Abstract
Objectives:
Evaluations from pharmacogenetics implementation programs at major U.S. medical centers have reported variability in the clinical adoption of pharmacogenetics across therapeutic areas. A potential cause for this variability may involve therapeutic area-specific differences in published pharmacogenetics recommendations to clinicians. To date, however, the potential for differences in clinical pharmacogenetics recommendations by therapeutic areas from prominent U.S. guidance sources has not been assessed. Accordingly, our objective was to comprehensively compare essential elements from clinical pharmacogenetics recommendations contained within Clinical Pharmacogenetics Implementation Consortium guidelines, U.S. Food and Drug Administration drug labels, and clinical practice guidelines from U.S. professional medical organizations across therapeutic areas.
Methods:
We analyzed clinical pharmacogenetics recommendation elements within Clinical Pharmacogenetics Implementation Consortium guidelines, U.S. Food and Drug Administration drug labels, and professional clinical practice guidelines through 05/24/19.
Results:
We identified 606 unique clinical pharmacogenetics recommendations, with the most recommendations involving oncology (217 recommendations), hematology (79), psychiatry (65), cardiovascular (43), and anesthetic (37) medications. Within our analyses, we observed considerable variability across therapeutic areas within the following essential pharmacogenetics recommendation elements: the recommended clinical management strategy; the relevant genetic biomarkers; the organizations providing pharmacogenetics recommendations; whether routine genetic screening was recommended; and the time since recommendations were published.
Conclusions:
Based on our results, we infer that observed differences in clinical pharmacogenetics recommendations across therapeutic areas may result from specific factors associated with individual disease states, the associated genetic biomarkers, and the characteristics of the organizations providing recommendations.
Keywords: Clinical Pharmacogenetics Implementation Consortium, Drug-gene interaction, FDA pharmacogenetics, Pharmacogenetic clinical practice guidelines, Pharmacogenetic recommendations, Therapeutic area
INTRODUCTION
Pharmacogenetics (PGx) holds tremendous promise as a means to inform optimal medication therapy based on patient-specific genetic factors. To date, genetic biomarkers have been discovered to predict the efficacy and/or toxicity of over 200 medications across a multitude of therapeutic areas.[1] PGx is typically viewed as a single field, but like many other areas of health care, PGx is highly specialized within different therapeutic areas (e.g., oncology, cardiology, psychiatry, etc.). Nuances that are specific to particular therapeutic areas may affect the clinical recommendations, use, and implementation of PGx.[2] For example, some PGx biomarkers, particularly in therapeutic areas like hematology, oncology, and infectious disease, involve genetic alterations apart from the human germline genome (i.e., genes inherited from parents). Genetic biomarkers in oncology include both germline biomarkers and somatic biomarkers that develop during oncogenesis.[3, 4] In contrast, antiviral pharmacotherapy is influenced by both germline biomarkers and biomarkers related to the genomes of human pathogens.[5] For most other PGx-relevant therapeutic areas, like cardiology and psychiatry, somatic and pathogen mutations are not applicable.
Formal evaluations of PGx implementation efforts have reported dramatic variability in the clinical adoption of PGx, including differences in adoption across therapeutic areas.[6–9] A 2017 survey conducted at the seven health centers within the Translational Pharmacogenomics Program of the NIH-led Pharmacogenomics Research Network revealed that certain germline drug-gene pairs involving cardiovascular drugs, including clopidogrel/CYP2C19, simvastatin/SLCO1B1, and warfarin/CYP2C9 & VKORC1, were among the most widely implemented.[6] In contrast, psychiatry drug-gene pairs, including tricyclic antidepressants/CYP2D6 & CYP2C19 and selective serotonin reuptake inhibitors/CYP2D6, were less widely implemented, with PGx testing only being performed at two sites and one site, respectively.[6] These observed differences in PGx implementation by therapeutic area are consistent with results from a nationwide PGx implementation survey that included responses from institutions within the Implementing Genomics in Practice (IGNITE), Clinical Sequencing Evidence-Generating Research (CSER), and Electronic Medical Record and Genomic (eMERGE) networks.[9]
One potential cause for the reported variability in the clinical PGx implementation could relate to differences in the clinical PGx recommendations published for different therapeutic areas. However, to our knowledge, clinical PGx recommendations have not been compared across therapeutic areas. Therefore, the objective of this study was to compare clinical PGx recommendations from prominent U.S. guidance sources, including Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines, U.S. Food and Drug Administration (FDA) drug labels, and clinical practice guidelines (CPGs) from professional medical organizations, across therapeutic areas. This investigation, which comprehensively compares PGx recommendation elements by therapeutic area, expands on our previous work which assessed inconsistencies among clinical PGx recommendations by the sources providing the recommendations (i.e., CPIC, FDA, or medical organizations).[10]
METHODS
Data Collection
Our data collection methods have been previously published in detail.[10] Briefly, we collected clinical PGx recommendations by reviewing CPIC guidelines, FDA drug labels, and official recommendations from U.S. professional medical organizations until 05/24/19. Collected PGx recommendation elements included the following: (1) the medication and (2) genetic biomarker (e.g., abacavir/HLA-B); (3) the recommended clinical strategy (e.g., contraindication); (4) whether routine genetic screening should be performed in all patients prescribed the drug; (5) the section of the FDA drug label containing the PGx recommendation, when applicable; (6) the publication date of the recommendation (i.e., when the article was first available online or in-print); and (7) the therapeutic area of the medication. Therapeutic area was assigned based on the indications listed in the FDA drug label and the FDA Division that approved the product, as found in the approval letter in the Drugs@FDA database. We analyzed genetic biomarkers contained in human germline, human somatic, and pathogen-related genomes, and we included genetic biomarkers that consisted of both genetic variants and alterations in gene expression.
CPIC guidelines and FDA drug labels were accessed from the CPIC website and the Drugs@FDA database, as previously described.[10] For drug-gene pairs identified in CPIC guidelines or FDA drug labels, we searched for PGx recommendations from U.S. professional organizations using the following procedures: (1) searching guidelines.gov, PubMed, and Google Scholar with keywords that included the specific drug and biomarker; (2) reviewing cited references in the most recent version of CPIC guidelines, when available; and (3) reviewing guidelines of prominent U.S. medical organizations in the therapeutic area of the drug in the drug-gene pair. CPGs intended for individual hospitals or health systems were excluded, and only recommendations from the most recently published professional CPG for each drug-gene pair was collected.
Categorization of PGx Clinical Management Strategies
Clinical management strategies within PGx recommendations were categorized as previously described,[10] and included the following: presence or absence of the biomarker required for drug Indication; Contraindication in the presence of the biomarker; administration Not Recommended in the presence of the biomarker; Dose Adjustment recommended in the presence of the biomarker; Use With Caution in the presence of the biomarker; No Dose Adjustment recommended in the presence of the biomarker; and Informational (None) when information related to the drug-gene interaction was provided but no explicit therapeutic management recommendation was given. Examples of this categorization method are provided in Supplementary Table 1. For FDA drug labels, the location of information was also used to assign the clinical management strategy when specific label language was not available (see Supplementary Table 2 for details). Clinical management strategy categories of Indication, Contraindication, Not Recommended, Dose Adjustment, and Use With Caution were defined as “clinically actionable”, while categories of No Dose Adjustment and Informational (None) were not.
Comparisons of PGx Recommendation Elements Across Therapeutic Areas
Information contained in PGx recommendations were compared based on the therapeutic areas of the medications involved. Specific recommendation elements that were tabulated and compared by therapeutic area included the following: (1) the clinical management strategy, including whether clinically actionable; (2) the sources providing PGx recommendations; (3) the genetic biomarkers, including whether biomarkers related to genetic variation or gene expression and whether biomarkers related to germline, somatic, or pathogenic genomes; (4) whether routine genetic screening was recommended; and (5) the elapsed time since the recommendations were published.
Statistical Analysis
We analyzed data using descriptive statistics in JMP v13.0.0 (SAS Institute®; Cary, NC). Data are expressed as counts and percentages or, for our time since publication analysis, median and interquartile range (IQR). We expressed time since publication dates in years with median and IQR as the measures of central tendency and variability, respectively, to account for data skewed towards the distant past.
RESULTS
We analyzed 302 unique PGx recommendation documents, yielding a total of 267 unique drugs, 86 unique genes, 433 unique drug-gene pairs, and 606 unique drug-gene pair recommendations. The complete dataset is available in the supplemental information of our previous publication.[10] The distribution of clinical management strategies within PGx recommendations, with stratification by therapeutic area, is shown in Table 1. Therapeutic areas with <15 clinical PGx recommendations were combined into the “all other therapeutic areas” group, with clinical management strategy categories for each therapeutic area comprising this group displayed in Supplementary Table 3. The therapeutic areas with the most PGx recommendations were oncology (217 drug-gene pairs; 62 unique drugs), hematology (80 drug-gene pairs; 24 unique drugs), and psychiatry (65 drug-gene pairs; 35 unique drugs). Indication was the most frequent recommendation for oncology (65.4%) and hematology (43.8%) drugs, while Informational (None) was the most common recommendation for drug-gene pairs in antivirals (61.5%), gastroenterology (47.4%), cardiovascular (30.2%), psychology (36.9%), and neurology (27.3%). Recommendations were actionable in 67.3% of cases overall, with the highest proportion of actionable recommendations in anesthetics (100%), anti-infectives (78.9%), and oncology (74.2%) and the lowest proportion in gastroenterology (47.4%) and antivirals (34.6%).
Table 1.
Distribution of Clinical Management Strategies Within Clinical Pharmacogenetic Recommendations by Therapeutic Area.
| Recommendation | Onc #, (%) | Heme #, (%) | Psych #, (%) | CV #, (%) | Anesth #, (%) | Neuro #, (%) | AV #, (%) | AI #, (%) | Gastro #, (%) | Others #, (%) | Total #, (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Indication | 142 (65.4) | 35 (43.8) | 0 (0) | 0 (0) | 0 (0) | 2 (6.1) | 2 (7.7) | 0 (0) | 3 (15.8) | 10 (14.9) | 194 (32.0) |
| Contraindication | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 14 (37.8) | 2 (6.1) | 1 (3.8) | 4 (21.1) | 0 (0) | 8 (11.9) | 29 (4.8) |
| Not Recommended | 9 (4.1) | 4 (5.1) | 17 (26.2) | 4 (9.3) | 14 (37.8) | 8 (24.2) | 5 (19.2) | 3 (15.8) | 3 (15.8) | 15 (22.4) | 82 (13.5) |
| Dose Adjustment | 2 (0.9) | 7 (8.9) | 10 (15.4) | 6 (14.0) | 0 (0) | 6 (18.2) | 1 (3.8) | 0 (0) | 1 (5.3) | 5 (7.5) | 38 (6.3) |
| Use With Caution | 8 (3.7) | 2 (2.5) | 11 (16.9) | 13 (30.2) | 9 (24.3) | 4 (12.1) | 0 (0) | 8 (42.1) | 2 (10.5) | 8 (11.9) | 65 (10.7) |
| Total Actionable | 161 (74.2) | 48 (60.0) | 38 (58.5) | 23 (53.5) | 37 (100) | 22 (66.7) | 9 (34.6) | 15 (78.9) | 9 (47.4) | 46 (68.7) | 408 (67.3) |
| No Dose Adjustment | 0 (0) | 0 (0) | 3 (4.6) | 7 (16.3) | 0 (0) | 2 (6.1) | 1 (3.8) | 0 (0) | 1 (5.3) | 3 (4.5) | 17 (2.8) |
| Informational (None) | 56 (25.8) | 32 (40.5) | 24 (36.9) | 13 (30.2) | 0 (0) | 9 (27.3) | 16 (61.5) | 4 (21.1) | 9 (47.4) | 18 (26.9) | 181 (29.9) |
| Total Drug-Gene Pairs | 217 | 80 | 65 | 43 | 37 | 33 | 26 | 19 | 19 | 67 | 606 |
| Unique Drugs | 62 | 24 | 35 | 17 | 14 | 23 | 19 | 13 | 14 | 46 | 267 |
Abbreviations: Anesth = Anesthetics; AI = anti-infectives; AV = antivirals; CV = cardiovascular; Gastro = gastroenterology; Heme = hematology; Neuro = neurology; Onc = oncology; Others = all other therapeutic areas, including pulmonary, rheumatology, endocrinology, analgesic, urology, immunosuppressant, medical countermeasure, metabolism, reproduction, addiction, and bone products; Psych = psychiatry.
The sources providing PGx recommendations varied by therapeutic area (Figure 1; numerical data available in Supplementary Table 4). CPIC guidelines most commonly provided recommendations for anesthetics (comprised 37.8% of all PGx recommendations for the therapeutic area), psychiatry (27.7%), and antivirals (23.1%); in contrast, CPIC guidelines least commonly provided recommendations for anti-infective (5.3%), hematology (5.0%), and oncology medications (1.4%). FDA drug labels were the most common source of PGx recommendations for all therapeutic areas except anesthetics (comprised 24.3% of all recommendations) and were most common for gastroenterology (89.5%), anti-infectives (78.9%), and neurology (78.8%). Professional CPGs most commonly provided recommendations for oncology (comprised 43.8% of all recommendations), hematology (40.0%), and anesthetics (37.8%), but they did not provide PGx recommendations for any gastroenterology medications. A list of the professional organizations providing PGx recommendations is shown in Supplementary Table 5.
Figure 1. Recommendation Sources Informing Clinical Pharmacogenetic Recommendations by Therapeutic Area.
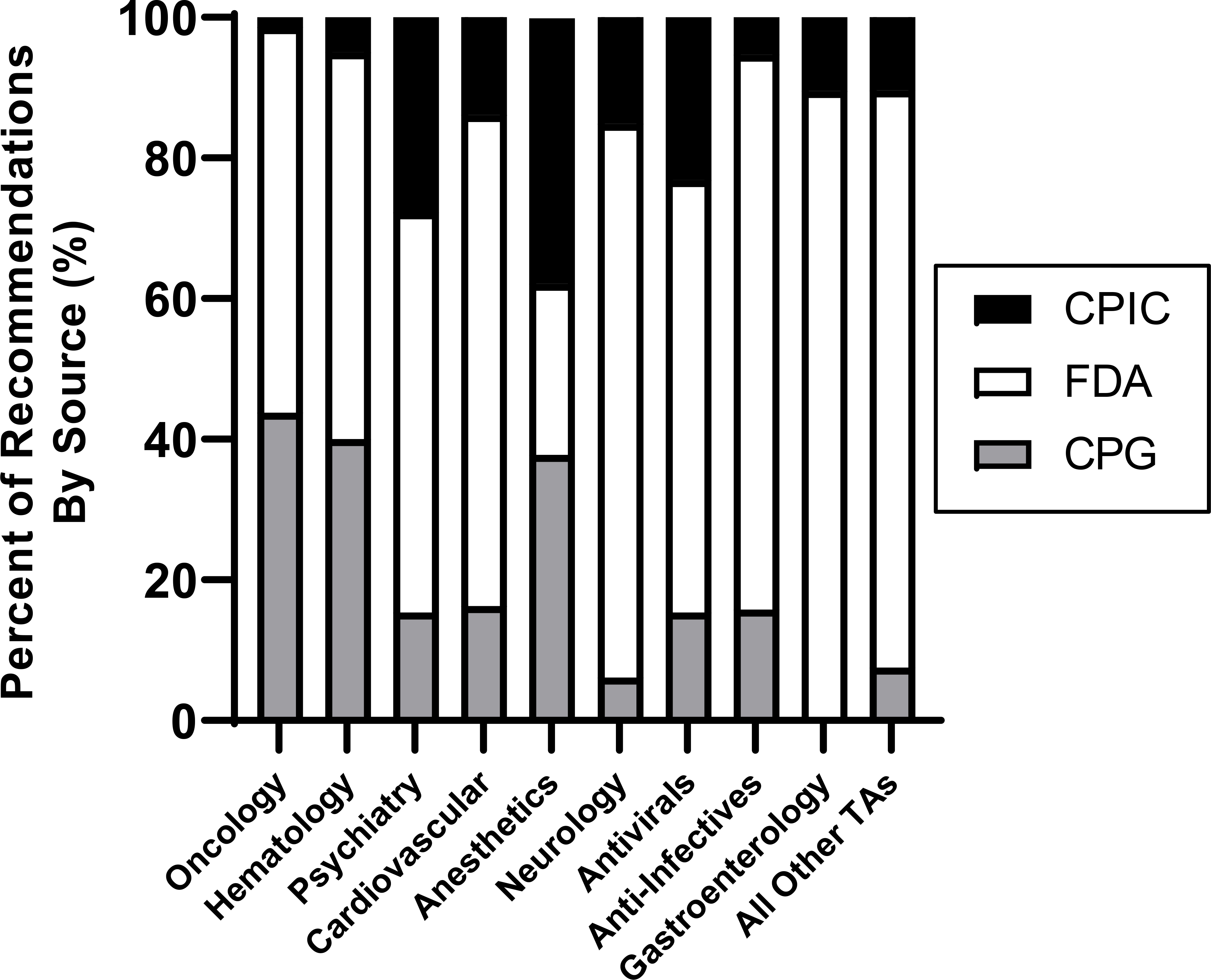
As collected from 302 pharmacogenetic recommendation documents, the total number of recommendations from each source (i.e., CPIC, FDA, or CPGs) are displayed as a percentage of the total number of recommendations for each therapeutic area. The “all other therapeutic areas” category includes pulmonary, rheumatology, endocrinology, analgesic, urology, immunosuppressant, medical countermeasure, metabolism, reproduction, addiction, and bone products.
Abbreviations: CPG = Clinical Practice Guidelines from U.S. Professional Organizations; CPIC = Clinical Pharmacogenetics Implementation Consortium; FDA = U.S. Food and Drug Administration; TA = therapeutic area
The most common genes contained in PGx recommendations by therapeutic area are displayed in Table 2. Broadly, these genes included (1) drug metabolism enzymes (e.g., CYP2D6), (2) drug targets or patient-related genetic predictors of drug response (e.g., ERBB2), (3) pathogen-related predictors of drug response (e.g., NS5A), and (4) pharmacodynamic genes that predispose to drug adverse events (e.g., HLA-A). Drug metabolizing enzymes comprised the most common biomarkers within drug-gene pairs for psychiatry, cardiovascular, and gastroenterology. In contrast, drug targets or predictors of drug response were the most common genetic biomarkers for oncology, hematology, and antivirals. Biomarkers related to pathogen-related predictors of drug response were only identified in antiviral PGx recommendations related to HCV therapy. Finally, pharmacodynamic genes that predispose to drug adverse events comprised the most common biomarkers for neurology, anesthetics, and anti-infectives.
Table 2.
Most Common Genetic Biomarkers Contained in Clinical Pharmacogenetic Recommendations by Therapeutic Area.
| Therapeutic Area | Most Common Genetic Biomarkers Within Pharmacogenetic Recommendations (Frequency, Percent) | ||||
|---|---|---|---|---|---|
| Oncology | ERBB2 (35, 16.1%) | ESR (33, 15.2%) | PGR (29, 13.4%) | EGFR (20, 9.2%) | ALK; BRAF (16, 7.4%) |
| Hematology | BCR-ABL1 (17, 21.5%) | TPMT (5, 6.3%) | 11q Del; 17p Del; FLT3; IDH1; IDH2; NUDT15; TP53 (4, 5.1%) | MS4A1; KIT; NPM1; SLC22A1 (3, 3.8%) | IGHV; TNFRSF8; UGT1A1 (2, 2.5%) |
| Psychiatry | CYP2D6 (47, 72.3%) | CYP2C19 (17, 26.2%) | CYP2C9 (1, 1.5%) | ||
| Cardiovascular | CYP2D6 (6, 14.0%) | CYP2C19 (5, 11.6%) | CYP2C9; F5; PROC; PROS1 (4, 9.3%) | F2; VKORC1 (3, 7.0%) | CYB5R; SERPINC1 (2, 4.7%) |
| Anesthetics | RYR1 (15, 40.5%) | CACNA1S (14, 37.8%) | G6PD (7, 18.9%) | BCHE (1, 2.7%) | |
| Neurology | HLA-B (7, 21.2%) | CYP2D6 (6, 18.2%) | CYP2C19 (5, 15.2%) | CYP2C9 (3, 9.1%) | HLA-A; SMN2; TTR (2, 6.1%) |
| Antivirals | IFNL3 (15, 57.7%) | NS5A; HLA-B; UGT1A1 (3, 11.5%) | CYP2B6 (2, 7.7%) | ||
| Anti-Infectives | G6PD (14, 73.7%) | CYP2C19 (3, 15.8%) | CYB5R; CYP2D6 (1, 5.3%) | ||
| Gastroenterology | CYP2C19; CYP2D6 (6, 31.6%) | G6PD (3, 15.8%) | ASS1; CPS1; CYB5R; OTC (1, 5.3%) | ||
| All Other TAs | CYP2D6 (14, 20.6%) | G6PD (10, 14.7%) | CYP2C9 (7, 10.3%) | CFTR (6, 8.8%) | CYP2C19; TPMT (3, 4.4%) |
Boxes containing multiple genes indicate that each gene individually has the designated frequency. “All Other therapeutic areas” category includes pulmonary, rheumatology, endocrinology, analgesic, urology, immunosuppressant, medical countermeasure, metabolism, reproduction, addiction, and bone products.
Abbreviations: 11q Del- chromosome 11q deletion; 17p Del- chromosome 17p deletion; ALK- anaplastic lymphoma kinase; ASS1- argininosuccinate synthase 1; BCHE- butyrylcholinesterase; BCR-ABL1- breakpoint cluster region- ABL proto-oncogene fusion; BRAF- B-Raf proto-oncogene; CACNA1S- calcium voltage-gated channel subunit alpha 1s; CFTR- cystic fibrosis transmembrane conductance regulator; CPS1- carbamoyl-phosphate synthase 1; CYB5R- cytochrome B5 reductase 1; CYP2B6- cytochrome P450 2B6; CYP2C19- cytochrome P450 2C19; CYP2C9- cytochrome P450 2C9; CYP2D6- cytochrome P450 2D6; EGFR- epidermal growth factor receptor; ERBB2- human epidermal growth factor receptor 2; ESR- estrogen receptor gene family; F2- coagulation factor II; F5- coagulation factor V; FLT3- fms-related tyrosine kinase 3; G6PD- glucose-6-phosphate dehydrogenase; HLA-A/B- major histocompatibility complex, class I, A/B; IGHV- immunoglobulin heavy chain variable region; IDH1/2- isocitrate dehydrogenase 1/2; IFNL3- interferon lambda-3; KIT- proto-oncogene c-KIT; MS4A1- membrane spanning 4-domains A1; NPM1- nucleophosmin 1; NS5A- non-structural gene 5A (hepatitis C virus); NUDT15- nudix hydrolase 15; OTC- ornithine carbamoyltransferase; PGR- progesterone receptor; PROC- vitamin K-dependent protein C; PROS1- vitamin K-dependent protein S; RYR1- ryanodine receptor 1; SERPINC1- serpin family C member 1; SLC22A1- solute carrier family 22 member 1; SMN2- survival Of motor neuron 2; TA = therapeutic area; TNFRSF8- tumor necrosis factor receptor superfamily, member 8; TP53- tumor protein P53; TPMT- thiopurine S-methyltransferase; TTR- transthyretin; UGT1A1- UDP-glucuronosyltransferase family 1 member A1; VKORC1- vitamin K epoxide reductase complex subunit 1.
Genetic variation, including both point mutations and structural variation (e.g., gene deletions), served as the genetic basis for 79.9% of clinical PGx recommendations overall. Recommendations based on gene expression comprised the remaining 20.1%. Gene expression formed the basis for 51.6% and 11.3% of PGx recommendations for oncology and hematology medications, respectively; all PGx recommendations for all other therapeutic areas were based on genetic variation. The majority of PGx recommendations were based on germline genetic biomarkers (58.3%), with fewer recommendations based on somatic (41.3%) and pathogen (0.5%) biomarkers. Somatic biomarkers were only present within PGx recommendations for oncology and hematology medications, forming the basis for 84.3% and 83.8% of recommendations, respectively. All genetic biomarkers related to pathogen genomes were contained within antiviral PGx recommendations (forming the genetic basis for 11.5% of recommendations), and the applicable pathogen in all of these cases was HCV. Germline genetic biomarkers formed the basis for all recommendations for the remaining therapeutic areas. Complete data for the genetic biomarker types (i.e., genetic variation or gene expression) and the relevant genome (i.e., germline, somatic, or pathogen) for all therapeutic areas are displayed in Table 3.
Table 3.
Distributions of the Types of Genetic Biomarkers and the Relevant Genomes for Genetic Biomarkers Contained in Clinical Pharmacogenetic Recommendations by Therapeutic Area.
| Therapeutic Area | Genetic Biomarker Type | Relevant Genome for Genetic Biomarker | |||
|---|---|---|---|---|---|
| Genetic Variation | Gene Expression | Germline | Somatic | Pathogen (Viral) | |
| Oncology | 105 (48.4%) | 112 (51.6%) | 34 (15.7%) | 183 (84.3%) | 0 (0%) |
| Hematology | 71 (88.8%) | 9 (11.3%) | 13 (16.3%) | 67 (83.8%) | 0 (0%) |
| Psychiatry | 65 (100%) | 0 (0%) | 65 (100%) | 0 (0%) | 0 (0%) |
| Cardiovascular | 43 (100%) | 0 (0%) | 43 (100%) | 0 (0%) | 0 (0%) |
| Anesthetics | 37 (100%) | 0 (0%) | 37 (100%) | 0 (0%) | 0 (0%) |
| Neurology | 33 (100%) | 0 (0%) | 33 (100%) | 0 (0%) | 0 (0%) |
| Antivirals | 26 (100%) | 0 (0%) | 23 (88.5%) | 0 (0%) | 3 (11.5%) |
| Anti-Infectives | 19 (100%) | 0 (0%) | 19 (100%) | 0 (0%) | 0 (0%) |
| Gastroenterology | 19 (100%) | 0 (0%) | 19 (100%) | 0 (0%) | 0 (0%) |
| All Other TAs | 66 (98.5%) | 1 (1.5%) | 67 (100%) | 0 (0%) | 0 (0%) |
| Total | 484 (79.9%) | 122 (20.1%) | 353 (58.3%) | 250 (41.3%) | 3 (0.5%) |
All data are presented as follows: counts (percentages). The “all other therapeutic areas” category includes pulmonary, rheumatology, endocrinology, analgesic, urology, immunosuppressant, medical countermeasure, metabolism, reproduction, addiction, and bone products.
Abbreviations: TA = therapeutic area
We next assessed whether recommendations for routine genetic screening were included within PGx recommendations from the FDA professional CPGs. CPIC does not provide recommendations for when genetic testing should be ordered;[11] therefore, routine screening recommendations were not assessed within CPIC guidelines. The rates of recommendations for routine screening by recommendation source are shown in Table 4 for all therapeutic areas with ≥15 recommendations and in Supplementary Table 6 for therapeutic areas comprising the “all other therapeutic areas” category. Overall, 38.2% of drug-gene pairs had recommendations for routine genetic screening, and the rate of routine screening was higher within professional CPGs (60.5%) relative to FDA drug labels (27.8%), a trend that was consistent across therapeutic areas except psychiatry and neurology. Routine screening was most commonly recommended for oncology (64.0%) and hematology (46.1%) drug-gene pairs. In contrast, psychiatry (4.3%), cardiovascular (0%), and anesthetics (0%) were among the therapeutic areas with the lowest rates of recommendations for routine screening.
Table 4. Whether Routine Screening for Genetic Biomarkers was Recommended by Therapeutic Area.
Counts represent instances wherein guidance sources recommended genetic screening in all patients being prescribed the applicable medication. CPIC guidelines were not included since they do not address whether genetic testing should be ordered.
| Therapeutic Area | Recommendation for Routine Screening | ||
|---|---|---|---|
| Overall | FDA | CPG | |
| Oncology | 137 (64.0%) | 60 (50.4%) | 77 (81.1%) |
| Hematology | 35 (46.1%) | 17 (38.6%) | 18 (56.3%) |
| Psychiatry | 2 (4.3%) | 2 (5.4%) | 0 (0%) |
| Cardiovascular | 0 (0%) | 0 (0%) | 0 (0%) |
| Anesthetics | 0 (0%) | 0 (0%) | 0 (0%) |
| Neurology | 8 (28.6%) | 8 (30.8%) | 0 (0%) |
| Antivirals | 3 (15.0%) | 1 (6.3%) | 2 (50.0%) |
| Anti-Infectives | 4 (23.5%) | 2 (14.3%) | 2 (66.7%) |
| Gastroenterology | 3 (17.6%) | 3 (17.6%) | N/A |
| All Other TAs | 14 (23.3%) | 9 (16.4%) | 5 (100%) |
| Total | 206 (38.2%) | 102 (27.8%) | 104 (60.5%) |
All data are presented as follows: counts (percentages). “All Other TAs” include pulmonary, rheumatology, endocrinology, analgesic, urology, immunosuppressant, medical countermeasure, metabolism, reproduction, addiction, and bone products.
Abbreviations: CPG = Clinical Practice Guidelines from U.S. Professional Organizations; CPIC = Clinical Pharmacogenetics Implementation Consortium; FDA = U.S. Food and Drug Administration; N/A = not applicable, indicates that no recommendations were provided by the designated source
The distribution of the times since publication for PGx recommendations across therapeutic areas is shown in Figure 2. For all recommendations (irrespective of therapeutic area), the average times since publication were 0.8 ± 1.6 [median ± IQR] years. PGx recommendations containing oncology (0.2 ± 0.6 years), hematology (0.5 ± 0.8), and anesthetic (0.6 ± 0.7) medications had median times since publication of under one year, while recommendations for psychiatry (2.5 ± 3.6) and anti-infective (2.5 ± 3.9) drugs averaged greater than two years old. All other therapeutic areas with ≥15 PGx recommendations had median times since publication between one and two years old. Average calculated times since publication for all therapeutic areas are shown in Supplementary Table 7.
Figure 2. Boxplots of Years Since Publication Date for the Most Recent Clinical Pharmacogenetic Recommendations by Therapeutic Area.
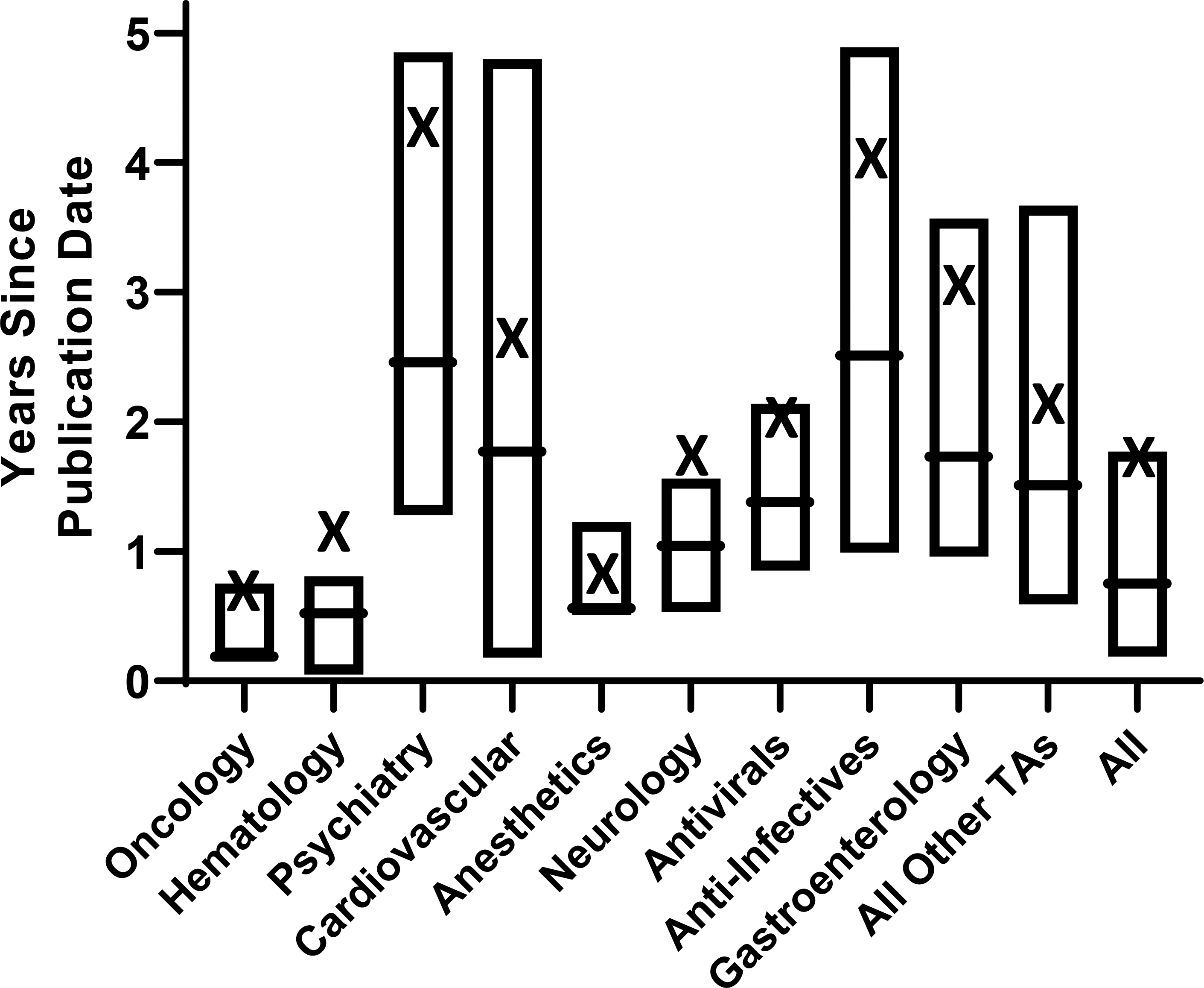
Horizontal box lines represent third quartiles, medians, and first quartiles (top to bottom), respectively. The X indicates the arithmetic mean. The “all other therapeutic areas” category includes pulmonary, rheumatology, endocrinology, analgesic, urology, immunosuppressant, medical countermeasure, metabolism, reproduction, addiction, and bone products.
Abbreviations: TA = therapeutic area
DISCUSSION
In this investigation, we comprehensively analyzed recommendation elements within clinical PGx recommendations from U.S. sources including CPIC guidelines, FDA drug labels, and professional organization CPGs by therapeutic area. PGx is typically viewed as a single field, but our results illustrate a vast amount of heterogeneity in PGx recommendations across different therapeutic areas. Specifically, we found notable differences in the following PGx recommendation elements by therapeutic area: the sources providing recommendations; the genetic biomarkers included; the recommended clinical management strategies; whether routine genetic screening was recommended; and how recently recommendations were published.
These findings likely reflect differences in the progress of clinical PGx adoption across therapeutic areas. Based on our findings, clinical adoption of PGx appears to be most strongly established within the fields of oncology and hematology. For example, nearly half (~49%) of all identified PGx recommendations involved oncology or hematology medications. PGx recommendations in oncology and hematology were published more recently than those from other therapeutic areas and were among those with the highest rates of both clinically actionable management strategies and recommendations for routine genetic screening. Perhaps most importantly, oncology and hematology were the two therapeutic areas with the most PGx recommendations from CPGs (~40% of recommendations for both), indicating the acceptance of PGx approaches by prominent professional organizations (e.g., the National Comprehensive Cancer Network) within these therapeutic areas. Our results align with the emerging role for PGx both within the fields of oncology and hematology. Given that genetic alterations are central to the pathophysiology of cancer, molecular tumor boards have been developed at major medical centers across the country to provide somatic genotype-guided treatment recommendations that have improved patient outcomes.[12, 13] Additionally, germline genetic biomarkers are increasingly being clinically implemented to ensure safe use of fluoropyrimidine and thiopurine chemotherapies.[14, 15]
In contrast, our findings also suggest the need for continued investigation to demonstrate the clinical utility of PGx approaches in other therapeutic areas in order to facilitate widespread adoption. For instance, psychiatry and cardiovascular medicine were the therapeutic areas with the third and fourth most PGx recommendations, respectively. However, our analyses revealed the following findings that reflect limited clinical PGx adoption within these fields: both therapeutic areas had a modest number of recommendations from CPGs (~15% for both); both were among the therapeutic areas with the longest time since publication for PGx recommendations; both had modest rates (~55% for both) of clinically actionable recommendations; and both had very low rates (<5% for both) of recommendations for routine genetic screening. These findings reflect current challenges towards PGx adoption within these fields. While CPIC guidelines currently provide clinically actionable PGx recommendations for 18 drug-gene pairs in psychiatry,[16–18] these same drug-gene pairs are either not mentioned (~28% of cases) or, if mentioned, are not given actionable recommendations (~47% of cases) within FDA drug labels or professional CPGs. In fact, the only identified professional CPG source for psychiatry is a 2007 statement from the Evaluation of Genomic Applications in Practice and Prevention Working Group that discourages PGx testing to guide prescribing of selective serotonin reuptake inhibitors in depression.[19] This finding is significant since CPGs, rather than CPIC guidelines or FDA drug labels, are likely the greatest indicator of PGx adoption within a field; CPGs are written by expert professional organizations within a therapeutic area and largely influence decisions related to clinical practice as well as medical policy and reimbursement.[20] Therefore, our findings support the need for continued PGx research and implementation efforts to better elucidate a clear role for PGx in psychiatry, an opinion that was recently voiced by PGx experts in the field.[21]
Similarly, our findings evidence limited clinical PGx adoption within the field of cardiovascular medicine. Although clinical PGx initiatives at major academic medical centers have successfully implemented genotype-guided approaches for medications with CPIC recommendations, including clopidogrel, simvastatin, and warfarin,[6] widespread adoption of these approaches remains limited. Currently there is no professional CPG for simvastatin/SLCO1B1, and guidelines from the American Heart Association-American College of Cardiology for clopidogrel and American College of Chest Physicians for warfarin do not make actionable PGx recommendations.[22, 23] CPGs in cardiology may include more PGx recommendations soon since randomized controlled trials of genotype-guided antiplatelet therapy have been recently published.[24, 25] Finally, more evidence is needed to conclusively define the potential role for PGx testing for other cardiovascular medications mentioned in FDA drug labels, including beta-blockers and thrombopoietin receptor agonists.
Previous studies have compared PGx recommendations from professional organizations and drug regulatory bodies both within the U.S. and abroad. An investigation by Shekhani, et al. compared recommendations from CPIC, the FDA, the Dutch Pharmacogenetics Working Group (DPWG), and the European Medicines Agency (EMA).[26] While the authors categorized drug therapeutic area based on the World Health Organization’s Anatomical Therapeutic Chemical (ATC) classification, they only compared whether there was overlap in actionable PGx recommendations between the EMA and FDA by ATC category. Bank, et al. compared PGx recommendations within guidelines from CPIC and DPWG, but they did not stratify their comparisons by therapeutic area.[27] Similarly, Abdullah-Koolmees, et al. did not stratify by therapeutic area in their comparison of recommendations from CPIC, DPWG, the Canadian Pharmacogenomics Network for Drug Safety (CPNDS), and the French National Network of Pharmacogenetics (RNPGx).[28] Filipski, et al. compared PGx recommendations for drug-metabolizing enzyme genes within CPIC guidelines, FDA drug labels, and professional CPGs but also did not assess differences across therapeutic areas.[29] We previously assessed the prevalence of inconsistencies within PGx recommendations from CPIC guidelines, FDA drug labels, and professional CPGs in the U.S.[10] Notably, we found significant differences across therapeutic areas in the rates of composite recommendation inconsistencies, which included inconsistencies related to the clinical management strategy, the group subject to the recommendation, and whether routine genetic screening was recommended. Within our analyses, psychiatry and cardiovascular medicine were the therapeutic areas with the highest rates of composite inconsistencies (100% for both) while oncology (42.3%) and hematology (50%) had lower inconsistency rates. Our past findings also likely reflect differences in the progress of PGx adoption by therapeutic area since inconsistent PGx recommendations to clinicians are an established barrier to PGx adoption.[26] In contrast to past studies, the current investigation assessed differences among therapeutic areas in novel and important PGx recommendation elements, including the sources providing recommendations, the rates of clinically actionable recommendations, whether routine genetic screening was recommended, and the time elapsed since publication. Accordingly, the present study builds upon past investigations by contributing novel insights into the observed variability in PGx adoption across therapeutic areas.
As evidenced by recent advances in oncology and hematology clinical practice, PGx approaches have the potential to enhance both medication outcomes when effectively implemented.[13, 30, 31] However, our findings along with those from PGx implementation efforts at academic medical centers have consistently demonstrated inconsistencies in PGx adoption across therapeutic areas that limit the potential benefit of PGx in many areas.[6–9] Furthering the widespread clinical adoption of PGx in these therapeutic areas will likely require multiple approaches. First, research is needed to demonstrate both the scientific validity and clinical utility of PGx approaches for new drug-gene pairs. As curated by CPIC, there are nearly 200 novel drugs (i.e., those not contained in current CPIC guidelines) with provisional PGx associations with genetic biomarkers that span many therapeutic areas.[32] Similarly, the FDA has released the “Table of Pharmacogenetic Associations” that lists nearly 100 drugs with PGx associations, the majority of which are not currently included in CPIC guidelines.[33] Research investigating both the biological underpinnings and clinical significance, including effects on important patient outcomes, is needed to establish the clinical potential of PGx-guided approaches for these drugs.[34] Second, implementation science efforts are needed to overcome common obstacles (e.g., challenges associated with embedding PGx clinical decision support into the electronic health record) that limit the clinical adoption of PGx approaches for drug-gene pairs with established clinical utility.[35, 36]
By comparing PGx recommendations from therapeutic areas that are leading and lagging behind in terms of PGx adoption, we believe our findings reveal critical barriers to PGx implementation. Specifically, we note two important differences when comparing oncology and hematology to other therapeutic areas. Most principally, the evidence base is stronger for many drug-gene pairs within oncology and hematology, particularly for those containing somatic genetic biomarkers for which drug sponsors have conducted rigorous clinical trials to establish the effectiveness of PGx-guided approaches during drug development. Next, and likely secondary to the stronger evidence base, oncology and hematology professional organizations publish far more PGx recommendations in CPGs than similar professional organizations in therapeutic areas like cardiology and psychiatry. Therefore, our findings suggest the need for stronger evidence, including findings from prospective clinical trials, and enhanced engagement with professional organizations in order to advance PGx adoption in lagging therapeutic areas.
We acknowledge several limitations of our investigation. First, our analyses focused on comparing PGx recommendations from U.S. organizations. By only analyzing U.S. sources, our findings may not accurately represent the progress of PGx adoption in other countries. However, PGx recommendations from both U.S. and non-U.S. sources, including pharmacogenetics organizations, drug regulatory bodies, and other professional medical societies are catalogued and accessible through the Pharmacogenetics Knowledge Base (PharmGKB).[37] In addition, our search strategy for identifying professional CPGs only focused on drug-gene pairs contained in CPIC guidelines or FDA drug labels. As a result, it is possible that our search strategy omitted professional CPGs for drug-gene pairs without CPIC or FDA recommendations. Given that PGx is a dynamic field, new PGx recommendations have been published by CPIC and the FDA since our data were collected and analyzed. However, at the time of manuscript preparation, our analyses included data from 23 of 25 published CPIC guidelines[32] and for ~88% of the drug-gene pairs included in the FDA’s “Table of Pharmacogenetic Associations” that was released in 2020.[33] Therefore, we do not expect that the new PGx recommendations from CPIC and the FDA would strongly influence our findings.
CONCLUSION
Our work provides the first comprehensive comparison of clinical PGx recommendations across therapeutic areas, demonstrating vast therapeutic area-specific differences in important recommendation elements. Our findings likely reflect differences in PGx adoption across therapeutic areas and identify therapeutic areas in which research is needed to advance the progress of PGx. In order to facilitate widespread PGx adoption, future research must demonstrate stronger evidence for the clinical benefit of PGx approaches in lagging therapeutic areas and identify best practices for PGx implementation when high-level evidence exists.
Supplementary Material
Acknowledgments
Sources of Funding:
J.A.L. is funded by a NIH K08 award from NHLBI (1K08HL146990-01) and the NIH Clinical Research Loan Repayment Program (L30 HL110279-03). The remaining authors declare no sources of funding.
Footnotes
Conflicts of Interest
All authors declare no conflicts of interest.
REFERENCES
- 1.U.S. Food and Drug Administration. Table of Pharmacogenomic Biomarkers in Drug Labeling. https://www.fda.gov/drugs/science-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling. Published 2019. Accessed February 24, 2021.
- 2.Daly AK and Cascorbi I. Opportunities and limitations: the value of pharmacogenetics in clinical practice. Br J Clin Pharmacol. 2014;77(4):583–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amstutz U, Henricks LM, Offer SM, Barbarino J, Schellens JHM, Swen JJ, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin Pharmacol Ther. 2018;103(2):210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanna N, Johnson D, Temin S, Baker S Jr., Brahmer J, Ellis PM, et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol. 2017;35(30):3484–3515. [DOI] [PubMed] [Google Scholar]
- 5.Hepatitis C Guidance 2018 Update: AASLD-IDSA Recommendations for Testing, Managing, and Treating Hepatitis C Virus Infection. Clin Infect Dis. 2018;67(10):1477–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luzum JA, Pakyz RE, Elsey AR, Haidar CE, Peterson JF, Whirl-Carrillo M, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Outcomes and Metrics of Pharmacogenetic Implementations Across Diverse Healthcare Systems. Clin Pharmacol Ther. 2017;102(3):502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owusu Obeng A, Fei K, Levy KD, Elsey AR, Pollin TI, Ramirez AH, et al. Physician-Reported Benefits and Barriers to Clinical Implementation of Genomic Medicine: A Multi-Site IGNITE-Network Survey. J Pers Med. 2018;8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sperber NR, Carpenter JS, Cavallari LH, L JD, Cooper-DeHoff RM, Denny JC, et al. Challenges and strategies for implementing genomic services in diverse settings: experiences from the Implementing GeNomics In pracTicE (IGNITE) network. BMC Med Genomics. 2017;10(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volpi S, Bult CJ, Chisholm RL, Deverka PA, Ginsburg GS, Jacob HJ, et al. Research Directions in the Clinical Implementation of Pharmacogenomics: An Overview of US Programs and Projects. Clin Pharmacol Ther. 2018;103(5):778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shugg T, Pasternak AL, London B and Luzum JA. Prevalence and types of inconsistencies in clinical pharmacogenetic recommendations among major U.S. sources. NPJ Genom Med. 2020;5:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Relling MV and Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89(3):464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Specchia ML, Frisicale EM, Carini E, Di Pilla A, Cappa D, Barbara A, et al. The impact of tumor board on cancer care: evidence from an umbrella review. BMC Health Serv Res. 2020;20(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato S, Kim KH, Lim HJ, Boichard A, Nikanjam M, Weihe E, et al. Real-world data from a molecular tumor board demonstrates improved outcomes with a precision N-of-One strategy. Nat Commun. 2020;11(1):4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weitzel KW, Smith DM, Elsey AR, Duong BQ, Burkley B, Clare-Salzler M, et al. Implementation of Standardized Clinical Processes for TPMT Testing in a Diverse Multidisciplinary Population: Challenges and Lessons Learned. Clin Transl Sci. 2018;11(2):175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hertz DL and Sahai V. Including DPYD on Cancer Genetic Panels to Prevent Fatal Fluoropyrimidine Toxicity. J Natl Compr Canc Netw. 2020;18(4):372–374. [DOI] [PubMed] [Google Scholar]
- 16.Brown JT, Bishop JR, Sangkuhl K, Nurmi EL, Mueller DJ, Dinh JC, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Cytochrome P450 (CYP)2D6 Genotype and Atomoxetine Therapy. Clin Pharmacol Ther. 2019;106(1):94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hicks JK, Sangkuhl K, Swen JJ, Ellingrod VL, Muller DJ, Shimoda K, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102(1):37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks JK, Bishop JR, Sangkuhl K, Muller DJ, Ji Y, Leckband SG, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharmacol Ther. 2015;98(2):127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Recommendations from the EGAPP Working Group: testing for cytochrome P450 polymorphisms in adults with nonpsychotic depression treated with selective serotonin reuptake inhibitors. Genet Med. 2007;9(12):819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woolf SH, Grol R, Hutchinson A, Eccles M and Grimshaw J . Clinical guidelines: potential benefits, limitations, and harms of clinical guidelines. Bmj. 1999;318(7182):527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicks JK, Bishop JR, Gammal RS, Sangkuhl K, Bousman CA, Leeder JS, et al. A Call for Clear and Consistent Communications Regarding the Role of Pharmacogenetics in Antidepressant Pharmacotherapy. Clin Pharmacol Ther. 2020;107(1):50–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holbrook A, Schulman S, Witt DM, Vandvik PO, Fish J, Kovacs MJ, et al. Evidence-based management of anticoagulant therapy: Antithrombotic Therapy and Prevention of Thrombosis, 9th ed: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest. 2012;141(2 Suppl):e152S–e184S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine GN, Bates ER, Bittl JA, Brindis RG, Fihn SD, Fleisher LA, et al. 2016 ACC/AHA Guideline Focused Update on Duration of Dual Antiplatelet Therapy in Patients With Coronary Artery Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines: An Update of the 2011 ACCF/AHA/SCAI Guideline for Percutaneous Coronary Intervention, 2011 ACCF/AHA Guideline for Coronary Artery Bypass Graft Surgery, 2012 ACC/AHA/ACP/AATS/PCNA/SCAI/STS Guideline for the Diagnosis and Management of Patients With Stable Ischemic Heart Disease, 2013 ACCF/AHA Guideline for the Management of ST-Elevation Myocardial Infarction, 2014 AHA/ACC Guideline for the Management of Patients With Non-ST-Elevation Acute Coronary Syndromes, and 2014 ACC/AHA Guideline on Perioperative Cardiovascular Evaluation and Management of Patients Undergoing Noncardiac Surgery. Circulation. 2016;134(10):e123–155. [DOI] [PubMed] [Google Scholar]
- 24.Claassens DMF, Vos GJA, Bergmeijer TO, Hermanides RS, van ‘t Hof AWJ, van der Harst P, et al. A Genotype-Guided Strategy for Oral P2Y(12) Inhibitors in Primary PCI. N Engl J Med. 2019;381(17):1621–1631. [DOI] [PubMed] [Google Scholar]
- 25.Pereira NL, Farkouh ME, So D, Lennon R, Geller N, Mathew V, et al. Effect of Genotype-Guided Oral P2Y12 Inhibitor Selection vs Conventional Clopidogrel Therapy on Ischemic Outcomes After Percutaneous Coronary Intervention: The TAILOR-PCI Randomized Clinical Trial. Jama. 2020;324(8):761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shekhani R, Steinacher L, Swen JJ and Ingelman-Sundberg M. Evaluation of Current Regulation and Guidelines of Pharmacogenomic Drug Labels: Opportunities for Improvements. Clin Pharmacol Ther. 2020;107(5):1240–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.PCD Bank, Caudle KE, Swen JJ, Gammal RS, Whirl-Carrillo M, Klein TE, et al. Comparison of the Guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin Pharmacol Ther. 2018;103(4):599–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdullah-Koolmees H, van Keulen AM, Nijenhuis M and Denser VHM. Pharmacogenetics Guidelines: Overview and Comparison of the DPWG, CPIC, CPNDS, and RNPGx Guidelines. Front Pharmacol. 2020;11:595219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filipski KK, Pacanowski MA, Ramamoorthy A, Feero WG and Freedman AN . Dosing recommendations for pharmacogenetic interactions related to drug metabolism. Pharmacogenet Genomics. 2016;26(7):334–339. [DOI] [PubMed] [Google Scholar]
- 30.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med. 2017;377(6):523–533. [DOI] [PubMed] [Google Scholar]
- 31.G Stocco, Cheok MH, Crews KR, Dervieux T, French D, Pei D, et al. Genetic polymorphism of inosine triphosphate pyrophosphatase is a determinant of mercaptopurine metabolism and toxicity during treatment for acute lymphoblastic leukemia. Clin Pharmacol Ther. 2009;85(2):164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clinical Pharmacogenetics Implementation Consortium. Guidelines. https://cpicpgx.org/guidelines/. Published 2020. Accessed December 16, 2021.
- 33.U.S. Food and Drug Administration. Table of Pharmacogenetic Associations. https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations. Published 2020. Accessed February 24, 2021.
- 34.Giri J, Moyer AM, Bielinski SJ and Caraballo PJ. Concepts Driving Pharmacogenomics Implementation Into Everyday Healthcare. Pharmgenomics Pers Med. 2019;12:305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy KD, Blake K, Fletcher-Hoppe C, Franciosi J, Goto D, Hicks JK, et al. Opportunities to implement a sustainable genomic medicine program: lessons learned from the IGNITE Network. Genet Med. 2019;21(3):743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts MC, Kennedy AE, Chambers DA and Khoury MJ. The current state of implementation science in genomic medicine: opportunities for improvement. Genet Med. 2017;19(8):858–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92(4):414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


