Abstract
Circulating HIV subtypes in the Philippines have increasingly diversified, potentially affecting treatment. We monitored outcomes of a treatment-naïve cohort and their virus subtype prevalence.
Retrospective/prospective study cohort.
HIV-I-REACT clinic patients co-enrolled in the Virology Quality Assurance Program (RUSH-VQA) from 7/2017-6/2019 were included. Relevant demographic and laboratory information were collected. The ViroSeq HIV-1 Genotyping System v.3 and HIV-1 Integrase Genotyping Kit identified protease-reverse transcriptase and integrase drug resistance mutations (DRM). Sequence subtyping followed using the Stanford University Drug Resistance Database and the REGA HIV-1 Subtyping Tool v.3. The jpHMM HIV-1 Tool and REGA HIV-1 Subtyping Tool provided additional subtype analysis of this cohort's 5’LTR-VIF regions after Sanger sequencing. One-year outcomes included virologic suppression, mortality, and follow-up.
86/88 patients were males. Median age was 30 (range 19–65) years; 61/88 were MSM. 15/85 carried baseline DRM. ViroSeq-generated sequences included subtypes CRF01_AE (66/85), B (14/85), and newer recombinants (4/85). Extensive sequencing (n = 71) of the 5’-LTR-GAG-Pol genes showed CRF01_AE (n = 50), subtype B (n = 7), and other recombinants (n = 13). Bootstrap analysis identified 7 pairs of highly related strains. Discordant DRM appeared in 2/7 pairs, where 1/2 strains displayed DRM. After 1 year, 87 individuals were alive, with 19 lost to care. Viral load (VL) was repeated for only 31/77 (40.2%). Follow-up CD4 testing for 39/77 (50.6%) showed an increase to a median of 327 cells/mm3.
Our cohort currently carries subtype CRF01_AE (∼68%–70%), followed by subtype B and CRF01_AE/B recombinants. Outcomes were favorable, regardless of subtype after 1 year on cART.
Keywords: HIV/AIDS, outcomes, sequencing, subtype
1. Introduction
More than 75 million people around the world have been affected by the HIV/AIDS pandemic,[1] with 32.7 million [24.8 million–42.2 million] people dying from an AIDS-related illness since the start of the epidemic. The overall prevalence is fairly low in the Asia-Pacific region, with Thailand and Cambodia the only countries with a prevalence above 1%.[2] In the Philippines, however, the prevalence has increased dramatically over the past decade, with 71% of the total 78,559 cases diagnosed in the last 5 years.[3] The number of daily reported cases has also exponentially increased from 6 per day in 2011 to 35 per day in 2019.
Although the Philippine Department of Health collects epidemiologic data for surveillance purposes, there are scant published data about the subtypes of HIV-1 virus circulating in the country[4–9] with only a few published recently.[5,9] Studies have shown that genotypic differences between subtypes influence the response to, and the emergence of resistance to anti-retroviral therapy.[10–12] As such, knowledge about circulating subtypes is imperative.
Historically, studies in the Philippines have reported that the majority of HIV infections were caused by subtypes B and E, with low numbers of subtypes A, C, D, and F sequenced via polymerase chain reaction (PCR) and heteroduplex assays,[8] or the env C2-V3 reqions from peripheral blood mononuclear cells.[4] However, a shift from subtype B to CRF01_AE was observed among female sex workers[6] in 1993. Validation by a later study among the MSM population (n = 63) revealed that 64% had subtype CRF01_AE and only 27% had subtype B.[7] A more recent study of 81 patients[5] in 2017, showed that the protease-RT genotype distribution was predominantly subtype CRF01_AE (62/81, 77%) followed by subtype B (18/81, 22%). The authors concluded that the molecular epidemiology of HIV in the Philippines had changed, with the more aggressive CRF01_AE currently being the predominant subtype. This study aimed to
-
1.
provide more detailed information regarding the genotypic characteristics of HIV in the Philippines in a treatment-naïve cohort in an urban setting, and
-
2.
correlate subtype with outcome data after 1 year.
The results of this study will help contribute important epidemiologic and clinical data regarding circulating viral subtypes among patients living with HIV (PLWHIV) in the Philippines.
2. Methods
2.1. Study setting
I-REACT, which stands for I Am Responsible thru Early Assessment, Consultation and Treatment, is an HIV Clinic located within the Medical City (TMC) hospital. It was established in February 2012 and offers HIV counseling and testing, multi-disciplinary needs assessment, clinical care, free anti-retroviral medicines, immunization and training workshops on HIV counseling. At the time of the study, there were about 1000 PLWHIV enrolled in the clinic.
2.2. Study participants
All newly diagnosed HIV-infected adult patients or treatment-experienced individuals failing anti-retroviral therapy (ART) enrolled in the TMC I-REACT HIV treatment hub from July 2017 until June 2019 were eligible to participate in the Rush Virology Quality Assurance (VQA) Program. This study was conducted in accordance with the Declaration of Helsinki and approved by TMC and Rush University Medical Center Institutional Review Boards.
The VQA is an international NIH-funded program that tests the accuracy and reliability of diagnostic laboratory tests currently being used or being developed for use in testing for markers HIV, and/or HIV-associated viruses in blood. For this project, the TMC/VQA cohort was screened for virus carrying DRM. These individuals were invited back for a second larger volume blood draw, before initiation of treatment, if genotyping of their screening samples showed the presence of drug resistant mutations (DRM). The larger volumes (∼100 mL) were processed and sent back de-identified to the VQA for repository storage for future use. Inclusion criteria into the TMC/VQA program incorporated the following Red Cross guidelines for larger volume blood collection:
-
1.
Weight of at least 50 kg;
-
2.
pulse rate of 50 to 100/minute
-
3.
blood pressure of 90/50 to 180/100 mm Hg,
-
4.
oral temperature of not more than 37.5°C, and
-
5.
normal hemoglobin level.
Written informed consent for screening was obtained from all patients before study enrolment; a separate consent was obtained for the second larger volume collection. Demographic, clinical, and laboratory data were collected from patient charts by study authors (CLA, JGB, AC) and samples were de-identified using code numbers, with a cohort number preceding the letters ph (for Philippines), before submission to the Rush VQA for genotyping and viral load (VL) determination. Of the individuals screened, 85 carried a detectable VL to allow genotyping for DRM.
2.3. Resistance testing/genomic sequencing/genomic analysis
2.3.1. Resistance testing
The FDA-approved ViroSeq HIV-1 Genotyping System v.3 and HIV-1 Integrase Genotyping Kit, with “research use only” approval were used to identify DRM. The kits generated sequence data for the entire protease, partial RT (codons 1–335), and entire integrase, respectively. Using kit instructions and reagents, plasma virions were concentrated to extract genomic viral RNA which was re-suspended in either 50 or 100 μL diluent defined by VL of the sample. For each kit, 10 μL of the re-suspended RNA was used in RT-PCR reactions to generate sequencing template for the protease-RT or integrase genes. The remainder was frozen at −80C for later use or for repeat analysis if needed. After sequencing, the individual sequencing primer files for each patient sample were aligned in software provided by the respective kits to evaluate sequence quality and allow resolution of ambiguous base calling[13] of mixed nucleotides. Each software uses a subtype B HIV strain as the reference strain for alignment. After evaluation, the software can generate a genotyping report of the observed DRM and a consensus FASTA file of the aligned sequences which can be saved and used for other bio-informatic activities.
2.3.2. Genome sequencing
An aliquot of the same extracted virus RNA from the ViroSeq DRM analysis was used to generate larger overlapping templates spanning the genome. Near full length cDNA was made with primers located in LTR sequences from which 4 overlapping larger-sized sequencing templates were produced by nested PCR based on modifications of the Novitsky protocol.[14,15] Sanger sequencing was performed with overlapping primers designed against HXB2 in both the 5’ and 3’ directions. For each patient sample, analysis, including alignment of the sequence chromatograms and generation of consensus FASTA data from the aligned sequences, was accomplished using SeqScape v2.7 (ABI, Applied Biosystems Inc).
2.3.3. Subtyping
The subtype designations of the ViroSeq-derived protease-RT and integrase sequences were made using the Stanford University Drug Resistance Database[16,17] and also the REGA HIV-1 Subtyping Tool v.3 (Katholieke Universiteit Leuven; HIV Bioinformatics, BioAfrica).[18] Subtype composition of the contiguous LTR-VIF region comprising the partial long terminal repeat (LTR)-entire group specific antigen (GAG)-polymerase (POL) and partial viral infectivity factor (VIF) regions of the genome, was determined for each patient sample with the jpHMM HIV-1 Tool (University of Göttingen, Department of Bioinformatics)[19] and REGA HIV-1 Subtyping Tool v.3. These programs also mapped the breakpoints of the recombinants. Consensus FASTA files of all the clinical samples yielding sequence data from LTR-VIF were analyzed by MEGA v.6[20] to generate neighbor- joining phylogenetic trees after bootstrapping analysis (500 iterations). The samples were gap-stripped prior to performing a clustal W alignment yielding a tree consisting of regions spanning the beginning of GAG through the end of integrase. Consensus strains from the Los Alamos HIV Sequence Database[21] of pure subtypes and selected older “original” recombinants, as well as the consensus O group and CPZ strains were used as references in the alignment.
2.4. Clinical profile and outcomes
Basic demographic data including gender, age, co-morbidities, co-infections (e.g., Hepatitis B or C) opportunistic infections or AIDS-defining illnesses, CD4 cell count, and VL were collected upon enrolment and during follow-up using the patient chart and a standardized case report form. Outcomes at 1 year included the following
-
1.
virologic suppression defined as VL <40copies/mL or undetectable at 1 year;
-
2.
mortality (e.g., death/alive), and
-
3.
retention in care defined as seen at least once on follow up within 1 year of enrollment.
2.5. Statistical analysis
We determined frequency distributions of demographic and clinical characteristics using mean or median, and range for quantitative variables. Missing variables was neither replaced nor estimated.
3. Results
3.1. Characteristics of the cohort
A total of 88 patients were included during the study period, enrolled by 6 different infectious disease providers within the institution. Of these, 82 were treatment naïve, 5 were treatment experienced and suspected to be failing ART (6.ph17, 16.ph17, 17.ph17 39.ph17, and 37.ph18), and one had a false positive HIV ELISA, confirmed by a negative Western Blot (4.ph19). All enrollees were Filipino, although one was half-European (31.ph18), and 2 had history of prolonged stays or travel abroad (34.ph17 and 27.ph17). Table 1 shows the majority were male (86/88, 97.7%), most of whom (61/88 [69.3%]) identified as men who had sex with men (MSM). Median age was 30 (range 19–65 years) and time from initial HIV diagnosis to first consult was 7.6 weeks (range 0–379.7). Of those who enrolled, 5 pairs acknowledged partnership at the time of the screening visit—32.ph17 and 33.ph17, 26.ph17 and 37.ph17, 35.ph17 and 36.ph17, 2.ph18 and 3.ph18, and 32.ph18 and 33.ph18.
Table 1.
Baseline characteristics of the cohort.
| Characteristic N = 88 | N (% or range) |
| Age, yr (median) | 30 (19–65) |
| Sex | |
| Male | 86 (97.7) |
| Female | 2 (2.3) |
| HIV status at enrollment | |
| Treatment naïve | 82 (93.2) |
| Treatment experienced | 5 (5.7) |
| HIV negative | 1 (1.1) |
| Sexual orientation | |
| MSM or same sex | 61 (69.3) |
| Heterosexual | 5 (5.7) |
| Bisexual | 8 (9.1) |
| Not reported | 14 (15.9) |
| Social history | |
| Smoking, any | 23 (26.1) |
| Alcohol intake, any | 42 (47.7) |
| Intravenous drug use (IVDU) | 2 (2.3) |
| Co-morbidity (n = 79) | |
| Hypertension | 2 |
| COPD | 1 |
| None | 76 |
| Not reported | 9 |
| Co-infection, untreated (n = 87) | |
| Hepatitis B | 9 (10.3) |
| Hepatitis C | 1 (1.1) |
| Syphilis | 15 (17.2) |
| Opportunistic infection | |
| Tuberculosis | 14 |
| PCP | 12 |
| Baseline laboratory result∗ | |
| CD4 cells/mm3 | 94 (0–2296) |
| VL, copies/mL | 168,473 (<20–1,729,378) |
| Subtype ViroSeq pro-RT (n = 85) | |
| CRF01_AE | 66 |
| B | 14 |
| Potential A/E or other recombinants | 5 |
| HAART† | |
| Yes | 83 |
| No data | 5 |
| HAART Regimen‡ | |
| TDF/3TC/EFV | 77 |
| TDF/3TC/LPV/r | 3 |
| AZT/3TC/LPV/r | 2 |
| AZT/3TC/NVP | 1 |
| Outcome, at 1 yr | |
| Alive | 87 |
| Lost to follow up§ | 19 |
| Dead | 1 |
| Repeat CD4 (n = 39) | 327 (58–1481) |
| Virologic suppression (VL <40; n = 31) | 24 (77.4) |
Viral load using Abbot RealTime HIV-1.
HAART initiated after 1st study visit.
HAART regimens were initiated prior to identification of drug resistant mutations.
Defined as not obtaining medications from the HIV clinic and/or with no consultation with the attending physician for >1 year.
LEGEND: TDF – Tenofovir, 3TC – Lamivudine, EFV – Efavirenz, LPV/r – Lopinavir-ritonavir, AZT – Zidovudine, NVP- Nevirapine.
At time of enrollment, the majority of patients had no known chronic co-morbid conditions (74/84, 88.1%). Only a few were co-infected with hepatitis B virus (HBV) (9/87, 10.3%) and only 1 had evidence of hepatitis C virus infection (1/87, 1.1%). Fifteen patients had serologic evidence of syphilis (15/87, 17.2%) with a reactive rapid plasma reagin test. A few had history of tuberculosis (14/88, 15.9%) and pneumocystis jirovecii pneumonia (12/88, 13.6%) (Table 1).
Baseline median CD4 count, and HIV viral load were 94 cells/mm3 (range 0–2296 cells/mm3) and 168,473 copies/mL (range 40–1,744,801 copies/mL) respectively. Two patients (16.ph17 and 37.ph18) who were treatment-experienced and thought to be failing ART based on self-reported poor compliance with medications had undetectable viral loads at time of enrollment. Most patients (78/83) began ART, with 77/83 starting the fixed-dose combination of tenofovir (TDF), lamivudine (3TC), and efavirenz (EFV).
3.1.1. DRM and subtyping in Pro-RT and integrase genes
Of the cohort, 85/88 DRM profiles were obtained prior to starting treatment. Table 2 shows that virus from 15/85 individuals carried recognized DRM in the protease-RT genes of which 7/15 had major DRM. The mutations observed were associated with both nucleoside inhibitors (NRTI) and non-nucleoside inhibitors (NNRTI) as well as protease inhibitors (PI). No mutations associated with integrase inhibitors (II) were observed.
Table 2.
Drug resistance mutations detected in screening samples.
| Mutations∗ | |||||
| Sample | Subtype† | Protease | NRTI | NNRTI | Integrase |
| 5.ph17 | CRF01_AE | None | None | V90I | |
| 6.ph17 | CRF01_AE | L10F; L10V | None | E138G | |
| 10.ph17 | B | L10V | None | none | |
| 11.ph17 | CRF01_AE | M46L | None | none | |
| 17.ph17 | CRF01_AE | None | D67N; K70R; M184V; T215F; T215I; T215S | A98G, G190A | |
| 21.ph17 | B | M46L; A71V | None | K101Q;E138K | |
| 27.ph17 | CRF01_AE | None | None | E138A | |
| 28.ph17 | CRF01_AE | None | None | V106I | none |
| 39.ph17 | CRF01_AE | None | K65R; K70T; L74I; M184I | V90I; K103N; V108I; Y181C; M230L | |
| 2.ph18∗ | B | L10I | None | V179D | |
| 17.ph18 | B | L10V | None | none | |
| 18.ph18 | CRF01_AE | None | None | V90I, V106I, E138G | |
| 32.ph18∗ | CRF01_AE | None | Y115F; T215Y | Y188C; G190V | |
| 34.ph18 | B | A71V; L90M | None | Y181C, H221Y | |
| 1.ph19 | CRF01_AE | None | None | V106I | |
Mutations associated with protease, reverse transcriptase (nucleoside – NRTI and non-nucleoside – NNRTI) and integrase inhibitors.
Subtype listed in the Stanford HIV Drug Resistance designation for Protease-RT FASTA submitted for that sample.
Bolded = treatment experienced, failing ART.
∗ Partners w/ another patient in the cohort.
There were no consistent patterns regarding the presence of mutations as 8/15 profiles displayed DRM for a single class of inhibitors and 7/15 showed DRM belonging to 2 classes of inhibitors. Samples 17.ph17 and 39.ph17 displayed more complex patterns, where each had several DRM to multiple drugs in both NRTI and NNRTI. DRM patterns appeared independent of subtype designation.
Table 3 shows subtyped ViroSeq-generated pro-RT sequences included CRF01_AE (77.6% 66/85), subtype B (16.5% 14/85), and potential recombinants (4.7% 4/85) including 2 between CRF01_AE and B (CRF-1_AE/B) and 2 other recombinant patterns. One sample displayed a subtype A classification. For the integrase gene, the subtype distribution was different where 72.9% (62/85), 22.4% (19/85), and 1.2% (1/85) were clearly CRF01_AE, subtype B and subtype A respectively. There were no CRF-1_AE/B recombinants, but 3 other recombinant patterns were recognized. Taken together, the pro-RT and integrase subtyping patterns suggested that recombination within the POL gene between CRF01_AE and subtype B is currently not a rare event.
Table 3.
Historical subtype prevalence of HIV patients in the Philippines based on area sequenced.
| First Author, yr [Ref] | Telan, 2013 [7] | Salvana, 2017[5] | Chen, 2019[9] | Current study[24] | ||
| Region Sequenced | Pro-RTA | Pro-RT | Near full- length | Pro-R (Stanford)D | Integrase (Stanford) | LTR-VIFB (jpHMM) |
| N | 163 | 81 | 23 | 85 | 85 | 71 |
| Years of Study Inclusion | 2008–2012C | Mar-Aug 2013 | 2015–2017 | 2017–2019 | ||
| Subtype (%) | ||||||
| CRF01_AE | 39.3∗ | 77 | 60.8 | 77.6 | 72.9 | 70.4 |
| B | 54 | 22 | 13 | 16.5 | 22.4 | 9.9 |
| CRF01_AE/BE | 21 | 2.3 | 0 | 14.1 | ||
| Other subtypes | 6.7 | 1 Subtype C | 1.2 Subtype A | 1.2 Subtype A | ||
| Other recombinantsF | – | 4.3 CRF01/07/B (n = 1) | 2.4 B/A1 (n = 1) G/CRF02_AG (n = 1) | 3.5 A/B (n = 1) B/CRF-3-AB (n = 1) CRF01_AG/G (n = 1) | 5.6 B/K (n = 1) CRF01/A1/B (n = 1) CRF01/A1 (n = 1) A1/G (n = 1) | |
HIV = human immunodeficiency virus.
A: region of HIV genome sequenced Pro-= protease, RT= reverse transcriptase, LTR=long terminal repeat, VIF = vial Infectivity factor
B: Average length ∼ 4500 nucleotides; partial LTR and VIF, entire GAG and POL genes. Consensus sequence incorporates the Pro-rt and Integrase data from VIroSeq analysis
C: Years that were sampled
D. Program used for these subtype designations
E: recombinant between CRF01_AE and subtype B
F. recombinants composed of other combinations other than just CRF01_AE and subtype B
includes CRF02.
3.1.2. Extended sequencing of the LTR-VIF regions
Extensive sequencing of these samples successfully covering LTR-VIF (n = 71) showed CRF01_AE (70.4%, 50/71), subtype B (9.9%, 7/71), recombinants between CRF01_AE and B (14.1%, 10/71), and recombinants between other subtypes (5.6%, 4/71). Collectively, acquisition of the longer contiguous sequence indicated the proportion of recombinants within the samples increased even though the shorter pro-RT and integrase regions may have presented as “true” subtypes for the established CRF01_AE recombinant.
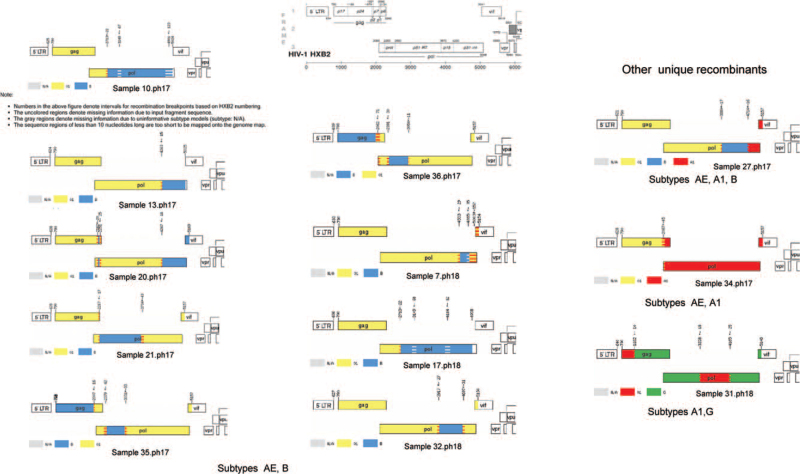
The breakpoint patterns of the CRF01_AE/ B, and other recombinants are visualized in Figure 1. The recombinant regions in the POL gene varied in size and position. The general pattern of 2 of these recombinants was very similar with slightly different breakpoints in 2 acknowledged partner pairs (Fig. 2). Surprisingly, another 2 strains displayed similar breakpoints and pattern of recombination with some discordance in composition. Here 1 sample, 32.ph18, showed CRF01_AE/B and the other, 27.ph17, appeared to have replaced an AE region with subtype A1 region to have a composite of 3 subtypes. Table 4 lists the proportions of the subtypes comprising the CRF01_AE/ B as well as other observed recombinants, 27.ph17 (AE/B/A1), 34.ph17 (AE/A1), and 31.ph18 (A1/G) for the regions encoding protease, RT, RNase H and Integrase. The data show diverse breakpoint patterns among protease (2/9), RT (6/9), RNAse H (3/9) and integrase (3/9) genes.
Figure 1.
Composite breakpoint maps of recombinant AE/B and other unique recombinant profiles spanning the GAG-integrase genes in the ph17-18 cohorts. Breakpoint maps were created by jpHMM analysis of the consensus sequence generated from each sample based on HXB2 genome numbering. The notes following sample 10.ph17 are applicable to all samples analysed by jpHMM and are identical on each map returned. The breakpoints patterns observed were generally different for each of the strains, although some patterns look similar. No unusual breakpoints were noted for the ph19 cohort.
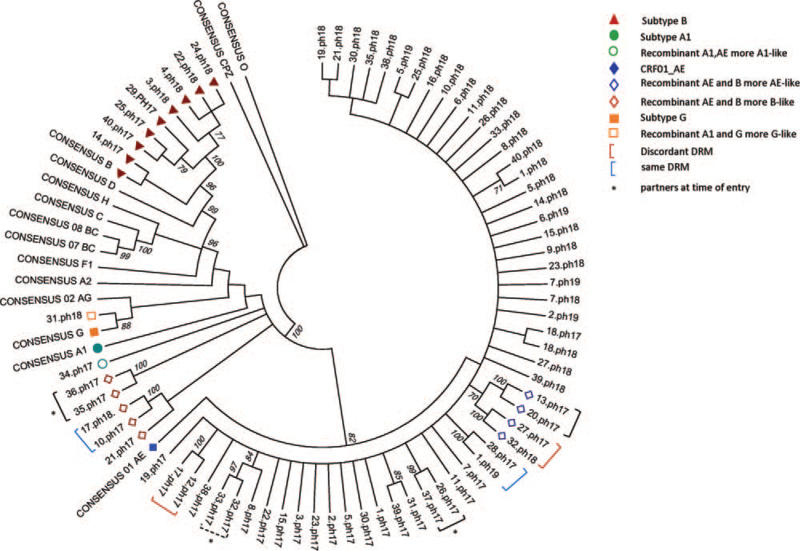
Figure 2.
Neighbor joining tree made after bootstrap analysis (500) using Mega 6. Sequences from cohorts ph17-18-19 span GAG-integrase and were gap-stripped prior to performing a clustal W alignment. Values >70 are shown on the branches. Subtypes are marked as described in the key. Pairs of strains at 99 to 100 values (solid brackets) are considered nearly identical. One pair may be closely related (hatched bracket). Discordant DRM appeared in 2 strain pairs (maroon bracket), where 1/2 strains carried DRM. Pairs showing the same DRM are marked with blue brackets. Strain pairs with high relatedness collected from long-term partners are marked by an asterisk.
Table 4.
POL gene subtype composition identified by jpHMM in recombinants (% of gene that is subtype B).
| Gene | ||||
| Sample | Protease∗ | RT† | Rnase H‡ | Integrase§ |
| 10.ph17 | AE | AE/B (87.6%)1 | B | B |
| 13.ph17 | AE | AE | AE | B (96%) |
| 20.ph17 | AE | AE | AE | B (95.7%) |
| 21.ph17 | B | B (∼86.7%)/AE | AE | AE |
| 35.ph17 | AE/B (∼57.5%) | B (∼36.4%)/AE | AE | AE |
| 36.ph17 | AE/B (∼53.5%) | B (38.2%)/AE | AE | AE |
| 7.ph18 | AE | AE | AE | AE/B (32.5%)/AE2 |
| 17.ph18 | AE | AE/B (87.6%)3 | B | B |
| 32.ph18 4 | AE | AE | AE/B (86.9%) | B (53.9%)/AE |
| 27.ph17 | AE | AE | AE/B (97.2%) | B (55.8%)/A1 |
| 34.ph17 | A1 | A1 | A1 | A1 |
| 31.ph18 | G | G/A1 (48.6%5 | A1 (80.6%)/G | G |
jpHMM POL location (HXB2 numbering) 2253–2550
jpHMM POL location (HXB2 numbering) 2550–3870
jpHMM POL location (HXB2 numbering) 3870–4230
jpHMM POL location (HXB2 numbering) 4230–5096
1subtype designation from 5’end - 3’end of gene; % calculated from breakpoints identified by jHMM
2breakpoints for subtype B portion in the the middle of the gene flanked by AE portions
3Stanford and Rega analysis classified p-RT together as B/A1
4breakpoints for subtype B portion in the middle of Rnase H and Integrase
5percentage of subtype in recombinants that do not have subytpe B.
A phylogenetic tree was generated from the 71 samples for which LTR-VIF sequence was obtained to observe the genetic relatedness of the strains (Fig. 2). Bootstrap analysis identified 7 pairs of strains with high relatedness (>99 confidence levels). One other pair appeared to be also highly related at a lesser level and showed no DRM (97 confidence level). Discordant DRM appeared in 2 pairs, where virus from 1/2 individuals displayed either no or a single DRM and the other carried a more complex pattern involving both NRTI and NNRTI. Two pairs carried the same single DRM, L10 V in protease, and V106I in the RT, respectively (Table 2).
3.2. Clinical outcomes
At 1 year after their initial enrollment, 87 are alive, although 19 (21.8%) were lost to follow-up. Of those who followed up, 27 (39.7%) were seen quarterly, 14 (20.6%) were seen at least twice, and 27 (39.7%) were seen only once since enrollment. HIV VL was repeated for only 31/77 (40.2%), with 24/31 (77.4%) being undetectable. Follow-up CD4 testing for 39/77 (50.6%) showed a median increase of 327 cells/mm3. By the end of the project period on September 2019, all except 1 were alive.
4. Discussion
Our study provides additional important information regarding the evolution of the HIV epidemic in the Philippines. First, the characterization of HIV strains found in our cohort of primarily young MSM reinforces that the HIV epidemic in the Philippines is shifting from predominantly subtype B to CRF01_AE and other diverse subtypes and recombinants. Our data show that transmission of similar subtypes and recombinants between partners may occur, and that strains carrying DRMs may not be uncommon prior to initiation of treatment. Second, we observe that regardless of the diversity of subtypes, patient outcomes, including virologic suppression, are favorable at 1 year.
Our TMC/VQA cohort was comprised mainly of young, male (97.75%), MSM (68%), with a median age of 30 (range 19–65) years. These numbers mirror the larger HIV epidemic in the country – among 82,865 total confirmed HIV cases reported to the HIV/AIDS Registry of the Philippines (HARP) as of December 2020, 77,821 (94%) were male, with a median age of 28 (range: 1 month–82 years), with MSM (86%,) reported as the predominant mode of transmission.[3]
We observe that the distribution of CRF01_AE, subtype B and recombinants has changed over time (Table 3). Our data for just the pro-RT are similar to the findings of Salvana et al.[5] Both our studies based on pro-RT gene sequence indicate that other subtypes and known subtypes are beginning to circulate, where CRF01_AE becomes increasingly prevalent over subtype B. But our data also suggests that recombinants in the pro-RT between these 2 subtypes are present. It was interesting to see within our data that the proportions of subtypes noted based on pro-RT were revised when the longer LTR-VIF region, which incorporated these pro-RT sequences, was analyzed. A higher proportion of the cohort appeared to carry CRF01_AE/B strains, as well as other recombinants, similar to the trend reported by Chen et al.[9] Hence, it is important to remember when reporting prevalence that the designation of subtype and recombinants may be influenced by the length of the sequence analyzed (i.e., just pro-RT vs partial full length vs near full-length).
Notably, 2 patients identified to have subtype A1 (34.ph17), and subtype G (31.ph18) in the Pol gene had history of travel and prolonged stays outside of the country. 34.ph17 was based in Japan working in a high-risk environment for several years before returning home, while 31.ph18 was foreign-born and had emigrated from Europe. The presence of these strains, less common for the area currently, in addition to their past travel history, indicates that their viruses were more likely imported strains, rather than endemic to the Philippines.
We generated a phylogenetic tree to characterize the genetic relatedness of the strains (Fig. 2) where bootstrap analysis identified 7 pairs of strains with high relatedness (>99 confidence levels). Of these, 2 pairs acknowledged they were long-term monogamous partners. Another partner pair also showed 97% relatedness. This implies that transmission of viral subtypes between these partners may have occurred, although the dynamics of transmission between them is beyond the scope of this study. Interestingly, one other set of partners identified upon enrollment did not appear to have genetically related strains, which may indicate that they were infected earlier, through other prior contacts. For the other partner pair, only the virus of one of the pair was able to yield the longer sequence data to be included in this tree. However, from their pro-RT DRM genotyping, both carried virus designated subtype B, but only one of the pair carried virus showing DRM. An alignment comparison of just their pro-RT sequences indicated a high homology level between 97% to 99% and placement on the same branch of a tree compared to the other subtype B pro-rt sequences in this cohort (data not shown).
In the Philippines, performing either a baseline genotype prior to ART initiation or when shifting regimens is not usual practice, since the test is not widely available, has a prolonged turn- around time, and entails additional out-of-pocket cost (e.g., 5,500P or 100$) to the patient. As such, little is known about DRMs. In our cohort, DRMs were observed in 15/85 (17.6%) samples, of which half (7/15) had major DRM. Proportionately for this cohort, 10/15 of the DRM were found in CRF01_AE and the other 5 in subtype B. The mutations observed were associated more with NRTI and NNRTI, than PI. No mutations associated with integrase were found. ART regimens are available only through the Department of Health treatment hubs and at the time of this study, the first line regimen based on local guidelines was the combination regimen TDF/3TC/EFV. The alternative or second line regimen often consisted of TDF/3TC and lopinavir-ritonavir (LPV/r), chosen if there was drug failure or severe side effects from the primary regimen. Integrase inhibitors were unavailable during this time period and were made accessible only in late 2020. It is unsurprising then, that the DRMs in this cohort reflect this selective drug pressure. 17.ph17 and 39.ph17 showed extensive DRMs, including multiple TAMs such as M184 V and K65R (17.ph17, 39.ph17) and key signature mutations from the NNRTI gene such as K013N, Y181C (39.ph17), reflective of their failed primary regimen. Both these patients had been on an EFV-based regimen for at least a year before failure was detected. Their regimens were adjusted after results of the genotypes were made available. Given these results, a genotype prior to ART initiation, or particularly when ART failure is suspected, should be recommended. Periodic viral load monitoring- at least twice yearly- is also advocated by World Health Organization guidelines,[22] but is often not performed because of the cost entailed.
Regardless of subtype, patient outcomes at 1-year were favorable. The majority of patients alive and maintained on ART show a substantial increase in median CD4. However, several observations warrant concern –19% of patients were lost to follow up, only 40.2% had a subsequent test for viral load, of whom only 24/31 (77.4%) were virologically suppressed. These are well below the targets of the HIV cascade of care,[23] which set the goal at 90-90-90-90% of PLWHIV should be linked to care, retained in care, and achieve virologic suppression. A similar study in a tertiary level government hospital treatment hub in the Philippines among 584 PLWHIV showed similar results, with only 79% of patients retained in care and only 49% with a timely viral load determination (unpublished, personal communication). The Philippine HIV health insurance outpatient package is meant to cover the costs of some laboratory tests, including the VL. However, there is tremendous paper work involved, a prolonged waiting period, and patients often choose not to avail of the package because they are hesitant to disclose their status. This may explain why only few in the cohort had a subsequent VL. Other underlying reasons why these goals could not be met within a relatively short period of time should merit further study.
Our study is limited by its small sample size, and short period of follow up. Details prior to enrollment were not captured, and information regarding follow-up may have been missed due to inadequate documentation. These details would have been useful to try to gain more specific insight into transmission patterns and contact tracing. However, all patients enrolled in the VQA program were included to minimize selection bias. To help minimize recall bias, patients were interviewed using a standardized case report form upon study enrollment, and on follow-up. Our genotypic data focused on the regions of the genome that covered at least the protease-RT and integrase genes in screening visit samples to link with subsequent treatment strategy. Samples from follow-up visits, if any, were not collected and might have added more molecular epidemiologic information. As discussed earlier, a caveat to our study is that our assessment of subtypes and recombinants circulating in the country is based on the partial genome sequence of strains comprising the front half of the genome (LTR-VIF, ∼4500 bases) and could be adjusted if more of the genome is sequenced. It is difficult to say whether the findings of this study are generalizable to other HIV-clinics, especially in public hospitals, although the demographic of our clinic is certainly similar. Nevertheless, this is one of the few studies in the Philippines that focused on circulating subtypes, baseline drug resistant mutations, and patient outcomes. For our cohort, the success of initiating ART, especially in the CRF01_AE/B recombinants does not seem to be influenced by genetic profile.
Acknowledgments
We would like to acknowledge the help of Dr. Raul V. Destura, who gave his permission to be named, with the initial protocol.
Author contributions
Conceptualization: Cybele Lara Rivera Abad, James W. Bremer, Diana D. Huang.
Data curation: Cybele Lara Rivera Abad, Jia An G. Bello, Angela Beatriz Cruz, Diana D. Huang.
Formal analysis: Diana D. Huang.
Funding acquisition: James W. Bremer, Diana D. Huang.
Investigation: Jia An G. Bello.
Methodology: Cybele Lara Rivera Abad, Aleksandra Danilovic, James W. Bremer, Diana D. Huang.
Project administration: Cybele Lara Rivera Abad, Juan Elias, James W. Bremer, Diana D. Huang.
Resources: Aleksandra Danilovic, Juan Elias.
Supervision: Cybele Lara Rivera Abad, Diana D. Huang, James W. Bremer.
Validation: Cybele Lara Rivera Abad, Angela Beatriz Cruz, Diana D. Huang, Aleksandra Danilovic, Juan Elias.
Writing – original draft: Cybele Lara Rivera Abad, Jia An G. Bello, Angela Beatriz Cruz, Juan Elias, Diana D. Huang.
Writing – review & editing: Cybele Lara Rivera Abad, Jia An G. Bello, Diana D. Huang.
Footnotes
Abbreviations: ART = antiretroviral therapy, DRM = drug resistant mutations, PCR = polymerase chain reaction, PLWHIV = people living with HIV, TMC = the medical city, VL = viral load, VQA = virologic quality assurance.
How to cite this article: Abad CL, Bello JA, Cruz AB, Danilovic A, Elias J, Bremer JW, Huang DD. Prevalent subtypes and one-year outcomes of an HIV-cohort from an urban Philippine center. Medicine. 2021;100:51(e28315).
This work was supported by NIAID (VQA contract # HHSN272201200023C).
Sequences of the partial genomes of the samples have been deposited into Genbank and have been given accession numbers OK663666-OK663736. They will be released upon publication.
The authors have no conflicts of interests to disclose.
The datasets generated during and/or analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
References
- [1].Global HIV & AIDS statistics — 2020 fact sheet. https://www.unaids.org/en/resources/fact-sheet. Accessed May 25, 2021. [Google Scholar]
- [2].Global Report. UNAIDS Report on the Global AIDS Epidemic. https://unaids-test.unaids.org/sites/default/files/unaids/contentassets/documents/unaidspublication/2010/20101123_globalreport_en%5b1%5d.pdf. 2010. Accessed May 2021. [Google Scholar]
- [3].HIV/AIDS and ART Registry of the Philippines. https://doh.gov.ph/sites/default/files/statistics/EB_HARP_December_AIDSreg2020.pdf Accessed May 25, 2021. [Google Scholar]
- [4].Paladin FJ, Monzon OT, Tsuchie H, Aplasca MR, Learn GH, Kurimura T. Genetic subtypes of HIV-1 in the Philippines. AIDS (London, England) 1998;12:291–300. [DOI] [PubMed] [Google Scholar]
- [5].Salvaña EMT, Schwem BE, Ching PR, Frost SDW, Ganchua SKC, Itable JR. The changing molecular epidemiology of HIV in the Philippines. Int J Infect Dis 2017;61:44–50. [DOI] [PubMed] [Google Scholar]
- [6].Santiago ML, Santiago EG, Hafalla JC, et al. Molecular epidemiology of HIV-1 infection in the Philippines, 1985 to 1997: transmission of subtypes B and E and potential emergence of subtypes C and F. Journal of acquired immune deficiency syndromes and human retrovirology: official publication of the International Retrovirology Association 1998;18:260–9. [DOI] [PubMed] [Google Scholar]
- [7].Telan EF, Samonte GM, Palaypayon N, et al. Possible HIV transmission modes among at-risk groups at an early epidemic stage in the Philippines. J Med Virol 2013;85:2057–64. [DOI] [PubMed] [Google Scholar]
- [8].Gloriani-Barzaga N, Graham RR, Santiago EG. Genotype analysis of HIV-1 isolates from various risk groups in Metro Manila. Phil J Microbiol Infect Dis 1998;27:137–42. [Google Scholar]
- [9].Chen Y, Hora B, DeMarco T, et al. Increased predominance of HIV-1 CRF01_AE and its recombinants in the Philippines. J Gen Virol 2019;100:511–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Holguín A, Ramirez de Arellano E, Rivas P, Soriano V. Efficacy of antiretroviral therapy in individuals infected with HIV-1 non-B subtypes. AIDS Rev 2006;8:98–107. [PubMed] [Google Scholar]
- [11].Kantor R. Impact of HIV-1 pol diversity on drug resistance and its clinical implications. Curr Opin Infect Dis 2006;19:594–606. [DOI] [PubMed] [Google Scholar]
- [12].Kiwanuka N, Laeyendecker O, Robb M, et al. Effect of human immunodeficiency virus Type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J Infect Dis 2008;197:707–13. [DOI] [PubMed] [Google Scholar]
- [13].Huang DD, Eshleman SH, Brambilla DJ, Palumbo PE, Bremer JW. Evaluation of the editing process in human immunodeficiency virus type 1 genotyping. J Clin Microbiol 2003;41:3265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gall A, Ferns B, Morris C, et al. Universal amplification, next-generation sequencing, and assembly of HIV-1 genomes. J Clin Microbiol 2012;50:3838–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Novitsky V, Zahralban-Steele M, McLane MF, et al. Long-range HIV genotyping using viral RNA and proviral DNA for analysis of HIV drug resistance and HIV clustering. J Clin Microbiol 2015;53:2581–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shafer RW. Rationale and uses of a public HIV drug-resistance database. J Infect Dis 2006;194:S51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shafer RW, Jung DR, Betts BJ. Human immunodeficiency virus type 1 reverse transcriptase and protease mutation search engine for queries. Nat Med 2000;6:1290–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pineda-Peña AC, Faria NR, Imbrechts S, et al. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: performance evaluation of the new REGA version 3 and seven other tools. Infection, Genetics Evolution 2013;19:337–48. [DOI] [PubMed] [Google Scholar]
- [19].Schultz AK, Zhang M, Bulla I, et al. jpHMM: improving the reliability of recombination prediction in HIV-1. Nucleic Acids Res 2009;37:W647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kuiken C, Korber B, Shafer RW. HIV sequence databases. AIDS Rev 2003;5:52–61. [PMC free article] [PubMed] [Google Scholar]
- [22].WHO. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Recommendations for a public health approach - Second edition. 2016. Accessed May 2021. [Google Scholar]
- [23].Joint United Nations Programme on HIV/AIDS (UNAIDS) 90-90-90 An ambitious treatment target to help end the AIDS epidemic. [Updated 2014]. [Accessed May 25, 2021]. https://www.unaids.org/sites/default/files/media_asset/90-90-90_en.pdf. [Google Scholar]