Abstract
The rapid spread of the SARS-CoV-2 Omicron variant suggests that the virus might become globally dominant. Further, the high number of mutations in the viral spike protein raised concerns that the virus might evade antibodies induced by infection or vaccination. Here, we report that the Omicron spike was resistant against most therapeutic antibodies but remained susceptible to inhibition by sotrovimab. Similarly, the Omicron spike evaded neutralization by antibodies from convalescent patients or individuals vaccinated with the BioNTech-Pfizer vaccine (BNT162b2) with 12- to 44-fold higher efficiency than the spike of the Delta variant. Neutralization of the Omicron spike by antibodies induced upon heterologous ChAdOx1 (Astra Zeneca-Oxford)/BNT162b2 vaccination or vaccination with three doses of BNT162b2 was more efficient, but the Omicron spike still evaded neutralization more efficiently than the Delta spike. These findings indicate that most therapeutic antibodies will be ineffective against the Omicron variant and that double immunization with BNT162b2 might not adequately protect against severe disease induced by this variant.
Keywords: SARS-CoV-2, Omicron, spike, antibody, neutralization, immune evasion, vaccine
Graphical abstract
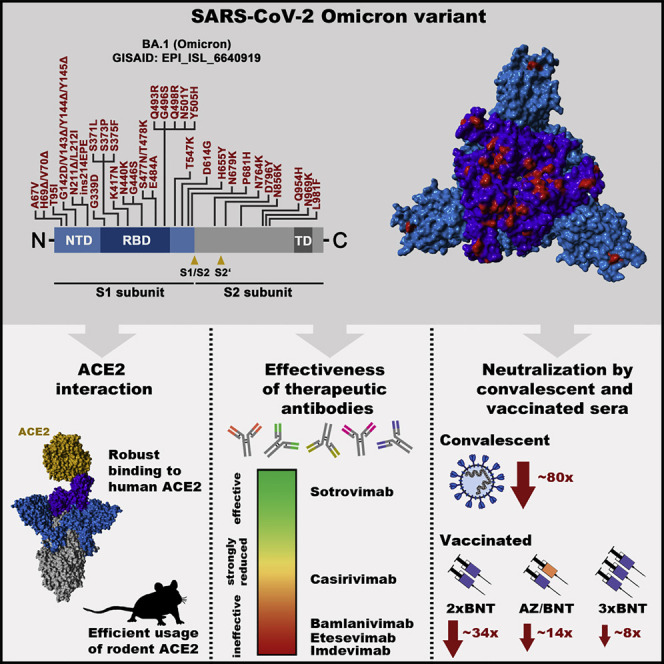
The SARS-CoV-2 Omicron variant is rapidly spreading worldwide and a public health concern. Experiments show that this variant is resistant against several therapeutic antibodies for COVID-19 and efficiently evades antibodies induced upon infection or double BNT162b2 vaccination, but not triple BNT162b2 or ChAdOx1/BNT162b2 vaccination.
Introduction
Vaccination is considered key to ending the devastating COVID-19 pandemic. However, inequities in vaccine distribution and the emergence of new SARS-CoV-2 variants threaten this approach. Several SARS-CoV-2 variants of concern (VOCs) have emerged in the recent year and the Delta variant (B.1.617.2) is currently dominating the pandemic (Harvey et al., 2021b; Tao et al., 2021). These VOCs exhibit increased transmissibility and/or immune evasion, traits that have been linked to mutations in the viral spike (S) protein (Harvey et al., 2021b; Tao et al., 2021).
The coronavirus S protein facilitates viral entry into host cells and constitutes the central target for antibodies that neutralize the virus. Mutations in the N-terminal domain (NTD), which contains an antigenic supersite (McCallum et al., 2021), and the receptor-binding domain (RBD), which binds to the angiotensin-converting enzyme 2 (ACE2) receptor (Hoffmann et al., 2020b; Zhou et al., 2020), can confer neutralization resistance by altering epitopes of neutralizing antibodies. In contrast, it is less well understood which mutations in the spike increase transmissibility and which mechanisms are responsible, although it is well established that mutation D614G increases viral transmission and promotes ACE2 engagement (Hou et al., 2020; Korber et al., 2020; Mansbach et al., 2021; Plante et al., 2021; Zhou et al., 2021).
A novel VOC, the Omicron variant (Pango lineages B.1.1.529, BA.1, BA.2, and BA.3), was recently identified in South Africa, and its emergence was associated with a steep increase in cases and hospitalizations (Abdullah, 2021). The Omicron variant was imported into several European, African and Asian countries, as well as the United States via infected air travelers (Abbasi, 2021; Graham, 2021; Gu et al., 2021; Petersen et al., 2021). In the United Kingdom, local transmission events were reported (Company, 2021) with case numbers doubling every 2 to 3 days (Torjesen, 2021). The S protein of the Omicron variant harbors an unusually high number of mutations, which might increase immune evasion and/or transmissibility. Indeed, a recent study suggested that the Omicron variant is more adept at infecting convalescent individuals as compared with previously circulating variants (Abdullah, 2021; Pulliam et al., 2021). Thus, the Omicron variant constitutes a rapidly emerging threat to public health and might undermine global efforts to control the COVID-19 pandemic. However, the susceptibility of the Omicron variant to antibody-mediated neutralization remains to be analyzed.
Here, we report that the Omicron S protein evades antibodies with up to 44-fold higher efficiency than the spike of the Delta variant, rendering therapeutic antibodies ineffective and likely compromising protection by antibodies induced upon infection or vaccination with two doses of BNT162b2 (BNT).
Results
The NTD and RBD of the Omicron spike are highly mutated
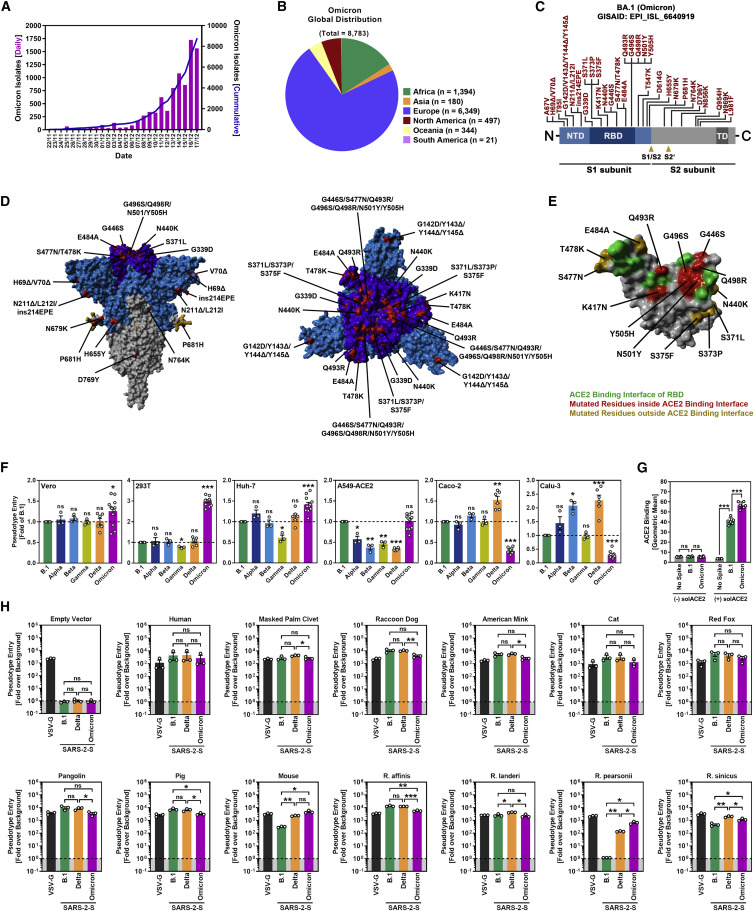
The first sequences of the Omicron variant were deposited into the GISAID (Global Initiative on Sharing All Influenza Data) database on November 22 and 23, 2021 (Figure 1 A). These sequences were obtained from patients in Botswana and South Africa as well as from a traveler returning to Hong Kong from South Africa. Subsequently, the number of deposited Omicron sequences rapidly increased (Figure 1A) and the virus was also detected in Europe, Asia, and the United States due to infected air travelers (Figure 1B). Analysis of the Omicron genomic sequence revealed striking differences as compared with other known SARS-CoV-2 variants, suggesting extensive, independent evolution, potentially in an isolated human population, immunocompromised patients, or an unknown animal species (Kupferschmidt, 2021).
Figure 1.
The Omicron spike mediates entry into cell lines with different efficiency as compared with B.1 and Delta spike, binds human ACE2 efficiently, and utilizes a broad range of animal ACE2 orthologues as receptor
(A) Epidemiology of SARS-CoV-2 Omicron variant. Purple bars indicate the number of newly reported isolates per day while the blue line shows the cumulative number of isolates as of November 17, 2021. (Based on data deposited in the GISAID database).
(B) Global distribution of SARS-CoV-2 Omicron variant. Sequences retrieved from the GISAID database were grouped based on the continent where they were detected (as of November 17, 2021).
(C) Schematic overview and domain organization of the S proteins of SARS-CoV-2 Omicron variant. Mutations compared with the SARS-CoV-2 Wuhan-Hu-1 isolate are highlighted in red. (NTD, N-terminal domain; RBD, receptor-binding domain; TD, transmembrane domain).
(D) Location of Omicron-specific mutations in the context of the trimeric spike protein. (S1 subunit, light blue; RBD, dark blue; S2 subunit, gray; S1/S2 and S2′ cleavage sites; mutated amino acid residues, red (compared with the S protein of the SARS-CoV-2 Wuhan-Hu-1 isolate).
(E) Locations of Omicron-specific mutations in the context of the RBD. (RBD, gray; RBD residues that interact with ACE2, green; ACE2-interacting RBD residues that are mutated in Omicron spike, red: non-ACE2-interacting RBD residues that are mutated in Omicron spike, orange).
(F) Particles bearing the indicated S proteins were inoculated onto different cell lines, and S-protein-driven cell entry was analyzed at 16–18 h postinoculation by measuring the activity of virus-encoded firefly luciferase in cell lysates. Presented are the mean from 3 to 12 biological replicates (each conducted with four technical replicates) in which S-protein-driven cell entry was normalized against B.1 (set as 1). (Error bars: SEM).
(G) Binding of soluble ACE2 to B.1 or Omicron spike proteins. 293T cells expressing the indicated S protein (or no S protein) following transfection were sequentially incubated with soluble ACE2 harboring a C-terminal Fc-tag derived from human IgG and AlexaFlour-488-conjugated antihuman antibody. Finally, ACE2 binding was analyzed by flow cytometry. Cells incubated with only secondary antibody served as controls. Presented is the mean geometric mean channel fluorescence from six biological replicates (each conducted with single samples). (Error bars: standard deviation).
(H) Particles bearing the indicated S proteins or VSV-G were inoculated on BHK-21 cells that were previously transfected to express the indicated ACE2 orthologues or empty vector. S protein-driven cell entry was analyzed as described in (F). Presented are the mean data from three biological replicates (each conducted with four technical replicates) in which signals obtained from particles bearing no viral glycoprotein (indicated by dashed line) were used for normalization (set as 1). (Error bars: SEM).
(F–H): statistical significance of differences between individual groups was assessed by two-tailed Students t test: p > 0.05, not significant (ns); ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
The Omicron spike (isolate hCoV-19/Botswana/R40B58_BHP_3321001245/2021; GISAID Accession ID: EPI_ISL_6640919) exhibits 37 mutations as compared with the Wuhan-Hu-1 spike (Figures 1C and 1D). Thirteen of these changes are unique, while the remaining changes are known from variants of interest or concern (Haseltine, 2021). Specifically, 11 mutations, including 6 deletions and 1 insertion, are located in the N-terminal domain (NTD) and mutations N211Δ and ins214EPE are unique. Some of the deletions are found in other VOCs and might reduce antibody binding or increase spike expression (Harvey et al., 2021b; Li et al., 2021b; Liu et al., 2021; Meng et al., 2021; Wang et al., 2021; Wheatley et al., 2021). Another 15 mutations are found in the receptor-binding domain (RBD) (Figures 1C–1E), of which G339D, S371L, S373P, and S375F are unique, and several were previously shown to modulate ACE2 binding and/or antibody evasion (Harvey et al., 2021b; Li et al., 2021b; Liu et al., 2021; Wang et al., 2021; Wheatley et al., 2021). Further, five mutations reside between the RBD and the S1/S2 site, including the unique mutation T547K and mutation P681H, which may modulate cleavage at the S1/S2 site by the host protease furin (Hoffmann et al., 2020a; Zhang et al., 2021a) (Figures 1C and 1D). Finally, five mutations are present in the S2 subunit, all of them unique (Haseltine, 2021).
Omicron spike binds human ACE2 and mediates efficient entry into cell lines
For analysis of host-cell entry, we employed vesicular stomatitis virus (VSV) particles pseudotyped with SARS-CoV-2 S proteins. These pseudotyped particles adequately mimic key features of SARS-CoV-2 entry into target cells, including receptor and protease choice and neutralization by antibodies (Hoffmann et al., 2020b; Riepler et al., 2020; Schmidt et al., 2020).
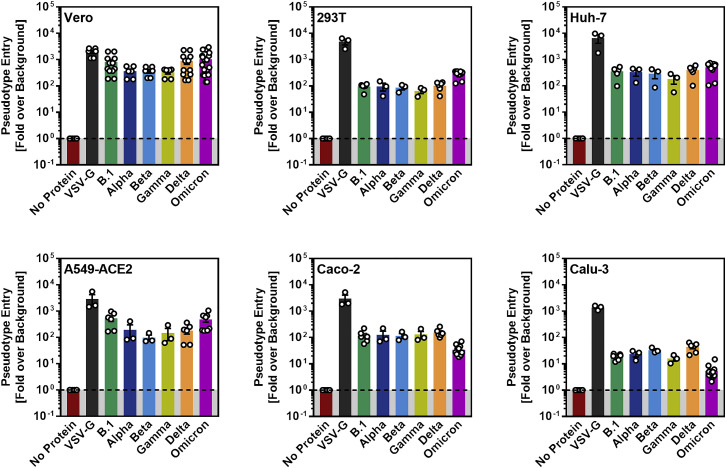
We first asked whether the Omicron spike differs from the spike of other VOCs regarding target cell choice and entry efficiency. The spike from SARS-CoV-2 B.1 (which is identical to the S protein of the Wuhan-Hu-1 isolate, except for the presence of mutation D614G) was analyzed in parallel, since this virus circulated early in the pandemic and does not harbor mutations found in the S proteins of VOCs. For the analysis of cell tropism, we employed the cell lines Vero (African green monkey, kidney), 293T (human kidney), and A549 (human lung), which were engineered to express high levels of ACE2 (A549-ACE2), Huh-7 (human liver), Caco-2 (human colon) and Calu-3 (human lung) cells. All cell lines were highly susceptible to entry driven by VSV-G and SARS-CoV-2 B.1 spike (Figures 1F and S1 ). Further, all VOC S proteins facilitated robust entry into the cell lines analyzed, but subtle differences were noted. Thus, the Delta spike mediated increased entry into Calu-3 and Caco-2 cells, in keeping with published data (Arora et al., 2021b), while the Omicron spike facilitated increased entry into Vero, Huh-7, and particularly 293T cells (Figure 1F). Further, B.1 and Omicron spike mediated comparable entry into A549-ACE2 cells while entry driven by the other S proteins was less efficient (Figure 1F). Finally, the Omicron spike bound efficiently to human ACE2 (Figure 1G) and used ACE2 for host-cell entry (see below), indicating that the mutations in the RBD do not compromise ACE2 interactions. In sum, the Omicron spike binds to ACE2 and mediates robust entry into diverse ACE2-positive cell lines.
Figure S1.
Cell entry driven by the spike proteins of VOC (related to Figure 1F)
Pseudotype entry data presented in Figure 1F normalized against the assay background. Pseudotype entry was normalized against signals obtained from cells inoculated with particles bearing no viral glycoprotein (background, set as 1), and data obtained for particles bearing VSV-G were included. Error bars indicate the SEM.
The Omicron spike can efficiently use ACE2 orthologues from different animal species for cell entry
We next asked whether the Omicron spike can use human ACE2 and ACE2 orthologues from different animal species, including horseshoe bat, masked palm civet, raccoon dog, and pangolin, for entry into target cells. Entry driven by the B.1 and Delta spikes as well as VSV-G served as controls. The expression of the ACE2 orthologues did not modulate entry driven by VSV-G but in most cases allowed robust and roughly comparable entry driven by the B.1, Delta, and Omicron spikes (Figure 1H). Two exceptions were noted. Murine ACE2 was used more efficiently by the Delta spike as compared with the B.1 spike and supported entry driven by the Omicron spike with the highest efficiency (Figure 1H). Further, the B.1 spike was unable to use ACE2 from Pearson’s horseshoe bat (Rhinolophus pearsonii) for entry while the Delta and particularly the Omicron spike used this receptor with high efficiency (Figure 1H). Collectively, these data reveal broad usage of ACE2 orthologues by Omicron spike for host-cell entry, which might hint toward high zoonotic potential.
Omicron spike is inhibited by soluble ACE2 but is resistant against several antibodies used for COVID-19 treatment
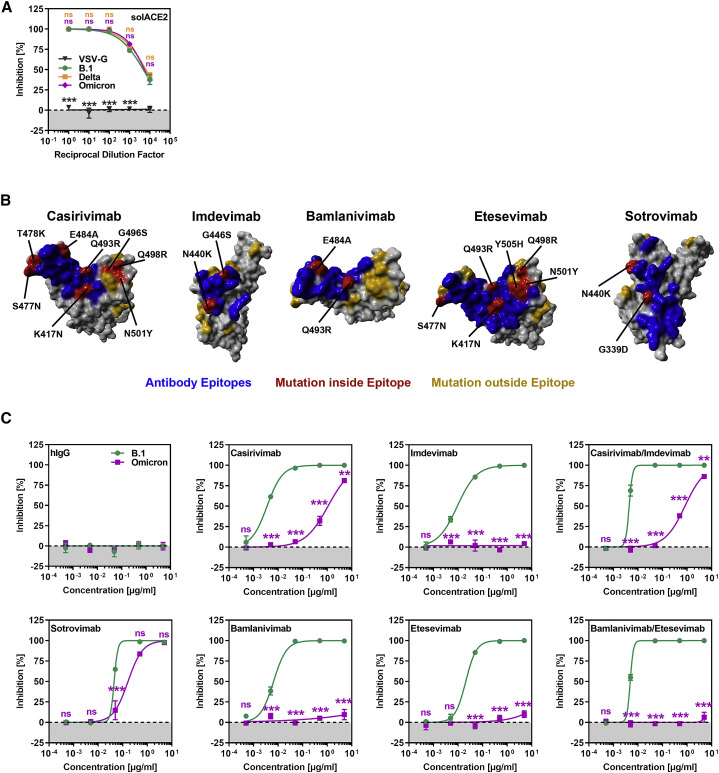
We next asked whether the Omicron spike can be inhibited by soluble ACE2, which binds to the RBD, blocks host-cell entry, and is currently being developed for COVID-19 therapy (Monteil et al., 2020). Soluble ACE2 did not modulate entry driven by VSV-G but robustly and comparably inhibited entry driven by B.1, Delta, and Omicron spikes (Figure 2 A), indicating that soluble ACE2 should be suitable for treatment of patients infected with the Omicron variant.
Figure 2.
The Omicron spike evades neutralization by four out of five monoclonal antibodies but is efficiently inhibited by soluble ACE2
(A) Inhibition of S-protein-driven cell entry by soluble ACE2. Particles harboring the indicated S proteins were preincubated (30 min, 37°C) with different dilutions of soluble ACE2 before being inoculated onto Vero cells. S-protein-driven cell entry was analyzed as described in Figure 1F. Presented are the mean data from three biological relocates (each conducted with four technical replicates).
(B) Locations of Omicron-specific mutations in the context of the RBD epitopes targeted by casirivimab, imdevimab, bamlanivimab, etesevimab, and sotrovimab. (RBD, gray; epitope targeted by the antibody, blue; Omicron-specific mutations within the epitope, red; Omicron-specific mutations outside the epitope, orange).
(C) Omicron spike is resistant against four out of five monoclonal antibodies used for treatment of COVID-19 patients. Particles harboring the indicated S proteins were preincubated (30 min, 37°C) with different concentrations of recombinant monoclonal antibody before being inoculated onto Vero cells. S-protein-driven cell entry was analyzed as described in Figure 1F. Presented are the mean data from three biological replicates (each conducted with four technical replicates).
(A and C): statistical significance of differences between individual groups was assessed by two-way analysis of variance with Dunnett’s (A) or Sidak’s (C) post hoc tests: p > 0.05, not significant (ns); ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Several recombinant, neutralizing monoclonal antibodies were identified that inhibit SARS-CoV-2 infection and cocktails of casirivimab and imdevimab (REGN-COV2, Regeneron) (Weinreich et al., 2021) and etesevimab and bamlanivimab (Eli Lilly) (Dougan et al., 2021) are currently used for COVID-19 therapy. In addition, the antibody sotrovimab was shown to inhibit SARS-CoV-2 and related viruses and was found to protect patients from COVID-19 (Gupta et al., 2021). Since the Omicron spike harbors several mutations within the structures that are recognized by these antibodies (Figure 2B), we investigated whether the antibodies were still able to neutralize the Omicron spike. All antibodies inhibited entry driven by the B.1 spike in a robust and concentration-dependent manner, while a control immunoglobulin was inactive (Figure 2C). In contrast, entry driven by the Omicron spike was fully resistant against bamlanivimab, etesevimab, and imdevimab and largely resistant against casirivimab. In agreement with these findings, a cocktail of bamlanivimab and etesevimab failed to inhibit entry mediated by the Omicron spike, while inhibition by a cocktail of casirivimab and imdevimab was inefficient (Figure 2C). In contrast, sotrovimab was active against Omicron spike, although inhibition was slightly less efficient than that measured for B.1 spike (Figure 2C). In sum, the Omicron spike is resistant against several antibodies used for COVID-19 treatment.
The Omicron spike evades neutralization by antibodies induced upon infection and BNT vaccination with high efficiency
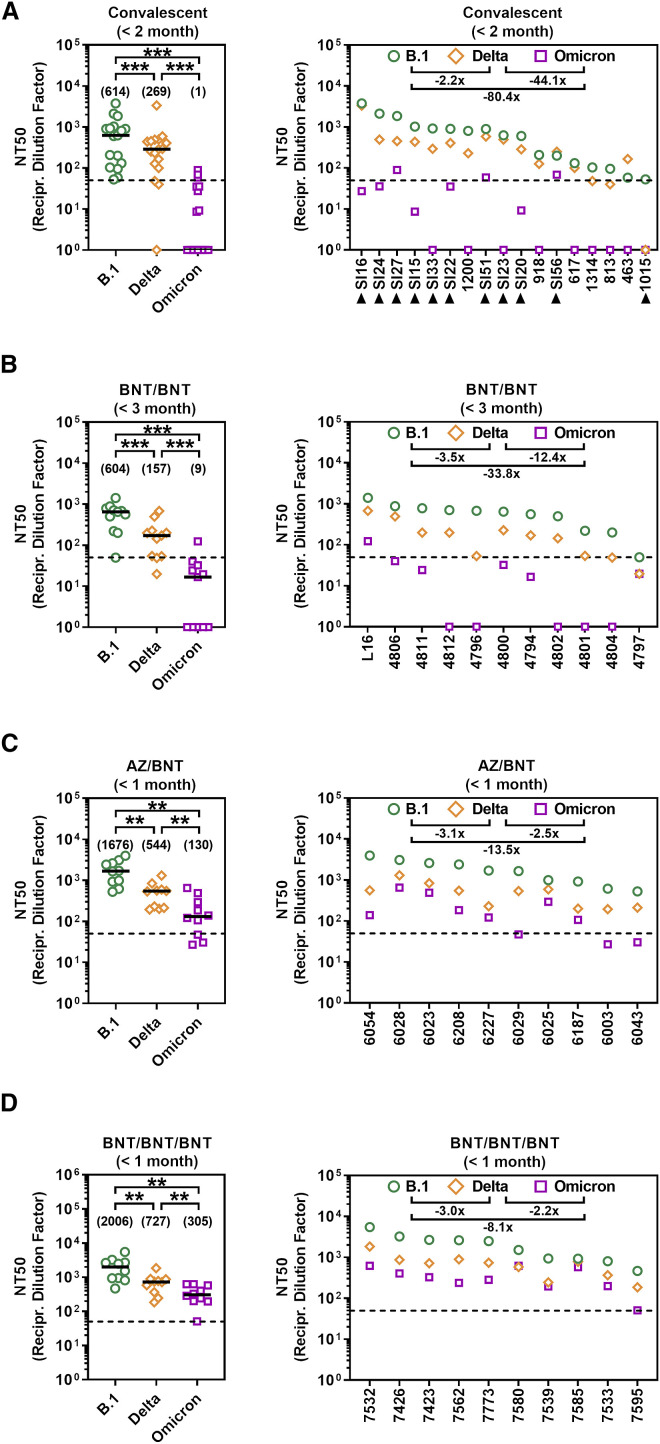
The resistance against several antibodies used for COVID-19 therapy suggested that the Omicron spike might also evade antibodies induced upon infection and vaccination. Indeed, sera/plasma collected within two months of convalescence from mild or severe COVID-19 inhibited entry driven by the Omicron spike 80-fold less efficiently as compared with the B.1 spike and 44-fold less efficiently as compared with the Delta spike, with 9 out of 17 sera tested being unable to neutralize particles bearing Omicron spike (Figures 3A and S2 ). The samples were collected in Germany during the first COVID-19 wave (Table S1), when neither the Alpha nor the Delta variant predominated, suggesting the antibodies raised against the virus circulating at the beginning of the pandemic offer little to no protection against the Omicron variant.
Figure 3.
The Omicron spike shows high resistance against antibodies elicited upon infection or vaccination
(A) Particles bearing the indicated S proteins were preincubated (30 min, 37°C) with different dilutions of convalescent sera/plasma (n = 17) before being inoculated onto Vero cells. S-protein-driven cell entry was analyzed as described in Figure 1F. Black triangles indicate patients with severe disease that required admission to the intensive care unit; all other patients showed mild disease.
(B) The experiment was performed as described in (A) but sera from BNT/BNT-vaccinated individuals were analyzed (n = 11).
(C) The experiment was performed as described in (A) but sera from AZ/BNT-vaccinated individuals were analyzed (n = 10).
(D) The experiment was performed as described in (A) but sera from BNT/BNT/BNT-vaccinated individuals were analyzed (n = 10).
(A–D) Patient identifiers are indicated on the x axes. The reciprocal serum/plasma dilution factors that caused a 50% reduction in S protein-driven cell entry (neutralization titer 50, NT50) are shown. Left panels show individual NT50 values clustered per SARS-CoV-2 variant. Black lines and numerical values in brackets indicate median NT50 values, whereas right panels show serum/plasma-specific NT50 values ranked from highest to lowest based on NT50 for B.1. Numerical values indicate the median fold change in neutralization between individual SARS-CoV-2 variants. Dashed lines indicate the lowest serum dilution tested. Statistical significance of differences between individual groups was assessed by Wilcoxon matched-pairs signed rank test: p > 0.05, not significant (ns); ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Figure S2.
Individual neutralization data (related to Figure 3)
Presented are the individual neutralization results for the data shown in Figure 3. Data represent the mean values of four technical replicates with error bars indicating the standard deviation. The curves were calculated based on a non-linear regression model with variable slope.
The mRNA-based vaccine BNT efficiently protects against COVID-19 (Polack et al., 2020) and is frequently used in Europe and the United States. Sera collected within one to three months after the second dose of BNT (10 sera were collected within 1 month, one serum was collected within 3 months; Table S2) inhibited entry by the Omicron spike with 34-fold lower efficiency as compared with the B.1. spike and with 12-fold lower efficiency as compared with the Delta spike (Figures 3B and S2). These results suggest that two immunizations with BNT, which can provide more than 90% protection from severe disease upon infection with the Delta variant (Chemaitelly et al., 2021), might be markedly less effective against the Omicron variant.
Evidence that heterologous and booster vaccination may provide improved protection against the Omicron variant
We next explored whether strategies known to increase production of neutralizing antibodies relative to BNT/BNT immunization might afford better protection against the Omicron variant. Heterologous vaccination with a first dose of ChAdO1-nCoV-19/AZD1222 (AZ) (Voysey et al., 2021) and a second dose of BNT was shown to induce higher neutralizing antibody titers as compared with the corresponding homologous vaccinations (Barros-Martins et al., 2021; Behrens et al., 2021). Sera collected within one month after heterologous AZ/BNT vaccination (Table S2) exhibited higher neutralizing activity as compared with sera collected within three months after BNT/BNT vaccination (Figure 3 C). Inhibition of Omicron-spike-driven entry by sera from AZ/BNT-vaccinated individuals was 14-fold less efficient as compared to B.1. spike but only 3-fold less efficient relative to Delta spike (Figure 3C). Further, inhibition of the Omicron spike by sera collected within one month of AZ/BNT vaccination was comparable with inhibition of the Delta spike by sera obtained within three months after BNT/BNT vaccination (Figures 3B and 3C), a time interval during which the vaccine provides more than 90% protection from severe COVID-19.
A third immunization with BNT increases protection against infection by 10-fold as compared to two doses of BNT (Bar-On et al., 2021). Sera from BNT/BNT/BNT-immunized donors collected within one month after the third dose (Table S2) had slightly higher neutralizing titers as sera obtained within the same time interval from donors immunized with AZ/BNT (Figures 3C and 3D). Sera from BNT/BNT/BNT-immunized individuals inhibited entry driven by the Omicron spike with 8-fold reduced efficiency as compared with the B.1 spike and 2-fold reduced efficiency as compared with the Delta spike (Figure 3D). Further, inhibition of Omicron spike by sera collected within one month after BNT/BNT/BNT immunization was moderately more efficient as compared with inhibition of the Delta spike by sera obtained within three months after BNT/BNT vaccination (Figures 3B and 3D). These results suggest that heterologous AZ/BNT as well as homologous BNT/BNT/BNT immunization might provide better protection against the Omicron variant as compared to BNT/BNT immunization.
Discussion
The emergence, rapid spread, and international dissemination of the highly mutated Omicron variant raised concerns that this variant might soon become globally dominant and that several therapeutic or preventive interventions might be ineffective against this variant. The present study indicates that several of these concerns are justified. The S protein of the Omicron variant evaded antibody-mediated neutralization with higher efficiency than any previously analyzed S proteins of variants of interest and VOCs. It was not appreciably inhibited by two antibody cocktails used for COVID-19 therapy and inhibition by antibodies induced by two immunizations with BNT was strongly reduced as compared with the spike of the Delta variant. Heterologous vaccinations and a BNT booster shot induced appreciable levels of neutralizing antibodies against the Omicron spike and might offer some protection against this variant. Nevertheless, our tools available to contain SARS-CoV-2 might require expansion and adaptation in order to efficiently combat the Omicron variant.
The findings that the Omicron spike facilitated efficient entry into several human cell lines and robustly bound human ACE2 suggest that the Omicron variant readily infects human cells. Nevertheless, some peculiarities of host cell entry driven by the Omicron spike relative to the B.1 spike were noted. The Omicron spike mediated slightly but significantly augmented entry into cell lines which only allow for cathepsin L-(293T, Huh-7, A549-ACE2) but not TMPRSS2-dependent entry, due to low or absent TMPRSS2 expression (Hoffmann et al., 2020b). We previously observed a similar phenotype for the S protein of another African SARS-CoV-2 lineage, A.30 (Arora et al., 2021a), and the mutations in the spike responsible for this phenotype remain to be determined. Further, the Omicron spike used ACE2 proteins from different animal species with high efficiency for entry. The efficient usage of murine ACE2 is in keeping with the presence of the mutations K417N and N501Y, which can also emerge upon SARS-CoV-2 adaptation to experimentally infected mice (Huang et al., 2021; Sun et al., 2021; Zhang et al., 2021b). In addition, the Omicron spike harbors mutations Q493R and Q498R, which are related to exchanges Q493K and Q498H, which were also detected in mouse-adapted SARS-CoV-2 (Huang et al., 2021; Sun et al., 2021; Zhang et al., 2021b). However, some of these mutations are present in other VOCs and cannot be taken as evidence that the Omicron variant may have evolved in infected mice in the wild. In addition, the Omicron spike was able to use ACE2 from the Pearson’s horseshoe bat (Rhinolophus pearsonii) with high efficiency. Again, this finding does not indicate that the Omicron variant infects these animals (which are found in Asia) in the wild but rather reflects the ability of the Omicron spike to use diverse ACE2 orthologues for entry. Collectively, our results suggest that the mutations in the Omicron spike are compatible with robust usage of diverse ACE2 orthologues for entry and might thus have broadened the ability of the Omicron variant to infect animal species.
Cocktails of the antibodies casirivimab and imdevimab as well as etesevimab and bamlanivimab are used for COVID-19 therapy. Entry driven by the Omicron spike was fully or largely resistant against each of these antibodies and against the antibody cocktails, most likely due to the mutations K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H, which are located within or close to the epitopes bound by these antibodies (Figure 2B). Thus, two frequently used COVID-19 treatment options will not be available to combat the Omicron variant. In contrast, sotrovimab, a pan-sarbecovirus-neutralizing antibody (Gupta et al., 2021), remained active against the Omicron spike, in keeping with sotrovimab recognizing an epitope not substantially altered by mutations found in the Omicron spike (Figure 2B). Finally, soluble ACE2 robustly blocked entry driven by the Omicron spike and might be an option for treatment of patients infected with the Omicron variant.
Studies conducted before the emergence of the Omicron variant indicated that convalescent COVID-19 patients are efficiently protected against reinfection and antibody responses likely play an important role in protection (Addetia et al., 2020; Hall et al., 2021; Hansen et al., 2021; Harvey et al., 2021a; Krammer, 2021; Lumley et al., 2021). Our findings with serum/plasma samples collected in Germany during the first wave of the pandemic indicate that this high level of protection might not apply to reinfection with the Omicron variant. Thus, neutralization of the Omicron spike was 80-fold less efficient as compared with controls and several sera did not exert neutralizing activity. Although neutralization by sera from patients infected with the Delta variant remains to be examined, it is likely that convalescent patients might not be adequately protected against symptomatic reinfection with the Omicron variant, in keeping with recent data (Pulliam et al., 2021).
Neutralizing antibody responses are also believed to be critical for protection against COVID-19 by BNT vaccination (Corbett et al., 2021; Feng et al., 2021; Gilbert et al., 2021). We found that antibodies induced upon BNT/BNT immunization neutralized the Omicron spike with 34-fold reduced efficiency as compared with B.1. Therefore, two doses of BNT might not provide robust protection against severe disease induced by the Omicron variant and adaptation of the vaccine to the new variant seems required. In the meantime, heterologous AZ/BNT vaccination or a BNT booster (BNT/BNT/BNT) might afford some protection against the Omicron variant, since sera from individuals who had received the respective vaccinations neutralized the Omicron spike with appreciable efficiency. However, it remains to be determined whether this protection is transient or long lasting.
Limitations of the study
Our study has several limitations, including the analysis of a limited number of sera/plasma collected within 3 months after vaccination and use of pseudotyped virus instead of authentic SARS-CoV-2 Omicron variant, which was not available to us. Further, T cell responses were not determined in our study. However, given the important role that antibodies play in immune protection against SARS-CoV-2, our results suggest that antibody cocktails used for therapy and likely also vaccines must be adapted for efficient protection against the Omicron variant. While such adaptations are in progress, heterologous or booster immunizations could help to limit the impact of the Omicron variant on public health, and conventional control measures like face masks and social distancing should be maintained.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Casirivimab | Laboratory of Hans-Martin Jäck | N/A |
| Imdevimab | Laboratory of Hans-Martin Jäck | N/A |
| Bamlanivimab | Laboratory of Hans-Martin Jäck | N/A |
| Etesevimab | Laboratory of Hans-Martin Jäck | N/A |
| Sotrovimab | Laboratory of Hans-Martin Jäck | N/A |
| hIgG | Laboratory of Hans-Martin Jäck | N/A |
| Goat anti-Human IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11013; RRID: AB_2534080 |
| Anti-VSV-G antibody (I1, produced from CRL-2700 mouse hybridoma cells) | ATCC | Cat# CRL-2700; RRID: CVCL_G654 |
| Bacterial and virus strains | ||
| VSV∗ΔG-FLuc | Laboratory of Gert Zimmer | N/A |
| One Shot™ OmniMAX™ 2 T1R Chemically Competent E. coli | Thermo Fisher Scientific | Cat# C854003 |
| Biological samples | ||
| Patient plasma (SI 15) | Laboratory of Martin Sebastian Winkler | N/A |
| Patient plasma (SI 16) | Laboratory of Martin Sebastian Winkler | N/A |
| Patient plasma (SI 20) | Laboratory of Martin Sebastian Winkler | N/A |
| Patient plasma (SI 22) | Laboratory of Martin Sebastian Winkler | N/A |
| Patient plasma (SI 23) | Laboratory of Martin Sebastian Winkler | N/A |
| Patient plasma (SI 24) | Laboratory of Martin Sebastian Winkler | N/A |
| Patient plasma (SI 27) | Laboratory of Martin Sebastian Winkler | N/A |
| Patient plasma (SI 33) | Laboratory of Martin Sebastian Winkler | N/A |
| Patient plasma (SI 51) | Laboratory of Martin Sebastian Winkler | N/A |
| Patient plasma (SI 56) | Laboratory of Martin Sebastian Winkler | N/A |
| Patient serum (463) | Laboratory of Georg M.N. Behrens | N/A |
| Patient serum (617) | Laboratory of Georg M.N. Behrens | N/A |
| Patient serum (813) | Laboratory of Georg M.N. Behrens | N/A |
| Patient serum (918) | Laboratory of Georg M.N. Behrens | N/A |
| Patient serum (1015) | Laboratory of Georg M.N. Behrens | N/A |
| Patient serum (1200) | Laboratory of Georg M.N. Behrens | N/A |
| Patient serum (1314) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (L16) | Laboratory of Martin Lier | N/A |
| Vaccinee serum (4796) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (4811) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (4801) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (4806) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (4804) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (4802) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (4812) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (4800) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (4799) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (4797) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (6187) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (6227) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (6208) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (6043) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (6029) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (6025) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (6028) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (6023) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (6054) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (6003) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (7539) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (7533) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (7423) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (7426) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (7532) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (7580) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (7773) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (7562) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (7585) | Laboratory of Georg M.N. Behrens | N/A |
| Vaccinee serum (7595) | Laboratory of Georg M.N. Behrens | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Soluble human ACE2 (sol-hACE2-Fc) | Laboratory of Stefan Pöhlmann | N/A |
| Critical commercial assays | ||
| Beetle-Juice Kit | PJK | Cat# 102511 |
| GeneArt Gibson Assembly HiFi Master Mix | Thermo Fisher Scientific | Cat# A46627 |
| Experimental models: Cell lines | ||
| 293T | DSMZ | Cat# ACC-635; RRID: CVCL_0063 |
| A549 (ACE2) | Laboratory of Stefan Pöhlmann | N/A |
| BHK-21 | Laboratory of Georg Herrler | ATCC Cat# CCL-10; RRID:CVCL_1915 |
| Caco-2 | Laboratory of Stefan Pöhlmann | ATCC Cat# HTB-37; RRID: CVCL_0025 |
| Calu-3 | Laboratory of Stephan Ludwig | ATCC Cat# HTB-55; RRID: CVCL_0609 |
| Huh-7 | Laboratory of Thomas Pietschmann | JCRB Cat# JCRB0403; RRID: CVCL_0336 |
| Vero | Laboratory of Andrea Maisner | ATCC Cat# CRL-1586; RRID: CVCL_0574 |
| Oligonucleotides | ||
| Human ACE2 (NotI) F (TGATCC GCGGCCGCATGTCAAGCTCT TCCTGGCTCC) |
Sigma-Aldrich | N/A |
| Human ACE2-cMYC (PacI) R (GATCCG TTAATTAACTACAGATCTTCTTCGCTA ATCAGTTTCTGTTCAAAGGAGGTCTG AACATCATCAG) |
Sigma-Aldrich | N/A |
| Masked Palm civet ACE2 (NotI) F (TGATCCGCGGCCGCATGTCA GGCTCTTTCTGGCTCC) |
Sigma-Aldrich | N/A |
| Masked Palm civet ACE2-cMYC (PacI) R (GATCCGTTAATTAACTACAGATCTT CTTCGCTAATCAGTTTCTGTTCAAATG AAGTCTGAACGTCATCAGC) |
Sigma-Aldrich | N/A |
| Raccoon dog ACE2 (NotI) F (TGATCC GCGGCCGCGCCACCATGTCAGGC TCTTCCTGGCTCC) |
Sigma-Aldrich | N/A |
| Raccoon dog ACE2-cMYC (PacI) R (GATC CGTTAATTAACTACAGATCTTCTTCGCTA ATCAGTTTCTGTTCAAATGAAGTCTGA GCATCATCCAC) |
Sigma-Aldrich | N/A |
| American mink ACE2 (NotI) F (TGATCC GCGGCCGCATGTTAGGCTCTTCCTG GCTCC) |
Sigma-Aldrich | N/A |
| American mink ACE2-cMYC (PacI) R (GATCCGTTAATTAACTACAGATCTTCTT CGCTAATCAGTTTCTGTTCAAATGACGT CTGAACATCATCGAC) |
Sigma-Aldrich | N/A |
| Cat ACE2 (NotI) F (TGATCCGCGGCCGC ATGTCAGGCTCTTTCTGGCTCC) |
Sigma-Aldrich | N/A |
| Cat ACE2-cMYC (PacI) R (GATCCG TTAATTAACTACAGATCTTCTTCGC TAATCAGTTTCTGTTCAAATGAAGT CTGAACATCATCAGC) |
Sigma-Aldrich | N/A |
| Red fox ACE2 (NotI) F (TGATCC GCGGCCGCATGAGCGGCTCC AGCTGGCTGC) |
Sigma-Aldrich | N/A |
| Red fox ACE2-cMYC (PacI) R (GATCCG TTAATTAACTACAGATCTTCTTCGCTAA TCAGTTTCTGTTCGAAGCTTGTCTG GGCGTCATCC) |
Sigma-Aldrich | N/A |
| Malayan pangolin ACE2 (NotI) F (TGATCC GCGGCCGCATGAGCGGCAGCAGCTG GCTGC) |
Sigma-Aldrich | N/A |
| Malayan pangolin ACE2-cMYC (PacI) R (GATCCGTTAATTAACTACAGATCTTCT TCGCTAATCAGTTTCTGTTCGAAGCT GGTCTGCACGTCGTCC) |
Sigma-Aldrich | N/A |
| Pig ACE2 (NotI) F (TGATCC GCGGCCGCATGTCAGGC TCTTTCTGGCTCC) |
Sigma-Aldrich | N/A |
| Pig ACE2-cMYC (PacI) R (GATCCG TTAATTAACTACAGATCTTCTTCG CTAATCAGTTTCTGTTCAAACGA AGTCTGAATGTCATCG) |
Sigma-Aldrich | N/A |
| Mouse ACE2 (NotI) F (TGATCC GCGGCCGCATGTCCAGCTCC TCCTGGCTCC) |
Sigma-Aldrich | N/A |
| Mouse ACE2-cMYC (PacI) R (GATCCG TTAATTAACTACAGATCTTCTTCGCTA ATCAGTTTCTGTTCAAAGGAAGTCTG AGCATCATCAC) |
Sigma-Aldrich | N/A |
| Bat (Rhinolophus affinis) ACE2 (NotI) F (TGATCCGCGGCCGCATGTCAGGCT CTTCCTGGCTCC) |
Sigma-Aldrich | N/A |
| Bat (Rhinolophus affinis) ACE2-cMYC (PacI) R (GATCCGTTAATTAACTACAGATCTTC TTCGCTAATCAGTTTCTGTTCAAAGGAG GTCTGAACATCATCACC) |
Sigma-Aldrich | N/A |
| Bat (Rhinolophus landeri) ACE2 (NotI) F (TGATCCGCGGCCGCATGTCAGGCT CTTCCTGGCTCTTTC) |
Sigma-Aldrich | N/A |
| Bat (Rhinolophus landeri) ACE2-cMYC (PacI) R (GATCCGTTAATTAACTACAG ATCTTCTTCGCTAATCAGTTTCTGTTC AAAGGAGGTCTGAACATCATCACC) |
Sigma-Aldrich | N/A |
| Bat (Rhinolophus pearsonii) ACE2 (NotI) F (TGATCCGCGGCCGCATGTCAGGCT CTTTCTGGTTCC) |
Sigma-Aldrich | N/A |
| Bat (Rhinolophus pearsonii) ACE2-cMYC (PacI) R (GATCCGTTAATTAACTACAGA TCTTCTTCGCTAATCAGTTTCTGTTC AAAGGAGGTCTGAACATCATCACC) |
Sigma-Aldrich | N/A |
| Bat (Rhinolophus sinicus) ACE2 (NotI) F (TGATCCGCGGCCGCATGTCAGGCTC TTCCTGGCTCC) |
Sigma-Aldrich | N/A |
| Bat (Rhinolophus sinicus) ACE2-cMYC (PacI) R (GATCCGTTAATTAACTACAG ATCTTCTTCGCTAATCAGTTTCTGTTCA AACGAAGTCTGAACATCATCACC) |
Sigma-Aldrich | N/A |
| SARS-2-S Seq-01 (CAAGATCTACAGCA AGCACACC) |
Sigma-Aldrich | N/A |
| SARS-2-S Seq-02 (GTCGGCGGCAACTA CAATTAC) |
Sigma-Aldrich | N/A |
| SARS-2-S Seq-03 (GCTGTCTGATCGGA GCCGAG) |
Sigma-Aldrich | N/A |
| SARS-2-S Seq-04 (TGAGATGATCGCCCA GTACAC) |
Sigma-Aldrich | N/A |
| SARS-2-S Seq-05 (GCCATCTGCCACGA CGGCAAAG) |
Sigma-Aldrich | N/A |
| pCG1 F (CCTGGGCAACGTGCTGGT) | Sigma-Aldrich | N/A |
| pCG1 R (GTCAGATGCTCAAGGGG CTTCA) |
Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| SARS-2-SΔ18 (Omicron), codon-optimized, DNA strings | Thermo Fisher Scientific | N/A |
| Plasmid: pCG1 | Laboratory of Roberto Cattaneo | N/A |
| Plasmid: pCAGGS-VSV-G | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCAGGS-DsRed | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (B.1), codon-optimized | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (Alpha), codon-optimized | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (Beta), codon-optimized | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (Gamma), codon-optimized | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (Delta), codon-optimized | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (Omicron), codon-optimized | This study | N/A |
| Plasmid: pQCXIP_Human ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_Masked palm civet ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_Raccoon dog ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_American Mink ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_Cat ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_Red fox ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_Pangolin ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_Pig ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_Mouse ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_Bat (Rhinolophus affinis) ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_Bat (Rhinolophus landeri) ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_Bat (Rhinolophus pearsonii) ACE2-cMYC | This study | N/A |
| Plasmid: pQCXIP_Bat (Rhinolophus sinicus) ACE2-cMYC | This study | N/A |
| Plasmid: pCG1-solACE2-Fc | Laboratory of Stefan Pöhlmann | N/A |
| Software and algorithms | ||
| Hidex Sense Microplate Reader Software | Hidex Deutschland Vertrieb GmbH | https://www.hidex.de |
| YASARA (version 19.1.27) | YASARA Biosciences GmbH | http://www.yasara.org |
| Adobe Photoshop CS5 Extended (version 12.0 x 32) | Adobe | https://www.adobe.com/ |
| GraphPad Prism (version 8.3.0(538)) | GraphPad Software | https://www.graphpad.com/ |
| SWISS-MODEL online tool | Protein Structure Bioinformatics Group, Swiss Institute of Bioinformatics Biozentrum, University of Basel |
https://swissmodel.expasy.org |
| Flowing software (version 2.5.1) | Turku Bioscience | https://bioscience.fi/services/cell-imaging/flowing-software/ |
| Microsoft Office Standard 2010 (version 14.0.7232.5000) | Microsoft Corporation | https://products.office.com/ |
| Other | ||
| Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies (PDB: 6XDG) | (Hansen et al., 2020) | https://www.rcsb.org/structure/6XDG |
| SARS-CoV 2 Spike Protein bound to LY-CoV555 (PDB: 7L3N) | (Jones et al., 2021) | https://www.rcsb.org/structure/7L3N |
| Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD (PDB: 7C01) | (Shi et al., 2020) | https://www.rcsb.org/structure/7C01 |
| Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment (open state) (PDB: 6WPT) | (Pinto et al., 2020) | https://www.rcsb.org/structure/6WPT |
| Distinct conformational states of SARS-CoV-2 spike protein (PDB: 6XR8) | (Cai et al., 2020) | https://www.rcsb.org/structure/6XR8 |
Resource availability
Lead contact
Requests for material can be directed to Markus Hoffmann (mhoffmann@dpz.eu) and the lead contact, Stefan Pöhlmann (spoehlmann@dpz.eu).
Materials availability
All materials and reagents will be made available upon installment of a material transfer agreement (MTA).
Experimental model and subject details
Cell culture
The following cells lines were used in the present study: 293T (human, female, kidney; ACC-635, DSMZ; RRID: CVCL_0063), A549-ACE2 ((Huang et al., 2021); based on parental A549 cells, human, male, lung; CRM-CCL-185, ATCC), BHK-21 (Syrian hamster, male, kidney; ATCC Cat# CCL-10; RRID: CVCL_1915, kindly provided by Georg Herrler, University of Veterinary Medicine Hannover, Germany), Vero (African green monkey kidney, female, kidney; CRL-1586, ATCC; RRID: CVCL_0574, kindly provided by Andrea Maisner), Huh-7 cells (human, male, liver; JCRB Cat# JCRB0403; RRID: CVCL_0336, kindly provided by Thomas Pietschmann), Calu-3 (human, male, lung; HTB-55, ATCC; RRID: CVCL_0609, kindly provided by Stephan Ludwig) and Caco-2 cells (human, male, colon; HTB-37, ATCC, RRID: CVCL_0025). 293T, BHK-21, Vero and Huh-7 cells were maintained in Dulbecco’s modified Eagle medium (DMEM, PAN-Biotech). Calu-3 and Caco-2 cells were cultured in minimum essential medium (GIBCO) while A549-ACE2 cells were maintained in DMEM/F-12 medium (GIBCO). All media were supplemented with 10% fetal bovine serum (Biochrom) and 100 U/ml penicillin and 0.1 mg/ml streptomycin (PAA). Further, media for Calu-3 and Caco-2 cells were supplemented with 1x non-essential amino acid solution (from 100x stock, PAA) and 1 mM sodium pyruvate (GIBCO). Cell lines were validated by STR-typing, amplification and sequencing of a fragment of the cytochrome c oxidase gene, microscopic examination and/or according to their growth characteristics. In addition, all cell lines were regularly tested for mycoplasma contamination. For transfection of 293T cells, the calcium-phosphate based method was applied, while BHK-21 cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific).
Method details
Expression plasmids
Plasmids encoding DsRed, VSV-G (vesicular stomatitis virus glycoprotein), soluble ACE2, SARS-CoV-2 S B.1 (codon optimized, contains C-terminal truncation of the last 18 amino acid) and SARS-CoV-2 S of VOCs Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1) and Delta (B.1.617.2) have been previously reported (Brinkmann et al., 2017; Hoffmann et al., 2021a, 2021b). The expression vector for the Omicron spike (based on isolate hCoV-19/Botswana/R40B58_BHP_3321001245/2021; GISAID Accession ID: EPI_ISL_6640919) was generated by Gibson assembly using five overlapping DNA strings (Thermo Fisher Scientific, sequences available upon request), linearized (BamHI/XbaI digest) pCG1 plasmid and GeneArt™ Gibson Assembly HiFi Master Mix (Thermo Fisher Scientific). Gibson assembly was performed according to manufacturer’s instructions. The pCG1 vector was kindly provided by Roberto Cattaneo (Mayo Clinic College of Medicine, Rochester, MN, USA). To generate expression plasmids for different ACE2 orthologues (harboring a C-terminal cMYC-epitope tag) the respective sequences were amplified from existing plasmids (human, pangolin, cat, masked palm civet and red fox, Rhinolophus landeri ACE2) (Hoffmann et al., 2021b; Krüger et al., 2021; Li et al., 2021a) or ACE2 sequences were obtained from a commercial gene synthesis service (raccoon dog, American mink, Pig, Mouse, Rhinolophus affinis, Rhinolophus sinicus, Rhinolophus pearsonii; GeneArt). Expression plasmids for pangolin, masked palm civet and red fox ACE2 used as templates for cloning were kindly provided by Zhaohui Qian (Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China). ACE2 open reading frames were inserted into the pQCXIP expression vector making use of NotI and PacI restriction sites. The integrity of all expression plasmids was confirmed using a commercial sequencing service (Microsynth SeqLab).
Sequence analysis and protein models
S protein sequences and information on collection date and global distribution of SARS-CoV-2 isolates belonging to the Omicron variant were obtained from the GISAID (Global Initiative on Sharing All Influenza Data) database (https://www.gisaid.org). Structural models of the S protein were generated using YASARA software (http://www.yasara.org/index.html) and are based on a template that was constructed by modelling the SARS-2 S sequence on PDB: 6XR8 (Cai et al., 2020) using the SWISS-MODEL online tool (https://swissmodel.expasy.org). Alternatively, the following published crystal structures were employed; PDB: 6XDG (Hansen et al., 2020), PDB: 7L3N (Jones et al., 2021), PDB: 7C01 (Shi et al., 2020) or PDB: 6WPT (Pinto et al., 2020).
Pseudotyping
Rhabdoviral pseudotypes harboring SARS-CoV-2 S protein were produced as described (Kleine-Weber et al., 2019). In brief, 293T cells were transfected with expression plasmids for SARS-CoV-2 S protein, VSV-G or empty plasmid (control). At 24 h posttransfection, cells were infected with at an MOI of 3 with a replication-deficient reporter VSV, termed VSV∗ΔG-FLuc (Berger Rentsch and Zimmer, 2011). This virus lacks the ORF for VSV-G and codes for enhanced green fluorescent protein and firefly luciferase (FLuc). After 1 h of incubation, the input virus was removed and cells were washed with phosphate-buffered saline (PBS). Thereafter, culture medium containing anti-VSV-G antibody (culture supernatant from I1-hybridoma cells; ATCC no. CRL-2700) was added in order to neutralize residual inpunt virus. For cells expressing VSV-G, culture medium without antibody was added. After 16-18 h, the culture supernatant was harvested, separated from cellular debris by centrifugation for 10 min at 4,000 x g at room temperature (RT), and the clarified supernatants were aliquoted and stored at -80 °C.
Analysis of spike protein-mediated cell entry
For analysis of S protein-driven cell entry, target cells were seeded in 96-well plates. In case of experiments addressing S protein usage of different ACE2 orthologues as receptor, cells were transfected with the respective ACE2 expression plasmids or empty vector. At 24 h post seeding (or transfection), the culture medium was removed and cells were inoculated with equal volumes of pseudotype preparations. At 16-18 h post inoculation, pseudotype entry was quantified by measuring the activity of virus-encoded luciferase in cell lysates. For this, cells were lysed using PBS supplemented with 0.5% triton X-100 (Carl Roth) for 30 min at RT. Subsequently, cell lysates were transferred into white 96-well plates, mixed with luciferase substrate (Beetle- Juice, PJK) and luminescence measured with a Hidex Sense Plate luminometer (Hidex).
Analysis of ACE2 binding
The production of soluble ACE2 fused to the Fc portion of human immunoglobulin has been previously described in detail (Hoffmann et al., 2021b). In order to test binding of soluble ACE2 to the S protein, 293T cells seeded in 6-well plates were transfected with S protein expression plasmids or empty plasmid as negative control. At 24 h posttransfection, the medium was replaced with fresh culture medium. At 48 h posttransfection, the culture medium was aspirated and cells resuspended in PBS and pelleted by centrifugation at 600 x g for 5 min. Subsequently, cells were washed with PBS containing 1 % bovine serum albumin (BSA, PBS-B) and pelleted again. Next, the cell pellets were resuspended in 250 μl PBS-B containing soluble ACE2-Fc (1:100 dilution of 100x concentrated stock) and incubated for 60 min at 4 °C, employing a Rotospin test tube rotator disk (IKA). Thereafter, the cells were pelleted, resuspended in 250 μl PBS-B containing anti-human AlexaFlour-488-conjugated antibody (1:200; Thermo Fisher Scientific) and incubated again for 60 min at 4 °C. After a final wash step with PBS-B, the cells were fixed with 4 % paraformaldehyde solution for 30 min at RT, washed, resuspended in 150 μl PBS-B and analyzed by flow cytometry, using an LSR II flow cytometer and the FACS Diva software (BD Biosciences). The collected data were then analyzed using the Flowing software version 2.5.1 (https://bioscience.fi/services/cell-imaging/flowing-software/).
Analysis of inhibition of S protein-driven cell entry by soluble ACE2
S protein (or VSV-G) bearing particles were pre-incubated for 30 min at 37 °C with different dilutions of soluble ACE2 (undiluted, 1:10, 1:100, 1:1,000, 1:10,000). After incubation, the mixtures were added to Vero cells. Particles exposed to medium without soluble ACE2 served as control. Transduction efficiency was determined at 16-18 h postinoculation by determining luciferase activities in cell lysates, as described above.
Collection of serum and plasma samples
Convalescent plasma/serum was obtained from COVID-19 patients treated at the University Medicine Göttingen (UMG) or Hannover Medical school (MHH). Sera from individuals vaccinated with BNT162b2/BNT162b2 (BNT/BNT), ChAdOx1 nCoV-19/BNT162b2 (AZ/BNT) or BNT162b2/BNT162b2/BNT162b2 (BNT/BNT/BNT) were collected 14-72 days after receiving the last (second or third) dose at UMG or MHH (specific details on the samples can be found in Table S1). All serum and plasma samples were heat-inactivated at 56 °C for 30 min. Collection of samples was approved by the ethic committee of the UMG (reference number: 8/9/20 and SeptImmun Study 25/4/19 Ü) and the Institutional Review Board of MHH (8973_BO_K_2020).
Neutralization assay
For neutralization experiments, S protein bearing particles were pre-incubated for 30 min at 37 °C with Casirivimab, Imdevimab, Bamlanivimab, Etesevimab, Casirivimab + Imdevimab, Bamlanivimab + Etesevimab, Sotrovimab or unrelated control human IgG (5, 0.5, 0.05, 0.005, 0.0005 μg/ml). Alternatively, pseudotyped particles were pre-incubated with different dilutions of convalescent or vaccinated plasma/serum (dilution range: 1:25 to 1:12,800). After incubation, the mixtures were added to Vero cells. Particles exposed to medium without antibodies served as control. Transduction efficiency was determined at 16-18 h postinoculation by determining luciferase activities in cell lysates, as described above.
Quantification and statistical analysis
The results on S protein-driven cell entry represent average (mean) data acquired from three to twelve biological replicates, each conducted with four technical replicates. The transduction was normalized against SARS-CoV-2 S B.1 (= 1). Alternatively, transduction was normalized against the background signal (luminescence measured for cells inoculated with particles bearing no viral glycoprotein; set as 1). For ACE2 binding, presented are the average (mean) geometric mean channel fluorescence data from six biological replicates, each conducted with single samples. The results on neutralization of S protein-driven cell entry by monoclonal antibodies and control IgG represent average (mean) data from three biological replicates (each conducted with technical quadruplicates) for which transduction was normalized against samples that did not contain any antibody (= 0% inhibition). The results on inhibition of S protein-driven cell entry by soluble ACE2 represent average (mean) data from three biological replicates (each conducted with technical quadruplicates) for which transduction was normalized against samples that did not contain soluble ACE2 (= 0% inhibition). The results on neutralization of spike protein-driven cell entry by convalescent plasma/serum or serum from vaccinated individuals are based on a single experiment, which was conducted with technical quadruplicates. For data normalization, the plasma/serum dilution factor that leads to 50% reduction in S protein-driven cell entry (neutralizing titer 50, NT50) was calculated. In addition, for each plasma/serum the fold change in NT50 between the indicated SARS-CoV-2 variants was calculated.
Error bars are defined as either standard deviation (SD) or standard error of the mean (SEM). Data were analyzed using Microsoft Excel (as part of the Microsoft Office software package, version 2019, Microsoft Corporation) and GraphPad Prism 8 version 8.4.3 (GraphPad Software). Statistical significance was analyzed by two-tailed Student’s t-test with Welch correction (pseudotype entry, ACE2 binding), two-way analysis of variance (ANOVA) with Dunnett’s or Sidak’s post-hoc test (soluble ACE2 inhibition, and monoclonal antibody neutralization) or Wilcoxon matched-pairs signed rank test (serum/plasma neutralization). Only p-values of 0.05 or lower were considered statistically significant (p > 0.05, not significant [ns]; p ≤ 0.05, ∗; p ≤ 0.01, ∗∗; p ≤ 0.001, ∗∗∗). Details on the statistical test and the error bars can be found in the figure legends.
Acknowledgments
We thank R. Cattaneo, G. Herrler, A. Maisner, S. Ludwig, T. Pietschmann, Z. Qian, and G. Zimmer for providing reagents and L. Čičin-Šain for discussion. We gratefully acknowledge the originating laboratories responsible for obtaining the specimens, as well as the submitting laboratories where the genome data were generated and shared via GISAID, on which this research is based. S.P. acknowledges funding by BMBF (01KI2006D, 01KI20328A, 01KX2021), the Ministry for Science and Culture of Lower Saxony (14-76103-184, MWK HZI COVID-19), and the German Research Foundation (DFG; PO 716/11-1, PO 716/14-1). N.K. received funding from BMBF (01KI2074A). M.S.W. received unrestricted funding from Sartorius AG, Lung research. H.-M.J. received funding from BMBF (01KI2043, NaFoUniMedCovid19-COVIM: 01KX2021), Bavarian State Ministry for Science, and the Arts and Deutsche Forschungsgemeinschaft (DFG) through the research training groups RTG1660 and TRR130. G.M.N.B. acknowledges funding by German Center for Infection Research (grant no 80018019238) and a European Regional Development Fund (Defeat Corona, ZW7-8515131, together with A.D.-J.).
Author contributions
Conceptualization, M.H. and S.P.; funding acquisition, H.M.J., G.M.N.B., and S.P.; investigation, M.H., N.K., C.R., A.K., I.N., L.G., and A.-S.M.; essential resources, S.S., A.C., M.S.W., M.L., A.D.-J., H.-M.J., and G.M.N.B.; writing, M.H. and S.P.; review and editing, all authors.
Declaration of interests
The authors declare no competing interests.
Published: December 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.12.032.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
References
- Abbasi J. Omicron has reached the US-here's what infectious disease experts know About the variant. JAMA. 2021 doi: 10.1001/jama.2021.22619. [DOI] [PubMed] [Google Scholar]
- Abdullah F. 2021. Tshwane District Omicron variant patient profile - early features.https://www.samrc.ac.za/news/tshwane-district-omicron-variant-patient-profile-early-features [Google Scholar]
- Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.-L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P., Rocha C., Kempf A., Nehlmeier I., Graichen L., Winkler M.S., Lier M., Schulz S., Jäck H.-M., Cossmann A., et al. The spike protein of SARS-CoV-2 variant A.30 is heavily mutated and evades vaccine-induced antibodies with high efficiency. Cell. Mol. Immunol. 2021;18:2673–2675. doi: 10.1038/s41423-021-00779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P., Sidarovich A., Krüger N., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.-S., Winkler M.S., Schulz S., Jäck H.-M., et al. B.1.617.2 enters and fuses lung cells with increased efficiency and evades antibodies induced by infection and vaccination. Cell Rep. 2021;37:109825. doi: 10.1016/j.celrep.2021.109825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On Y.M., Goldberg Y., Mandel M., Bodenheimer O., Freedman L., Alroy-Preis S., Ash N., Huppert A., Milo R. Protection against Covid-19 by BNT162b2 booster across age groups. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2115926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros-Martins J., Hammerschmidt S.I., Cossmann A., Odak I., Stankov M.V., Morillas Ramos G., Dopfer-Jablonka A., Heidemann A., Ritter C., Friedrichsen M., et al. Immune responses against SARS-CoV-2 variants after heterologous and homologous ChAdOx1 nCoV-19/BNT162b2 vaccination. Nat. Med. 2021;27:1525–1529. doi: 10.1038/s41591-021-01449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens G.M., Cossmann A., Stankov M.V., Nehlmeier I., Kempf A., Hoffmann M., Pöhlmann S. SARS-CoV-2 delta variant neutralisation after heterologous ChAdOx1-S/BNT162b2 vaccination. Lancet. 2021;398:1041–1042. doi: 10.1016/S0140-6736(21)01891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Rentsch M., Zimmer G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS One. 2011;6:e25858. doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann C., Hoffmann M., Lübke A., Nehlmeier I., Krämer-Kühl A., Winkler M., Pöhlmann S. The glycoprotein of vesicular stomatitis virus promotes release of virus-like particles from tetherin-positive cells. PLoS One. 2017;12:e0189073. doi: 10.1371/journal.pone.0189073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemaitelly H., Tang P., Hasan M.R., AlMukdad S., Yassine H.M., Benslimane F.M., Al Khatib H.A., Coyle P., Ayoub H.H., Al Kanaani Z., et al. Waning of BNT162b2 vaccine protection against SARS-CoV-2 infection in Qatar. N. Engl. J. Med. 2021;385:e83. doi: 10.1056/NEJMoa2114114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company B.B. 2021. Covid: Omicron spreading in the community, Javid confirms.https://www.bbc.com/news/uk-59553460 [Google Scholar]
- Corbett K.S., Nason M.C., Flach B., Gagne M., O'Connell S., Johnston T.S., Shah S.N., Edara V.V., Floyd K., Lai L., et al. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science. 2021;373:eabj0299. doi: 10.1126/science.abj0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan Michael, Nirula Ajay, Azizad Masoud, Mocherla Bharat, Gottlieb Robert, Chen Peter, Hebert Corey, Perry Russell, Boscia Joseph, Heller Barry, et al. Bamlanivimab plus Etesevimab in Mild or Moderate Covid-19. N Engl J Med. 2021;385(15):1382–1392. doi: 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2021:eab3435. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. Daily briefing: Omicron was already spreading in Europe. Nature. 2021 doi: 10.1038/d41586-021-03610-3. [DOI] [PubMed] [Google Scholar]
- Gu H., Krishnan P., Ng D.Y.M., Chang L.D.J., Liu G.Y.Z., Cheng S.S.M., Hui M.M.Y., Fan M.C.Y., Wan J.H.L., Lau L.H.K., et al. Probable transmission of SARS-CoV-2 omicron variant in quarantine hotel, Hong Kong, China, November 2021. Emerg. Infect. Dis. 2021;28 doi: 10.3201/eid2802.212422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Gonzalez-Rojas Y., Juarez E., Crespo Casal M., Moya J., Falci D.R., Sarkis E., Solis J., Zheng H., Scott N., et al. Early treatment for Covid-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N. Engl. J. Med. 2021;385:1941–1950. doi: 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- Hall V.J., Foulkes S., Charlett A., Atti A., Monk E.J.M., Simmons R., Wellington E., Cole M.J., Saei A., Oguti B., et al. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: a large, multicentre, prospective cohort study (SIREN) Lancet. 2021;397:1459–1469. doi: 10.1016/S0140-6736(21)00675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C.H., Michlmayr D., Gubbels S.M., Mølbak K., Ethelberg S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. 2021;397:1204–1212. doi: 10.1016/S0140-6736(21)00575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R.A., Rassen J.A., Kabelac C.A., Turenne W., Leonard S., Klesh R., Meyer W.A., 3rd, Kaufman H.W., Anderson S., Cohen O., et al. Association of SARS-CoV-2 seropositive antibody test With risk of future infection. JAMA Intern. Med. 2021;181:672–679. doi: 10.1001/jamainternmed.2021.0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Robertson D.L., COVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine W. 2021. Understanding Omicron: changes in the spike protein and beyond and what they portend.https://www.forbes.com/sites/williamhaseltine/2021/12/08/omicron-the-sum-of-all-fears/?sh=2ef4182d5b51 [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Zhang L., Krüger N., Graichen L., Kleine-Weber H., Hofmann-Winkler H., Kempf A., Nessler S., Riggert J., Winkler M.S., et al. SARS-CoV-2 mutations acquired in mink reduce antibody-mediated neutralization. Cell Rep. 2021;35:109017. doi: 10.1016/j.celrep.2021.109017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Zhang Y., Hui X., Zhao Y., Gong W., Wang T., Zhang S., Yang Y., Deng F., Zhang Q., et al. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine. 2021;67:103381. doi: 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Weber H., Elzayat M.T., Wang L., Graham B.S., Müller M.A., Drosten C., Pöhlmann S., Hoffmann M. Mutations in the spike protein of Middle East respiratory syndrome coronavirus transmitted in Korea increase resistance to antibody-mediated neutralization. J. Virol. 2019;93 doi: 10.1128/JVI.01381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. Correlates of protection from SARS-CoV-2 infection. Lancet. 2021;397:1421–1423. doi: 10.1016/S0140-6736(21)00782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger N., Rocha C., Runft S., Krüger J., Färber I., Armando F., Leitzen E., Brogden G., Gerold G., Pöhlmann S., et al. The upper respiratory tract of felids is highly susceptible to SARS-CoV-2 infection. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms221910636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K. Where did 'weird' Omicron come from? Science. 2021;374:1179. doi: 10.1126/science.acx9738. [DOI] [PubMed] [Google Scholar]
- Li P., Guo R., Liu Y., Zhang Y., Hu J., Ou X., Mi D., Chen T., Mu Z., Han Y., et al. The Rhinolophus affinis bat ACE2 and multiple animal orthologs are functional receptors for bat coronavirus RaTG13 and SARS-CoV-2. Sci. Bull. (Beijing) 2021;66:1215–1227. doi: 10.1016/j.scib.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371.e9. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.-M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumley S.F., O'Donnell D., Stoesser N.E., Matthews P.C., Howarth A., Hatch S.B., Marsden B.D., Cox S., James T., Warren F., et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N. Engl. J. Med. 2021;384:533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansbach R.A., Chakraborty S., Nguyen K., Montefiori D.C., Korber B., Gnanakaran S. The SARS-CoV-2 Spike variant D614G favors an open conformational state. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B., Kemp S.A., Papa G., Datir R., Ferreira I.A.T.M., Marelli S., Harvey W.T., Lytras S., Mohamed A., Gallo G., et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021;35:109292. doi: 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E., Ntoumi F., Hui D.S., Abubakar A., Kramer L.D., Obiero C., Tambyah P.A., Blumberg L., Yapi R., Al-Abri S., et al. Emergence of new SARS-CoV-2 Variant of Concern Omicron (B.1.1.529) - highlights Africa's research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID-19 response and control efforts. Int. J. Infect. Dis. 2021;114:268–272. doi: 10.1016/j.ijid.2021.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam J.R.C., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M.J., Dushoff J., Mlisana K., Moultrie H. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv. 2021 doi: 10.1101/2021.11.11.21266068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riepler L., Rössler A., Falch A., Volland A., Borena W., von Laer D., Kimpel J. Comparison of four SARS-CoV-2 neutralization assays. Vaccines (Basel) 2020;9 doi: 10.3390/vaccines9010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.-H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric virusesSARS-CoV-2. J. Exp. Med. 2020;217 doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Sun S., Gu H., Cao L., Chen Q., Ye Q., Yang G., Li R.T., Fan H., Deng Y.-Q., Song X., et al. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat. Commun. 2021;12:5654. doi: 10.1038/s41467-021-25903-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torjesen I. Covid restrictions tighten as omicron cases double every two to three days. BMJ. 2021;375:n3051. doi: 10.1136/bmj.n3051. [DOI] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley A.K., Pymm P., Esterbauer R., Dietrich M.H., Lee W.S., Drew D., Kelly H.G., Chan L.-J., Mordant F.L., Black K.A., et al. Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain. Cell Rep. 2021;37:109822. doi: 10.1016/j.celrep.2021.109822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Mann M., Syed Z.A., Reynolds H.M., Tian E., Samara N.L., Zeldin D.C., Tabak L.A., Ten Hagen K.G. Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2109905118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang K., Wang T., Deng F., Gong W., Hui X., Zhao Y., He X., Li C., Zhang Q., et al. SARS-CoV-2 rapidly adapts in aged BALB/c mice and induces typical pneumonia. J. Virol. 2021;95 doi: 10.1128/JVI.02477-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Pohlmann A., King J., Steiner S., Kelly J.N., et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592:122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request