Supplemental Digital Content is available in the text
Keywords: biochemical tumor marker, gene expression regulation, prognosis, uterine cervical neoplasms
Abstract
Background:
DNA damage is a fundamental process that plays a considerable role in generating protein diversity. FANCI, loaded on the altered chromatin, plays a vital role in DNA damage. Abnormal FANCI expression is potentially associated with carcinogenesis.However, the biological role of FANCI in cervical cancer is yet to be determined.
Methods:
We analyzed FANCI expression via multiple gene expression databases. Genes co-expressed with FANCI and its regulators were identified using LinkedOmics. The correlations between FANCI and cancer immune infiltrates were investigated via Tumor Immune Estimation Resource (TIMER).
Results:
FANCI was found upregulated with amplification in tumor tissues of multiple cervical cancer cohorts. High FANCI expression was associated with poorer overall survival (OS). Functional network analysis suggested that FANCI regulates spliceosome, DNA replication, and cell cycle signaling via pathways involving several cancer-related kinases and the E2F family. In additional, FANCI expression was positively correlated with infiltrating levels of CD4+ T and CD8+ T cells, and neutrophils. FANCI expression also showed strong correlations with diverse immune marker sets in cervical cancer.
Conclusion:
These findings suggested that FANCI is correlated with prognosis of and immune infiltration in cervical cancer, laying a foundation for further study of the immune regulatory role of FANCI in cervical cancer.
1. Introduction
Cervical cancer is one of the most common malignancies of the female genital tract. It is the fourth leading cause of cancer-related death in females, with an estimated 311,365 deaths worldwide in 2018.[1] The risk of death in females with cervical cancer is higher in low-income countries (0.9%) than in high-income countries (0.3%).[2]
Despite a series of advances in the prevention, screening, and treatment of cervical cancer (e.g., modern radiotherapy techniques and targeted therapy), treatment against cervical cancer has not significantly improved.[3,4] In China, the overall morbidity and mortality associated with cervical cancer have steadily increased from 1991 to 2013, and it is predicted to rise continually in future.[5] In addition to metastasis or recurrence, the disease is linked to poor prognosis, with a 5-year relative survival rate of 66.3% (Based on data from 2011–2017), and distant stage with a 5-year relative overall survival (OS) rate of only 17.6%.[6] Hence, identifying novel therapeutic targets and survival-associated biomarkers is essential to enhance the therapeutic effect in cervical cancer.
DNA damage is a fundamental process that plays a considerable role in generating protein diversity. DNA damage is also the key to the pathology of numerous diseases, especially cancers.[7] The connection between cancer biology and DNA damage is of primary importance to understand the mechanisms leading to disease and also to improve the development of therapeutic approaches.[8] Upon DNA damage, FANCI and FANCD2 can be phosphorylated by ATM and ATR kinases. This complex holds a E3 ubiquitin ligase activity via FANCI, which collaborates with the UBE2T ubiquitin-conjugating enzyme.[9] After DNA damage, FANCI and FANCD2 proteins are loaded on the altered chromatin.[10] Both partners are required for their reciprocal mono-ubiquitination by the FA core complex.[10–12]
It is also known that FANCI, complexed with FANCD2, suppresses the fanconi anemia pathway in the absence of DNA damage.[13] One study indicated that the expression of FANCI is changed in prostate cancer.[14] However, the biological function of FANCI in cervical cancer remains to be determined. Here, we investigated FANCI expression and mutations in data from patients with cervical cancer in The Cancer Genome Atlas (TCGA) and various public databases. Using multi-dimensional analysis, we evaluated genomic alterations and functional networks related to FANCI in cervical cancer and explored its role in tumor immunity. Our results could potentially reveal new targets and strategies for cervical cancer diagnosis and treatment.
2. Materials and methods
2.1. Databases description
2.1.1. Oncomine database analysis
The expression level of the FANCI gene in cervical cancers was examined in the Oncomine 4.5 database (https://www.oncomine.org/).[15] Oncomine is a cancer microarray database and web-based data-mining platform. The threshold was determined according to the following values: P value of .05, fold change of 1.5, and gene ranking of all.
2.1.2. UALCAN database analysis
UALCAN (http://ualcan.path.uab.edu) uses TCGA level 3 RNA-seq and clinical data from 31 cancer types,[16] allowing analysis of relative expression of genes across tumor and normal samples, as well as in various tumor sub-groups based on individual cancer stages, tumor grade, or other clinicopathological features.
2.2. Kaplan–Meier survival curve analysis
Kaplan–Meier Plotter (http://kmplot.com/analysis), an online survival analysis tool, was used to assess the effects of genes on the survival rates in cancers, including 371 CERVICAL CANCER.[17] The correlation between FANCI expression and cervical cancer patients OS was analyzed using Kaplan–Meier Plotter.
2.2.1. LinkedOmics database analysis
The LinkedOmics database (http://www.linkedomics.org/admin.php) is a web-based platform for analyzing 32 TCGA cancer-associated multi-dimensional datasets.[18]
FANCI co-expression was analyzed statistically using Pearson's correlation coefficient, presenting in volcano plots, heat maps, or scatter plots. The Function module of LinkedOmics analyzes Gene Ontology (GO) biological process (GO_BP), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, kinase-target enrichment, miRNA-target enrichment, and transcription factor-target enrichment by the gene set enrichment analysis (GSEA). The rank criterion was FDR < 0.05, and 1000 simulations were performed.
2.2.2. TIMER database analysis
TIMER is a comprehensive resource for systematic ally analyzing immune infiltrates across diverse cancer types from TCGA (https://cistrome.shinyapps.io/timer/), which includes 10,897 samples across 32 cancer types.[19] TIMER applies a deconvolution method to infer the abundance of tumor-infiltrating immune cells (TIICs) from gene expression profiles. We analyzed FANCI expression in cervical cancer, and the correlation of FANCI expression with the abundance of immune infiltrates, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells, as well as the tumor purity.
2.3. Statistical analysis
The t test P < .05 was utilized to determine the statistical significance between groups with different expression levels of FANCI. We compared the OS of cervical cancer patients separated by the median expression level of specific genes. Kaplan–Meier curves were used to compare the survival time differences. The log-rank test P < .05 was used to indicate the significance of survival time differences.
3. Results
3.1. Elevated expression of FANCI in cervical cancer
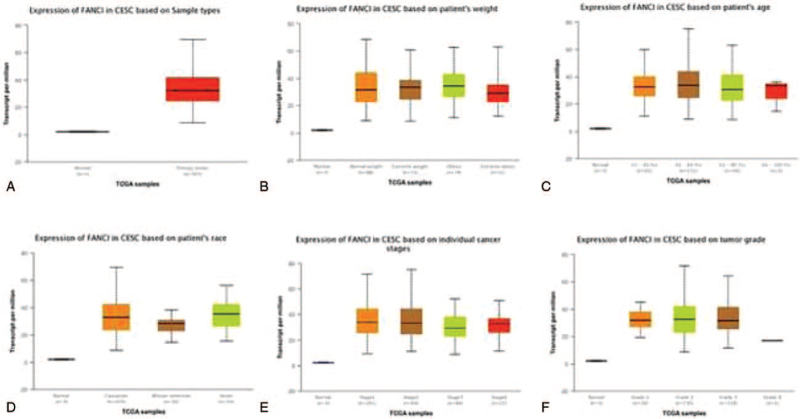
We initially evaluated the FANCI transcription levels in multiple cervical cancer studies from TCGA. Data in the Oncomine database indicated that the mRNA expression of FANCI was significantly higher in cervical cancer tissues than in the adjacent normal tissues (Fig. 1). In addition, results from the UALCAN database showed that the FANCI expression was significantly higher in tumor tissues than in the normal tissue (Fig. 2A). Further sub-group analysis of multiple clinic-pathological features of TCGA cervical cancer samples in UALCAN database consistently showed elevated transcription levels of FANCI. The expression of FANCI was significantly higher in cervical cancer patients than normal controls in subgroup analysis based on weight, age, ethnicity, disease stages, and tumor grade (Fig. 2B–F). Thus, FANCI expression may serve as a potential diagnostic indicator in cervical cancer.
Figure 1.
FANCI expression in cervical cancer according to public databases. (A,B) Box plot showing FANCI mRNA levels in cervical cancer specimens (dark blue) and their normal controls (light blue) according to datasets from Biewenga Cervix and Scotto Cervix 2, respectively.
Figure 2.
FANCI transcription in cervical cancer based on TCGA database. (A) Boxplot showing relative expression of FANCI in normal individuals or cervical cancer patients. (B) Boxplot showing relative expression of FANCI in normal individuals or cervical cancer patients with average weight, extreme weight, obesity, or extreme obesity. (C) Boxplot showing relative expression of FANCI in normal individuals of any age or cervical cancer patients aged 21–40, 41–60, 61–80, or 81– 100 yr. (D) Boxplot showing relative expression of FANCI in normal individuals of any ethnicity or cervical cancer patients of Caucasian, African-American, or Asian ethnicity. (E) Boxplot showing relative expression of FANCI in normal individuals or cervical cancer patients with stage 1, 2, 3, or 4 tumors. (F) Boxplot showing relative expression of FANCI in normal individuals or cervical cancer patients with grade 1, 2, 3, or 4 tumors. Figures (A–F) conducted using UALCAN.
3.2. FANCI expression is survival-associated
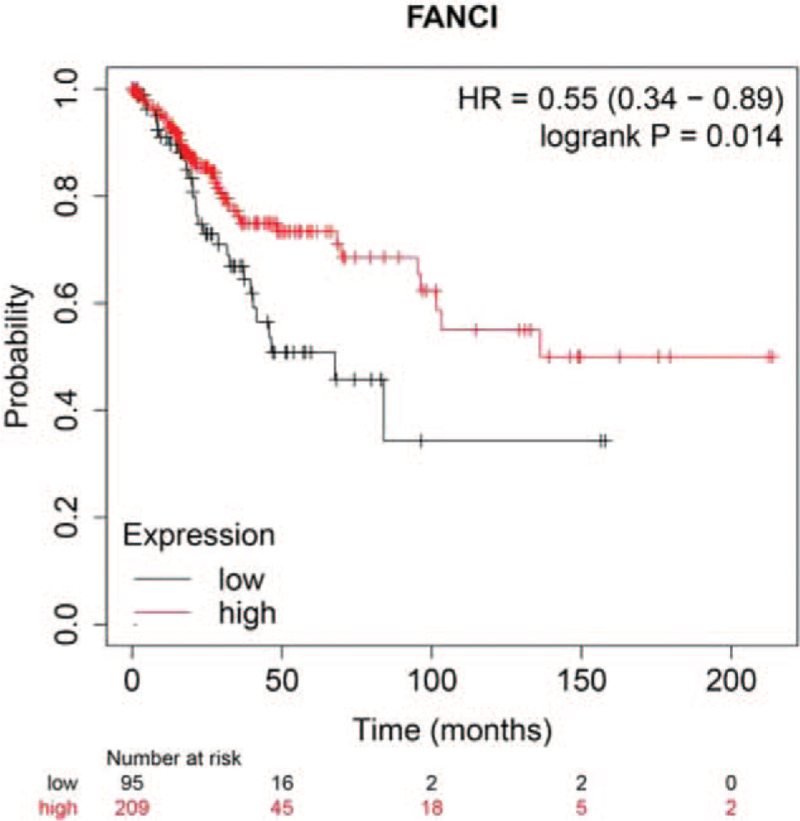
Kaplan–Meier survival curves were used to assess the association between FANCI expression and the survival outcomes of cervical cancer cohorts with survival information available. The patients were separated into 2 groups according to the median value of FANCI expression level in each cohort. Generally, the high FANCI expression group had significantly shorter OS (log-rank test, P < .05), compared with the low expression group in the cervical cancer cohort (Fig. 3).
Figure 3.
Survival curve analyses showing the correlation between FANCI mRNA expression and overall survival in patients with cervical cancer.
3.3. FANCI co-expression networks in cervical cancer
To gain an insight into the biological function of FANCI in cervical cancer, the function modu of LinkedOmics was used to examine FANCI co-expression mode in cervical cancer cohort. As shown in Figure 4A, 3558 genes (dark red dots) were shown significant positive correlations with FANCI, whereas 1891 genes (dark green dots) were shown to have significant negative correlations (false discovery rate, FDR < 0.01). The top 50 significant genes positively and negatively correlated with FANCI were shown in the heat map (Fig. 4B,C). A total description of the co-expressed genes was detailed in Supplementary Table 1, http://links.lww.com/MD/G558.
Figure 4.
Genes differentially expressed in correlation with FANCI in cervical cancer (LinkedOmics). (A) Pearson test used to analyze correlations between FANCI and genes differentially expressed in cervical cancer. (B–C) Heat maps showing genes negatively and positively correlated with FANCI in cervical cancer (TOP 50).
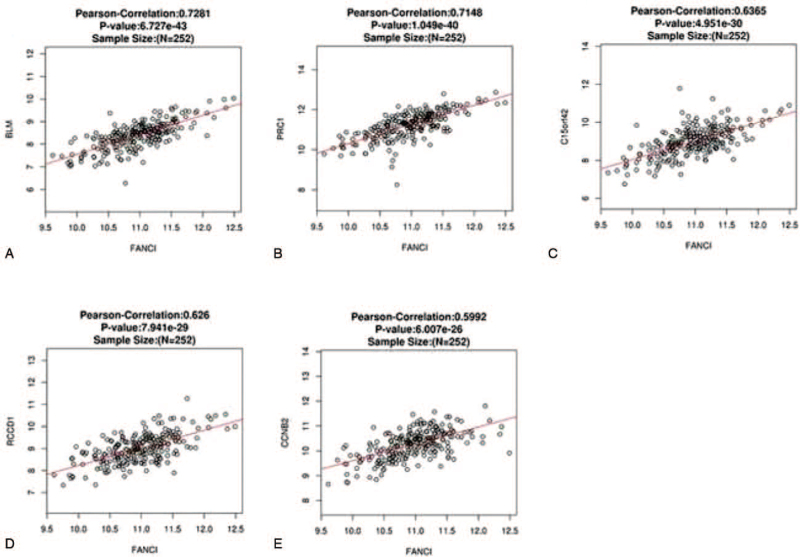
FANCI expression showed a strong positive association with expression of BLM (r = 0.7281, P = 6.727E-43), PRC1 (r = 0.7148, P = 1.049E-40), and C15orf42 (r = 0.6365, P = 4.951E-30), RCCD1 (r = 0.626, P = 7.941E-29), CCNB2 (r = 0.5992, P = 6.007E-26), etc (Fig. 5).
Figure 5.
Overall survival (OS) of the top 5 positive genes correlated with FANCI. (A-E) All of these genes were significantly correlated with OS in cervical cancer.
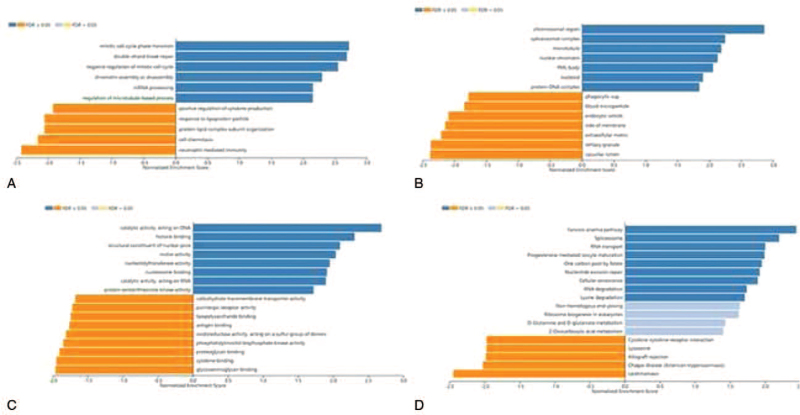
Significant Gene Ontology (GO) term annotation by gene set enrichment analysis (GSEA) showed that FANCI co-expressed genes participate primarily in chromosome segregation, mitotic cell cycle phase transition, double-strand break repair, and mRNA processing, while the activities like fatty acid metabolic process, peroxisomal transport, and multiple metabolic processes were inhibited (Fig. 6 and Supplementary Table 2, http://links.lww.com/MD/G559). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed enrichment in the spliceosome, fanconi anemia pathway, RNA transport, and nucleotide excision repair pathways, etc (Fig. 6 and Supplementary Table 3, http://links.lww.com/MD/G560). These results suggest that a widespread impact of FANCI on the global transcriptome.
Figure 6.
Genes co-expressed with FANCI in cervical cancer (LinkedOmics). Significantly enriched GO annotations and KEGG pathways of FANCI in cervical cancer cohort.
3.4. Regulators of FANCI networks in cervical cancer
To further explore the regulators of FANCI in cervical cancer, we analyzed the kinases, miRNAs, and transcription factors’ (TF) enrichment of FANCI co-expressed genes. The top 5 most significant kinases related primarily to the cyclin-dependent kinase 1 (CDK1), polo like kinase 1 (PLK1), ATM serine/threonine kinase (ATM), cyclin- dependent kinase 2 (CDK2), and checkpoint kinase 1 (CHEK1) (Table 1 and Supplementary Table 4, http://links.lww.com/MD/G561). In addition, the top 5 correlated miRNA-target networks were (CACGTTT) MIR-302A, (TATCTGG) MIR-488, (CCTGAGT) MIR-510,
Table 1.
The kinase, miRNA, and transcription factor-target networks of FANCI in cervical cancer (LinkedOmics).
| Enriched category | Geneset | LeadingEdgeNum | P |
| Kinase Target | Kinase_ CDK1 | 88 | 0 |
| Kinase_ PLK1 | 34 | 0 | |
| Kinase_ATM | 42 | 0 | |
| Kinase_CDK2 | 88 | 0 | |
| Kinase_CHEK1 | 44 | 0 | |
| MiRNA Target | ATAGGAA, MIR-302A | 8 | 0 |
| ATGTACA, MIR-488 | 3 | 0 | |
| ATAAGCT, MIR-510 | 6 | .020548 | |
| ATAACCT, MIR-492 | 11 | .003413 | |
| TTGGAGA, MIR-154 | 25 | .02027 | |
| Transcription Factor Target | V$E2F1_Q4 | 104 | 0 |
| V$E2F1_02 | 85 | 0 | |
| V$E2F1_Q6 | 104 | 0 | |
| V$E2F1DP1_01 | 84 | 0 | |
| V$E2F4DP2_01 | 84 | 0 |
(CAGGTCC) MIR-492, and (ATAACCT) MIR-154 (Table 1 and Supplementary Table 5, http://links.lww.com/MD/G562). Moreover, the enrichment of transcription factors was related mainly to the E2F transcription factor family (Supplementary Table 6, http://links.lww.com/MD/G563), including V$E2F_Q4, V$E2F_02, V$E2F_Q6, V$E2F1DP1_01, and V$E2F4DP2_01. One recent study, using combinatorial mapping of chromatin occupancy and transcriptome profiling, identified an E2F-driven transcriptional program that was associated with cervical cancer development and progression.
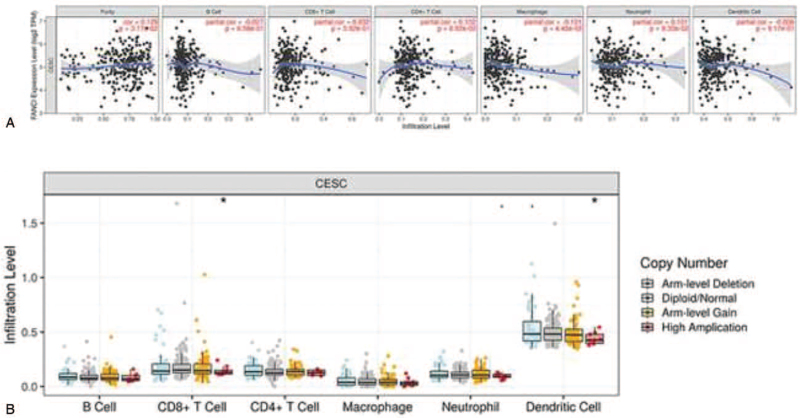
ANCI is related with tumor purity and immune infiltration level in cervical cancer. Therefore, we investigated whether FANCI expression was correlated with immune infiltration levels in cervical cancer from TIMER database. The results show that FANCI expression has significant correlations with tumor purity (r = 0.129, P = 3.17E-02) and significant correlations with the dominant immune cells infiltration levels (Fig. 7A). Particularly, FANCI CNV has significant correlations with infiltrating levels of CD8+ T cells and dendritic cells (Fig. 7B). Moreover, multivariable hazards models were used to evaluate the impacts of FANCI expression in the presence of varying immune cells.
Figure 7.
Correlations of FANCI expression with immune infiltration level in cervical cancer. (A) FANCI expression is significantly related to tumor purity and has significant positive correlations with infiltrating levels of CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in cervical cancer. (B) FANCI CNV affects the infiltrating levels of CD8+ T cells, macrophages, neutrophils, and dendritic cells in cervical cancer.
4. Discussion
DNA damage is emerging as a critical step in abnormal phenotypic heterogeneity. Moreover, DNA damage is expected to be a major potential factor for untapped molecular targets in precision oncology and cancer disparities.[20] FANCI, a core component of the FANCD2-FANCI complex, is involved in multiple steps of DNA damage. To gain more detailed insights into the potential functions of FANCI in cervical cancer and its regulatory network, we performed the bioinformatics analysis of public data to guide future research in cervical cancer.
Analysis of transcriptome from more than 3400 clinical samples comprising 6 geographic regions and ethnic cervical cancer studies confirmed that FANCI mRNA levels are significantly higher in cervical cancer than in normal tissue. In addition, high expression of FANCI was significantly to improve poor survival in cervical cancer patients. Thus, our results suggested that FANCI upregulation occurs in many cases of cervical cancer and deserves further clinical validation as a potential diagnostic and prognostic marker.
For mining regulators potentially responsible for FANCI dysregulation, we found that FANCI in cervical cancer is associated with a network of kinases, including CDK1, PLK1, ATM, CDK2, and CHEK1. These kinases regulated genomic stability, mitosis, and the cell cycle, and showed differential expression and survival prognosis in cervical cancer. CDK1 participates in the regulation of mitosis, self-renewal, differentiation, and somatic reprogramming. Various inhibitors of CDK1 have been developed, and some have entered phase I and II clinical trials to treat a variety of solid tumors and hematologic malignancies.[21] As a critical driver gene, a causal link has recently been established between PLK1 and cervical cancer.[22]FANCI may regulate DNA replication, repair, and cell cycle progression via interacted kinases in cervical cancer.
Next, the E2F family constitutes the main transcription factors for FANCI dysregulation. E2F1 is one of the critical links in the cell cycle regulatory network. Activated E2F oncogenic signaling was always seen in the progression of typical cancer, and studies have shown that dosage-dependent copy number gains in E2F1 and E2F3 drive cervical cancer.[23] Our results suggest that E2F1 is an essential regulator of FANCI and that FANCI might act through this factor to regulate the cell cycle and proliferation capacity of cervical cancer. Further studies are needed to test this hypothesis. Our study did not identify any miRNA that was significantly associated with FANCI, possibly because FANCI is involved in mRNA splicesome and has no role in regulation of miRNA cellular machinery.
To probe the signaling events in controlling abnormal FANCI expression, we tested the FANCI co-expression network. Our results suggested that the functional consequence of FANCI mainly include spliceosome, DNA repair, DNA replication, and cell cycle, while it inhibits the metabolic processes, such as fatty acid, lipid, antibiotic, nucleoside bisphosphate, and cellular modified amino acid metabolic process. These findings are consistent with the molecular pathways implicated in cervical cancer.[24]
The tumor microenvironment is the non-cancerous cells present in and around a tumor, have a strong influence on the genomic analysis of tumor samples.[25] As gene dynamics are known to influence belowground genetic diversity and microenvironment processes, co-occurrence analysis was performed. Most genes co-occurring with FANCI CNV were distributed in the 1q21 locus. Further, a gene-level network representing the co-occurrence of genes across cervical cancer genomes was built, giving the clues of the role of FANCI in regulating the immune response.
Herein, by tumor purity analysis, the network of FANCI alterations is involved in the tumor purity and tumor immunity. Our findings provide a detailed characterization of the association between FANCI and immune marker sets in cervical cancer patients. Further studies need to be done to elucidate whether FANCI is a crucial factor in mediating T-cell therapy.
In conclusion, this study provides multilevel evidence for the role of FANCI in immune response and its potential as a biomarker in cervical cancer. Our results suggest that FANCI upregulation in cervical cancer may likely have far-reaching effects in RNA splicing and genomic stability, and at multiple cell cycle steps. Further, our results suggest a potential novel immune regulatory role of FANCI in tumor immunity. These findings call for large-scale cervical cancer genomics research and subsequent functional studies. And further clinical research, as well as convincing validations in cell lines and animal models are necessary.
Author contributions
Data curation: Xiaoling Liu, Xiqin Liu, Xia Han.
Formal analysis: Xiaoling Liu, Xiqin Liu.
Methodology: Xiaoling Liu.
Project administration: Xiaoling Liu.
Software: Xiaoling Liu.
Supervision: Xiaoling Liu, Xia Han.
Validation: Xia Han.
Writing – original draft: Xiaoling Liu, Xia Han.
Writing – review & editing: Xiaoling Liu.
Footnotes
Abbreviations: OS = overall survival, TCGA = the cancer genome atlas, TIMER = tumor immune estimation resource, GSEA = gene set enrichment analysis, GSEA = gene set enrichment analysis.
How to cite this article: Liu X, Liu X, Han X. FANCI may serve as a prognostic biomarker for cervical cancer: A systematic review and meta-analysis. Medicine. 2021;100:51(e27690).
The authors report no conflicts of interest.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet (London, England) 2019;393:169–82. [DOI] [PubMed] [Google Scholar]
- [3].Chargari C, Soria JC, Deutsch E. Controversies and challenges regarding the impact of radiation therapy on survival. Ann Oncol Off J Eur 2013;24:38–46. [DOI] [PubMed] [Google Scholar]
- [4].Zeng H, Chen W, Zheng R, et al. Changing cancer survival in China during 2003–15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health 2018;6:e555–67. [DOI] [PubMed] [Google Scholar]
- [5].Du P-L, Wu K-S, Fang J-Y, et al. Cervical cancer mortality trends in China, 1991–2013, and predictions for the future. Asian Pac J Cancer Prev 2015;16:6391. [DOI] [PubMed] [Google Scholar]
- [6].Centers for Disease Control and Prevention. Cervical Cancer Statistics. Available at: www.cdc.gov/cancer/cervical/statistics. Accessed June 29, 2021. [Google Scholar]
- [7].O’Connor MJ. Targeting the DNA damage response in cancer. Mol Cell 2015;60:547–60. [DOI] [PubMed] [Google Scholar]
- [8].Carrassa L, Damia G. DNA damage response inhibitors: mechanisms and potential applications in cancer therapy. Cancer Treat Rev 2017;60:139–51. [DOI] [PubMed] [Google Scholar]
- [9].Longerich S, San Filippo J, Liu D, Sung P. FANCI binds branched DNA and is monoubiquitinated by UBE2T-FANCL. J Biol Chem 2009;284:23182–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Smogorzewska A, Matsuoka S, Vinciguerra P, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 2007;129:289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Knipscheer P, Räschle M, Smogorzewska A, et al. The fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science (New York, NY) 2009;326:1698–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ishiai M, Kitao H, Smogorzewska A, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol 2008;15:1138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lopez-Martinez D, Kupculak M, Yang D, et al. Phosphorylation of FANCD2 inhibits the FANCD2/FANCI complex and suppresses the Fanconi anemia pathway in the absence of DNA damage. Cell Rep 2019;27:2990–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Isaacsson Velho P, Qazi F, Hassan S, et al. Efficacy of Radium-223 in bone- metastatic castration-resistant prostate cancer with and without homologous repair gene defects. Eur Urol 2019;76:170–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia (New York, NY) 2007;9:166–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (New York, NY) 2017;19:649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lánczky A, Nagy Á, Bottai G, et al. miRpower: a web-tool to validate survivalassociated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat 2016;160:439–46. [DOI] [PubMed] [Google Scholar]
- [18].Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017;77:e108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer 2016;16:20–33. [DOI] [PubMed] [Google Scholar]
- [21].Wang Q, Su L, Liu N, et al. Cyclin dependent kinase 1 inhibitors: a review of recent progress. Curr Med Chem 2011;18:2025–43. [DOI] [PubMed] [Google Scholar]
- [22].Yang X, Chen G, Li W, et al. Cervical cancer growth is regulated by a c-ABL- PLK1 signaling axis. Cancer Res 2017;77:1142–54. [DOI] [PubMed] [Google Scholar]
- [23].Chen J, Deng Y, Ao L, et al. The high-risk HPV oncogene E7 upregulates miR-182 expression through the TGF-β/Smad pathway in cervical cancer. Cancer Lett 2019;460:75–85. [DOI] [PubMed] [Google Scholar]
- [24].Abbas M, Srivastava K, Imran M, Banerjee M. Genetic polymorphisms in DNA repair genes and their association with cervical cancer. Br J Biomed Sci 2019;76:117–21. [DOI] [PubMed] [Google Scholar]
- [25].Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun 2015;6:8971. [DOI] [PMC free article] [PubMed] [Google Scholar]