Abstract
To differentiate the Borrelia burgdorferi sensu lato genospecies, LightCycler real-time PCR was used for the fluorescence (SYBR Green I) melting curve analysis of borrelial recA gene PCR products. The specific melting temperature analyzed is a function of the GC/AT ratio, length, and nucleotide sequence of the amplified product. A total of 32 DNA samples were tested. Of them three were isolated from B. burgdorferi reference strains and 16 were isolated from B. burgdorferi strains cultured from Ixodes ricinus ticks; 13 were directly isolated from nine human biopsy specimens and four I. ricinus tick midguts. The melting temperature of B. garinii was 2°C lower than that of B. burgdorferi sensu stricto and B. afzelii. Melting curve analysis offers a rapid alternative for identification and detection of B. burgdorferi sensu lato genospecies.
Borrelia burgdorferi sensu stricto, B. garinii, and B. afzelii are the genospecies of B. burgdorferi sensu lato proven to be responsible for human Lyme borreliosis (19). In Europe, B. garinii and B. afzelii are the most prevalent genospecies, whereas B. burgdorferi sensu stricto is the only genospecies encountered in North America (19). For the genotypic identification of B. burgdorferi sensu lato genospecies, PCR and PCR-based assays, such as species-specific PCR (8, 10), randomly amplified polymorphic DNA analysis (20, 21), PCR-based sequencing (4–6, 11, 18), and restriction fragment length polymorphism (9, 13, 14), are commonly used. All of these methods, although they have the advantage of high discriminative power, are considered to be relatively laborious and expensive. Recently, Morrison et al. (12) described a continuous fluorescence-monitoring PCR for the recA gene of B. burgdorferi sensu stricto. The recA gene belongs to a set of genes responsible for homologous recombination in bacteria (3). The gene is located on the chromosome of the spirochete and thought to be evolutionally conserved (3, 12). Using primers described by Morrison et al. (12), we analyzed the melting curves of PCR products derived from the different genospecies of B. burgdorferi sensu lato.
A total of 32 DNA samples were tested. Of them 3 were isolated from B. burgdorferi reference strains and 16 were isolated from B. burgdorferi strains cultured from Ixodes ricinus tick midguts; 13 were directly isolated from nine human biopsy specimens punched from marginal areas of erythema migrans lesions and four I. ricinus ticks from which the midgut was removed (Table 1). The human biopsy specimens obtained were first cut in half by scissors. One half was cultured and from the other half DNA was extracted directly and used for PCR. The midgut of each tick was removed by tweezers under a microscope and cultured. DNA was extracted directly from the ticks from which the midgut was removed. The specimens submitted to culture were inoculated into tubes containing Barbour-Stoenner-Kelly (BSK-II) medium (1) and incubated at 30°C for 4 to 6 weeks. The tubes were observed macroscopically twice a week. Dark-field microscopy was carried out if the color indicated growth. The final identification of cultured spirochetes was based on PCR and sequencing (7, 17). The DNA was extracted from cultured bacteria and directly from human and tick tissues using InstaGene matrix (Bio-Rad, Hercules, Calif.), according to the manufacturer's instructions.
TABLE 1.
Origins and Tm analysis of strains used in this studya
| Species and sample (n) | Location (yr) of isolation | Mean Tm ± SD (°C) |
|---|---|---|
| B. burgdorferi sensu stricto ATCC 35210 (1) | United States | 84.02 |
| B. garinii | ||
| 387 CSF (1) | Germany | 81.62 |
| Human samples (8) | Finland (1996–1999) | 80.62 ± 0.33b |
| Tick samples (10) | Finland (1996–1998) | 81.30 ± 0.40 |
| B. afzelii | ||
| Bo 23 (1) | Germany | 83.71 |
| Human samples (1) | Finland (1999) | 83.84 |
| Tick samples (10) | Finland (1998–1999) | 83.42 ± 0.27 |
DNA was extracted from the Borrelia reference strain, human skin biopsy samples, and tick midguts and was subjected to real-time PCR for the recA gene; melting curve analysis was also performed.
P = 0.0014 when compared to Tm of recA PCR products amplified from tick samples.
PCR was performed using a fluorescence temperature cycler (LightCycler; Roche Molecular Biochemicals, Mannheim, Germany). The primers were the same as those used by Morrison et al. (12) and flanked a product 222 bp in length. The primers were nTM17.F (5′ GTG GAT CTA TTG TAT TAG ATG AGG CTC TCG 3′) and nTM17.R (5′GCC AAA GTT CTG CAA CAT TAA CAC CTA AAG 3′). Amplification was done according to the general guidelines provided by the manufacturer of the LightCycler after optimization of the various reaction parameters. The 20-μl reaction volume in a glass capillary contained 2 μl of LightCycler-DNA Master SYBR Green I mix (Taq DNA polymerase, reaction buffer, deoxynucleoside triphosphate mix, and SYBR Green I dye), 3 mM MgCl2, 20 ng of bovine serum albumin [BSA], an 8 μM concentration of each primer, 220 ng of TaqStart antibody (ClonTech, Palo Alto, Calif.), and 4 μl of extracted DNA. DNA preparations (concentration, ∼2 μg/ml) extracted from the reference strains of B. burgdorferi sensu stricto, B. afzelii, and B. garinii were used as positive controls, and mixtures of all reagents, devoid of added DNA, were used as negative controls. The three positive controls and one negative control were included in each PCR run.
The amplification program included the initial denaturation step at 95°C for 40 s and 50 cycles of denaturation at 95°C for 1 s, annealing at 59°C for 5 s, and extension at 72°C for 11 s. The temperature transition rate was 20°C/s. Fluorescence was measured at the end of each extension step. After amplification, a melting curve was acquired by heating the product at 20°C/s to 95°C, cooling it at 20°C/s to 55°C, keeping it at 55°C for 20 s, and then slowly heating it at 0.1°C/s to 94°C. Fluorescence was measured through the slow heating phase. For improved visualization of the melting temperatures (Tm), melting peaks were derived from the initial melting curves (fluorescence [F] versus temperature [T]) by plotting the negative derivative of fluorescence over temperature versus temperature (−dF/dT versus T). Melting curves were used to determine the specific PCR products (15), which were further confirmed using conventional gel electrophoresis.
The Student t test was used to analyze statistical significance. All P values corresponded to two-tailed tests, and a P of <0.05 was considered statistically significant.
The mean Tm values for the reference strains of B. burgdorferi sensu stricto, B. afzelii, and B. garinii, obtained by testing aliquots of the same sample in five reaction capillaries during the same run, were 84.02, 83.71, and 81.62°C, respectively (Table 1 and Fig. 1). The corresponding intra-assay coefficients of variation were 0.08, 0.05, and 0.06%. The mean Tm values for the reference strains of B. burgdorferi sensu stricto, B. afzelii, and B. garinii, obtained by testing aliquots of the same sample during six separate runs, were 83.57, 83.54, and 81.42°C, respectively (Table 1). The corresponding interassay coefficients of variation were 0.4, 0.4, and 0.5%. To test the effect of target DNA concentration on Tm, six fourfold dilutions of B. garinii DNA (starting concentration, ∼2 μg/ml) were tested during the same run, and the mean Tm was 81.77°C (standard deviation [SD], 0.41).
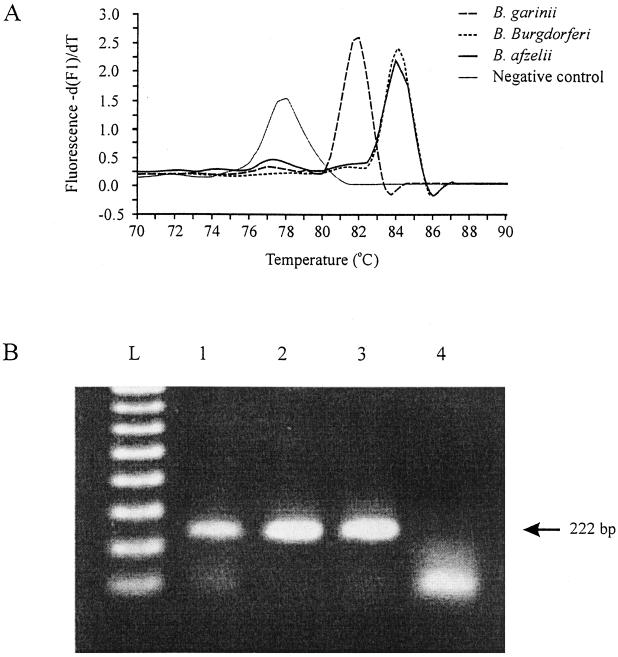
FIG. 1.
Using the real-time PCR, a fragment of the recA gene was amplified from the genospecies of B. burgdorferi sensu lato, and fluorescence melting curve analyses as well as gel electrophoresis of the products were done. (A) The Tm was 81.62°C for B. garinii, 84.02°C for B. burgdorferi sensu stricto, and 83.71°C for B. afzelii. Primer-dimers in the negative control melted at temperatures below 80°C. (B) Lane L contains molecular weight markers (100 bp), and lanes 1 to 4 show PCR products obtained from B. garinii, B. burgdorferi sensu stricto, B. afzelii, and the negative control, respectively.
The mean Tm values obtained from the DNA derived from the tick samples harboring B. garinii (n = 10) or B. afzelii (n = 10) were 81.30°C (SD, 0.40) and 83.42°C (SD, 0.27), respectively (Table 1). The mean Tm obtained from human samples (n = 8) harboring B. garinii was 80.62°C (SD, 0.33). A statistically significant difference was found between the Tm of B. garinii DNA isolated directly from human clinical specimens and the Tm of borrelial DNA of tick origin (P = 0.0014). The Tm of one human specimen positive for B. afzelii was 83.84°C.
To our knowledge, this is the first report on the use of the fluorescence melting curve analysis of PCR products to identify the genospecies of B. burgdorferi sensu lato. Compared with conventional PCR and PCR-based assays, this analysis system is less complex and decisively more rapid, with results obtained within 1 hour. Our preliminary results show that using the present real-time PCR we can reach the same sensitivity (about five organisms of B. burgdorferi including all three genospecies of B. burgdorferi sensu lato) as using the nested PCR used in our laboratory (7, 17). This suggests that the LightCycler PCR can be used for the detection and differentiation of B. burgdorferi directly in clinical samples.
It is known that the DNA extracted from clinical samples, such as blood and tissue, contains inhibitors. These inhibitors can interfere with the PCR-based detection of microbial infection and cause false-negative results. It has been reported that pretreatment of DNA with or addition of BSA into the PCR mixture can remove inhibitors (2, 16). This is why in this study BSA was added to all reaction mixtures. Further, a relatively small volume of DNA was used for the PCR to minimize the inhibitory action of the sample.
The intra- and interassay variation coefficients of the Tm values were very low, showing that the technical principles of the LightCycler allow consistent temperature conditions for the reaction capillaries. The difference between the Tm values of the reference strains B. garinii and B. afzelii or B. burgdorferi sensu stricto was about 2°C. A similar difference between B. garinii and the other genospecies was found when DNA isolated from clinical samples was tested. The shape and position of the DNA melting curve are functions of the GC/AT ratio, length, and sequence (15). Ririe et al. (15) used the LightCycler to analyze the melting curves of a mixture of a 180-bp fragment of the hepatitis B surface antigen gene and a 536-bp fragment of the human β-globin gene. They concluded that PCR products with Tm differences of even less than 2°C can be distinguished (15).
The Tm values of B. garinii DNA isolated directly from human clinical specimens and borrelial DNA of tick origin showed differences which were statistically significant. This may reflect the change that the bacterium has to undergo during its adaptation to different environments. However, the effect of the matrix from which the DNA was extracted cannot be totally excluded.
During PCR amplification, unspecific products, even primer-dimers that are produced when little or no template is present, can be formed. Because these products are double stranded, they also bind SYBR Green I dye and give a signal. The analysis of melting curves can differentiate the unspecific products from the specific product, because the Tm of unspecific products will be lower. In our PCR, all unspecific products melted at temperatures below 80°C.
The difference in Tm between B. burgdorferi sensu stricto and B. afzelii reference strains was so small (84.02 versus 83.71°C) that it cannot be used for distinguishing these two species. However, these two species could possibly be differentiated if a gene with a wider heterogeneity is used for the LightCycler PCR and melting curve analysis. Rapid differentiation of B. garinii and B. afzelii is highly valuable in Europe, where these genospecies are prevalent and B. burgdorferi sensu stricto is rarely encountered.
Our results indicate that the fluorescence melting curve analysis of recA PCR products can be used for differentiation of B. garinii from B. afzelii and B. burgdorferi sensu stricto genospecies. Studies are under way to sequence the amplified fragments for determination of factors underlying Tm differences. Further, the fluorescence melting curve analysis of PCR products could be applied to differentiation between other bacterial species, provided that gene segments with suitable variation are available.
Acknowledgments
We thank Harri Marttila and Miikka Peltomaa for providing part of the sample material, Tuula Lehtonen and Ulla Toivonen for technical assistance, and Erkki Nieminen for help in preparation of the figure. Simo Merne revised the language of the manuscript.
The Academy of Finland and the Special Governmental Fund for University Hospitals (EVO) supported this work.
REFERENCES
- 1.Barbour A G, Burgdorfer W, Hayes S F, Peter O, Aeschlimann A. Isolation of a cultivable spirochete from Ixodes ricinus ticks of Switzerland. Curr Microbiol. 1983;8:123–126. [Google Scholar]
- 2.Forbes B A, Hicks K E. Substances interfering with direct detection of Mycobacterium tuberculosis in clinical specimens by PCR: effects of bovine serum albumin. J Clin Microbiol. 1996;34:2125–2128. doi: 10.1128/jcm.34.9.2125-2128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 4.Fukunaga M, Hamase A. Outer surface protein C gene sequence analysis of Borrelia burgdorferi sensu lato isolates from Japan. J Clin Microbiol. 1995;33:2415–2420. doi: 10.1128/jcm.33.9.2415-2420.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukunaga M, Koreki Y. A phylogenetic analysis of Borrelia burgdorferi sensu lato isolates associated with Lyme disease in Japan by flagellin gene sequence determination. Int J Syst Bacteriol. 1996;46:416–421. doi: 10.1099/00207713-46-2-416. [DOI] [PubMed] [Google Scholar]
- 6.Gassmann G S, Jacobs E, Deutzmann R, Gobel U B. Analysis of the Borrelia burgdorferi GeHo fla gene and antigenic characterization of its gene product. J Bacteriol. 1991;173:1452–1459. doi: 10.1128/jb.173.4.1452-1459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Junttila J, Peltomaa M, Soini H, Marjamäki M, Viljanen M K. Prevalence of Borrelia burgdorferi in Ixodes ricinus ticks in urban recreational areas of Helsinki. J Clin Microbiol. 1999;37:1361–1365. doi: 10.1128/jcm.37.5.1361-1365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liebisch G, Sohns B, Bautsch W. Detection and typing of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks attached to human skin by PCR. J Clin Microbiol. 1998;36:3355–3358. doi: 10.1128/jcm.36.11.3355-3358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liveris D, Wormser G P, Nowakowski J, Nadelman R, Bittker S, Cooper D, Varde S, Moy F H, Forseter G, Pavia C S, Schwartz I. Molecular typing of Borrelia burgdorferi from Lyme disease patients by PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 1996;34:1306–1309. doi: 10.1128/jcm.34.5.1306-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marconi R T, Garon C F. Development of polymerase chain reaction primer sets for diagnosis of Lyme disease and for species-specific identification of Lyme disease isolates by 16S rRNA signature nucleotide analysis. J Clin Microbiol. 1992;30:2830–2834. doi: 10.1128/jcm.30.11.2830-2834.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misonne M C, Hoet P P. Species-specific plasmid sequences for PCR identification of the three species of Borrelia burgdorferi sensu lato involved in Lyme disease. J Clin Microbiol. 1998;36:269–272. doi: 10.1128/jcm.36.1.269-272.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison T B, Ma Y, Weis J H, Weis J J. Rapid and sensitive quantification of Borrelia burgdorferi-infected mouse tissues by continuous fluorescence monitoring of PCR. J Clin Microbiol. 1999;37:987–992. doi: 10.1128/jcm.37.4.987-992.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Postic D, Assous M V, Grimont P A D, Baranton G. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf(5S)-rrl(23S) intergenic spacer amplicons. Int J Syst Bacteriol. 1994;44:743–752. doi: 10.1099/00207713-44-4-743. [DOI] [PubMed] [Google Scholar]
- 14.Rijpkema S G, Herbes R G, Verbeek-De Kruif N, Schellekens J F. Detection of four species of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks collected from roe deer (Capreolus capreolus) in the Netherlands. Epidemiol Infect. 1996;117:563–566. doi: 10.1017/s0950268800059252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ririe K M, Rasmussen R P, Wittwer C T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 16.Satoh Y, Takasaka N, Hoshikawa Y, Osaki M, Ohfuji S, Ito H, Kaibara N, Kurata T, Sairenji T. Pretreatment with restriction enzyme or bovine serum albumin for effective PCR amplification of Epstein-Barr virus DNA in DNA extracted from paraffin-embedded gastric carcinoma tissue. J Clin Microbiol. 1998;36:3423–3425. doi: 10.1128/jcm.36.11.3423-3425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt B, Muellegger R P, Stockenhuber C, Soyer H P, Hoedl S, Luger A, Kerl H. Detection of Borrelia burgdorferi-specific DNA in urine specimens from patients with erythema migrans before and after antibiotic therapy. J Clin Microbiol. 1996;34:1359–1363. doi: 10.1128/jcm.34.6.1359-1363.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valsangiacomo C, Balmelli T, Piffaretti J C. A phylogenetic analysis of Borrelia burgdorferi sensu lato based on sequence information from the hbb gene, coding for a histone-like protein. Int J Syst Bacteriol. 1997;47:1–10. doi: 10.1099/00207713-47-1-1. [DOI] [PubMed] [Google Scholar]
- 19.Wang G, Van Dam A P, Schwartz I, Dankert J. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin Microbiol Rev. 1999;12:633–653. doi: 10.1128/cmr.12.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang G, van Dam A P, Spanjaard L, Dankert J. Molecular typing of Borrelia burgdorferi sensu lato by randomly amplified polymorphic DNA fingerprinting analysis. J Clin Microbiol. 1998;36:768–776. doi: 10.1128/jcm.36.3.768-776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welsh J, Pretzman C, Postic D, Saint Girons I, Baranton G, McClelland M. Genomic fingerprinting by arbitarily primed polymerase chain reaction resolves Borrelia burgdorferi into three distinct phyletic groups. Int J Syst Bacteriol. 1992;42:370–377. doi: 10.1099/00207713-42-3-370. [DOI] [PubMed] [Google Scholar]