcinoma, lung squamous cell carcinoma, pheochromocytoma, and paraganglioma, stomach adenocarcinoma. Furthermore, according to a survival analysis performed by using the GEPIA database, we found that high level of AGAP2-AS1 expression was significantly with unfavorable OS (HR = 1.6, P < .01) and DFS (HR = 1.3, P < .01) in cancer patients expressing a low level of AGAP2-AS1, as shown in Figure 6C and D.
Figure 6.
Validation of the role of lncRNA AGAP2-AS1 in human cancers in the GEPIA dataset: (A and B) plot of SNHG3AGAP2-AS1 expression in different types of human cancers and normal tissues, (C) overall survival plot of AGAP2-AS1, and (D) disease-free survival plot of AGAP2-AS1. AGAP2-AS1 = ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 antisense 1, GEPIA = gene expression profiling interactive analysis, lncRNA = long noncoding RNA.
3.5. Publication bias and sensitivity analysis
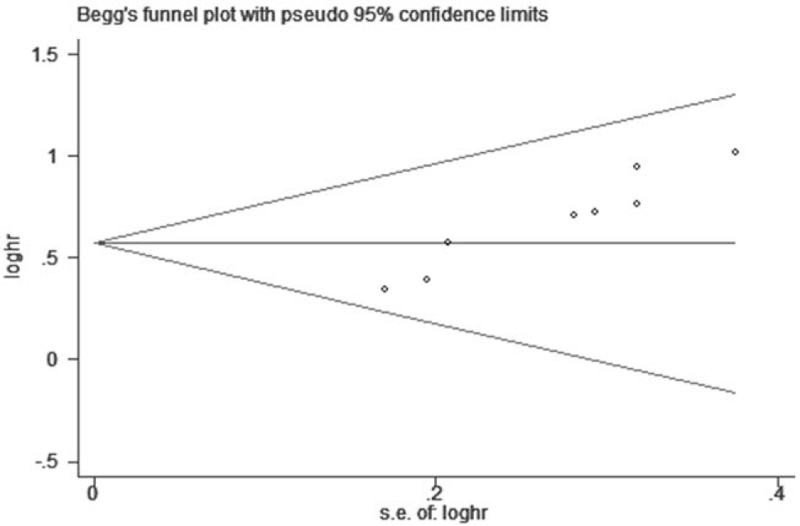
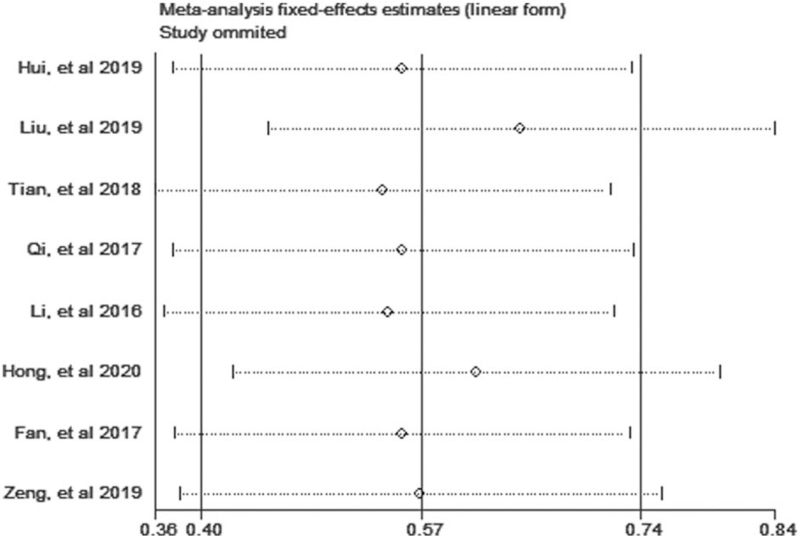
The Begg test was used to evaluate the publication bias in this meta-analysis. No significant publication bias for OS and independent factor for OS was found in this meta-analysis (Fig. 7). As illustrated in Figure 8, we performed the sensitivity analysis to prove that the results were robust, and the summary HRs were not affected after removal of study one by one.
Figure 7.
Begg funnel plot of publication bias on the correlation between AGAP2-AS1 expression and OS in this meta-analysis. AGAP2-AS1 = ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 antisense 1, OS = overall survival.
Figure 8.
Sensitivity analysis for the correlation between AGAP2-AS1 expression and OS in this meta-analysis. AGAP2-AS1 = ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 antisense 1, OS = overall survival.
4. Discussion
While only 2% of human genomic sequences are found to encode proteins, most of the genome are transcribed into noncoding RNA that has no known biological function.[22] LncRNAs, a class of noncoding RNAs with more than 200 nucleotides in length but by no means encode protein,[23] have been shown to be significantly involved in various essential cellular processes including cell cycle regulation, immune regulation, stem cells differentiation,[4] insensitivity to radiation and drugs,[24] and energy metabolism[25] through interacting with DNA, RNA, or proteins. A growing number of studies have shown that the abnormal expression of lncRNAs plays an important role in the clinicopathological features and prognosis of cancers.[26] Furthermore, lncRNAs, which are easily detected in body fluids, have the potential to be accurate prognosis for cancer patients.[27]
AGAP2-AS1 is a member of a cancer-associated lncRNA family, located at 12q14.1 and 1567 nt in length. The upregulation of AGAP2-AS1 expression is detected in numerous cancer types and promotes the progression of cancers.[28] Recently, accumulating evidence demonstrated that AGAP2-AS1 overexpression was highly related to the poor prognosis of clear cell renal cell cancer patients and strongly promoted cell proliferation.[29] It has been confirmed that the up-regulation of AGAP2-AS1 could cause the proliferation of ovarian carcinoma cells by downregulating MEG3, implicating the link between high AGAP2-AS1 expression and the progression of cancer cells invasion.[30] In esophageal carcinoma, Shen et al[31] determined that FOSL1 had a significant connection with the cellular growth, proliferation, and invasion, which was attributed to the upregulation of the miR-195-5p by AGAP2-AS1. Also, a study by Xu et al[32] demonstrated that the knockdown of AGAP2-AS1 prevented the occurrence of pre-eclampsia by via inhibition of JDP2 at the post-transcriptional level by competing for miR-574. In another study, AGAP2-AS1 was up-regulated in prostate carcinoma tissues compared with normal tissues and regulated the miR-195-5p expression, which was important for the development of prostate cancer including proliferation, migration, and invasion.[32] Furthermore, Tao et al[33] proposed that lncRNA AGAP2-AS1 combined with TBILA could serve as potential targets for the diagnosis and treatment of NSCLC. AGAP2-AS1 was also shown to be a crucial lncRNA expressed during the migration and invasion of NSCLC cells, suggesting AGAP2-AS1 could help to identify effective treatment strategies for NSCLC patients.[34] Meanwhile, Li et al[35] also found a similar function for AGAP2-AS1 in facilitating CRC cell growth via the regulation of the targeting the miR-4,668-3p/SRSF1 axis, indicating that AGAP2-AS1 had the high possibility of being a novel prognostic and therapeutic biomarker for CRC. It is well-known that Target drug chemotherapy is an effective strategy to treat advance tumors, and there is evidence that AGAP2-AS1 is involved in drug resistance. The latest research found that knockdown of AGAP2-AS1 sensitize breast cancer cells to trastuzumab by regulating MyD88 expression via g the NF-κB signaling pathway, which imply that designing drugs to lower the AGAP2-AS1 expression could boost the value of chemotherapy in the treatment of breast cancer, further regarding AGAP2-AS1 as a therapeutic target for breast cancer patients.[36,37] Despite the well-identified link between AGAP2-AS1 and cancer, further studies are needed to validate the function of AGAP2-AS1 in cancer.
To further define the role of AGAP2-AS1 in different cancers, we conducted the first meta-analysis to elucidate the impact of abnormal AGAP2-AS1 expression levels on the prognostic value and clinicopathological characteristic of cancer patients. From merged results, we found that the patients with a high level of expression of AGAP2-AS1 had worse outcomes in terms of OS, PFS, and DFS when in contrast to those with low AGAP2-AS1 expression, suggesting that elevated AGAP2-AS1 expression was highly related to poor prognosis and could act as an unfavorable prognostic predictor for patients with cancers. Also, the merged results suggest that the AGAP2-AS1 expression could be investigated as an independent predictive factor for OS in cancers. Moreover, the inferiority of high AGAP2-AS1 expression on LNM, tumor size and advanced tumor node metastasis stage was also exhibited, clearly indicating that the overexpression of AGAP2-AS1 had a connection with worse clinicopathological characteristics. However, no relationship was found between AGAP2-AS1 and age, gender, DM, and histological grade. The GEPIA analysis validated that high AGAP2-AS1 expression was frequently appeared in multiple cancers, and cancer patients with increased tissue AGAP2-AS1 expression had worse OS and DFS.
Some limitations should be clearly delineated. The shortcomings of this meta-analysis are as follows: First, most studies were from China, which might be potentially suitable for China or Asia. Second, the included studies were only from China. Consequently, the results might only capture the clinical characteristics of Asian populations. Third, the tumor types and number of patients and other prognostic indicators, such as PFS, were insufficient for a more comprehensive analysis. Therefore larger sample studies should be conducted to sustain the results. Fourth, the HRs were determined indirectly from survival curves by using available software, which might contribute to a calculation bias. Thus, more relevant high-quality studies that contain a large number of samples are needed to verify the findings.
5. Conclusion
In conclusion, our results provided novel insights into the correlation between AGAP2-AS1 expression, prognosis, and clinical outcomes in cancer patients. In the present meta-analysis, the results indicated that cancer patients with a high expression level of AGAP2-AS1 were at higher risk for poor OS compared with those with low AGAP2-AS1 expression. Our data strongly suggest that lncRNA AGAP2-AS1 might be capable of predicting poor prognosis of cancer patients as a novel biomarker. Taking the limitations of this study into account, more high-quality researches are needed to confirm the prognostic value of AGAP2-AS1 in tumors.
Acknowledgments
We would like to thank Dr. Xie for his guidance on this article and for his editing and proofreading of this English manuscript
Author contributions
Conceptualization: Hao Hua.
Data curation: Pingyong Zhong.
Formal analysis: Shun Chen, Zhidan Zhu.
Funding acquisition: Fei Xie.
Investigation: Hao Hua, Pingyong Zhong.
Project administration: Shun Chen.
Software: Hao Hua.
Supervision: Fei Xie.
Writing – original draft: Hao Hua, Pingyong Zhong.
Writing – review & editing: Fei Xie.
Footnotes
Abbreviations: AGAP2-AS1 = ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 antisense 1, CI = confidence interval, CRC = colorectal cancer, DFS = disease-free survival, DM = distant metastasis, GEPIA = gene expression profiling interactive analysis, HR = hazard ratios, lncRNA = long noncoding RNA, LNM = lymph node metastasis, NSCLC = nonsmall cell lung cancer, OR = odds ratio, OS = overall survival, PFS = progression-free survival.
How to cite this article: Zhong P, Hua H, Chen S, Zhu Z, Xie F. The prognostic value of lncRNA AGAP2-AS1 in cancer patients: a meta-analysis. Medicine. 2021;100:51(e28425).
PZ and HH contributed equally to this work.
The authors have no funding and conflicts of interest to disclose.
The study was approved by the Human Research Ethics Committees of the First People's Hospital of Neijiang, Neijiang, Sichuan.
Consent for publication is not applicable.
All data used to support the findings of this study are included within the article.
All data generated or analyzed during this study are included in this published article.
References
- [1].Zadeh HG, Haddadnia J, Ahmadinejad N, Baghdadi MR. Assessing the potential of thermal imaging in recognition of breast cancer. Asian Pac J Cancer Prev 2015;16:8619–23. [DOI] [PubMed] [Google Scholar]
- [2].Chen W, Zhang W, Wu R, Cai Y, Xue X, Cheng J. Identification of biomarkers associated with histological grade and prognosis of gastric cancer by co-expression network analysis. Oncol Lett 2019;18:5499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yan Y, Wu Q, Li ZY, Bu ZD, Ji JF. Endoscopic ultrasonography for pretreatment T-staging of gastric cancer: an in vitro accuracy and discrepancy analysis. Oncol Lett 2019;17:2849–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155–9. [DOI] [PubMed] [Google Scholar]
- [5].Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell 2016;29:452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yamashita A, Shichino Y, Yamamoto M. The long non-coding RNA world in yeasts. Biochim Biophys Acta 2016;1859:147–54. [DOI] [PubMed] [Google Scholar]
- [7].Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol 2017;18:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xu YC, Liang CJ, Zhang DX, et al. LncSHRG promotes hepatocellular carcinoma progression by activating HES6. Oncotarget 2017;8:70630–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fan KJ, Liu Y, Yang B, Tian XD, Li CR, Wang B. Prognostic and diagnostic significance of long non-coding RNA AGAP2-AS1 levels in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2017;21:2392–6. [PubMed] [Google Scholar]
- [10].Wang W, Yang F, Zhang L, et al. LncRNA profile study reveals four-lncRNA signature associated with the prognosis of patients with anaplastic gliomas. Oncotarget 2016;7:77225–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hui B, Ji H, Xu Y, et al. RREB1-induced upregulation of the lncRNA AGAP2-AS1 regulates the proliferation and migration of pancreatic cancer partly through suppressing ANKRD1 and ANGPTL4. Cell Death Dis 2019;10:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Qi F, Liu X, Wu H, et al. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J Hematol Oncol 2017;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Oremus M, Oremus C, Hall GB, McKinnon MC. Inter-rater and test-retest reliability of quality assessments by novice student raters using the Jadad and Newcastle-Ottawa Scales. BMJ Open 2012;2: doi:10.1136/bmjopen-2012-001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu Z, Wang Y, Wang L, et al. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res 2019;38:194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [16].Tian Y, Zheng Y, Dong X. AGAP2-AS1 serves as an oncogenic lncRNA and prognostic biomarker in glioblastoma multiforme. J Cell Biochem 2019;120:9056–62. [DOI] [PubMed] [Google Scholar]
- [17].Li W, Sun M, Zang C, et al. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis 2016;7:e2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hong S, Yan Z, Song Y, Bi M, Li S. LncRNA AGAP2-AS1 augments cell viability and mobility, and confers gemcitabine resistance by inhibiting miR-497 in colorectal cancer. Aging 2020;12:5183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tingting Z, Xiaojing L, Xiaoyan T, Keqin H, Junjun Q. The antisense long noncoding RNA AGAP2-AS1 regulates cell proliferation and metastasis in epithelial ovarian cancer. J Cancer 2020;11:5318–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shao L, Sun W, Zhang H, et al. Long non-coding RNA AGAP2-AS1 increases the invasiveness of papillary thyroid cancer. Aging 2020;12:18019–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zheng Y, Lu S, Xu Y, Zheng J. Long non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells by sponging miR-15a/b-5p to upregulate the expression of HDGF and activating Wnt/β-catenin signaling pathway. Int J Biol Macromol 2019;128:521–30. [DOI] [PubMed] [Google Scholar]
- [22].Santer BD, Wigley TM, Mears C, et al. Amplification of surface temperature trends and variability in the tropical atmosphere. Science (New York, NY) 2005;309:1551–6. [DOI] [PubMed] [Google Scholar]
- [23].Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zheng R, Yao Q, Ren C, et al. Upregulation of long noncoding RNA small nucleolar RNA host gene 18 promotes radioresistance of glioma by repressing semaphorin 5A. Int J Radiat Oncol Biol Phys 2016;96:877–87. [DOI] [PubMed] [Google Scholar]
- [25].Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol 2013;45:1895–910. [DOI] [PubMed] [Google Scholar]
- [26].Zhang J, Feng S, Su W, et al. Overexpression of FAM83H-AS1 indicates poor patient survival and knockdown impairs cell proliferation and invasion via MET/EGFR signaling in lung cancer. Sci Rep 2017;7:42819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xie Y, Zhang Y, Du L, et al. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol Oncol 2018;12:648–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Luo W, Li X, Song Z, Zhu X, Zhao S. Long non-coding RNA AGAP2-AS1 exerts oncogenic properties in glioblastoma by epigenetically silencing TFPI2 through EZH2 and LSD1. Aging 2019;11:3811–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gao L, Zhao A, Wang X. Upregulation of lncRNA AGAP2-AS1 is an independent predictor of poor survival in patients with clear cell renal carcinoma. Oncol Lett 2020;19:3993–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chen J, Peng X, Dai Y. The long non-coding RNA (lncRNA) AGAP2-AS1 is upregulated in ovarian carcinoma and negatively regulates lncRNA MEG3. Med Sci Monit 2019;25:4699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shen S, Li K, Liu Y, et al. Silencing lncRNA AGAP2-AS1 upregulates miR-195-5p to repress migration and invasion of EC cells via the decrease of FOSL1 expression. Mol Therap Nucleic Acids 2020;20:331–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xu Y, Xia X, Jiang Y, et al. Down-regulated lncRNA AGAP2-AS1 contributes to pre-eclampsia as a competing endogenous RNA for JDP2 by impairing trophoblastic phenotype. J Cell Mol Med 2020;24:4557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tao Y, Tang Y, Yang Z, et al. Exploration of serum exosomal LncRNA TBILA and AGAP2-AS1 as promising biomarkers for diagnosis of non-small cell lung cancer. Int J Biol Sci 2020;16:471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Poulet C, Njock MS, Moermans C, et al. Exosomal long non-coding RNAs in lung diseases. Int Mol Sci 2020;21: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li H, Guo S, Zhang M, Li L, Wang F, Song B. Long non-coding RNA AGAP2-AS1 accelerates cell proliferation, migration, invasion and the EMT process in colorectal cancer via regulating the miR-4,668-3p/SRSF1 axis. J Gene Med 2020;22:e3250. [DOI] [PubMed] [Google Scholar]
- [36].Dong H, Wang W, Mo S, et al. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res 2018;37:202. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [37].Zheng Z, Chen M, Xing P, Yan X, Xie B. Increased expression of exosomal AGAP2-AS1 (AGAP2 antisense RNA 1) in breast cancer cells inhibits trastuzumab-induced cell cytotoxicity. Med Sci Monit 2019;25:2211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]