Abstract
The rapid accumulation of mutations in the SARS-CoV-2 Omicron variant that enabled its outbreak raises questions as to whether its proximal origin occurred in humans or another mammalian host. Here, we identified 45 point mutations that Omicron acquired since divergence from the B.1.1 lineage. We found that the Omicron spike protein sequence was subjected to stronger positive selection than that of any reported SARS-CoV-2 variants known to evolve persistently in human hosts, suggesting a possibility of host-jumping. The molecular spectrum of mutations (i.e., the relative frequency of the 12 types of base substitutions) acquired by the progenitor of Omicron was significantly different from the spectrum for viruses that evolved in human patients but resembled the spectra associated with virus evolution in a mouse cellular environment. Furthermore, mutations in the Omicron spike protein significantly overlapped with SARS-CoV-2 mutations known to promote adaptation to mouse hosts, particularly through enhanced spike protein binding affinity for the mouse cell entry receptor. Collectively, our results suggest that the progenitor of Omicron jumped from humans to mice, rapidly accumulated mutations conducive to infecting that host, then jumped back into humans, indicating an inter-species evolutionary trajectory for the Omicron outbreak.
Keywords: SARS-CoV-2, Omicron, Evolutionary origins, Molecular spectrum of mutations, Spike-ACE2 interaction, Receptor-binding domain
Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by the SARS-CoV-2 RNA virus, has led to significant illness and death worldwide. The SARS-CoV-2 Omicron variant was first reported in South Africa on November 24th, 2021, and was designated as a variant of concern (VOC) within two days by the World Health Organization (WHO) based on the increase in infections attributable to this variant in South Africa (i.e., Omicron outbreak). In addition, the open reading frame encoding the spike protein (ORF S) of Omicron harbors an exceptionally high number of mutations. These mutations are particularly relevant to infection characteristics because the SARS-CoV-2 spike protein is well-known to mediate viral entry into the host cell by interacting with angiotensin-converting enzyme 2 (ACE2) on the cell surface (Zhou et al., 2020). In addition, the spike protein is also a target for vaccine development and antibody-blocking therapy (Huang et al., 2020; Martinez-Flores et al., 2021).
The proximal origins of Omicron have quickly become a controversial topic of heated debate in the scientific and public health communities (Callaway, 2021; Kupferschmidt, 2021). Many mutations detected in Omicron were rarely reported among previously sequenced SARS-CoV-2 variants (Shu and McCauley, 2017; Hadfield et al., 2018), leading to three prevalent hypotheses regarding its evolutionary history. The first hypothesis is that Omicron could have ‘cryptically spread’ and circulated in a population with insufficient viral surveillance and sequencing. Second, Omicron could have evolved in a chronically infected COVID-19 patient, such as an immunocompromised individual who provided a suitable host environment conducive to long-term intra-host virus adaptation. The third possibility is that Omicron could have accumulated mutations in a nonhuman host and then jumped into humans. Currently, the second scenario represents the most popular hypothesis regarding the proximal origins of Omicron (Callaway, 2021; Kupferschmidt, 2021).
The first two hypotheses assume that Omicron acquired these mutations in humans (collectively referred to as ‘human origin hypothesis’ hereafter), while the third assumes that Omicron acquired mutations in a nonhuman species. Based on our previous work in viral evolution (Shan et al., 2021), we hypothesized that the host species in which Omicron acquired its particular set of mutations could be determined by analyzing the molecular spectra of mutations (i.e., the relative frequency of the 12 types of base substitutions). In previous work, we showed that many de novo mutations in RNA virus genomes are generated in a replication-independent manner and are highly dependent on mutagenic mechanisms specific to the host cellular environment, resulting in overrepresentation with specific mutation types. For example, reactive oxygen species (ROS) can oxidize guanine to 8-oxoguanine and thereby induce the G>U transversion (Li et al., 2006; Kong and Lin, 2010), while cytidine deaminases can induce RNA editing such as C>U transitions (Blanc and Davidson, 2010; Harris and Dudley, 2015). Consistent with this phenomenon, viruses belonging to different orders (e.g., poliovirus, Ebola virus, and SARS-CoV-2) were found to exhibit similar molecular spectra of mutations when evolving in the same host species, while members of the same virus species exhibit divergent molecular spectra when evolving in different host species (Shan et al., 2021). Since de novo mutations can thus strongly influence the molecular spectrum of mutations that accumulate during virus evolution in a host-specific manner, the host species in which Omicron acquired its mutations could be determined by analyzing information carried by the mutations themselves.
In this study, we identified mutations acquired by Omicron before its outbreak and tested whether the molecular spectrum of these mutations was consistent with the cellular environment of human hosts. Prominent dissimilarities were observed between the molecular spectrum of Omicron and a relatively comprehensive set of molecular spectra from variants known to have evolved in humans, including those of three isolates from chronic COVID-19 patients. Therefore, we next examined the molecular spectra of mutations obtained from a wide range of host mammals for comparison with that of Omicron. Finally, we used molecular docking-based analyses to investigate whether the mutations in the Omicron spike protein could be associated with adaptation to the host species inferred from molecular spectrum analysis. Our study provides insight into the evolutionary trajectory and proximal origins of Omicron through careful scrutiny of its mutations and suggests strategies for avoiding future outbreaks caused by SARS-CoV-2 variants proliferating in wild animals.
Results
Over-representation of nonsynonymous mutations in Omicron ORF S suggests strong positive selection
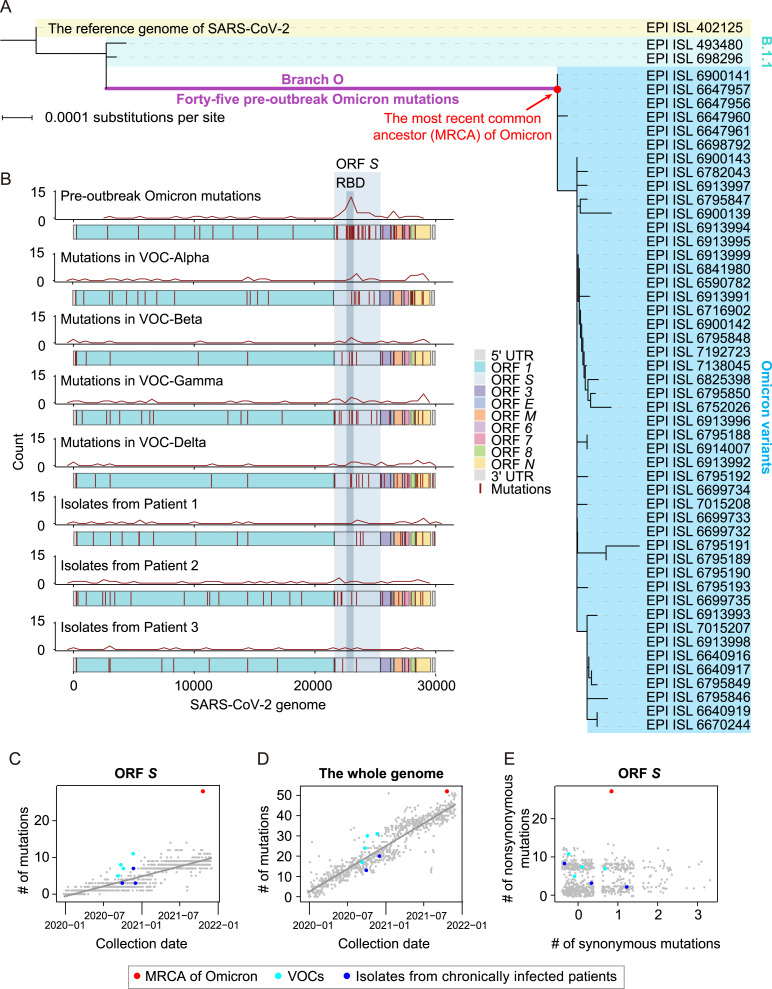
To first identify mutations that accumulated in the SARS-CoV-2 genome prior to the Omicron outbreak, we constructed a phylogenetic tree that included the genomic sequences of the reference SARS-CoV-2 (Wu et al., 2020a), two variants in the B.1.1 lineage, which were genetically close to Omicron (based on the results of BLASTn), and 48 Omicron variants sampled before November 15th, 2021 (Fig. 1 A). These two B.1.1 variants were sampled during April 22nd–May 5th, 2020, which suggested that the progenitor of Omicron diverged from the B.1.1 lineage roughly in mid-2020. Intermediate versions have gone largely undetected, thus resulting in an exceptionally long branch leading to the most recent common ancestor (MRCA) of Omicron in the phylogenetic tree (Fig. 1A). We hereafter refer to this long branch as Branch O.
Fig. 1.
The characterization of pre-outbreak Omicron mutations. A: The phylogenetic tree of Omicron variants, including the reference genome of SARS-CoV-2, two B.1.1 variants, and 48 Omicron variants. A total of 45 pre-outbreak Omicron point mutations in the long branch (Branch O, labeled in purple) leading to the MRCA of Omicron (dot in red) in the phylogenetic tree are shown in Fig. S1. B: The genomic distribution of the 45 pre-outbreak Omicron mutations, the mutations detected in each progenitor of the other four VOCs (i.e., Alpha, Beta, Gamma, and Delta), and the mutations identified from the SARS-CoV-2 isolates of three chronically infected patients. The density curves for mutations were generated by the geom_freqpoly function in R. C: Number of mutations that accumulated in ORF S of the MRCA of Omicron (red), the other four VOCs (cyan), and three SARS-CoV-2 isolates from chronically infected patients (blue), against the date of sample collection. SARS-CoV-2 variants randomly sampled (one variant per day) are shown in grey, and the grey line represents their linear regression. D: Similar to (C), for the whole genome. The dot for a variant isolated from a chronically infected patient is overlapped with a VOC. E: A scatterplot shows the numbers of synonymous and nonsynonymous mutations in ORF S (jittered in order to reduce overplotting). UTR, untranslated region; RBD, receptor-binding domain.
We identified 45 point mutations that were introduced in Branch O (hereafter referred to as ‘pre-outbreak Omicron mutations’; Fig. S1). We observed that the pre-outbreak Omicron mutations were over-represented in ORF S (P = 1.2 × 10−13, binomial test with the expected probability equal to the length of ORF S relative to the SARS-CoV-2 genome; Fig. 1B), especially in the coding region of the receptor-binding domain (RBD) (P = 1.1 × 10−13, Fig. 1B). We further identified mutations in the other four SARS-CoV-2 VOCs (i.e., Alpha, Beta, Gamma, and Delta) as well as those in the SARS-CoV-2 variants isolated from three chronically infected patients (Kemp et al., 2021; Truong et al., 2021), but did not observe such a level of over-representation of mutations in ORF S or RBD region as in the pre-outbreak Omicron mutations (Fig. 1B).
To test if the rate at which mutations accumulated in ORF S was accelerated in Branch O, we randomly sampled one SARS-CoV-2 variant per day since December 24th, 2019, from the Global Initiative on Sharing All Influenza Data (GISAID) (Shu and McCauley, 2017) to compare mutation accumulation rates among different variants. We found that mutations accumulated in ORF S at a rate of ∼0.45 mutations per month on average. In sharp contrast, 27 mutations accumulated in ORF S in Branch O during the 18 months spanning May 2020–November 2021, equivalent to ∼1.5 mutations per month, or ∼3.3 times faster than the average rate of other variants (Fig. 1C).
Counting mutations across the whole SARS-CoV-2 genome indicated that Omicron acquired mutations in the genome at a similar rate to other variants (Fig. 1D), suggesting that the accelerated evolutionary rate of ORF S could not be explained by an overall elevated mutation rate in Omicron progenitors. In light of these findings, we hypothesized that positive selection could have helped accelerate the evolutionary rate of ORF S. To test this hypothesis, we sought to infer the strength of positive selection by estimating the ratio of nonsynonymous to synonymous mutations. Twenty-six of the 27 pre-outbreak mutations in the ORF S of Omicron were nonsynonymous (Fig. 1E), resulting in a d N/d S ratio of 6.64, significantly greater than a d N/d S of 1.00 (P = 0.03, Fisher’s exact test). These results indicated that positive selection contributed to increasing the mutation rate in ORF S in Branch O.
To test if such a level of positive selection is common among SARS-CoV-2 variants, we counted the number of nonsynonymous and synonymous mutations in ORF S in the other four VOCs as well as in the variants isolated from three chronically infected patients (Kemp et al., 2021; Truong et al., 2021). None of these other VOCs or isolates exhibited comparable numbers of nonsynonymous mutations as that of mutations in Branch O (Fig. 1E). These observations strongly suggested that the Omicron variant had undergone a strong positive selection for the spike protein that no other known SARS-CoV-2 variants evolved in humans had been subjected to. Considering that the spike protein determines the host range of a coronavirus (i.e., which organisms it can infect), we therefore hypothesized that the progenitor of Omicron might host-jump from humans to a nonhuman species because this process would require substantial mutations in the spike protein for rapid adaptation to a new host.
The molecular spectrum of pre-outbreak Omicron mutations is inconsistent with an evolutionary history in humans
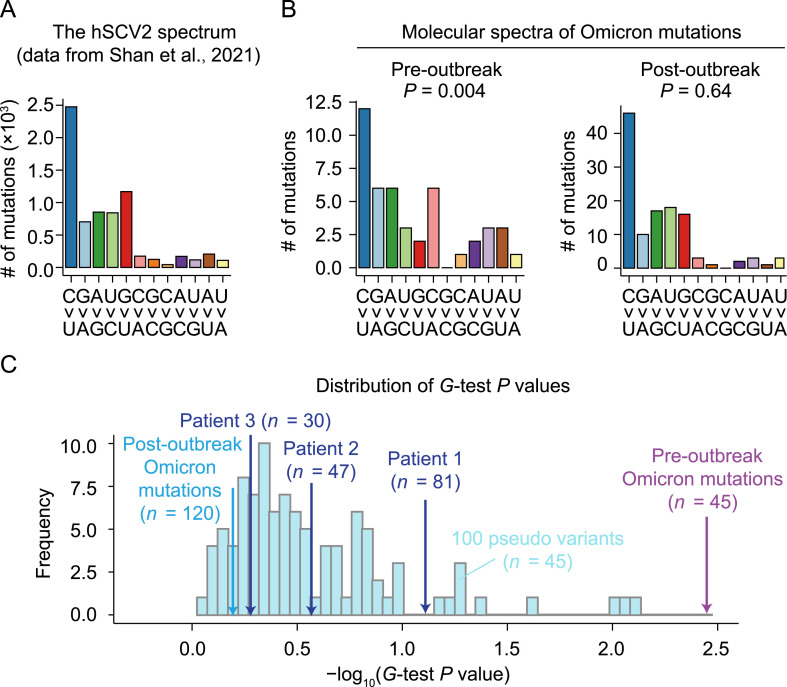
Previous studies showed that the molecular spectrum of mutations that accumulate in a viral genome reflects a host-specific cellular environment (Deng et al., 2021; Shan et al., 2021). To test the human origin hypothesis of Omicron, we compared the molecular spectrum of the 45 pre-outbreak Omicron mutations with the ‘standard’ molecular spectrum for SARS-CoV-2 variants known to have evolved strictly in humans (hereafter referred to as ‘the hSCV2 spectrum’; Fig. 2 A). The hSCV2 spectrum included 6986 point mutations that were compiled from 34,853 high-quality sequences of SARS-CoV-2 variants isolated from patients worldwide (Shan et al., 2021). We found that the molecular spectrum of the pre-outbreak Omicron mutations was significantly different from the hSCV2 spectrum (P = 0.004, G-test; Fig. 2B). In particular, as in the hSCV2 spectrum, transitions were more abundant than transversions, and C>U mutation was more abundant than its complementary mutation G>A. However, a hallmark of RNA virus mutations when evolving in humans—a higher rate of G>U mutation than its complementary mutation C>A (Panchin and Panchin, 2020; De Maio et al., 2021; Deng et al., 2021; Shan et al., 2021) that is likely caused by cellular ROS—was absent in the pre-outbreak Omicron mutations.
Fig. 2.
Comparison of the molecular spectrum of pre-outbreak Omicron mutations and spectra of mutations known to accumulate in humans. A: The molecular spectrum of viral mutations that accumulated in humans (the hSCV2 spectrum). B: The molecular spectra of pre-outbreak and post-outbreak Omicron mutations. P values were given by G-tests against the hSCV2 spectrum. C: The distribution of P values (given by G-tests) of 100 pseudo samples that were downsampled from the hSCV2 spectrum. The number of mutations (n) of each pseudo sample was equal to 45. SARS-CoV-2 data of Patient 1 were retrieved from Kemp et al. (2021), and those of Patients 2 and 3 were retrieved from Truong et al. (2021).
To exclude the possibility that this apparent difference in the molecular spectrum was caused by the relatively small number of pre-outbreak Omicron mutations, we generated 100 ‘pseudo’ variants in silico by randomly down sampling 45 mutations from the hSCV2 spectrum. None of the pseudo variants showed smaller P values (based on G-tests) than that obtained using the pre-outbreak Omicron mutations (Fig. 2C), nor did the SARS-CoV-2 isolates with mutations known to be acquired in the three chronically infected patients (# of mutations are 30, 47, and 81; Fig. 2C). These observations indicated that the difference between the molecular spectrum of pre-outbreak Omicron mutations and the hSCV2 spectrum could not be strictly attributed to statistical randomness.
To exclude the possibility that some mutations which occurred early in the evolution of Omicron (e.g., mutations in the RNA-dependent RNA polymerase) distorted the molecular spectrum of mutations that accumulated afterward, we identified 120 point mutations that occurred after the Omicron outbreak by screening 695 Omicron variants collected spanning November 8th–December 7th, 2021 (hereafter referred to as ‘post-outbreak Omicron mutations’). The molecular spectrum of these post-outbreak Omicron mutations was not significantly different from the hSCV2 spectrum (P = 0.64, G-test; Fig. 2B and 2C). This finding indicated that Omicron would acquire mutations following the same molecular spectrum as other SARS-CoV-2 variants during its evolution in human hosts. Collectively, these molecular spectrum analyses revealed that pre-outbreak Omicron mutations were unlikely to have been acquired in humans.
The molecular spectrum of pre-outbreak Omicron mutations is consistent with an evolutionary history in mice
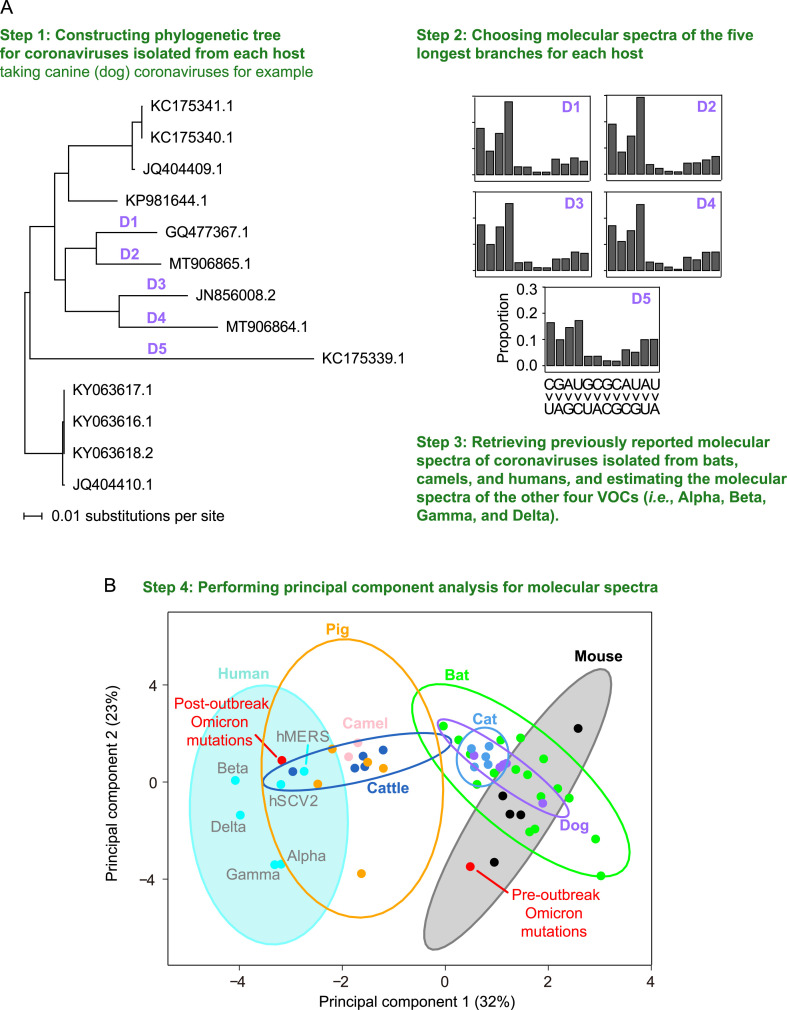
In light of our findings that Omicron may have evolved in another host before its outbreak, we next sought to determine the nonhuman host species in which these mutations accumulated. To this end, we first characterized the molecular spectra of coronaviruses that evolved in different host species for comparison with that of Omicron. Specifically, we retrieved 17 sequences of murine hepatitis viruses, 13 canine coronaviruses, 54 feline coronaviruses, 23 bovine coronaviruses, and 110 porcine deltacoronaviruses (Table S1), constructed the phylogenetic tree for the coronaviruses isolated from each host species (canine coronavirus as an example shown in Fig. 3 A and the rest are shown in Fig. S2), and identified the mutations that accumulated in each branch (Fig. 3A). The longest five external branches of each host species were used for the subsequent analysis (see Materials and methods). We also included some previously reported molecular spectra (Shan et al., 2021), including 17 spectra of mutations acquired by SARS-CoV-, SARS-CoV-2-, and MERS-CoV-related coronaviruses during their evolution in bats, two spectra of camel MERS-CoV, one spectrum estimated from 807 MERS-CoV mutations accumulated in human (the hMERS spectrum), as well as the hSCV2 spectrum. Furthermore, we also included the molecular spectrum of mutations identified in an early variant of each of the other four VOCs.
Fig. 3.
The similarity in molecular spectra between Omicron and coronaviruses isolated from various mammalian species. A: A schematic shows the workflow for analyzing the similarity in molecular spectra across various hosts. B: The principal component analysis plot depicts the molecular spectra of virus mutations that accumulated in humans and various host species. Dots were colored according to the corresponding host species. The 95% confidence ellipses are shown for each host species. VOC, variant of concern.
We performed principal component analysis to reduce the dimensionality of the molecular spectrum of mutations and subsequently visualized the data using the first two principal components (Fig. 3B). Consistent with the results of our previous study (Shan et al., 2021), drawing 95% confidence ellipses for each host species showed that the molecular spectra clustered according to their respective hosts (Fig. 3B), likely because viruses evolving in the same host species share the mutagens specific to that host’s cellular environment. In supporting this point, the molecular spectrum of post-outbreak Omicron mutations (which are known to have accumulated in humans) was located within the human 95% confidence ellipse. In contrast, the molecular spectrum of pre-outbreak Omicron mutations was within the mouse ellipse, suggesting that the pre-outbreak mutations accumulated in a rodent (in particular a mouse) host.
Pre-outbreak Omicron mutations in the spike protein significantly overlap with mutations in mouse-adapted SARS-CoV-2
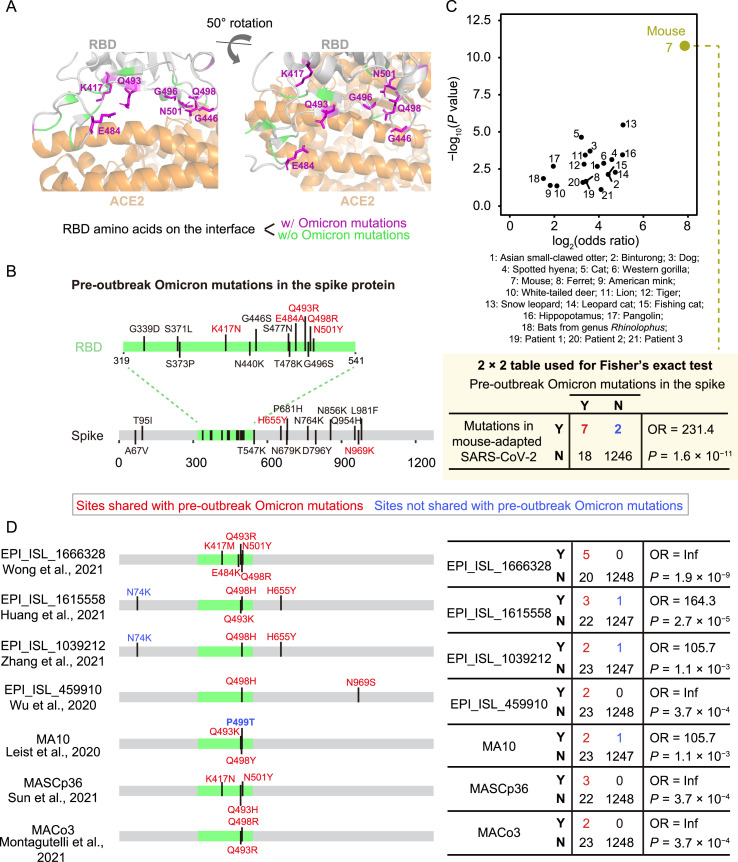
Mice were previously reported to serve as poor hosts for SARS-CoV-2 because the spike protein of early SARS-CoV-2 variants exhibited low-affinity interactions with mouse ACE2 (Lam et al., 2020; Zhou et al., 2020; Ren et al., 2021; Wong et al., 2021). However, over the course of the pandemic, SARS-CoV-2 variants emerged that could infect mice. For example, variants harboring the spike mutation N501Y, which are relatively common in human patients (24.7%, CoV-GLUE-Viz, accessed on November 23rd, 2021), could infect mice (Gu et al., 2020; Leist et al., 2020; Sun et al., 2021). If the progenitor of Omicron indeed evolved in a mouse species before the Omicron outbreak, we postulated that its spike protein likely adapted through increased binding affinity for mouse ACE2. To test this possibility, we projected the pre-outbreak Omicron mutations in the spike protein onto a three-dimensional structure of the spike:ACE2 complex (Lan et al., 2020). Seven mutations (i.e., K417N, G446S, E484A, Q493R, G496S, Q498R, and N501Y) were located at the interface of ACE2 and the spike protein RBD, and could potentially affect their interactions (Fig. 4 A).
Fig. 4.
The similarity in the spike protein sequence between Omicron and SARS-CoV-2 variants isolated from various hosts. A: The interface structure between SARS-CoV-2 spike protein and human ACE2 (PDB: 6M0J). RBD residues on the interface (with a distance cut-off of 5 Å) were labeled. B: The pre-outbreak Omicron mutations in the spike protein. C: The statistical assessment on the overlapping in mutated positions of the spike protein between Omicron and SARS-CoV-2 variants isolated from chronically infected patients or nonhuman mammals using Fisher’s exact tests. The 2 × 2 contingency table for mice is shown. D: Comparison between pre-outbreak Omicron mutations and mutations detected in seven mouse-adapted SARS-CoV-2 variants in the spike protein. OR, odds ratio; Y, with a mutation at a particular site; N, without a mutation at the site.
Previous studies reported specific amino acid mutations that allow SARS-CoV-2 variants (mouse-adapted SARS-CoV-2) to use mouse ACE2 more efficiently for entry into cells (Leist et al., 2020; Wu et al., 2020b; Huang et al., 2021; Montagutelli et al., 2021; Sun et al., 2021; Wong et al., 2021; Zhang et al., 2021). In addition, previous studies have described some reverse zoonotic events (i.e., host-jumping from humans to other mammals such as mink and white-tailed deer) for SARS-CoV-2 (Chandler et al., 2021; Oude Munnink et al., 2021), and the variants isolated from these mammalian hosts presumably harbored amino acid mutations that could potentially participate in their adaptation to these hosts (Telenti et al., 2021). Thus, if the progenitor of Omicron evolved in mice and adapted to mouse ACE2, we predicted that the pre-outbreak Omicron mutations should share considerable overlap with the mutations identified in these mouse-adapted SARS-CoV-2 variants, but not those of isolates from other mammalian species.
To test this prediction, we identified the mutations in ORF S of SARS-CoV-2 variants isolated from 18 mammalian species (e.g., mice, cats, dogs, minks, and deer; Tables S2 and S3) and found that pre-outbreak Omicron mutations tended to share the same positions as the ORF S mutations identified in mice (odds ratio = 231.4, P = 1.6 × 10−11, Fisher’s exact test; Fig. 4B and 4C). In contrast, the same statistical test showed much lower odds ratios and significance levels for overlap in these mutations with other species (Fig. 4C). Pre-outbreak Omicron mutations also overlapped with some mutations detected in isolates from chronically infected patients (Kemp et al., 2021; Truong et al., 2021); however, they too showed substantially lower odds ratios and significance levels than those isolated from mice (Fig. 4C). These observations implied that the pre-outbreak Omicron mutations in ORF S promoted its adaptation to a mouse host.
We then conducted enrichment analysis for each of the seven mouse-adapted SARS-CoV-2 variants and observed statistical significance for all these variants (Fig. 4D). In particular, we observed amino acid mutations at residues 493 and 498 in five and six of the seven mouse-adapted SARS-CoV-2 variants, respectively (Fig. 4D). Identical amino acid mutations (i.e., Q493R and Q498R) were both observed in two variants (Montagutelli et al., 2021; Wong et al., 2021). Considering that these two amino acid mutations are uncommon in human patients infected by non-Omicron SARS-CoV-2 variants (0.005% and 0.002%, respectively, CoV-GLUE-Viz, accessed on November 23rd, 2021), we proposed the hypothesis that the progenitor of Omicron evolved in mice.
Pre-outbreak Omicron mutations in the RBD significantly enhance binding affinity with mouse ACE2
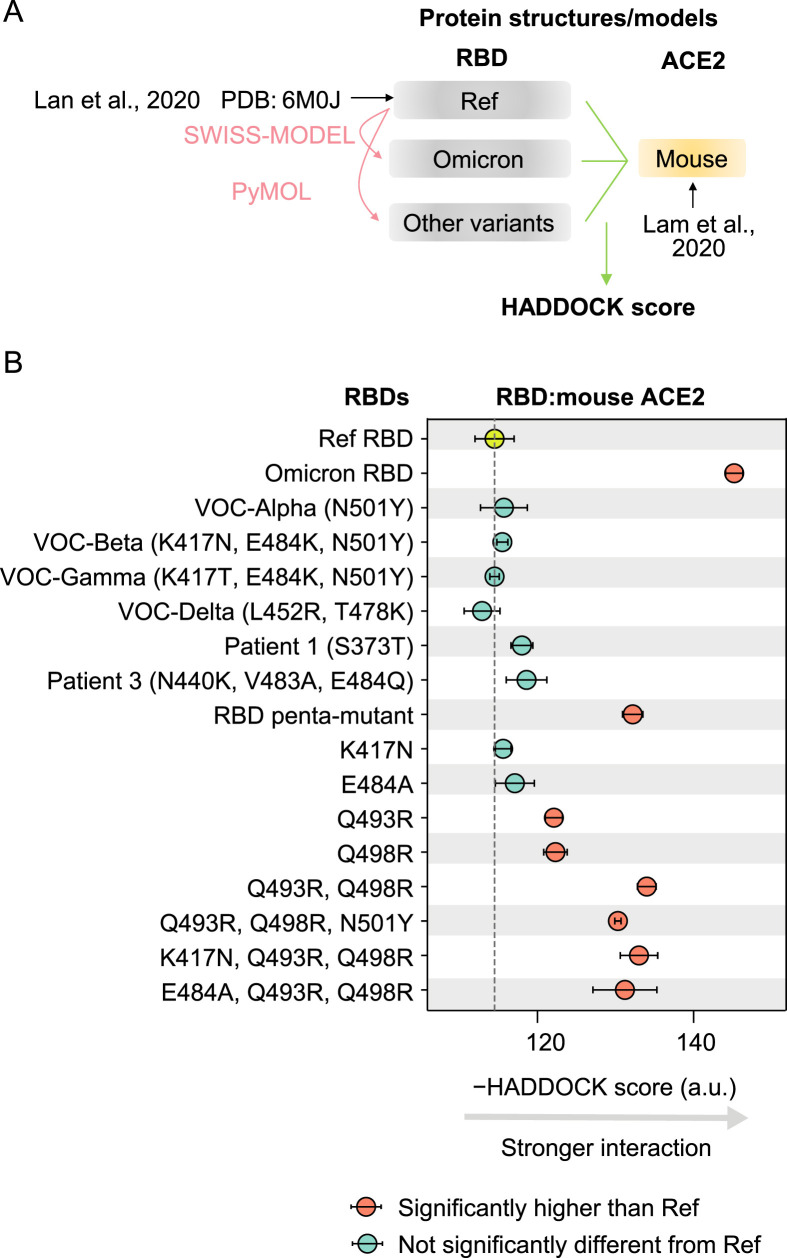
To investigate the mechanisms by which the pre-outbreak Omicron mutations in the spike protein could have contributed to its adaptation to a mouse host, we examined their interaction through molecular docking analysis of the spike protein RBD and mouse ACE2 (Fig. 5 A). Following previous studies (Lam et al., 2020; Rodrigues et al., 2020), we estimated the HADDOCK score (van Zundert et al., 2016), which is positively associated with the dissociation constant (K D, with smaller K D indicating stronger binding) of protein interactions (Kastritis and Bonvin, 2010), and can be used to predict the susceptibility of a mammalian species to infection with SARS-CoV-2 (Rodrigues et al., 2020).
Fig. 5.
Predicted binding affinities between RBD variants and mouse ACE2. A: A schematic shows the workflow to estimate the HADDOCK scores between RBD variants and mouse ACE2. B: The HADDOCK scores for the interaction of various RBD variants with the mouse ACE2. The error bars represent standard errors. Penta-mutant of RBD harbored five mutations (K417N, E484A, Q493R, Q498R, and N501Y). The result of Patient 2, who did not harbor any amino acid mutations in RBD, was not shown.
To confirm the accuracy of molecular docking-based inferences regarding the binding affinity between spike protein RBD and ACE2, we estimated HADDOCK scores for the interaction between the reference RBD and ACE2 of various mammalian species that have experimental evidence about the susceptibility to infection with the reference SARS-CoV-2. The susceptible mammalian species indeed exhibited lower HADDOCK scores (P = 0.001, t-test; Fig. S3). Furthermore, we calculated the HADDOCK score for eight experimentally determined K D values between four RBD variants and human (or mouse) ACE2 (Sun et al., 2021). The HADDOCK scores were positively correlated with the K D values in the analysis (Pearson’s correlation coefficient r = 0.93, P = 0.002; Fig. S4A–S4C). In addition, the binding affinity with mouse ACE2 was elevated in all seven mouse-adapted SARS-CoV-2 variants (five of them were statistically significant; Fig. S4D). All these observations supported the validity of molecular docking-based predictions of ACE2-binding affinity for other RBD variants.
The molecular docking-based predictions suggested that the RBD of Omicron exhibited higher binding affinity for mouse ACE2 than that of RBD encoded in the reference SARS-CoV-2 genome, further suggesting an evolutionary history in mice (Fig. 5B). And as expected, the mutations detected in the RBD of the other four VOCs of SARS-CoV-2 as well as those of variants isolated from chronically infected human patients, showed no apparent changes in their binding affinity for mouse ACE2 compared with the reference RBD (Fig. 5B).
Since five amino acid mutations were shared between Omicron and mouse-adapted SARS-CoV-2 variants in RBD (i.e., K417N, E484A, Q493R, Q498R, and N501Y; Fig. 4B), and that they together enhanced RBD binding affinity for mouse ACE2 (Fig. 5B), we next determined the individual effects of each of these five mutations. Notably, only Q493R and Q498R significantly increased the binding affinity with mouse ACE2, which was consistent with their repeated detection in mouse-adapted SARS-CoV-2 variants (Montagutelli et al., 2021; Wong et al., 2021). Indeed, docking analysis showed that Q493R/Q498R double mutation could further increase the RBD binding affinity for mouse ACE2 (Fig. 5B). By contrast, the other three mutations showed no significant effects on the binding affinity between RBD and mouse ACE2, neither in the reference RBD nor in the Q493R/Q498R double mutant (Fig. 5B), suggesting that they did not contribute to the enhanced interaction between Omicron RBD and mouse ACE2. We speculated that these mutations (K417N, E484K, and N501Y) were acquired in Omicron because they were related to escape from neutralizing antibodies, as indicated by previous studies (Li et al., 2021; Nelson et al., 2021).
The pre-outbreak Omicron mutations in the RBD showed the greatest enhanced binding affinity for mouse ACE2 among 32 mammals
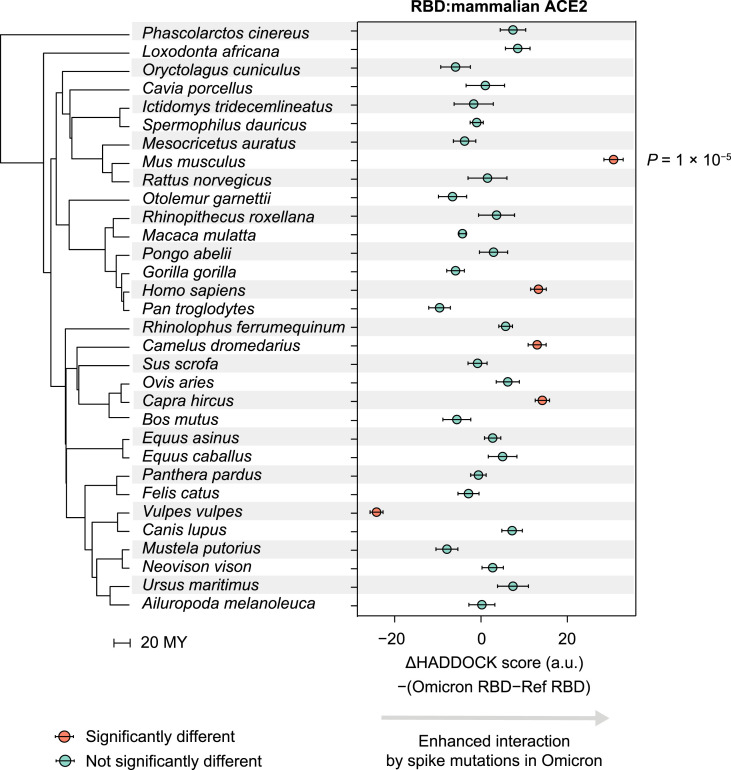
Our characterization of the molecular spectrum of mutations and observations of RBD-ACE2 interactions both suggested that mice were the most likely host species in which the progenitor of Omicron evolved. However, it remained plausible that Omicron could have evolved in another species with a similar cellular mutagen environment and ACE2 structure to that of mice. We therefore postulated that if Omicron evolved in another species, the pre-outbreak Omicron mutations in the RBD should enhance its interactions with the ACE2 of that host. To test this prediction, we applied molecular docking analysis to ACE2 from 31 other species, representing markedly different mammalian lineages (Kumar et al., 2017). We found that, compared with the RBD encoded in the reference genome, the Omicron RBD showed the highest ACE2-interaction enhancement with mice among all these mammals (Fig. 6 ), suggesting that mice were the most likely host species to influence the evolution of the progenitor of Omicron.
Fig. 6.
Predicted interaction enhancement with ACE2 of various mammalian species caused by pre-outbreak Omicron mutations in the spike protein. P values were given by two-tailed t-tests. The phylogenetic tree was constructed using TimeTree, in the unit of MY. MY, million years.
Discussion
In this study, we used the molecular spectrum of mutations of the SARS-CoV-2 Omicron variant to trace its proximal host origins. We found that the molecular spectrum of pre-outbreak Omicron mutations was inconsistent with the rapid accumulation of mutations in humans but rather suggested a trajectory in which the progenitor of Omicron experienced a reverse zoonotic event from humans to mice sometime during the pandemic (most likely in mid-2020) and accumulated mutations in a mouse host for more than one year before jumping back to humans in late-2021. While evolving in mice, the progenitor of Omicron adapted to the mouse host by acquiring amino acid mutations in the spike protein that increased its binding affinity with mouse ACE2 (this is also recently reported by another study, Cameroni et al., 2021). In addition, mutations associated with immune escape also accumulated, which may also be a contributing factor in its rapid spread in humans.
The B.1.1 variants showed the highest sequence similarities to Omicron in the GISAID database (where SARS-CoV-2-related viruses such as those isolated from bats were also deposited), strongly suggesting that the progenitor of Omicron jumped from humans, instead of another animal (such as bats), to mice. Nevertheless, it in principle remains plausible that the MRCA of Omicron was an evolutionary product of recombination between a human variant (that provided the genomic sequence for the non-RBD region, or the ‘backbone’) and a variant from another species (that provided the RBD region). Although not highlighted in our results, note that we did test this possibility by BLAST searching against the GISAID database using Omicron’s backbone sequence. The top hits were again from the B.1.1 lineage, which differed from Omicron by 31 mutations, indicating that human SARS-CoV-2 variants reported to date could not provide a backbone for Omicron. Furthermore, the molecular spectrum of these 31 mutations in Omicron was also significantly different from the hSCV2 spectrum (P = 0.008, G-test; Fig. S5), suggesting that these backbone mutations were not acquired in humans.
While we show a phylogenetically long branch leading to the MRCA of current Omicron variants (i.e., Branch O), it is worth noting that intermediate versions of Omicron were occasionally reported. For example, a SARS-CoV-2 variant (EPI_ISL_7136300) was collected by the Utah Public Health Laboratory on December 1st, 2021, which harbored 32 of the 45 pre-outbreak Omicron mutations. However, the 13 mutations absent in this variant clustered within residues 371–501 of the spike protein (Fig. S6). The absence of these spike protein mutations thus suggested that this variant was a product of recombination between an Omicron variant and another SARS-CoV-2 variant rather than a direct progenitor of Omicron. Considering the large number of pre-outbreak Omicron mutations (45) combined with the sparsity of intermediate versions identified to date, this long branch leading to Omicron in our phylogenetic reconstruction remains valid.
Although we primarily focused on point mutations because the molecular spectrum of these mutations can reflect the host cellular environment (Deng et al., 2021; Shan et al., 2021), we also realized that the information of deletions and insertions could be used to infer the evolutionary trajectory of Omicron. For example, it was noted that Omicron harbored a nine-nucleotide insertion (GAGCCAGAA, encoding the peptide EPE) after residue 214 in the spike protein. This insertion is identical to the sequence of TMEM245 in the human genome or that of ORF S in the human coronavirus hCoV-229E, which was used as evidence to support a human origin for Omicron (Venkatakrishnan et al., 2021). However, we provide a simpler explanation for this insertion, namely that it was derived from an RNA fragment of ORF N in the SARS-CoV-2 genome (Fig. S7) because the RNA abundance of ORF N is much higher than that of mRNA encoded by the human genome (Wei et al., 2021). And this is especially so for ORF N due to the nested nature of the coronavirus genome and subgenomes (Kim et al., 2020).
The molecular docking-based predictions showed that the adaptation of Omicron to mice also promoted its adaptation to other species, such as humans, camels, and goats, via stronger RBD-ACE2 interaction (Fig. 6). Such a ‘pleiotropic effect’ of mutations was likely caused by structural similarity of ACE2 across species, and indicates that once a SARS-CoV-2 variant acquires the capacity to infect a new host, it can accumulate mutations in this new animal reservoir and becomes transmittable to another host. This ‘chain reaction’ of host jumping could potentially lead to remarkably high diversity in the adaptation to ACE2 from various host species. Consistent with this possibility, numerous mutations were identified in the spike protein of SARS-CoV-2 RNA fragment amplified from wastewater samples (Smyth et al., 2021).
Humans represent the largest known reservoir of SARS-CoV-2, and frequently come in contact with other animals, including livestock animals, pets, or wild animals that invade homes searching for food and shelter. Given the ability of SARS-CoV-2 to jump across various species, it appears likely that global populations will face additional animal-derived variants until the pandemic is well under control. Our study thus emphasizes the need for viral surveillance and sequencing in animals, especially those in close contact with humans. Furthermore, computational characterization of the spike RBD in animals and identification of their potentials to interact with human ACE2 will likely help to prevent future outbreaks of dangerous SARS-CoV-2 variants.
Materials and methods
Identification of pre-outbreak and post-outbreak Omicron mutations
Genomic sequences of 695 SARS-CoV-2 Omicron variants were downloaded from GISAID (https://www.gisaid.org/) on December 7th, 2021. The reference genome of SARS-CoV-2 (EPI_ISL_402125) and two variants in the B.1.1 lineage (EPI_ISL_698296 and EPI_ISL_493480) were also downloaded from GISAID. The variants from the B.1.1 lineage were chosen because they showed the highest sequence similarities to the early Omicron samples in a BLASTn search.
The genomes of SARS-CoV-2 variants were aligned by MUSCLE v3.8.1551 (Edgar, 2004). The phylogenetic tree and ancestral sequences were reconstructed using FastML v3.11 (Ashkenazy et al., 2012) with default parameters. The single-nucleotide substitutions obtained by the most recent common ancestor (MRCA) of Omicron variants after its divergence from the B.1.1 lineage were defined as pre-outbreak Omicron mutations. To detect the post-outbreak Omicron mutations, the sequences of 695 Omicron variants were aligned to the Omicron’s MRCA sequence, and sequences with >10 single-nucleotide substitutions were discarded. The single-nucleotide substitutions detected in at least two variants were defined as the post-outbreak Omicron mutations.
The numbers of synonymous and nonsynonymous sites in ORF S of SARS-CoV-2 were estimated by PAML in a previous study (Wei et al., 2021). Briefly, d N was calculated as the ratio between the number of nonsynonymous mutations and the number of nonsynonymous sites, while d S was calculated as the ratio between the number of synonymous mutations and the number of synonymous sites.
Comparison between the sequence evolutionary rate of Omicron and other SARS-CoV-2 variants
A total of 764 variant sequences were randomly sampled from patient-related SARS-CoV-2 genomic sequences deposited at GISAID, one variant each day since the COVID-19 outbreak. The progenitors of the other four VOCs (Alpha, Beta, Gamma, and Delta) were retrieved from Nextstrain (https://nextstrain.org/) (Hadfield et al., 2018). Single-nucleotide substitutions (relative to the reference genome) of each variant were defined as the mutations acquired by the SARS-CoV-2 variant. The single-nucleotide base substitutions of three chronically infected patients were retrieved from two previous studies (Kemp et al., 2021; Truong et al., 2021). The mutations with allele frequency >50% on the final monitored day were used to count mutations that accumulated in a chronically infected patient.
We performed a resampling test to estimate the statistical significance. Specifically, we randomly sampled 45 mutations from the 6986 point mutations identified in a previous study from the 34,853 high-quality sequences of SARS-CoV-2 variants isolated from patients worldwide (Shan et al., 2021). This operation was repeated 100 times in silico.
Characterization of molecular spectra of mutations
Complete genomic sequences of 23 bovine coronaviruses (Betacoronavirus 1), 13 canine coronaviruses (Alphacoronavirus 1), 54 feline coronaviruses (Alphacoronavirus 1), 17 murine hepatitis viruses (Murine coronavirus), and 110 porcine deltacoronaviruses (Coronavirus HKU15) were downloaded from National Center for Biotechnology Information (NCBI) Virus database (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/) (Hatcher et al., 2017), querying the hosts as Bos taurus (cattle), Canis lupus familiaris (dogs), Felis catus (cats), Mus musculus (mice), and Sus scrofa (pigs), respectively (Table S1). These coronaviruses were chosen from coronaviruses recorded in the International Committee on Taxonomy of Viruses (https://talk.ictvonline.org/taxonomy/). All mammalian coronaviruses were retrieved, and only those with at least ten reported sequences were used to ensure accurate estimation of the molecular spectrum. If there were multiple coronaviruses infecting the same host species, we randomly chose one for the subsequent analyses. The molecular spectra of accumulated mutations in the coronaviruses that infected bats, camels, or humans were retrieved from a previous study (Shan et al., 2021).
The virus genome sequences were aligned by MUSCLE, and the phylogenetic trees and ancestral sequences were reconstructed using FastML. Since the roots of these phylogenetic trees were not readily identified, we kept only external branches to ensure the correction direction of base substitutions (e.g., C>U vs. U>C). For the sake of clarity, we showed the molecular spectra for five branches with the largest number of mutations for each coronavirus species in the main text. The full data set is available in Table S1.
We characterized the molecular spectra of mutations accumulated in chronically infected patients, in which single-nucleotide base substitutions that ever occurred during the monitored period were counted. We downloaded the genomic sequences of four variants (EPI_ISL_5803018, EPI_ISL_3730369, EPI_ISL_4003132, and EPI_ISL_6260720), each from one of the other four VOCs (Alpha, Beta, Gamma, and Delta, respectively), to estimate the molecular spectra of mutations accumulated in VOCs.
Principal component analyses
We performed principal component analysis (prcomp function in R) with the proportions of the 12 base-substitution types as the input and then projected molecular spectra into a two-dimensional space according to the first two principal components. To define the borderlines of molecular spectra for each host species (i.e., cattle, bats, dogs, cats, mice, pigs, or humans), we estimated the 95% confidence ellipses (stat_ellipse function in R) from the molecular spectra of these host species. The spectra of pre and post-outbreak Omicron mutations were further projected into the same two-dimensional space.
Comparison of pre-outbreak Omicron mutations with mutations detected in SARS-CoV-2 variants isolated from various mammalian hosts
We downloaded from GISIAD the genomic sequences of SARS-CoV-2 variants isolated from 21 mammalian hosts (Tables S2 and S3): Aonyx cinereus (Asian small-clawed otter), Arctictis binturong (binturong), Canis lupus familiaris (dog), Crocuta crocuta (spotted hyena), Felis catus (cat), Gorilla gorilla (western gorilla), Mus musculus (mouse), Mustela furo (ferret), Neovison vison (American mink), Odocoileus virginianus (white-tailed deer), Panthera leo (lion), Panthera tigris (tiger), Panthera uncia (snow leopard), Prionailurus bengalensis (leopard cat), Prionailurus viverrinus (fishing cat), Hippopotamus amphibius (hippopotamus), Manis javanica (pangolin), Mesocricetus auratus (golden hamster), Chlorocebus sabaeus (green monkey), Puma concolor (puma), and the bats from genus Rhinolophus. BLASTx was performed to identify ORF S in each variant, and mutations relative to the reference SARS-CoV-2 genome were identified at the same time. Three species (Mesocricetus auratus, Chlorocebus sabaeus, and Puma concolor) were discarded because they harbored less than three single amino acid mutations. Amino acid mutation data from three additional viruses isolated from mice were retrieved from three studies (Leist et al., 2020; Montagutelli et al., 2021; Sun et al., 2021).
Estimation of the binding affinity of RBD-ACE2 interaction by molecular docking
We extracted three-dimensional structures of the spike RBD and human ACE2 from the crystal structure (PDB: 6M0J) reported in a previous study (Lan et al., 2020), and those of other representative mammalian ACE2 from the predicted models reported in a previous study (Lam et al., 2020). The structure models of the Omicron RBD were generated using SWISS-MODEL (Waterhouse et al., 2018), and those of other RBD variants were generated using PyMOL ‘mutagenesis’ (https://pymol.org/). The structure models of the RBD:ACE2 complex were generated by aligning against the reported complex structure of the corresponding species using PyMOL (Lam et al., 2020; Lan et al., 2020).
We performed molecular docking following previous studies (Lam et al., 2020; Rodrigues et al., 2020). Briefly, we refined the three-dimensional models using default refinement protocols and then estimated the HADDOCK scores for each RBD:ACE2 complex using the HADDOCKv2.4 webserver (van Zundert et al., 2016). Docking results of each RBD-ACE2 variant pair were clustered, and the average HADDOCK score of the top cluster was reported for the RBD:ACE2 complex.
Data availability
All scripts used to analyze the data and to generate the figures are available at github (https://github.com/ChangshuoWei/Omicron_origin) and Zenodo (DOI: 10.5281/zenodo.5778199). All data that were used to support the findings of this study are available in the public databases.
CRediT authorship contribution statement
Changshuo Wei: Data curation, Investigation, Writing - Original draft, Writing - Review & Editing. Ke-Jia Shan: Data curation, Investigation, Writing - Original draft, Writing - Review & Editing. Weiguang Wang: Data curation, Investigation, Writing - Review & Editing. Shuya Zhang: Investigation. Qing Huan: Writing - Original draft, Writing - Review & Editing, Supervision. Wenfeng Qian: Conceptualization, Writing - Original draft, Writing - Review & Editing, Supervision, Funding acquisition.
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgments
We thank Dr. Xionglei He from Sun Yat-sen University and Dr. Mingkun Li from Beijing Institute of Genomics CAS for discussion. We acknowledge the authors and laboratories for generating and submitting the sequences to GISAID Database on which this research is based. The list is detailed in Table S3. This work was supported by grants from the National Natural Science Foundation of China (31922014).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgg.2021.12.003.
Supplementary data
The following are the supplementary data to this article:
References
- Ashkenazy H., Penn O., Doron-Faigenboim A., Cohen O., Cannarozzi G., Zomer O., Pupko T. FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 2012;40:W580–W584. doi: 10.1093/nar/gks498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V., Davidson N.O. APOBEC-1-mediated RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010;2:594–602. doi: 10.1002/wsbm.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021 doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., Marco A.D., Di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021 doi: 10.1038/d41586-021-03825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J.C., Bevins S.N., Ellis J.W., Linder T.J., Tell R.M., Jenkins-Moore M., Root J.J., Lenoch J.B., Robbe-Austerman S., DeLiberto T.J., et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus) Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2114828118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio N., Walker C.R., Turakhia Y., Lanfear R., Corbett-Detig R., Goldman N. Mutation rates and selection on synonymous mutations in SARS-CoV-2. Genome Biol. Evol. 2021;13 doi: 10.1093/gbe/evab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S., Xing K., He X. Mutation signatures inform the natural host of SARS-CoV-2. Natl. Sci. Rev. 2021 doi: 10.1093/nsr/nwab220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S., Dudley J.P. APOBECs and virus restriction. Virology. 2015;479–480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., Schaffer A.A., Brister J.R. Virus variation resource – improved response to emergent viral outbreaks. Nucleic Acids Res. 2017;45:D482–D490. doi: 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Zhang Y., Hui X., Zhao Y., Gong W., Wang T., Zhang S., Yang Y., Deng F., Zhang Q., et al. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine. 2021;67:103381. doi: 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastritis P.L., Bonvin A.M. Are scoring functions in protein-protein docking ready to predict interactomes? Clues from a novel binding affinity benchmark. J. Proteome Res. 2010;9:2216–2225. doi: 10.1021/pr9009854. [DOI] [PubMed] [Google Scholar]
- Kemp S.A., Collier D.A., Datir R.P., Ferreira I., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q., Lin C.L. Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell. Mol. Life Sci. 2010;67:1817–1829. doi: 10.1007/s00018-010-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K. Where did ‘weird’ Omicron come from? Science. 2021;374:1179. doi: 10.1126/science.acx9738. [DOI] [PubMed] [Google Scholar]
- Lam S.D., Bordin N., Waman V.P., Scholes H.M., Ashford P., Sen N., van Dorp L., Rauer C., Dawson N.L., Pang C.S.M., et al. SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals. Sci. Rep. 2020;10:16471. doi: 10.1038/s41598-020-71936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Leist S.R., Dinnon K.H., 3rd, Schafer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183:1070–1085. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wu J., Deleo C.J. RNA damage and surveillance under oxidative stress. IUBMB Life. 2006;58:581–588. doi: 10.1080/15216540600946456. [DOI] [PubMed] [Google Scholar]
- Martinez-Flores D., Zepeda-Cervantes J., Cruz-Resendiz A., Aguirre-Sampieri S., Sampieri A., Vaca L. SARS-CoV-2 vaccines based on the spike glycoprotein and implications of new viral variants. Front. Immunol. 2021;12:701501. doi: 10.3389/fimmu.2021.701501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagutelli X., Prot M., Jouvion G., Levillayer L., Conquet L., Reyes-Gomez E., Donati F., Albert M., van der Werf S., Jaubert J., et al. A mouse-adapted SARS-CoV-2 strain replicating in standard laboratory mice. bioRxiv. 2021 doi: 10.1101/2021.07.10.451880. [DOI] [Google Scholar]
- Nelson G., Buzko O., Spilman P., Niazi K., Rabizadeh S., Soon-Shiong P. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. bioRxiv. 2021 doi: 10.1101/2021.01.13.426558. [DOI] [Google Scholar]
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchin A.Y., Panchin Y.V. Excessive G-U transversions in novel allele variants in SARS-CoV-2 genomes. PeerJ. 2020;8 doi: 10.7717/peerj.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Zhu Y., Wang Y., Shi H., Yu Y., Hu G., Feng F., Zhao X., Lan J., Wu J., et al. Comparative analysis reveals the species-specific genetic determinants of ACE2 required for SARS-CoV-2 entry. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J., Barrera-Vilarmau S., J M.C.T., Sorokina M., Seckel E., Kastritis P.L., Levitt M. Insights on cross-species transmission of SARS-CoV-2 from structural modeling. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1008449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan K.J., Wei C., Wang Y., Huan Q., Qian W. Host-specific asymmetric accumulation of mutation types reveals that the origin of SARS-CoV-2 is consistent with a natural process. Innovation (N Y). 2021;2:100159. doi: 10.1016/j.xinn.2021.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.S., Trujillo M., Gregory D.A., Cheung K., Gao A., Graham M., Guan Y., Guldenpfennig C., Hoxie I., Kannoly S., et al. Tracking cryptic SARS-CoV-2 lineages detected in NYC wastewater. medRxiv. 2021 doi: 10.1101/2021.07.26.21261142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Gu H., Cao L., Chen Q., Ye Q., Yang G., Li R.T., Fan H., Deng Y.Q., Song X., et al. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat. Commun. 2021;12:5654. doi: 10.1038/s41467-021-25903-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenti A., Arvin A., Corey L., Corti D., Diamond M.S., Garcia-Sastre A., Garry R.F., Holmes E.C., Pang P.S., Virgin H.W. After the pandemic: perspectives on the future trajectory of COVID-19. Nature. 2021;596:495–504. doi: 10.1038/s41586-021-03792-w. [DOI] [PubMed] [Google Scholar]
- Truong T.T., Ryutov A., Pandey U., Yee R., Goldberg L., Bhojwani D., Aguayo-Hiraldo P., Pinsky B.A., Pekosz A., Shen L., et al. Increased viral variants in children and young adults with impaired humoral immunity and persistent SARS-CoV-2 infection: a consecutive case series. EBioMedicine. 2021;67:103355. doi: 10.1016/j.ebiom.2021.103355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zundert G.C.P., Rodrigues J., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan A., Anand P., Lenehan P., Suratekar R., Raghunathan B., Niesen M.J., Soundararajan V. Omicron variant of SARS-CoV-2 harbors a unique insertion mutation of putative viral or human genomic origin. OSF Preprints. 2021 doi: 10.31219/osf.io/f7txy. [DOI] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Chen Y.M., Chen Y., Qian W. The missing expression level-evolutionary rate anticorrelation in viruses does not support protein function as a main constraint on sequence evolution. Genome Biol. Evol. 2021;13 doi: 10.1093/gbe/evab049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.Y.R., Zheng J., Wilhelmsen K., Li K., Ortiz M.E., Schnicker N.J., Pezzulo A.A., Szachowicz P.J., Klumpp K., Aswad F., et al. Eicosanoid signaling as a therapeutic target in middle-aged mice with severe COVID-19. bioRxiv. 2021 doi: 10.1101/2021.04.20.440676. [DOI] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Zhong G., Zhang J., Shuai L., Zhang Z., Wen Z., Wang B., Zhao Z., Song X., Chen Y., et al. A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020;11:4081. doi: 10.1038/s41467-020-17972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang K., Wang T., Deng F., Gong W., Hui X., Zhao Y., He X., Li C., Zhang Q., et al. SARS-CoV-2 rapidly adapts in aged BALB/c mice and induces typical pneumonia. J. Virol. 2021;95 doi: 10.1128/JVI.02477-20. e02477–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts used to analyze the data and to generate the figures are available at github (https://github.com/ChangshuoWei/Omicron_origin) and Zenodo (DOI: 10.5281/zenodo.5778199). All data that were used to support the findings of this study are available in the public databases.