Abstract
Main conclusion
The free bases of cytokinins are the biologically active forms of the hormone while cytokinin ribosides become active only upon removal of the ribose residue.
Abstract
Cytokinins (CKs) belong to the classical plant hormones. They were discovered more than 65 years ago, but which molecular forms possess genuine CK activity is still matter of debate. Numerous studies support the view that only the free bases are the biologically active molecules. This standpoint has been challenged in a recent review (Nguyen et al. in Planta 254: 45, 2021) proposing that also CK ribosides may have genuine own CK activity. Here we critically discuss the pros and cons of this viewpoint considering the results of biological assays, CK binding studies, 3D structural data of CK-receptor interaction and mutant analyses. It is concluded that all types of study provide clear and convincing evidence only for biological activity of free bases and not ribosides; the latter are rather a transport form of the hormone without their own biological activity.
Keywords: 3D protein structure, Cytokinin receptor, Cytokinin riboside, Cytokinin signaling, Plant hormone, Structure–activity relation
Introduction
Intrinsic hormonal activity of a compound implies its ability to affect the receptor directly, switching it from one state to another. It is important to distinguish the biologically active compound of a given hormone from related compounds, which might be metabolic intermediates, storage or transport forms of the hormone. This is also true for cytokinins (CKs), which belong to the classical plant hormones with a broad spectrum of activity in planta. Chemically they are adenine derivatives carrying N6-linked side chain. The main forms are isopentenyladenine (iP), trans- and cis-zeatin (tZ and cZ). They typically occur as a mixture in plants, including free bases (or nucleobases), their ribosides and ribotides, as well as O-, N7- and N9-linked sugar conjugates. Recently, a review appeared in Planta focusing on CK ribosides and describing their history, metabolism, transport and putative functions (Nguyen et al. 2021). Importantly, the authors propose that the long-standing view that the active CKs are their free base forms should be expanded to include respective ribosides (Nguyen et al. 2021). The main arguments put forward by the authors has been the correlation of high concentrations of CK ribosides with CK activities in planta and their apparent ability to bind to CK receptors in certain heterologous test systems mentioned further below. The question whether or not CK ribosides are active CKs has been matter of debate during the history of CK studies. Therefore, this question and the arguments put forward by Nguyen et al. (2021) in favor of a genuine CK activity of CK ribosides need to be considered carefully. In this commentary, we are going to describe the current knowledge and give an oversight of the pros and cons. It will be concluded that most if not all of the available data do not support genuine CK activity of the ribosides.
Historical outline
CKs were first discovered in the form of kinetin, an artificial N6-modified nucleobase (Miller et al. 1955). Almost 10 years later the first natural CK–nucleobase termed zeatin was isolated from Zea mays seeds (Letham 1963). Afterwards various additional CKs and their derivatives were discovered as free molecules and constituents of tRNA, among them CK nucleosides with a ribose residue at the N9 of the adenine heterocycle (Skoog and Armstrong 1970). Because CK ribosides are often abundant and display CK activity in many bioassays, the question arose whether they possess genuine CK activity. Already at that time there were indications that they lack own CK activity. Addition of a non-hydrolyzable ribose group to N9 of a CK analog practically eliminated its CK activity (Hecht et al. 1971). It was also demonstrated that typical N9-substituted CKs can rapidly (within a few minutes of incubation) loose the N9-substitution in contact with living tissue (Fox et al. 1971). Unlike CK bases, the ribosides had no significant CK activity in the Funaria biotest (Spiess 1975). All this led to the widespread viewpoint that ribosides have no own activity (Sakakibara 2006).
Ligand binding to CK receptors
The question which CK compounds have biological activity emerged again after the discovery of CK receptors (Inoue et al. 2001; Suzuki et al. 2001). The first studies were limited to the three Arabidopsis CK receptors, termed AHK2, AHK3 and CRE1/AHK4. Checking the binding activities of free bases and ribosides using transgenic E. coli expressing the CRE1/AHK4 receptor gene yielded positive results for both compounds (Yamada et al. 2001). At the same time, evaluation of the ligand binding capabilities of the CRE1/AHK4 receptor residing in membranes isolated from yeast expressing the CRE1/AHK4 gene revealed the inability of ribosides, exemplified by iPR, to bind to the receptor (Yamada et al. 2001). However, subsequently the E. coli test system was developed as a convenient tool for quantitative analysis of hormone-receptor interactions (Romanov et al. 2005), and in this (Romanov et al. 2005; Stolz et al. 2011; Kuderová et al. 2015) and similar (Spíchal et al. 2004) test systems the CK ribosides usually exhibited binding/activity with rather high affinity for different CK receptors. Therefore, at that time many researchers believed that ribosides might possess their own CK activity.
CK receptor 3D structure
Ten years ago, Hothorn et al. (2011) succeeded in crystallizing the sensory module of the CRE1/AHK4 receptor, which allowed to readdress the question of “true ligands” with a novel approach. Analysis of the 3D structure of the binding site showed a space deficiency to accommodate the ribose residue in the correct nucleobase orientation. Hothorn et al. (2011) stated: "Although the structure strongly suggests that N3-, N7-, and N9-substituted cytokinins should not bind effectively to AHK4, previous studies using hormone binding assays in live E. coli cells have shown that, for example, trans-zeatin riboside (tZR) N9-substituted cytokinin binds to AHK4 with high affinity. To resolve this discrepancy, we purified AHK4 in the presence of tZR and crystallized the complex. However, we cannot detect any density differences for the ribose portion of tZR, suggesting that the ligand may have undergone hydrolysis during purification, a reaction that can also occur in hormone binding assays in living cells." Modeling of the 3D structures of the sensory modules of CK receptors from different species (Arabidopsis, maize, tobacco, potato, rice) pointed to their high structural conservation and especially of their binding sites, adjusted to bind specifically CK nucleobases and not ribosides (Lomin et al. 2015, 2018b; Savelieva et al. 2018; Arkhipov et al. 2019; and data not published). Notably, also iP9G and tZ9G, which carry glucosides at the N9 position of iP and tZ, did not inhibit binding of the respective free bases on AHK3 and CRE1/AHK4 receptors (Šmehilová et al. 2016). This result supports the viewpoint that attachment of groups at the N9 position interferes with receptor binding.
Plant membrane assay system
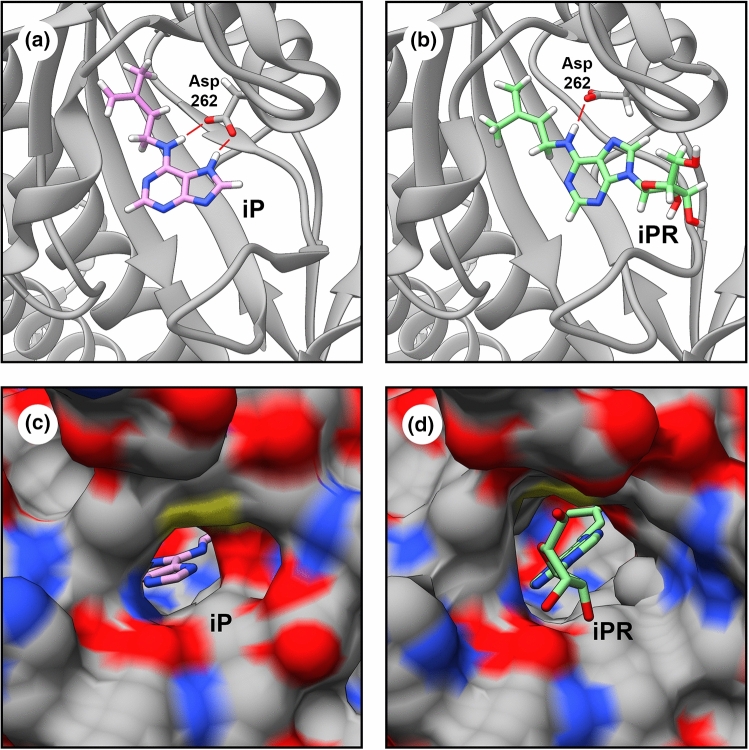
The apparent conflict between the results obtained in the E. coli test system and those obtained by other methods was resolved by Lomin et al. (2015). They described a new method for quantitative analysis of the CK receptor–ligand interaction in microsomes isolated from tobacco leaves transiently expressing one or the other individual CK receptor. In comparative experiments, it was found that the apparent activity of CK ribosides, in contrast to nucleobases, is strongly dependent on the test system. In the E. coli system, ribosides repeatedly showed relatively high affinity to the receptors, but in the system with plant microsomes, the apparent affinity of ribosides for receptors (Ka) dropped by orders of magnitude. The binding of CK ribosides to receptors in the plant test system was in most cases so small that the ligand–receptor affinity (Ka or Kd) could be not be determined (Lomin et al. 2015). In subsequent experiments using the plant test system, it turned out that not only addition of ribose but also of other substituents at the N9-position of the adenine heterocycle dramatically decreases the ligand affinity to receptors, turning biologically active adenine derivatives into inactive compounds (Savelieva et al. 2018). A detailed structural analysis showed that due to N9 substitution, a H-bond donor cannot emerge at N7, this H-bond with Asp262 in the ligand-binding domain playing a key role in the ligand-specific recognition (Fig. 1). Moreover, hydrogen bonding of CK ribosides with Leu194 is realized through N3 instead of N9, leading to the elimination of a favorable interaction with a water molecule. Furthermore, a ribose residue in the ligand molecule prevents the closure of the cavity by the β6-β7 loop (Fig. 1). Without site closure, specific high-affinity ligand binding by the CK receptor leading to receptor activation is hardly possible (Lomin et al. 2015).
Fig. 1.
Schematic presentation of the CK binding site in the sensory module of the CRE1/AHK4 receptor (AHK4sm) of A. thaliana. a and c Crystal structure of the AHK4sm–iP complex (PDB ID: 3T4J) (Hothorn et al. 2011). b and d Protein–ligand docking of the iPR molecule into AHK4sm, performed using the Rosetta software (Rosie web service) (DeLuca et al. 2015). a and b 3D presentations showing the binding of iP and iPR to the ligand binding site of CRE1/AHK4. Numbering of the Asp residue is according to the crystal structure of CRE1/AHK4 (Hothorn et al. 2011). c and d Comparison of iP and iPR positions in the ligand-binding pocket of CRE1/AHK4sm. The protruding "tail" of the ribose of iPR does not fit inside the ligand-binding pocket. Visualization was performed using UCSF Chimera software (Pettersen et al. 2004)
Receptor binding vs. receptor activation
Unfortunately, the commented review (Nguyen et al. 2021) considers neither the information obtained by 3D modeling of the receptor ligand-binding domain nor the results from binding studies performed with plant membranes. The opinion put forward was based mainly on the correlation of high concentrations of CK ribosides with biological activity and binding studies performed with the E. coli test system, including data of Romanov et al. (2006), Stolz et al. (2011) and Kuderová et al. (2015). But as it was mentioned above, the E. coli assay was shown to be not appropriate for CK riboside investigation. A further work by Daudu et al. (2017), which was cited to prove CK riboside activity but not discussed, assessed semiquantitatively the ligand preference of CK receptors of apple tree using a yeast test system. It was concluded that of the five tested CK receptors of apple, four do not respond to CK ribosides at all. Only one receptor, MaCHK2, was apparently able to react when exposed to CK ribosides, but its activation by ribosides required 500–1000 fold higher concentrations compared to the cognate tZ– and iP–nucleobases (Daudu et al. 2017). Taking into account that some tiny contamination (~ 0.1%) of the ribosides with cognate nucleobases can be hardly ruled out, it is doubtful that this particular apple tree receptor recognizes CK ribosides as activating ligands in planta. Another recent article by Jaworek et al. (2020) showing that CK ribosides are able to bind with high affinity to poplar CK receptors was not mentioned by Nguyen et al. either. In this article, the conversion of iPR to iP during the bacterial binding assay was indeed recorded but found to be low, which argued for relatively strong binding of iPR themselves to poplar CK receptors. To explain this result, Jaworek et al. (2020) suggested an induced fit, rearrangement of the loop close to the nucleobase or eventually a nucleobase flip that may occur in planta leading to CK riboside binding by the receptor. However, despite this binding, the activation of the poplar CK receptors by iPR was not reported (Jaworek et al. 2020). Indeed, high affinity binding of a compound to the receptor is not necessarily associated with the induction of hormonal activity. In a study of numerous CK derivatives recognized by CK receptors, inert ones or even compounds with opposite (inhibitory) action were quite common (Savelieva et al. 2018).
N9-bound fluorescent probe of CK
Lack of activation of a receptor by a binding compound was also found in a study using a fluorescently labeled CK that interacts with the receptor CK binding site. This fluorescent probe contains 7-nitro-2,1,3-benzoxadiazole (NBD), a small fluorophore comparable in size to a ribose residue and attached to the N9 position of iP (Kubiasová et al. 2020). Docking simulations using the CRE1/AHK4–iP crystal structure suggested that iP-NBD may be fully embedded into the active sites of AHK receptors. Competitive binding assays with E. coli expressing either AHK3 or CRE1/AHK4 showed that iP-NBD competes for receptor binding with radiolabelled trans-zeatin (tZ), with Ki ~ 37 μM in AHK3 and Ki ~ 1.4 μM in CRE1/AHK4 assays. This means that the attached N9 residue decreased the affinity of modified iP to CK receptors by about three to four orders of magnitude. Despite iP-NBD fitted into the CK-binding pockets of the receptors, it failed to trigger a CK response in an E. coli receptor activation assay with recombinant AHK3 and CRE1/AHK4 receptors, and it failed as well to induce the CK reporter genes TCS::GFP and ARR5::GUS in planta (Kubiasová et al. 2020). This strongly suggested that similar to CK ribosides and other N9 derivatives, iP-NBD binds to CK receptors but does not activate them, thus working as a partial CK receptor antagonist. Consistent with its highest affinity to CRE1/AHK4, iP-NBD had a pronounced inhibitory effect on the induction of a CK response gene (ARR5) in the ahk2ahk3 double mutant with only the CRE1/AHK4 receptor functional when compared to either wild type or the other CK receptor double mutants (ahk3ahk4 and ahk2ahk4) (Kubiasová et al. 2020).
CK mutants
Additional results supporting CK activity of the free bases but not of the ribosides come from the analysis of CK mutants of Arabidopsis, however these publications were not considered either by Nguyen et al. (2021). In the LOG sextuple Arabidopsis mutants, which are deficient in the production of iP and tZ from the nucleotides iPRP and tZRP, the iPR and tZR content was about six- and threefold higher compared to wild-type plants. However, this excess of iPR and tZR could not compensate for a much lower deficiency in iP and tZ bases (Tokunaga et al. 2012). Furthermore, mutant analyses and grafting experiments have shown that the activity of root-derived tZR depends on its metabolic conversion to the free base through the LONELY GUY(LOG)-mediated pathway (Osugi et al. 2017), possibly involving an adenosine kinase activity as a first step to form intracellular ZRMP as an intermediate compound (Lomin et al. 2018a; Romanov et al. 2018; Sakakibara 2021).
Concluding remarks
In summary, structural data, receptor-ligand interaction studies and mutant analyses argue, in our opinion quite convincingly, against a genuine biological activity of CK ribosides. The hormonal activity of CK ribosides found in experimental systems is most likely due to a rapid hydrolytic removal of the ribose moiety thus releasing the active CK base. The prevailing viewpoint that CK nucleobases are the only active forms is documented in more recent fundamental reviews (Kieber and Schaller 2018; Oschepkov et al. 2020). Nevertheless, despite the lack of own CK activity the ribosides are an important cellular source of active CK corresponding with their high concentrations in tissues with high CK activity. Therefore, it would be important to explore the metabolism of CK ribosides and their role as a transport form, as well as active CK precursors, in more detail. In particular, identification of the genes coding for specific enzymes catalyzing the interconversion between CK free bases and ribosides will be of great value and open up new avenues of research.
Author contribution statement
GAR and TS designed the project, GAR wrote the draft, TS and GAR prepared the final version.
Acknowledgements
We thank Dmitry Arkhipov for the figure preparation. The work was partly supported by the Ministry of Science and Higher Education of the Russian Federation (theme No. 121033000137-1).
Abbreviations
- CK
Cytokinin
- cZR
cis-Zeatin riboside
- iP
Isopentenyladenine
- iPR
Isopentenyladenine riboside
- tZ
trans-Zeatin
- tZR
trans-Zeatin riboside
Funding
Open Access funding enabled and organized by Projekt DEAL.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Georgy A. Romanov, Email: gromanov@yahoo.com, Email: gar@ippras.ru
Thomas Schmülling, Email: tschmue@zedat.fu-berlin.de.
References
- Arkhipov DV, Lomin SN, Myakushina YuA, et al. Modeling of protein–protein interactions in cytokinin signal transduction. Int J Mol Sci. 2019;20:2096. doi: 10.3390/ijms20092096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudu D, Allion E, Liesecke F, et al. CHASE-containing histidine kinase receptors in apple tree: from a common receptor structure to divergent cytokinin binding properties and specific functions. Front Plant Sci. 2017;8:1614. doi: 10.3389/fpls.2017.01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca S, Khar K, Meiler J. Fully flexible docking of medium sized ligand libraries with Rosetta ligand. PLoS ONE. 2015;10:e0132508. doi: 10.1371/journal.pone.0132508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JE, Sood CK, Buckwalter B, McChesney JD. The metabolism and biological activity of a 9-substituted cytokinin. Plant Physiol. 1971;47:275–281. doi: 10.1104/pp.47.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht SM, Bock RM, Schmitz RY, et al. Question of the ribosyl moiety in the promotion of callus growth by exogenously added cytokinins. Biochemistry. 1971;10:4224–4228. doi: 10.1021/bi00799a011. [DOI] [PubMed] [Google Scholar]
- Hothorn M, Dabi T, Chory J. Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase AHK4. Nature Chem Biol. 2011;7:766–768. doi: 10.1038/nchembio.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, et al. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Jaworek P, Tarkowski P, Hluska T, et al. Characterization of five CHASE-containing histidine kinase receptors from Populus × canadensis cv. Robusta sensing isoprenoid and aromatic cytokinins. Planta. 2020;251:1. doi: 10.1007/s00425-019-03297-x. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE. Cytokinin signaling in plant development. Development. 2018;145:dev149344. doi: 10.1242/dev.149344. [DOI] [PubMed] [Google Scholar]
- Kubiasová K, Montesinos JC, Šamajová O, et al. Cytokinin fluoroprobe reveals multiple sites of cytokinin perception at plasma membrane and endoplasmic reticulum. Nature Comm. 2020;11:4285. doi: 10.1038/s41467-020-17949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuderová A, Gallová L, Kuricová K, et al. Identification of AHK2- and AHK3-like cytokinin receptors in Brassica napus reveals two subfamilies of AHK2 orthologues. J Exp Bot. 2015;66:339–353. doi: 10.1093/jxb/eru422. [DOI] [PubMed] [Google Scholar]
- Letham DS. Zeatin, a factor inducing cell division, isolated from Zea mays. Life Sci. 1963;2:569–573. doi: 10.1016/0024-3205(63)90108-5. [DOI] [PubMed] [Google Scholar]
- Lomin SN, Krivosheev DM, Steklov MY, et al. Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J Exp Bot. 2015;66:1851–1863. doi: 10.1093/jxb/eru522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomin SN, Myakushina YA, Arkhipov DV, et al. Studies of cytokinin receptor–phosphotransmitter interaction provide evidence for the initiation of cytokinin signalling in the endoplasmic reticulum. Funct Plant Biol. 2018;45:192–202. doi: 10.1071/FP16292. [DOI] [PubMed] [Google Scholar]
- Lomin SN, Myakushina YA, Kolachevskaya OO, et al. Cytokinin perception in potato: new features of canonical players. J Exp Bot. 2018;69:3839–3853. doi: 10.1093/jxb/ery199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CO, Skoog F, von Saltza NM, Strong FM. Kinetin, a cell division factor from deoxyribonucleic acid. J Am Chem Soc. 1955;77:1329–1334. doi: 10.1021/ja01610a105. [DOI] [Google Scholar]
- Nguyen HN, Nguyen TQ, Kisiala AB, Emery RJN. Beyond transport: cytokinin ribosides are translocated and active in regulating the development and environmental responses of plants. Planta. 2021;254:45. doi: 10.1007/s00425-021-03693-2. [DOI] [PubMed] [Google Scholar]
- Oshchepkov MS, Kalistratova AV, Savelieva EM, et al. Natural and synthetic cytokinins and their applications in biotechnology, agrochemistry and medicine. Russian Chem Rev. 2020;89:787–810. doi: 10.1070/RCR4921. [DOI] [Google Scholar]
- Osugi A, Kojima M, Takebayashi Y, et al. Systemic transport of trans-zeatin and its precursor have differing roles in Arabidopsis shoots. Nature Plants. 2017;3:17112. doi: 10.1038/nplants.2017.112. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Romanov GA, Spíchal L, Lomin SN, et al. A live cell hormone-binding assay on transgenic bacteria expressing a eukaryotic receptor protein. Anal Biochem. 2005;347:129–134. doi: 10.1016/j.ab.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Romanov GA, Lomin SN, Schmülling T. Biochemical characteristics and ligand-properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J Exp Bot. 2006;57:4051–4058. doi: 10.1093/jxb/erl179. [DOI] [PubMed] [Google Scholar]
- Romanov GA, Lomin SN, Schmülling T. Cytokinin signaling: from the ER or from the PM? That is the question! New Phytol. 2018;218:41–53. doi: 10.1111/nph.14991. [DOI] [PubMed] [Google Scholar]
- Sakakibara H. Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- Sakakibara H. Cytokinin biosynthesis and transport for systemic nitrogen signaling. Plant J. 2021;105:421–430. doi: 10.1111/tpj.15011. [DOI] [PubMed] [Google Scholar]
- Savelieva EM, Oslovsky VE, Karlov DS, et al. Cytokinin activity of N6-benzyladenine derivatives assayed by interaction with the receptors in planta, in vitro, and in silico. Phytochemistry. 2018;149:161–177. doi: 10.1016/j.phytochem.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Skoog F, Armstrong DJ. Cytokinins. Annu Rev Plant Physiol. 1970;21:359–384. doi: 10.1146/annurev.pp.21.060170.002043. [DOI] [Google Scholar]
- Šmehilová M, Dobrůšková J, Novák O, et al. Cytokinin-specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front Plant Sci. 2016;7:1264. doi: 10.3389/fpls.2016.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spíchal L, Rakova NYu, Riefler M, et al. Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol. 2004;45:1299–1305. doi: 10.1093/pcp/pch132. [DOI] [PubMed] [Google Scholar]
- Spiess LD. Comparative activity of isomers of zeatin and ribosyl-zeatin on Funaria hygrometrica. Plant Physiol. 1975;55:583–585. doi: 10.1104/pp.55.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Riefler M, Lomin SN, et al. The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J. 2011;67:157–168. doi: 10.1111/j.1365-313X.2011.04584.x. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, et al. The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 2001;42:107–113. doi: 10.1093/pcp/pce037. [DOI] [PubMed] [Google Scholar]
- Tokunaga H, Kojima M, Kuroha T, et al. Arabidopsis LONELY GUY (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 2012;69:355–365. doi: 10.1111/j.1365-313X.2011.04795.x. [DOI] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, et al. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 2001;42:1017–1023. doi: 10.1093/pcp/pce127. [DOI] [PubMed] [Google Scholar]