Abstract
Non melanoma skin cancer (NMSC) is one of the most common types of skin cancer. It has a number of subtypes, which include basal cell carcinoma, cutaneous squamous cell carcinoma and Merkel cell carcinoma. MicroRNAs are short, non-coding RNA (ribonucleic acid) molecules, capable of regulating gene expression at a post transcriptional level. They play a pivotal role in a variety of physiologic cellular functions and pathologies, including malignant diseases. The development of miRNAs represents an important study field, which has been extensively exploited in melanoma for almost a decade with promising results, therefore we consider it a stepstone for further research projects also in non-melanoma skin cancers. The aim of our study was to explore the current literature in order to present the role of the different miRNAs in some of the most frequent types of NMSC pertaining to oncogenesis, evolution and therapy. The most relevant and accurate available data from the literature were evaluated. Our study concluded that there are almost 100 miRNAs which can be upregulated or downregulated and can play a role in oncogenesis. They can be easily identified in circulation, are stable and they can be important diagnosis/prognosis and therapy monitoring markers.
Keywords: NMSC, miRNA, skin, cancer, basal, squamous, Merkel, cell, carcinoma, oncogenesis
1. Introduction
The skin is the body’s largest organ and one of the first defense lines against various external factors. The maintenance of cellular integrity and underlying complex physiology of the skin are crucial for the overall health of organisms. Exposure to environmental agents including UV(ultraviolet)-radiation or carcinogens could lead to multiple pathological manifestations and compromised cellular integrity. The worldwide incidence of skin cancer is increasing rapidly due to the damage of the ozonosphere [1,2,3,4,5,6,7].
Malignant skin tumors are largely divided into melanoma and non-melanoma cancers. Non melanoma skin cancer (NMSC) is one of the most common types of skin cancer. According to the Globocan statistical data, there were over 1 million new cases worldwide in 2020, placing NMSC on the 4th place among all malignancies. Although the mortality is lower than other types of cancer, there were more than 60,000 cases reported in 2020 [8]. It has a number of subtypes, which include basal cell carcinoma (BCC), cutaneous squamous cell carcinoma (SCC) and Merkel cell carcinoma (MCC). BCC is the most frequent cutaneous carcinoma, followed by SCC and accounts for more than 20% of all skin cancer cases all over the world [9,10,11,12].
MicroRNAs (miRNAs) are short, non-coding RNA molecules formed by 17 to 23 nucleotides (nt). These molecules have the ability of regulating gene expression in post transcriptional phase. Being encoded within exons and introns, they play a major role in various physiological cellular functions and pathologies, including cancer [13,14,15,16,17,18].
Their discovery by two research groups in 1993 was one of the greatest achievements in molecular biology [19,20]. MiRNA biogenesis is a progressive process beginning with the production of primary miRNAs (pri-miRNAs) by specific DNA (deoxyribonucleic acid) transcripts which are transformed into precursor miRNAs (pre-miRNAs) followed by mature miRNAs. MiRNAs are able to regulate not only gene expression, but also protein translation. They are actively secreted into the bloodstream by a variety of cancer cells. MiRNAs can be identified in tissue samples and biological fluids such as cerebrospinal fluid, serum, plasma, saliva and urine. Usually, the circulating miRNAs are the first choice in the clinical setting due to the simplicity of technique but the result can be less concludent than the ones from tissue samples [21,22,23,24,25,26,27].
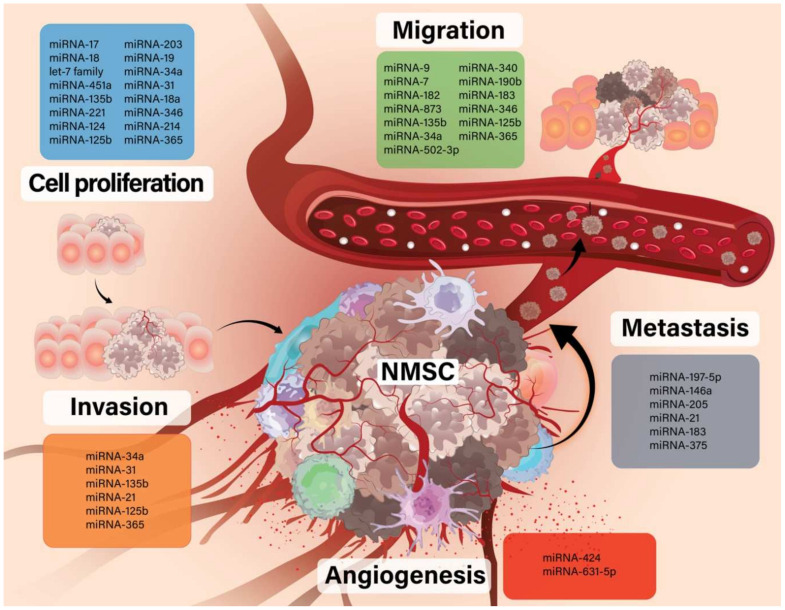
The aim of our study was to explore the current literature in order to present the role of the different miRNAs in some of the most frequent types of NMSC pertaining to oncogenesis, evolution and therapy (Figure 1).
Figure 1.
The role of different miRNA in NMSC evolution.
2. Materials and Methods
In the present review we encompassed the most relevant and accurate available data from the literature regarding the role of miRNAs mechanisms in oncogenesis, tumor progression, therapy and drug response in NMSC. The following keywords were searched: “miRNA”, “non-melanoma”, “skin”, “cancer”. Our process of selection included two of the most reliable databases in the medical field, EMBASE and PubMed, under specified criteria such as 10-years filter English language. The final article selection remained subjective. After the selection of the articles we presented the miRNAs function and their implications of miRNAs in different types of NMSC.
3. miRNA Function in Oncogenesis, Evolution and Therapy
miRNAs are small noncoding RNAs (17–23 nucleotides) expressed endogenously that occur both intra- and intergenically. The primary miRNAs are transcribed by RNA polymerase II resulting in long transcript forms which are nuclear cleaved by the Drosha RNase III endonuclease in a 60–70 nt precursor miRNA (pre-miRNA). The pre-miRNA is further processed by the RNase III endonucleproliferationase Dicer, resulting in a miRNA: miRNA duplex. One strand of the miRNA: miRNA duplex, the mature miRNA, is loaded into the RNA-induced silencing complex (RISC), while the other strand is degraded. The binding of the miRNA-RISC complex induces a translational repression or destabilization of mRNA.
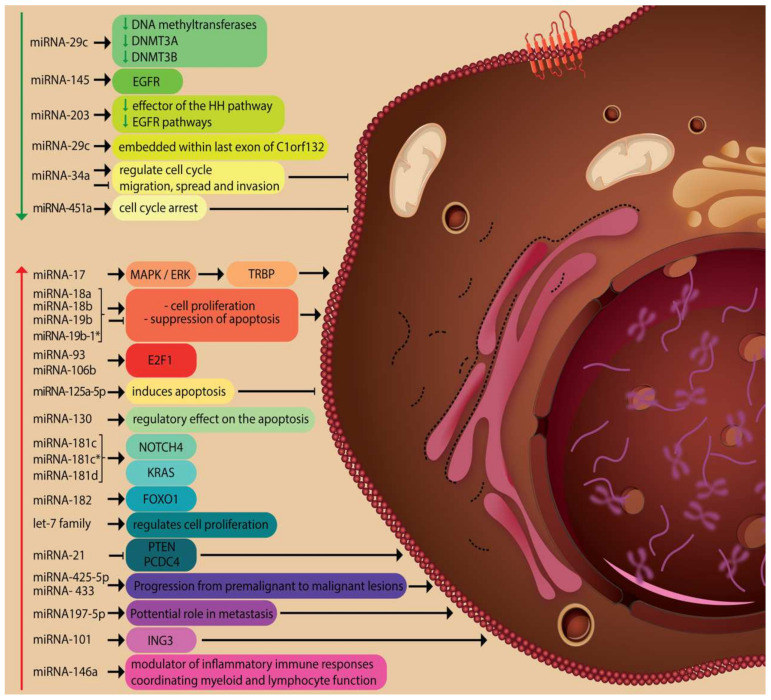
Through this action, miRNAs are involved in many physiological and pathological processes being recognized as potential therapeutic targets as well as possible biomarkers with diagnostic and/or prognostic potential (Figure 2). There is also a series of unconventional roles attributed to these molecules. Thus, miRNAs can activate Toll-like receptors displaying a pro-inflammatory/pro-metastatic potential, and hence, specific miRNAs can become future therapy targets. Also, miRNAs can act on protein expression in a cell cycle-dependent manner [28,29,30,31,32,33].
Figure 2.
Physiological and pathological processes involving miRNAs.
3.1. The Role of miRNAs in Basal Cell Carcinoma (BCC)
BCC is the most commonly encountered invasive skin cancer worldwide, arising from the basal skin cell layer. Metastasis of BCC is very rare, but the metastasized disease has a poor prognosis [34,35].
Regarding the role of miRNA in BCC, relevant contributions were brought by Sand M. et al. [13,14], as well as Heffelfinger et al. [36]. According to these authors, miRNA-21, miRNA-143, miRNA-148a, miRNA-378, miRNA-182, and let-7 family members were the most highly expressed in these tumors. 21 miRNAs have shown particular difference in expression between nodular and infiltrative BCCs. Sixteen significantly up-regulated miRNAs (miRNA-17, miRNA-18a, miRNA-18b, miRNA-19b, miRNA-19b-1*, miRNA-93, miRNA-106b, miRNA-125a-5p, miRNA-130a, miRNA-181c, miRNA-181c*, miRNA-181d, miRNA-182, miRNA-455-3p, miRNA-455-5p and miRNA-542-5p) and ten significantly down-regulated miRNAs (miRNA-29c, miRNA-29c*, miRNA-139-5p, miRNA-140-3p, miRNA-145, miRNA-378, miRNA-572, miRNA-638, miRNA-2861 and miRNA-3196) were detected in BCC compared with normal skin.
Sonkoly E et al. found in 2012 that miRNA-203 functions as a tumor suppressor miRNA in BCC. Reduced levels of miRNA-203 expression in BCCs are likely to sustain a high proliferative rate. It is known that upregulation of the Hedgehog (HH) pathway is the main molecular anomaly in all BCCs. Evidence suggests that aberrant activation of HH signaling is sufficient to initiate BCC carcinogenesis. MiRNA-203 functions as a downstream effector of the HH pathway and the epidermal growth factor (EGFR) pathways. They also mentioned that miRNA-203 may be a potential therapeutic target for the treatment of BCC [37].
Another study of Sand and colleagues identified 33 upregulated miRNAs in BCC by next-generation sequencing under neoadjuvant vismodegib therapy [38].
Al-Eryani L and collaborators noted that miRNA-425-5p and miRNA-433 were elevated in BCCs and other six miRNAs (miRNA-29c, miRNA-381, miRNA-452, miRNA-487b, miRNA-494 and miRNA-590-5p) were specifically inhibited [1,39].
Hu P et al. described the role of miRNA-34a present in serum samples of BCC patients. They observed low levels of miRNA-34a expression in patients having BCC relative to the healthy population and they established a correlation between miRNA expression with tumor cell diameter, lymph node metastasis and histological types of BCC. In the low expression group, median progression-free survival, overall survival time and rate were significantly improved. Also, the prognosis of basal cell carcinoma patients with low expression levels of miRNA-34a was poor [1,40].
Sun and Jiang confirmed miRNA-451a as a BCC tumor suppressor miRNA. In their study, miRNA-451a expression was reduced and the overexpression of miRNA-451a suppressed BCC cell growth through G1 cell cycle arrest, suggesting a therapeutic target of miRNA-451a in BCC [41].
Chang et al. studied the role of miR197-5p and they found that the miRNA was expressed in all patients with metastatic basal cell carcinoma (MBCC) and non-metastatic basal cell carcinoma (non-MBCC). Addition of a synthetic inhibitor of miRNA-197 significantly reduced fibroblast migration but not invasion. This means that miR197-5p is a potential candidate for future research into mechanisms of BCC metastasis [1,42].
Wan and Li found many dysregulated miRNAs, such as miRNA-29b, miRNA-7b, miRNA-141 miRNA-9, miRNA-203, miRNA-200a, miRNA-7c and miRNA-132, and that these miRNAs were significantly associated with BCC. MiRNA-203, miRNA-495, miRNA-385, miRNA-220a and miRNA-30e target PTCH1, which is an important component of tumor deriving signaling in BCC pathophysiology. Their network analysis identified some miRNAs such as miRNA-101, miRNA-103, miRNA-130a, and that miRNA-144 directly targeted the inhibitor of growth protein 3 (ING3), a tumor suppressor protein that interacts with tumor suppressor TP53 [1,43].
MiRNA-146a is of particular interest in the etiology of NMSCs, as it is an important modulator of inflammatory immune responses, coordinating myeloid and lymphocyte function to impact aspects of immunity. MiRNA-146a has 224 potential mRNA binding targets on the cancer susceptibility gene RNASEL (Ribonuclease L). In their study, Farzan et al. found that miR146a may play a role in the development of NMSC, both BCC and SCC [44] (Table 1).
Table 1.
Modified miRNAs in BCC.
| Nr | Ref | Expression | miRNA | Probe | Species | Role |
|---|---|---|---|---|---|---|
| 1 | Sand M. et al. [13,14] | upregulated | miRNA-17 | tissue | human | pro-growth miRNA regulated in vitro by MAPK (mitogen-activated protein kinase)/ ERK-induced phosphorylation of TRBP (TAR-RNA binding protein) |
| miRNA-18a miRNA-18b |
cell proliferation and the suppression of apoptosis | |||||
| miRNA-19b miRNA-19b-1* |
responsible for enhanced cell pro- liferation and the suppression of apoptosis | |||||
| miRNA-93 | transcription factor E2F1 (E2 promoter binding factor 1) is a target gene of miRNA-93 |
|||||
| miRNA-106b | transcription factor E2F1 is a target gene of miRNA-106b | |||||
| miRNA-125a-5p | induces apoptosis | |||||
| miRNA-130a | regulatory effect on the apoptosis | |||||
| miRNA-181c miRNA-181c* miRNA-181d |
targets NOTCH4( neurogenic locus notch homolog-4) and KRAS (Kirsten rat sarcoma virus) |
|||||
| miRNA-182 | negatively regulate human Forkheadbox
O1 (FOXO1) |
|||||
| miRNA-455-3p miRNA-455-5p miRNA-542-5p |
not mentioned | |||||
| downregulated | miRNA-29c | downregulates DNA methyltransferases DNMT3A and DNMT3B | ||||
| miRNA-29c* miRNA-139-5p miRNA-140-3p |
not mentioned | |||||
| miRNA-145 | targets EGFR | |||||
| miRNA-572 miRNA-638 miRNA-2861 miRNA-3196 |
not mentioned | |||||
| 2 | Heffelfinger et al. [36] | upregulated | let-7 family | tissue | human | involved in regulating cell proliferation |
| miRNA-21 | represses a variety of tumor suppressors such as PTEN (Phosphatase And Tensin Homolog) and PCDC4 (Programmed cell death protein 4) |
|||||
| miRNA-148a miRNA-143 miRNA-378 |
not mentioned | |||||
| 3 | Sonkoly E et al. [37] | downregulated | miRNA-203 | tissue | human | downstream effector of the HH pathway and EGFR pathways. Potential therapeutic target for the treatment of BCC |
| 4 | Al-Eryani L et al. [39] | upregulated | miRNA-425-5p | tissue (arsenic induced lession) | human | Premalignant lesions progression to malignancy |
| miRNA- 433 | ||||||
| downregulated | miRNA-29c | encoded in the last exon of C1orf132 (chromosome 1 open reading frame 132), and the transcript converts an unknown open reading frame |
||||
| miRNA-381 miRNA-452 miRNA487b miRNA-494 miRNA-590-5p |
not mentioned | |||||
| 5 | Hu P et al. [40] | downregulated | miRNA-34a | blood | human | can regulate cell cycle and inhibit the migration, spread and invasion of tumor cells |
| 6 | Sun H, Jiang P [41] | downregulated | miRNA-451a | tissue | human & mouse | limits cell proliferation by cell cycle arrest induction, suggesting the potential therapeutic target of miRNA-451a in BCC |
| 7 | Chang J et al. [42] | upregulated | miRNA197-5p | blood | human | Potential role in metastasis process |
| 8 | Wan C, Li Y [43] | miRNA-101 | tissue | human | targets ING3 | |
| miRNA-7b miRNA-141 miRNA-9 miRNA-200a miRNA-203 miRNA-7c miRNA-132 miRNA-203 miRNA-495 miRNA-385 miRNA-220a miRNA-30e miRNA-29b miRNA-103 miRNA-130a miRNA-144 |
not mentioned | |||||
| 9 | Farzan SF et al. [44] | upregulated | miRNA-146a | blood | human | modulator of inflammatory immune responses, coordinating myeloid and lymphocyte function to impact aspects of both innate and adaptive immunity |
3.2. The Implication of miRNAs in Squamous Cell Carcinoma (SCC)
The cutaneous SCC is a skin tumor originated from epidermal keratinocytes and the second most common skin cancer with arising incidence [34,45].
Exposure to the ultraviolet radiation is the main risk factor for SCC development. UVR induces a significant accumulation of DNA damage in the cutaneous cells, providing a wide mutational landscape which drives to SCC carcinogenesis. Other risk factors are old age, immunosuppression, smoking and genetic factors [46,47].
In the latest years, there have been major improvements in the recognition of target genes and the functions of numerous miRNAs [48].
Sand et al. published a series of works regarding the role of miRNA in SCC. They found in 2010 that the expression levels of the miRNA machinery, namely Drosha, DGCR8, AGO1, AGO2, PACT, and TARBP1, were significantly higher compared to healthy controls and Dicer levels were significantly higher compared to intra-individual controls [49,50]. They also performed a comparative miRNA analysis between SCC biopsies and adjacent healthy skin. Their result identified 13 particularly elevated and 18 downregulated miRNAs in SCC tumors. MiRNA-31 was expressed as higher upregulated miRNA in SCC in comparison with normal skin [34,51].
Dziunycz et al. identified in SCC the elevated expression of miRNA-21 and miRNA-184 and the lower expression of miRNA-203. Also, they described the increased expression of miRNA-21, hsamiR-203, and miRNA-205 by UVA exposure, in comparison with UVB exposure, which increased miRNA-203 and decreased miRNA-205 [14,52].
Yamane et al. identified the downregulation in SCC both in vitro and in vivo of miRNA-124 and miRNA-214. They are regulators of extracellular-signal-regulated kinase 1, 2 and 3 [14,53].
MiRNA-205 was associated with particular pathological characteristics of poor prognosis, including desmoplasia, perineural invasion and infiltrative patterns, clinically correlated with local recurrence. On the other side, miRNA-203 expression was linked to a favorable prognosis due to its identification mainly in well-differentiated areas and rarely in the invasion site [27,54,55].
Zhang et al. correlated low levels of miRNA-20a with poorer overall survival, proving the reduced expression of miRNA-20a in association with worse TNM (tumor node metastasis) staging [27,56].
Gong et al. demonstrated that miRNA-221 plays an oncogenic function in SCC. miRNA-221 expression was significantly higher in SCC tissues than in normal tissues. miRNA-221 knockdown inhibited the proliferation and cell cycle, while upregulation of miRNA-221 presented the opposite role. PTEN is a direct target gene of miRNA-221 [57].
Mutations to the TP53 gene are common in UV-exposed keratinocytes and contribute to apoptotic resistance in skin cancer. MiRNA-34a influences TP53 action on several cell processes such as growth arrest, senescence and apoptosis, also recognized as a target of P53. Therefore, in non-melanoma skin tumors, the key of overcoming resistance to cell death signaling could be represented by targeting TP53 gain-of-function mutations. Lefort et al. found that in SCC miRNA-34a expression was downregulated both in vivo and in vitro [58,59,60,61,62,63].
Kanitz et al. associated lower levels of miRNA-361-5p in SCC in comparison with normal skin and identified VEGFA (Vascular Endothelial Growth Factor A) as a direct target of miRNA-361-5p [64].
Chen et al. demonstrated that miRNA-346 is downregulated and can function as an onco- miRNA in SCC [65].
MiRNA-125b has been found by Xu et al. to be downregulated in premalignant lesions (actinic keratosis) and in SCC in comparison with normal tissue. It has a potential role of therapeutic biomarker. Matrix metalloproteinase (MMP)13 was identified as a direct target of miRNA-125b [66].
Olasz et al. identified in SCC an oncogenic role of miRNA-135b, with leucine zipper tumor suppressor 1 (LZTS1) as its direct target. In early stages of SCC progression, MiRNA-135b can influence cell migration and tumor invasiveness, acting as an oncogenic miRNA in human keratinocytes. By in vitro analysis, downregulation of LZTS1 mRNA was associated with miRNA-135b overexpression and also with increased mobility and aggressivity of malignant cells [67].
In the study of Zhou M. et al. the authors demonstrated that miRNA-365 was overexpressed both in fresh SCC samples and paraffin-embedded tissues and that miRNA-365 may act as an onco-miRNA in cutaneous SCC both in vitro and in vivo. These findings demonstrate that miRNA-365 may be a carcinogenic factor in cutaneous SCC and a potential target in cutaneous SCC therapy. The overexpression of miRNA-365 in cutaneous SCC can be used as a potential indicator both in the clinical diagnosis and treatment [68] (Table 2).
Table 2.
Modified miRNAs in SCC.
| Nr | Ref | Expression | miRNA | Probe | Species | Role |
|---|---|---|---|---|---|---|
| 1 | Sand M. et al. [51] | upregulated | miRNA-31 | tissue | human | downregulate the tumor suppressor RhoBTB1 (Rho Related BTB Domain Containing 1) in the cSCC cell line A-431, determing cell proliferation and invasion |
| miRNA-135b | miRNA-135b can regulate cell migration and tumor invasiveness in early stages of SCC progression and can act as an oncogenic miRNA in human keratinocytes | |||||
| miRNA-424 | determines angiogenesis, regulates cell-autonomous angiogenic functions | |||||
| miRNA-21* miRNA-374a miRNA-196a |
not mentioned | |||||
| miRNA-18a | associated with the Sonic Hedgehog pathway, correlated with molecular pathogenesis of cSCC | |||||
| miRNA-766 miRNA-128 |
not mentioned | |||||
| miRNA-130b | downregulate the tumor suppressor protein 53-induced nuclear protein 1 (TP53INP1) | |||||
| miRNA-455-5p | not mentioned | |||||
| miRNA-21 | targets phosphatase and tensin homolog (PTEN), PDC4 (Programed Cell Death 4) and BTG2 (B-cell translocation gene 2) | |||||
| downregulated | miRNA-30a* miRNA-133b miRNA-101 miRNA-4324 miRNA-136 |
not mentioned | ||||
| miRNA-378 | targets insulin-like growth factor 1 receptor (IGF1R) and caspase 3; reduced expression in basal cell carcinoma | |||||
| miRNA-204 miRNA-497 miRNA-29c miRNA-214 |
not mentioned | |||||
| miRNA-145 | inhibits actin-binding protein Fascin homolog 1 (FSCN1) in esophageal squamous cell carcinoma; down-regulated in basal cell carcinoma | |||||
| miRNA-199a-5p miRNA-125b |
not mentioned | |||||
| miRNA-140-3p | targets CD38; down-regulated in basal cell carcinoma | |||||
| miRNA-26a | downregulation of oncogene Histone-lysine N-methyltransferase (EZH2) | |||||
| 2 | P Dziunycz et al. [52] | upregulated | miRNA-21 | tissue | human | essential role in the development or maintenance of SCC of the skin |
| miRNA-184 miRNA-205 |
not mentioned | |||||
| downregulated | miRNA-203 | unleash p63 expression, leading to decreased cell senescence and supporting SCC formation | ||||
| miRNA-378 | not mentioned | |||||
| 3 | Yamane et al. [53] | downregulated | miRNA-124 miRNA-214 |
Tissue and serum | human | lead to overexpression of ERK1/2. May lead to the development of useful biomarkers for early detection of this tumor and to new treatments using miRNA |
| 4 | Zhang L et al. [56] | downregulated | miRNA-20a | tissue | human | might play important roles in the tumorigenesis and progression of CSCC patients, may serve as a novel molecular marker to predict the tumor progression and inferior prognosis of CSCC patients |
| 5 | Gong et al. [57] | upregulated | miRNA-221 | blood | human | significantly promotes cell proliferation |
| 6 | Kanitz et al. [64] | downregulated | miRNA-361-5p | tissue | human | regulator of VEGFA expression |
| 7 | Chen et al. [65] | downregulated | miRNA-346 | tissue | human | promotes the cSCC cell proliferation and migration through directly targeting SRCIN1 (SRC Kinase Signaling Inhibitor 1). This study may provide a new therapeutic target for cSCC. |
| 8 | Xu et al. [66] | downregulated | miRNA-125b | tissue | human | potential therapeutic biomarker. Matrix metalloproteinase (MMP)13 was considered a direct target of miRNA-125b |
| 9 | Olasz et al. [67] | upregulated | miRNA-135b | tissue | human | miRNA-135b can regulate cell migration and tumor invasiveness in early stages of SCC progression and can act as an oncogenic miRNA in human keratinocytes |
| 9 | Zhou M et al. [68] | upregulated | miRNA-365 | tissue | human | downregulates NFIB (nuclear Factor I B) and inhibits the expression of cyclin-dependent kinase CDK6 and CDK4 |
3.3. Modified miRNAs in Merkel Cell Carcinoma (MCC)
Merkel cell carcinoma (MCC) is a highly malignant non-melanoma skin tumor which usually arises in the sun-exposed skin [69,70]. A very high percent of the MCC is also associated with immune deficiencies and the presence of Merkel cell polyomavirus (MCPyV) [71,72].
The miRNAs role in MCC was studied by several authors. As compared to other variants of NMSCs, Ning et al. have found the overexpression of eight miRNAs (miRNA-502-3p, miRNA-9, miRNA-7, miRNA-340, miRNA-182, miRNA-190b, miRNA-873, and miRNA-183) and the suppression of three (miRNA-3170, miRNA-125b, and miRNA-374c), with the abundance of miRNA-182 in the malignant cells. By concomitent evaluating the expression of four miRNAs (miRNA-182, miRNA-183, miRNA 190b, and miRNA-340) in the MCPyV-negative cell line noted their downregulation. The authors concluded that these miRNA have a possible diagnostic role in cases of MCPyV-positive MCC [73].
Renwick et al. have correctly discerned BCC from MCC, due to overexpression of miRNA-205 and miRNA-375 [74]. In 2019 it was found that Atonal homolog 1 (ATOH1) expression, a tumor suppressor gene, binds and further activates miRNA-375, the highest abundant miRNA in MCC. In experimental models, it was shown that ATOH1 knockdown in cell lines reduced miRNA-375 expression. ATOH1 overexpression increases the risk of metastasis. The infection with MCPyV promotes carcinogenesis by ATOH1 stimulation [75].
Vieja et al. described the miRs related to positivity and negativity of MCPyV in MCCs. Therefore, miRNA-30a, miRNA-34a, miRNA-142-3p, and miRNA-1539 were overexpressed in MCPyV-positive MCCs, while miRNA-181d was overexpressed in MCPyV-negative MCCs [76].
Another important finding was the clinical possibility of associating circulating miRNAs as biomarkers for MCC. Using RT-PCR method, Moens et al. evaluated the expression of several miRNAs in exosomes and identified the presence of miRNA-30a, miRNA-125b, miRNA-183, miRNA-190b, and miRNA-375 [77]. Tuaeva et al. translated 11 of 22 circulating tumor DNA and circulating miRNAs into clinical practice to predict the clinical-pathological features of tumors thus ameliorating the diagnostic and therapeutic strategies available for cancer patients [78] (Table 3).
Table 3.
Modified miRNAs in MCC.
| Nr | Ref | Expression | miRNA | Probe | Species | Role |
|---|---|---|---|---|---|---|
| 1 | Ning et al. [73] | upregulated | miRNA-9 miRNA-502-3p miRNA-7 miRNA-340 miRNA-182 miRNA-190b miRNA-873 miRNA-183 |
tissue | human | increasing tumor motility and colony formation. Potential diagnostic and therapeutic applications in cases of MCPyV-positive MCC |
| downregulated | miRNA-3170 miRNA-125b miRNA-374c |
not mentioned | ||||
| 2 | Renwick et al. [74] | upregulated | miRNA-205 miRNA-375 | tissue | human | microRNAs downregulate the expression of gene targets through interaction with their three prime untranslated region (3′ UTR). miRNA-375 targets MTPN(myotrophin) gene, which encodes the myotrophin protein, further regulating hormone release and exocytosis. Potential diagnostic role by discerning BCC from MCC |
| 3 | Veija et al. [76] | downregulated | miR-34a, miR-1539, miR-30a, miR-142-3p | tissue | human | may play a role in the oncogenesis of MCV-negative tumors. |
| upregulated | miR-181d |
4. Discussion
Despite the advances in the prevention, diagnosis and treatment of NMSC worldwide, the exact molecular basis of NMSC remains unclear. miRNAs are considered to be important genetic regulators of various biological processes, including cell proliferation, development, invasion and apoptosis and can exhibit a dual function of either pro- or antitumoral action.
A number of previous studies have demonstrated that miRNAs are involved in several pathologies, especially in different types of cancer. MiRNAs were brought in the spotlight by the intense research made in this domain due to their implication in the tumorigenesis mechanism. For example, miRNA-17-92 with its subtype miRNA-17-15p expresses an oncogene function involved in tumor aggressiveness in several malignancies such as colorectal, liver and gastric cancer. On the other hand, the same miRNA can exert a tumor-suppressive effect in breast, prostate and lung cancer [79,80].
Lately, the Next-Generation Sequencing technologies (NGS) have offered a new opportunity to identify different miRNAs sequences that play a crucial role in the regulation of several cellular processes, such as transcription, post-transcriptional modifications, and signal transduction. Several published reviews have shown a wide range of miRNAs that play an important role in tumorigenesis, tumor progression and in drug response [81,82,83,84].
In the latest years, miRNAs have been under the spotlight due to their potential role of therapeutic targets. In vivo studies on different mice cell disease lines have proved the efficacy of miRNA targeting therapeutic strategies. Nowadays, RNA molecules are being clinically studied as therapeutic molecules, mostly for systemic application. For in vivo applications, multiple modifications are used in combination to achieve the best functional efficiency and stability [34,85,86,87].
In the field of skin cancers, miRNA have been broadly studied mostly in melanoma but also in other tumor types [88]. Due to the tumoral heterogeneity which determines highly aggressive characteristics and negative prognostic factors, melanoma has represented an intense study field for miRNAs. According to the miRNA expression in tumor tissue or in body fluids, a few years ago the role of miRNA-221 as prognostic factor was one of the first identified [89]. Furthermore, the expression of other miRNAs such as miRNA-203, miRNA-10b, miRNA-200b, miRNA-155 have been associated with metastasis and negative outcome. Potential therapeutic targets such as miRNA-675, miRNA-204, the combination between miRNA-135 and large tumor suppressor kinase 2 (LATS2) have also been suggested [90,91,92]. As for prognostic biomarker panel of shorter survival, miRNA-125b, miRNA-200c and miRNA-205 have been proposed [93]. Larger studies involving the analysis of over 25 miRNAs have correlated the presence of miRNA-10b, miRNA-16 and miRNA-21 with poor prognosis [94]. Another role for miRNAs in melanoma patients is the involvement in the therapy resistance process and particular miRNAs such as miRNA-92a-1-5p, miRNA-708-5p in association with specific genes have been identified [95].
In patients who undergo targeted therapy with BRAF (v-raf murine sarcoma viral oncogene homolog B1) and MEK (mitogen-activated protein kinase) inhibitors the expression of downregulated miRNA-579-3p is associated with treatment resistance, but miRNA-126-3p is responsible for increasing the sensitivity in dabrafenib-resistant patients [96,97]. Chemosensitivity of the malignant cells was correlated with the expression of miRNA-211 but the sensitivity to MAPK inhibitors is decreased when miRNA-214 is overexpressed [98,99]. Referring to immunotherapy in melanoma, the most innovative treatment option implemented so far, miRNAs can be further used as blood markers for efficacy by measuring their plasmatic levels before starting the treatment [100].
Increasing numbers of miRNAs have been identified as critical regulators in the initiation and progression of NMSC. In this study we have selected three of the most frequent types of NMSC and the role of miRNA was presented individually. Regarding BCC, studies have shown that more than 60 miRNAs were identified and can play a role in diagnosis or treatment.
The BCC remains the most frequent type of NMSC. The literature data identified numerous types of miRNAs associated with the malignancy and more particularly, with the variation between the nodular and infiltrative BCCs. A particular type of miRNA, miRNA-203 is involved in the carcinogenesis process by exerting a downstream effectory function upon the Hedgehog pathway and EGFR (epidermal growth factor receptor) and could represent a possible therapeutic target in this setting. Also, numerous types of miRNAs were associated with other mechanisms of the malignant cycle such as PTCH1 (patched 1) and ING3 (inhibitor of growth factor family 3). Studies have evidenced a possible role upon tumor progression and metastasis and miR197-5p was the main type identified. Another possible biomarker was miRNA-34a, associated with poor prognosis characteristics in BCC patients. MiRNA-146a has gained particular attention due to its effect on the inflammatory immune response and its common role in both BCC and SCC tumor development. Even if the prognosis is not severe and metastases are rare, some cases of advanced BCC or subtypes of BCC like the morpheus type are associated with a very poor prognosis. In our opinion, a further direction of research could be represented by the differentiation of BCC subtypes according to the miRNAs expression. The results could provide a better treatment selection and improve patients care delivery.
Many miRNAs are upregulated or downregulated in SCC and MCC and they play a role in the evolution of these pathologies. As we already know, the presence of regional metastasis in SCC and MCC is more frequent than in BCC and can have a poor prognosis. In SCC, distinct types of miRNAs were influenced and associated with either UVA or UVB exposure and several specific types were correlated to the malignant process in comparison with the expression in the normal skin, out of which miRNA-31 was the most common. Apart from their negative predictive role due to carcinogenesis and tumor progression, particular types were linked to a favorable prognosis such as miRNA-203. MiRNAs can also express their inhibiting or stimulating role in the malignant process by targeting specific genes such as PTEN, TP53, VEGFA, MMP13 and LZTS1 suggesting a synergic mechanism of action. Another miRNA with oncogene role intense studied in vivo and in vitro was miRNA-365 with a potential role in the clinical setting.
In MCC, the study of miRNA has shown the association between particular types of miRNA and their correlation with the MCPyV-positive subtype of MCC. Also, previous studies highlighted distinct types of miRNAs linked to MCPyV-positive and MCPyV-negative MCC, suggesting their different prognostic role in this particular setting. The influence of miRNA in MCC carcinogenesis was integrated either by downregulation, overexpression or correlation to ATOH1 tumor suppression gene. The potential role of miRNA as a biomarker in MCC was evidenced by RT-PCR (reverse transcription polymerase chain reaction) of exosomes and by circulating DNA and miRNA and could provide a key to personalized therapy in this category of patients.
Understanding the role of miRNAs in the diagnosis and metastasis process could increase the survival rate in this category of patients. The method of identification can vary from tissue sample, which can present disadvantages due to the biopsy failure, to circulating miRNAs from body fluids. The latter mechanism involves a non-invasive technique, which can be easily repeated and provides real-time monitoring, but its sensitivity can vary. Still, this method can be used for early detection as in comparison with the tissue sample, which is characterized by a higher sensitivity because the miRNAs are originated only from the tumor cells [101,102]. Introducing the liquid biopsy in the clinical daily practice can become a preferred diagnosis tool due to the less invasive characteristic and in combination with dermoscopy could differentiate the aggressivity profile types of NMSC.
Regardless the type of sample, miRNAs can be identified by the following technologies: microarray platforms which provide the characteristics of the detected miRNAs, quantitative real-time polymerase chain reaction (qPCR), quantitative reverse transcription real-time PCR (RT-qPCR) with different versions, used mainly for malignant biomarker identification. Nevertheless, RT-PCR is the gold standard tool for diagnosis and research in this field [103,104,105]. Furthermore, apart from the traditional techniques mentioned above, recent studies suggest the use of next-generation sequencing (NGS), miRNA enzyme immunoassay (miREIA) and multiplexed miRNA analysis which need further investigation in skin cancer patients in order to be broadly validated for clinical use and replace the actual gold standard method of detection [106,107,108].
The development of miRNAs represents an important study field, which has been extensively exploited in melanoma for almost a decade with promising results, therefore we consider it a stepstone for further research projects also in non-melanoma skin cancers which provide so far limited results.
Probably the highest impact upon treatment and prognosis could be represented by the early identification of the patients with high risk of developing metastasis. This work-up could offer a personalized treatment approach with a better prognosis and improved overall survival.
5. Conclusions
Non melanoma skin cancers need constant attention due to their increasing incidence and rapid evolution. Discovering molecular mechanisms helps us understand tumorigenesis, the process of metastasis and evolution while also leading to development of new therapeutic strategies. The miRNAs, these small non-coding RNAs that control gene expression at the post transcriptional level, are gaining increasing attention. There are almost 100 miRNAs which can be upregulated or downregulated and can play a role in oncogenesis. They can be easily identified in circulation, are stable and they can be important indicators of therapeutic evolution, diagnosis and prognosis. A reliable technology for assessing miRNAs, especially in the clinical workflow, is also mandatory. The miRNAs impact on the chemosensitivity and chemoresistance in tumors can be a pathway for new researches and could provide new strategies to optimize therapeutic approaches.
Even if immunotherapy has revolutionized the therapeutic approach of advanced NMSC, a high percent of the patients do not respond to this therapy, and their prognosis is severe with a very low survival rate.
Unfortunately, selecting patients who have a real clinical benefit from this therapy from those who do not improve is not yet currently available. Using miRNAs could allow for individualized treatments and better responses to chemotherapy treatments and immunotherapy.
Acknowledgments
Not applicable.
Author Contributions
Conceptualization, T.T. and M.B. Methodology, S.S. and A.T.; Software, A.N., L.C. and B.C.; Validation, M.B., C.D. and S.B.; Resources, D.O., H.O., A.M., B.B.; Writing—original draft preparation, T.T., M.B., C.D.; Writing—review and editing, G.A. and F.O.; Supervision, G.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kashyap M.P., Sinha R., Mukhtar M.S., Athar M. Epigenetic regulation in the pathogenesis of non-melanoma skin cancer. Semin. Cancer Biol. 2020 doi: 10.1016/j.semcancer.2020.11.009. in press. [DOI] [PubMed] [Google Scholar]
- 2.Di Meglio P., Perera G.K., Nestle F.O. The multitasking organ: Recent insights into skin immune function. Immunity. 2011;23:857–869. doi: 10.1016/j.immuni.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal R., Woodfolk J.A. Skin barrier defects in atopic dermatitis. Curr. Allergy Asthma Rep. 2014;14:433. doi: 10.1007/s11882-014-0433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li C., Athar M. Ionizing Radiation Exposure and Basal Cell Carcinoma Pathogenesis. Radiat. Res. 2016;185:217–228. doi: 10.1667/RR4284.S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Errico M., Calcagnile A., Canzona F., Didona B., Posteraro P., Cavalieri R., Corona R., Vorechovsky I., Nardo T., Stefanini M., et al. UV mutation signature in tumor suppressor genes involved in skin carcinogenesis in xeroderma pigmentosum patients. Oncogene. 2000;19:463–467. doi: 10.1038/sj.onc.1203313. [DOI] [PubMed] [Google Scholar]
- 6.Moodycliffe A.M., Nghiem D., Clydesdale G., Ullrich S.E. Immune suppression and skin cancer development: Regulation by NKT cells. Nat. Immunol. 2000;1:521–525. doi: 10.1038/82782. [DOI] [PubMed] [Google Scholar]
- 7.Gordon R. Skin Cancer: An Overview of Epidemiology and Risk Factors. Semin. Oncol. Nurs. 2013;29:160–169. doi: 10.1016/j.soncn.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 9.Glass A.G., Hoover R.N. The emerging epidemic of melanoma and squamous cell skin cancer. JAMA. 1989;262:2097–2100. doi: 10.1001/jama.1989.03430150065027. [DOI] [PubMed] [Google Scholar]
- 10.Dubas L.E., Ingraffea A. Nonmelanoma skin cancer. Facial. Plast. Surg. Clin. N. Am. 2013;21:43–53. doi: 10.1016/j.fsc.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Parekh V., Seykora J.T. Cutaneous Squamous Cell Carcinoma. Clin. Lab. Med. 2017;37:503–525. doi: 10.1016/j.cll.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Green A.C., Olsen C.M. Cutaneous squamous cell carcinoma: An epidemiological review. Br. J. Dermatol. 2017;177:373–381. doi: 10.1111/bjd.15324. [DOI] [PubMed] [Google Scholar]
- 13.Sand M., Skrygan M., Sand D., Georgas D., Hahn S.A., Gambichler T., Altmeyer P., Bechara F.G. Expression of microRNAs in basal cell carcinoma. Br. J. Dermatol. 2012;167:847–855. doi: 10.1111/j.1365-2133.2012.11022.x. [DOI] [PubMed] [Google Scholar]
- 14.Sand M., Sand D., Altmeyer P., Bechara F.G. MicroRNA in non-melanoma skin cancer. Cancer Biomark. 2012;11:253–257. doi: 10.3233/CBM-2012-0274. [DOI] [PubMed] [Google Scholar]
- 15.Sand M., Gambichler T., Sand D., Skrygan M., Altmeyer P., Bechara F.G. MicroRNAs and the skin: Tiny players in the body’s largest organ. J. Dermatol. Sci. 2009;53:169–175. doi: 10.1016/j.jdermsci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Tian J., Shen R., Yan Y., Deng L. miR-186 promotes tumor growth in cutaneous squamous cell carcinoma by inhibiting apoptotic protease activating factor-1. Exp. Ther. Med. 2018;16:4010–4018. doi: 10.3892/etm.2018.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han C., Seebacher N.A., Hornicek F.J., Kan Q., Duan Z. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget. 2017;8:64622–64637. doi: 10.18632/oncotarget.19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Self-Fordham J.B., Naqvi A.R., Uttamani J.R., Kulkarni V., Nares S. MicroRNA: Dynamic Regulators of Macrophage Polarization and Plasticity. Front. Immunol. 2017;8:1062. doi: 10.3389/fimmu.2017.01062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 20.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 21.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 22.Broughton J.P., Lovci M.T., Huang J.L., Yeo G.W., Pasquinelli A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell. 2016;64:320–333. doi: 10.1016/j.molcel.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip. Rev. RNA. 2012;3:311–330. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- 24.Makarova J.A., Shkurnikov M.U., Wicklein D., Lange T., Samatov T.R., Turchinovich A.A., Tonevitsky A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016;51:33–49. doi: 10.1016/j.proghi.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 25.O’Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018;3:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creemers E.E., Tijsen A.J., Pinto Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 27.Nikolouzakis T.K., Falzone L., Lasithiotakis K., Krüger-Krasagakis S., Kalogeraki A., Sifaki M., Spandidos D.A., Chrysos E., Tsatsakis A., Tsiaoussis J. Current and Future Trends in Molecular Biomarkers for Diagnostic, Prognostic, and Predictive Purposes in Non-Melanoma Skin Cancer. J. Clin. Med. 2020;9:2868. doi: 10.3390/jcm9092868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 31.Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Horsham J.L., Ganda C., Kalinowski F.C., Brown R.A., Epis M.R., Leedman P.J. MicroRNA-7: A miRNA with expanding roles in development and disease. Int. J. Biochem. Cell Biol. 2015;69:215–224. doi: 10.1016/j.biocel.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Gerloff D., Sunderkötter C., Wohlrab J. Importance of microRNAs in Skin Oncogenesis and Their Suitability as Agents and Targets for Topical Therapy. Skin Pharmacol. Physiol. 2020;33:270–279. doi: 10.1159/000509879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neagu M., Constantin C., Cretoiu S.M., Zurac S. miRNAs in the Diagnosis and Prognosis of Skin Cancer. Front. Cell Dev. Biol. 2020;8:71. doi: 10.3389/fcell.2020.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walling H.W., Fosko S.W., Geraminejad P.A., Whitaker D.C., Arpey C.J. Aggressive basal cell carcinoma: Presentation, pathogenesis, and management. Cancer Metastasis Rev. 2004;23:389–402. doi: 10.1023/B:CANC.0000031775.04618.30. [DOI] [PubMed] [Google Scholar]
- 36.Heffelfinger C., Ouyang Z., Engberg A., Leffell D.J., Hanlon A.M., Gordon P.B., Zheng W., Zhao H., Snyder M.P., Bale A.E. Correlation of Global MicroRNA Expression with Basal Cell Carcinoma Subtype. G3 (Bethesda) 2012;2:279–286. doi: 10.1534/g3.111.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonkoly E., Lov J., Xu N., Meisgen F., Wei T., Brodin P., Jacks V., Kasper M., Shimokawa T., Harada M., et al. MicroRNA-203 functions as a tumor suppressor in basal cell carcinoma. Oncogenesis. 2012;1:e3. doi: 10.1038/oncsis.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sand M., Bechara F.G., Gambichler T., Sand D., Friedländer M.R., Bromba M., Schnabel R., Hessam S. Next-generation sequencing of the basal cell carcinoma miRNome and a description of novel microRNA candidates under neoadjuvant vismodegib therapy: An integrative molecular and surgical case study. Ann. Oncol. 2016;27:332–338. doi: 10.1093/annonc/mdv551. [DOI] [PubMed] [Google Scholar]
- 39.Al-Eryani L., Jenkins S.F., States V.A., Pan J., Malone J.C., Rai S.N., Galandiuk S., Giri A.K., States J.C. miRNA expression profiles of premalignant and malignant arsenic-induced skin lesions. PLoS ONE. 2018;13:e0202579. doi: 10.1371/journal.pone.0202579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu P., Ma L., Wu Z., Zheng G., Li J. Expression of miR-34a in basal cell carcinoma patients and its relationship with prognosis. J. Buon. 2019;24:1283–1288. [PubMed] [Google Scholar]
- 41.Sun H., Jiang P. MicroRNA-451a acts as tumor suppressor in cutaneous basal cell carcinoma. Mol. Genet. Genom. Med. 2018;6:1001–1009. doi: 10.1002/mgg3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang J., Tran D.C., Zhu G.A., Li R., Whitson R., Kim Y.H., Gupta A., Afshari A., Antes T., Spitale R.C., et al. Initial in vitro functional characterization of serum exosomal microRNAs from patients with metastatic basal cell carcinoma. Br. J. Dermatol. 2017;177:e187–e190. doi: 10.1111/bjd.15508. [DOI] [PubMed] [Google Scholar]
- 43.Wan C., Li Y. Integrative analysis of mRNA-miRNA-TFs reveals the key regulatory connections involved in basal cell carcinoma. Arch. Dermatol. Res. 2020;312:133–143. doi: 10.1007/s00403-019-02002-y. [DOI] [PubMed] [Google Scholar]
- 44.Farzan S.F., Karagas M.R., Christensen B.C., Li Z., Kuriger J.K., Nelson H.H. New Hampshire Skin Cancer Study. RNASEL and MIR146A SNP-SNP interaction as a susceptibility factor for non-melanoma skin cancer. PLoS ONE. 2014;9:e93602. doi: 10.1371/journal.pone.0093602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alam M., Ratner D. Cutaneous squamous-cell carcinoma. N. Engl. J. Med. 2001;344:975–983. doi: 10.1056/NEJM200103293441306. [DOI] [PubMed] [Google Scholar]
- 46.Inman G.J., Wang J., Nagano A., Alexandrov L.B., Purdie K.J., Taylor R.G., Sherwood V., Thomson J., Hogan S., Spender L.C., et al. The genomic landscape of cutaneous SCC reveals drivers and a novel azathioprine associated mutational signature. Nat. Commun. 2018;9:3667. doi: 10.1038/s41467-018-06027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y., Sun B., Wen X., Hao D., Du D., He G., Jiang X. The Roles of lncRNA in Cutaneous Squamous Cell Carcinoma. Front. Oncol. 2020;10:158. doi: 10.3389/fonc.2020.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Konicke K., López-Luna A., Muñoz-Carrillo J.L., Servín-González L.S., Flores-de la Torre A., Olasz E., Lazarova Z. The microRNA landscape of cutaneous squamous cell carcinoma. Drug Discov. Today. 2018;23:864–870. doi: 10.1016/j.drudis.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 49.Sand M., Gambichler T., Skrygan M., Sand D., Scola N., Altmeyer P., Bechara F.G. Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Investig. 2010;28:649–653. doi: 10.3109/07357901003630918. [DOI] [PubMed] [Google Scholar]
- 50.Sand M., Skrygan M., Georgas D., Arenz C., Gambichler T., Sand D., Altmeyer P., Bechara F.G. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol. Carcinog. 2012;51:916–922. doi: 10.1002/mc.20861. [DOI] [PubMed] [Google Scholar]
- 51.Sand M., Skrygan M., Georgas D., Sand D., Hahn S.A., Gambichler T., Altmeyer P., Bechara F.G. Microarray analysis of microRNA expression in cutaneous squamous cell carcinoma. J. Dermatol. Sci. 2012;68:119–126. doi: 10.1016/j.jdermsci.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 52.Dziunycz P., Iotzova-Weiss G., Eloranta J.J., Lauchli S., Hafner J., French L.E., Altmeyer P., Bechara F.G. Squamous cell carcinoma of the skin shows a distinct microRNA profile modulated by UV radiation. J. Investig. Dermatol. 2010;130:2686–2689. doi: 10.1038/jid.2010.169. [DOI] [PubMed] [Google Scholar]
- 53.Yamane K., Jinnin M., Etoh T., Kobayashi Y., Shimozono N., Fukushima S., Masuguchi S., Maruo K., Inoue Y., Ishihara T., et al. Down-regulation of miR-124/-214 in cutaneous squamous cell carcinoma mediates abnormal cell proliferation via the induction of ERK. J. Mol. Med. 2013;91:69–81. doi: 10.1007/s00109-012-0935-7. [DOI] [PubMed] [Google Scholar]
- 54.Cañueto J., Cardeñoso-Álvarez E., García-Hernández J.L., Galindo-Villardón P., Vicente-Galindo P., Vicente-Villardón J.L., Alonso-López D., De Las Rivas J., Valero J., Moyano-Sanz E., et al. MicroRNA (miR)-203 and miR-205 expression patterns identify subgroups of prognosis in cutaneous squamous cell carcinoma. Br. J. Dermatol. 2017;177:168–178. doi: 10.1111/bjd.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stojadinovic O., Ramirez H., Pastar I., Gordon K.A., Stone R., Choudhary S., Badiavas E., Nouri K., Tomic-Canic M. MiR-21 and miR-205 are induced in invasive cutaneous squamous cell carcinomas. Arch. Dermatol. Res. 2017;309:133–139. doi: 10.1007/s00403-016-1705-0. [DOI] [PubMed] [Google Scholar]
- 56.Zhang L., Xiang P., Han X., Wu L., Li X., Xiong Z. Decreased expression of microRNA-20a promotes tumor progression and predicts poor prognosis of cutaneous squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2015;8:11446–11451. [PMC free article] [PubMed] [Google Scholar]
- 57.Gong Z.H., Zhou F., Shi C., Xiang T., Zhou C.K., Wang Q.Q., Jiang Y.S., Gao S.F. miRNA-221 promotes cutaneous squamous cell carcinoma progression by targeting PTEN. Cell Mol. Biol. Lett. 2019;24:9. doi: 10.1186/s11658-018-0131-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gal-Yam E.N., Saito Y., Egger G., Jones P.A. Cancer epigenetics: Modifications, screening, and therapy. Annu. Rev. Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 59.Lodygin D., Tarasov V., Epanchintsev A., Berking C., Knyazeva T., Körner H., Knyazev P., Diebold J., Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 60.Vousden K.H., Lu X. Live or let die: The cell’s response to p53. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 61.Rivlin N., Brosh R., Oren M., Rotter V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer. 2011;2:466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodust P.M., Stockfleth E., Ulrich C., Leverkus M., Eberle J. UV-induced squamous cell carcinoma—A role for antiapoptotic signalling pathways. Br. J. Dermatol. 2009;161((Suppl. 3)):107–115. doi: 10.1111/j.1365-2133.2009.09458.x. [DOI] [PubMed] [Google Scholar]
- 63.Lefort K., Brooks Y., Ostano P., Cario-André M., Calpini V., Guinea-Viniegra J., Albinger-Hegyi A., Hoetzenecker W., Kolfschoten I., Wagner E.F., et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 2013;32:2248–2263. doi: 10.1038/emboj.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanitz A., Imig J., Dziunycz P.J., Primorac A., Galgano A., Hofbauer G.F.L., Gerber A.P., Detmar M. The expression levels of microRNA-361-5p and its target VEGFA are inversely correlated in human cutaneous squamous cell carcinoma. PLoS ONE. 2012;7:e49568. doi: 10.1371/journal.pone.0049568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen B., Pan W., Lin X., Hu Z., Jin Y., Chen H., Ma G., Qiu Y., Chang L., Hua C., et al. MicroRNA-346 functions as an oncogene in cutaneous squamous cell carcinoma. Tumour Biol. 2016;37:2765–2771. doi: 10.1007/s13277-015-4046-2. [DOI] [PubMed] [Google Scholar]
- 66.Xu N., Zhang L., Meisgen F., Harada M., Heilborn J., Homey B., Grandér D., Ståhle M., Sonkoly E., Pivarcsi A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J. Biol. Chem. 2012;287:29899–29908. doi: 10.1074/jbc.M112.391243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olasz E.B., Seline L.N., Schock A.M., Duncan N.E., Lopez A., Lazar J., Flister M.J., Lu Y., Liu P., Sokumbi O., et al. MicroRNA-135b Regulates Leucine Zipper Tumor Suppressor 1 in Cutaneous Squamous Cell Carcinoma. PLoS ONE. 2015;10:e0125412. doi: 10.1371/journal.pone.0125412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou M., Liu W., Ma S., Cao H., Peng X., Guo L., Zhou X., Zheng L., Guo L., Wan M., et al. A novel onco-miR-365 induces cutaneous squamous cell carcinoma. Carcinogenesis. 2013;34:1653–1659. doi: 10.1093/carcin/bgt097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Calder K.B., Smoller B.R. New insights into merkel cell carcinoma. Adv. Anat. Pathol. 2010;17:155–161. doi: 10.1097/PAP.0b013e3181d97836. [DOI] [PubMed] [Google Scholar]
- 70.Albores-Saavedra J., Batich K., Chable-Montero F., Sagy N., Schwartz A.M., Henson D.E. Merkel cell carcinoma demographics, morphology, and survival based on 3870 cases: A population based study. J. Cutan. Pathol. 2010;37:20–27. doi: 10.1111/j.1600-0560.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 71.Hughes M.P., Hardee M.E., Cornelius L.A., Hutchins L.F., Becker J.C., Gao L. Merkel Cell Carcinoma: Epidemiology, Target, and Therapy. Curr. Dermatol. Rep. 2014;3:46–53. doi: 10.1007/s13671-014-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ning M.S., Kim A.S., Prasad N., Levy S.E., Zhang H., Andl T. Characterization of the Merkel Cell Carcinoma miRNome. J. Skin Cancer. 2014;2014:289548. doi: 10.1155/2014/289548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Renwick N., Cekan P., Masry P.A., McGeary S.E., Miller J.B., Hafner M., Li Z., Mihailovic A., Morozov P., Brown M., et al. Multicolor microRNA FISH effectively differentiates tumor types. J. Clin. Investig. 2013;123:2694–2702. doi: 10.1172/JCI68760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fan K., Gravemeyer J., Ritter C., Rasheed K., Gambichler T., Moens U., Shuda M., Schrama D., Becker J.C. MCPyV Large T Antigen-Induced Atonal Homolog 1 Is a Lineage-Dependency Oncogene in Merkel Cell Carcinoma. J. Investig. Dermatol. 2020;140:56–65.e3. doi: 10.1016/j.jid.2019.06.135. [DOI] [PubMed] [Google Scholar]
- 76.Veija T., Sahi H., Koljonen V., Bohling T., Knuutila S., Mosakhani N. miRNA-34a underexpressed in Merkel cell polyomavirus-negative Merkel cell carcinoma. Virchows Arch. 2015;466:289–295. doi: 10.1007/s00428-014-1700-9. [DOI] [PubMed] [Google Scholar]
- 77.Konstatinell A., Coucheron D.H., Sveinbjørnsson B., Moens U. MicroRNAs as Potential Biomarkers in Merkel Cell Carcinoma. Int. J. Mol. Sci. 2018;19:1873. doi: 10.3390/ijms19071873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tuaeva N.O., Falzone L., Porozov Y.B., Nosyrev A.E., Trukhan V.M., Kovatsi L., Spandidos D.A., Drakoulis N., Kalogeraki A., Mamoulakis C., et al. Translational Application of Circulating DNA in Oncology: Review of the Last Decades Achievements. Cells. 2019;8:1251. doi: 10.3390/cells8101251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu F., Zhang F., Li X., Liu Q., Liu W., Song P., Qiu Z., Dong Y., Xiang H. Prognostic role of miR-17-92 family in human cancers: Evaluation of multiple prognostic outcomes. Oncotarget. 2017;8:69125–69138. doi: 10.18632/oncotarget.19096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bobbili M.R., Mader R.M., Grillari J., Dellago H. OncomiR-17-5p: Alarm signal in cancer? Oncotarget. 2017;8:71206–71222. doi: 10.18632/oncotarget.19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Volpicella M., Leoni C., Costanza A., Fanizza I., Placido A., Ceci L.R. Genome walking by next generation sequencing approaches. Biology. 2012;1:495–507. doi: 10.3390/biology1030495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X., Liang Z., Wang S., Lu S., Song Y., Cheng Y., Ying J., Liu W., Hou W., Liu Y., et al. Application of next-generation sequencing technology to precision medicine in cancer: Joint consensus of the Tumor Biomarker Committee of the Chinese Society of Clinical Oncology. Cancer Biol. Med. 2019;16:189–204. doi: 10.20892/j.issn.2095-3941.2018.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martinez-Gutierrez A.D., Catalan O.M., Vázquez-Romo R., Porras Reyes F.I., Alvarado-Miranda A., Lara Medina F., Bargallo-Rocha J.E., Orozco Moreno L.T., De León D.C., Herrera L.A., et al. miRNA profile obtained by next generation sequencing in metastatic breast cancer patients is able to predict the response to systemic treatments. Int. J. Mol. Med. 2019;44:1267–1280. doi: 10.3892/ijmm.2019.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garofoli M., Volpicella M., Guida M., Porcelli L., Azzariti A. The Role of Non-Coding RNAs as Prognostic Factor, Predictor of Drug Response or Resistance and Pharmacological Targets, in the Cutaneous Squamous Cell Carcinoma. Cancers. 2020;12:2552. doi: 10.3390/cancers12092552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Corsten M.F., Miranda R., Kasmieh R., Krichevsky A.M., Weissleder R., Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 86.Tivnan A., Tracey L., Buckley P.G., Alcock L.C., Davidoff A.M., Stallings R.L. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer. 2011;11:33. doi: 10.1186/1471-2407-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Babar I.A., Cheng C.J., Booth C.J., Liang X., Weidhaas J.B., Saltzman W.M. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc. Natl. Acad. Sci. USA. 2012;109:E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Skourti E., Logotheti S., Kontos C.K., Pavlopoulou A., Dimoragka P.T., Trougakos I.P., Gorgoulis V., Scorilas A., Michalopoulos I., Zoumpourlis V. Progression of mouse skin carcinogenesis is associated with the orchestrated deregulation of mir-200 family members, mir-205 and their common targets. Mol. Carcinog. 2016;55:1229–1242. doi: 10.1002/mc.22365. [DOI] [PubMed] [Google Scholar]
- 89.Li P., He Q.Y., Luo C.Q., Qian L.Y. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med. Sci. Monit. 2014;20:2472–2477. doi: 10.12659/MSM.891327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu K., Jin J., Rong K., Zhuo L., Li P. MicroRNA 675 inhibits cell proliferation and invasion in melanoma by directly targeting metadherin. Mol. Med. Rep. 2018;17:3372–3379. doi: 10.3892/mmr.2017.8264. [DOI] [PubMed] [Google Scholar]
- 91.Hu Y., Wang Q., Zhu X.H. MiR-135b is a novel oncogenic factor in cutaneous melanoma by targeting LATS2. Melanoma Res. 2019;29:119–125. doi: 10.1097/CMR.0000000000000524. [DOI] [PubMed] [Google Scholar]
- 92.D’Aguanno S., Valentini E., Tupone M.G., Desideri M., Di Martile M., Spagnuolo M., Buglioni S., Ercolani C., Falcone I., De Dominici M., et al. Semaphorin 5A drives melanoma progression: Role of Bcl-2, miR-204 and c-Myb. J. Exp. Clin. Cancer Res. 2018;37:278. doi: 10.1186/s13046-018-0933-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sánchez-Sendra B., Martinez-Ciarpaglini C., González-Muñoz J.F., Murgui A., Terrádez L., Monteagudo C. Downregulation of intratumoral expression of miR-205, miR-200c and miR-125b in primary human cutaneous melanomas predicts shorter survival. Sci. Rep. 2018;8:17076. doi: 10.1038/s41598-018-35317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sabarimurugan S., Madurantakam Royam M., Das A., Das S., Gothandam K.M., Jayaraj R. Systematic Review and Meta-analysis of the Prognostic Significance of miRNAs in Melanoma Patients. Mol. Diagn. Ther. 2018;22:653–669. doi: 10.1007/s40291-018-0357-5. [DOI] [PubMed] [Google Scholar]
- 95.Kozar I., Cesi G., Margue C., Philippidou D., Kreis S. Impact of BRAF kinase inhibitors on the miRNomes and transcriptomes of melanoma cells. Pt 11Biochim. Biophys. Acta Gen. Subj. 2017;1861:2980–2992. doi: 10.1016/j.bbagen.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 96.Fattore L., Mancini R., Acunzo M., Romano G., Laganà A., Pisanu M.E. miR-579-3p controls melanoma progression and resistance to target therapy. Proc. Natl. Acad. Sci. USA. 2016;113:E5005–E5013. doi: 10.1073/pnas.1607753113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Caporali S., Amaro A., Levati L., Alvino E., Lacal P.M., Mastroeni S., Ruffini F., Bonmassar L., Antonini Cappellini G.C., Felli N., et al. miR-126-3p down-regulation contributes to dabrafenib acquired resistance in melanoma by up-regulating ADAM9 and VEGF-A. J. Exp. Clin. Cancer Res. 2019;38:272. doi: 10.1186/s13046-019-1238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li N., Liu Y., Pang H., Lee D., Zhou Y., Xiao Z. Methylation-Mediated Silencing of MicroRNA-211 Decreases the Sensitivity of Melanoma Cells to Cisplatin. Med. Sci. Monit. 2019;25:1590–1599. doi: 10.12659/MSM.911862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Prabhakar K., Rodrίguez C.I., Jayanthy A.S., Mikheil D.M., Bhasker A.I., Perera R.J., Setaluri V. Role of miR-214 in regulation of β-catenin and the malignant phenotype of melanoma. Mol. Carcinog. 2019;58:1974–1984. doi: 10.1002/mc.23089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huber V., Vallacchi V., Fleming V., Hu X., Cova A., Dugo M. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J. Clin. Investig. 2018;128:5505–5516. doi: 10.1172/JCI98060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.de Planell-Saguer M., Rodicio M.C. Analytical aspects of microRNA in diagnostics: A review. Anal. Chim. Acta. 2011;699:134–152. doi: 10.1016/j.aca.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 102.Greenberg E., Hershkovitz L., Itzhaki O., Hajdu S., Nemlich Y., Ortenberg R., Gefen N., Edry L., Modai S., Keisari Y., et al. Regulation of cancer aggressive features in melanoma cells by microRNAs. PLoS ONE. 2011;6:e18936. doi: 10.1371/journal.pone.0018936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang K., Zhang Z.W. Expression of miR-203 is decreased and associated with the prognosis of melanoma patients. Int. J. Clin. Exp. Pathol. 2015;8:13249–13254. [PMC free article] [PubMed] [Google Scholar]
- 104.Hunt E.A., Broyles D., Head T., Deo S.K. MicroRNA Detection: Current Technology and Research Strategies. Annu. Rev. Anal. Chem. 2015;8:217–237. doi: 10.1146/annurev-anchem-071114-040343. [DOI] [PubMed] [Google Scholar]
- 105.Ouyang T., Liu Z., Han Z., Ge Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019;91:3179–3186. doi: 10.1021/acs.analchem.8b05909. [DOI] [PubMed] [Google Scholar]
- 106.Kappel A., Backes C., Huang Y., Zafari S., Leidinger P., Meder B., Schwarz H., Gumbrecht W., Meese E., Staehler C.F., et al. MicroRNA in vitro diagnostics using immunoassay analyzers. Clin. Chem. 2015;61:600–607. doi: 10.1373/clinchem.2014.232165. [DOI] [PubMed] [Google Scholar]
- 107.Ozsolak F., Milos P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tanase C.P., Albulescu R., Neagu M. Application of 3D hydrogel microarrays in molecular diagnostics: Advantages and limitations. Expert Rev. Mol. Diagn. 2011;11:461–464. doi: 10.1586/erm.11.30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.