Abstract
Stress-associated proteins (SAPs), a group of zinc-finger-type proteins, have been identified as novel regulators of plant abiotic and biotic stresses. However, although they have been discovered in different plant species, their precise functional roles remain unclear. Here, we identified 14 SAP subfamily genes in the pepper genome. An investigation of the promoter regions of these genes for cis-regulatory elements associated with abiotic stress responses revealed the presence of multiple stress-related elements. Domain and phylogenetic analyses using the corresponding protein sequences revealed that the CaSAP genes can be classified into six groups (I–VI) and sorted into two broad types. Expression levels of the CaSAP genes were found to be differentially induced by low temperature, the dehydration stress, or exogenous abscisic acid. Group II and IV genes were highly induced by the low temperature and dehydration treatments, respectively. Moreover, subcellular localization analysis indicated that the proteins in these two groups are distributed in the nucleus, cytoplasm, and plasma membrane. Among the pepper plants silenced with the three identified group II CaSAP genes, the CA02g10410-silenced plants showed tolerance to low temperature, whereas the CA03g17080-silenced plants were found to have temperature-sensitive phenotypes. Interestingly, group IV CaSAP-silenced pepper plants showed drought-tolerant phenotypes. These findings contribute to a preliminary characterization of CaSAP genes and provide directions for future research on the biological role of CaSAPs in response to different abiotic stresses.
Keywords: ABA, drought, low temperature, pepper, virus-induced gene silencing
Introduction
With the ever-increasing severity of environmental problems, plants are becoming more frequently subjected to adverse abiotic stresses, such as extreme temperatures, high salinity, and dehydration, which contribute to limiting plant growth and crop productivity (Hasegawa et al., 2000; Park et al., 2016; He et al., 2018; Waqas et al., 2019). To survive under stress conditions, plants accordingly need to adapt to these stresses by mounting appropriate defense responses. In this regard, signaling molecules, such as different types of protein kinases, protein phosphatases, transcription factors, and other regulatory proteins, participate in several stress signaling pathways to maintain homeostasis (Shinozaki and Yamaguchi-Shinozaki, 2000; Schroeder et al., 2001; Zhu, 2002; Ding et al., 2020). In response to the transduction of such signals, downstream target genes are up- or downregulated (Chinnusamy et al., 2005; Tuteja, 2007; Ding et al., 2020), thereby enabling plants to adaptively modify their physiological or morphological status (Parihar et al., 2015; Volkov, 2015; Liu et al., 2018).
Exposure to the low temperature stress, also referred to as cold stress, can enable plants to cold acclimate under temperate climatic conditions (Guo et al., 2018); it can be divided into chilling (0–15°C) and freezing (under 0°C) stresses. Chilling stress is the primary stress to which subtropical crops, such as rice, are subjected; to adapt to chilling stress conditions, plants have evolved the ability to perceive cold signals and the transduction of related signals. In this context, the C-repeat (CRT)-binding factors/dehydration-responsive element binding protein 1 (CBF/DREB1)-dependent signaling pathway has been established to be the main regulatory mechanism activated in response to the low temperature stress (Thomashow, 1999). Recent studies have reported that a range of different proteins, including kinases, transcription factors, and regulator proteins, are involved in this mechanism, which leads to the activation of cold-regulated (COR) genes and enhanced chilling tolerance (Shi et al., 2018; Ding et al., 2020). Moreover, water deficit and elevated temperature are severe issues that contribute to promoting the dehydration stress in plants (Park et al., 2016; Lim et al., 2018a; Zhang et al., 2021). In this regard, plants can alter their physiological or morphological patterns by controlling changes in the balance of certain hormones (Waqas et al., 2019), among which the phytohormone abscisic acid (ABA) is known to play key roles in the responses to the dehydration stress (Cutler et al., 2010). The main ABA signaling pathway is the PYR/PYL/RCAR-PP2Cs-SnRK2s cascade (Shinozaki and Yamaguchi-Shinozaki, 2000; Nakashima and Yamaguchi-Shinozaki, 2013; Sah et al., 2016). Nevertheless, although the responses of plants to abiotic stress have been generally well characterized, much is still unknown regarding the underlying mechanisms, and studies on stress responses are accordingly still being actively conducted.
Stress-associated proteins (SAPs) are a group of zinc-finger-type proteins that are reportedly associated with abiotic stress responses, immunity, and development (Mukhopadhyay et al., 2004; Dixit et al., 2018; Dong et al., 2018; Liu et al., 2019c; Wang et al., 2019; Zhao et al., 2020). Since the discovery of the first SAP gene, the OsiSAP1 (OsSAP1) gene in rice, others have been characterized in various plant species as genome sequencing technology has advanced (Mukhopadhyay et al., 2004; Vij and Tyagi, 2006; Solanke et al., 2009; Giri et al., 2013; Dong et al., 2018; Liu et al., 2019a; Zhang et al., 2019; Ben Saad et al., 2020; Lai et al., 2020; Wang et al., 2021). However, no SAP genes have been isolated from pepper plant. Relatively well-characterized SAPs have been found to contain an A20 domain at the N terminus and/or an AN1 domain at the C terminus, which are highly conserved across species (Vij and Tyagi, 2008; Lai et al., 2020). In animals, the A20 zinc-finger proteins function as negative regulators of inflammation and have de-ubiquitinating activity (Lademann et al., 2001; Evans et al., 2004). Although the precise function of the AN1 zinc-finger proteins is unclear, recent studies have reported that they may regulate protein–protein interactions (Linnen et al., 1993; Chang et al., 2011). In plants, several SAPs containing A20/AN1 zinc-finger domains have been reported to confer tolerance to multiple abiotic stresses (Huang et al., 2008; Kanneganti and Gupta, 2008; Giri et al., 2011; Lloret et al., 2017; Yoon et al., 2018; He et al., 2019) and play a role in immunity (Liu et al., 2019b,c). Moreover, Arabidopsis SAP5 has been reported to have E3 ubiquitin ligase activity (Kang et al., 2011). A further type of SAP consists of an N-terminal AN1 zinc-finger domain and/or a C-terminal Cys2-His2 (C2H2) zinc-finger domain, and over the past two decades, C2H2 zinc-finger proteins have been studied and reported to have diverse functions in plant growth, development, and biotic/abiotic stress resistance (Han et al., 2020).
Pepper (Capsicum annuum) is one of the most important vegetable crops cultivated globally that has economic value as a spice, medicine, vegetable, and biopesticide (Lim et al., 2018b). Although the demand for peppers is increasing worldwide, their productivity can be limited to varying extents by adverse environmental conditions, such as dehydration, high salinity, and extreme temperatures. To solve this problem, numerous studies have focused on the defense mechanisms activated in response to such environmental stresses (Chen et al., 2014; Guo et al., 2014; Park et al., 2016; Lim et al., 2018a,2020; Kang et al., 2020; Wu et al., 2020). In recent decades, numerous stress-related genes have been discovered in the pepper plants. Furthermore, the recent whole-genome sequencing of pepper has accelerated these ongoing research efforts (Kim et al., 2014; Qin et al., 2014; Hulse-Kemp et al., 2018). Nonetheless, to our knowledge, SAP gene family and their functional role, especially in response to dehydration and low temperature, have been not yet identified from pepper plants.
In the present study, we performed a genome-wide analysis of SAP family genes in C. annuum and identified 14 SAP genes, the expression patterns of which were investigated in response to different abiotic stresses and exogenous ABA. In addition, we selected six SAP genes for conducting phenotypic assays. It is anticipated that the findings of this study will make a significant contribution to advancing our current understanding of plant SAPs and the mechanisms underlying the responses of plants to abiotic stress.
Materials and Methods
Plant Material and Growth Conditions
In the present study, we used pepper (Capsicum annuum cv. Nockwang) and tobacco (Nicotiana benthamiana) as experimental plants. Pepper seeds were soaked in a growth chamber at 28°C under dark conditions for 4 days. The germinated seeds were then planted in a mixture of steam-sterilized soil (peat moss, perlite, and vermiculite, 5:3:2, v/v/v), sand, and loam soil (1:1:1, v/v/v). The seeds of tobacco plants were sown in the same soil mixture. Both plant types were grown in a growth room at 25 ± 1°C and 60% relative humidity under white fluorescent light (130 μmol photons⋅m–2⋅s–1) on a 16 h light/8 h dark cycle.
Abiotic Stress Treatments
Plants were subjected to different abiotic stresses, the effects of which were subsequently assessed based on quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and phenotypic analyses. Pepper plants were subjected to dehydration by detaching the shoots and subsequently harvesting the leaves at designated time points (0, 6, and 12 h). For low temperature treatments, pepper plants were placed in an unilluminated growth chamber at 10°C, and leaves were harvested after 0, 6, and 12 h. Other plants were treated with ABA (100 μM) or mannitol (600 mM), with leaves again being collected after 0, 6, and 12 h as described by previous studies (Lim et al., 2019; Lim C. W. et al., 2021; Lim J. et al., 2021).
RNA Isolation and Quantitative Reverse Transcription-Polymerase Chain Reaction
Total RNA was isolated from the leaves of pepper plants at the 6th-leaf stage. The RNA thus obtained was quantified using spectrophotometer, and 1 (μg of the quantified RNA was used as a template to synthesize cDNA using the iScriptTM cDNA synthesis kit (Bio-Rad, Hercules, CAalifornia, United StatesA). The synthesized cDNA was amplified in a CFX96 TouchTM Real-Time PCR detection system (Bio-Rad) using the iQTMSYBR Green Supermix (Bio-Rad) and specific primers (Supplementary Table 1). Pepper Actin1 (CaACT1 and CA12g08730) was used as an internal control.
Subcellular Localization
The coding regions of the CaSAP genes (groups II and IV), minus stop codons, were inserted into a bar 35S-GFP vector containing the cauliflower mosaic virus 35S promoter and a green fluorescent protein (GFP) tag at the C-terminal end of the insert. Agrobacterium tumefaciens strain GV3101 harboring the GFP-tagged CaSAP (group II and IV) constructs was mixed with Agrobacterium strain p19 (1:1; OD600 = 0.5) and co-infiltrated into the epidermal cells of 5-week-old Nicotiana benthamiana leaves to induce transient expression. The 35S-GFP empty vector was used as a positive control. Two days after infiltration, we examined GFP signals using an LSM700 confocal microscope (Carl Zeiss, Jena, Germany) and analyzed the signals using the ZEN 3.1 software. DAPI (1 μg/ml) was used as a nucleus marker and FM4-64 (50 μM) as a plasma membrane marker.
A Virus-Induced Gene Silencing System
A tobacco rattle virus (TRV)-based virus-induced gene silencing system was used to generate a CaSAP (group II and IV) knock-down model in pepper plants, as described in previous studies (Pflieger et al., 2013; Jeong et al., 2020). We designed 300 bp fragments for each cDNA of CaSAP (groups II and IV), which were validated as regions for gene silencing using the VIGS tool1. Agrobacterium tumefaciens strain GV3101 carrying pTRV1 and pTRV2:CaSAP (groups II and IV) or pTRV2:00 (negative control) was co-infiltrated into the cotyledons of 2-week-old pepper plants (OD600 = 0.2 for each construct). The infected pepper plants were cultivated under the aforementioned growth conditions, and 2 weeks later, we measured the expression of each CaSAP gene using qRT-PCR (Supplementary Figure 1).
Results
Identification of SAP Family Genes in Capsicum annuum
In recent decades, SAP family genes have been identified as novel regulators involved in the responses of plants to different stresses, such as drought, high salinity, and extreme temperatures; additionally, they are known to play crucial roles in plant immunity (Mukhopadhyay et al., 2004; Kanneganti and Gupta, 2008; Ströher et al., 2009; Dixit and Dhankher, 2011; Tyagi et al., 2014; Liu et al., 2019b). To investigate whether members of the SAP family are present in pepper and function similarly to those of other species, we obtained the amino acid sequences of 14, 27, and 13 SAP family genes from Arabidopsis, soybean, and tomato, respectively, and then searched for pepper SAP family genes using these sequences as a query in BLASTP searches. We identified 14 SAP genes in pepper containing the typical SAP conserved domains, including the AN1, A20, and C2H2 zinc-finger domains (Table 1 and Figure 1A). On the basis of the location of the AN1 domain (the N or C terminus), the 14 pepper SAP genes can be divided into two types (Type I and II, respectively). Moreover, the predicted subcellular localization indicated that these genes are primarily distributed in the nucleus (Table 1). Using the amino acid sequences of SAP genes in pepper, Arabidopsis, soybean, and tomato, we conducted phylogenetic analysis based on the neighbor-joining method implemented using the MEGA X software (Kumar et al., 2018). We found that the pepper SAP genes were clustered into six clades (designated groups I to VI), each of which contained one to three members of the 14 pepper genes (Figure 1B). As several previous studies have reported that the functions of SAP proteins are primarily associated with responses to abiotic stresses, we focused on these responses in the present study.
TABLE 1.
Information on the CaSAP subfamily genes.
| Group | Gene locus | Chromosome | Location | Strand direction | CDS (bp) | Intron | Predicted protein (aa) | pI | MW (kDa) | Zing finger domain | Predicted subcellular location |
| I | CA01g04040 | 01 | 6602752–6603225 | – | 474 | 0 | 157 | 8.59 | 16.91 | A20-AN1 | Chloroplast, Nucleus, Cytoplasm |
| II | CA02g10410 | 02 | 129918896–129919300 | – | 405 | 0 | 134 | 8.88 | 15.28 | A20-AN1 | Nucleus, Chloroplast |
| CA03g17080 | 03 | 196705800–196706183 | – | 384 | 0 | 127 | 7.95 | 14.21 | AN1 | Chloroplast, Nucleus, Cytoplasm | |
| CA00g44110 | nd | nd | 426 | 0 | 141 | 8.88 | 15.76 | A20-AN1 | Nucleus | ||
| III | CA10g16510 | 10 | 220169266–220169757 | – | 492 | 0 | 163 | 7.53 | 18.09 | A20-AN1 | Chloroplast |
| CA12g06430 | 12 | 22845946–22846587 | + | 642 | 0 | 213 | 8.25 | 23.52 | A20-AN1 | Mitochondria, Chloroplast, Cytosol | |
| IV | CA01g16220 | 01 | 101708816–101709355 | + | 540 | 0 | 179 | 7.94 | 19.16 | A20-AN1 | Chloroplast |
| CA01g19950 | 01 | 160637657–160638169 | + | 513 | 0 | 170 | 8.43 | 18.33 | A20-AN1 | Chloroplast | |
| CA10g20690 | 10 | 230827282–230827800 | + | 519 | 0 | 172 | 8.62 | 18.23 | A20-AN1 | Nucleus | |
| V | CA01g19980 | 01 | 160706592–160707140 | + | 555 | 0 | 184 | 9.12 | 20.26 | A20-AN1 | Nucleus |
| CA12g22410 | 12 | 234718042–234718554 | – | 513 | 0 | 170 | 9.25 | 19.19 | AN1 | Nucleus | |
| VI | CA02g29220 | 02 | 168266440–168267317 | – | 570 | 1 | 189 | 8.96 | 20.81 | AN1-AN1 | Nucleus |
| CA05g01100 | 05 | 1965718–1969104 | – | 834 | 1 | 277 | 8.70 | 31.25 | AN1-AN1-C2H2-C2H2 | Chloroplast, Nucleus | |
| CA10g19590 | 10 | 228970650–228973903 | + | 822 | 1 | 273 | 8.79 | 30.28 | AN1-AN1-C2H2-C2H2 | Nucleus |
Gene locus and location are from Capsicum annuum cv ‘CM334’ genome (release 1.55). Domain analysis is performed using a web tool SMART (Simple Modular Architecture Research Tool; http://smart.embl.de/) Subcellular localization of protein is predicted using a web tool WOLF PSORT (http://www.genscript.com/wolf-psort.html). nd, not detected due to unsequenced portion of gene.
FIGURE 1.
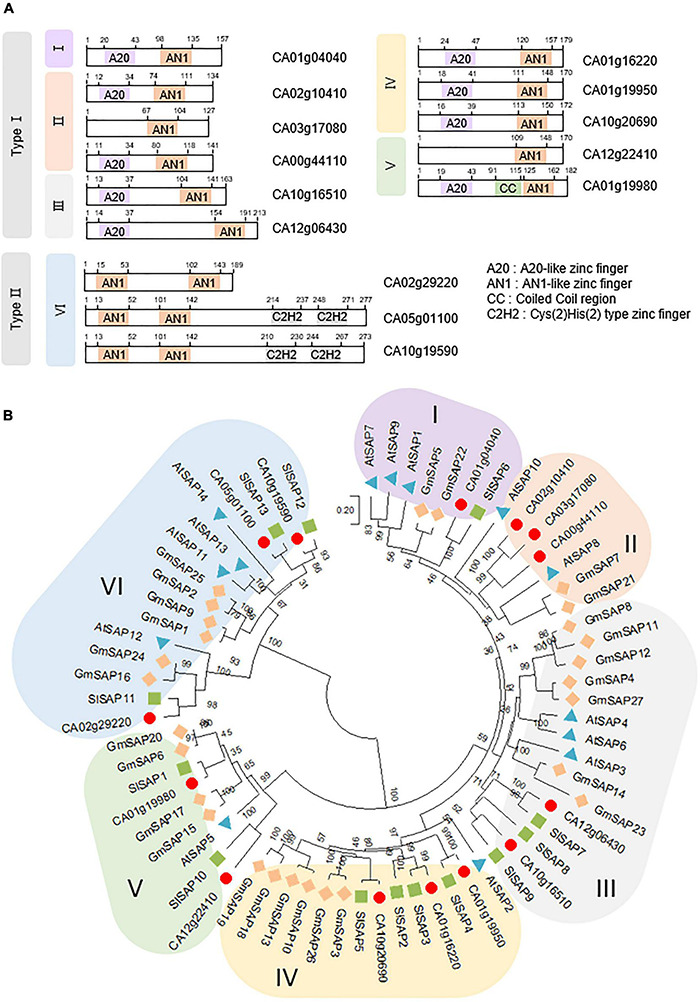
Pepper stress-associated protein (SAP) subfamily genes. (A) Domain organization of the pepper SAP subfamily genes. The amino acid sequences were obtained from SGN (https://solgenomics.net/), and domains were predicted using SMART (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/). (B) Phylogenetic tree analysis of the pepper SAP subfamily genes. The amino acid sequences were deduced from the full-length coding sequences of SAP genes in pepper (red circles), Arabidopsis (blue triangles), soybean (orange diamonds), and tomato (green squares) and used for comparisons. The phylogenetic tree was generated based on the neighbor-joining method implemented in MEGA X software.21. Bootstrap values shown at branch points were calculated from 1,000 bootstrap replications. The scale bar denotes the evolutionary distance computed using the Poisson correction method.
Expression Patterns of the Pepper SAP Family
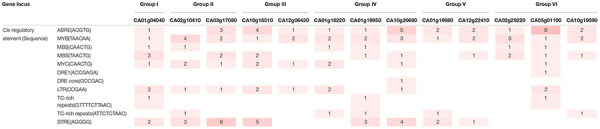
To investigate the tissue-specific expression of pepper SAP genes, we conducted qRT-PCR using cDNA templates derived from different tissues (leaf, stem, flower, and root) (Figure 2A). In common, group II and VI CaSAP genes showed low expression levels in all tissues. The expression level of group V CA01g19980 was higher in the leaf and stem than other CaSAPs. In the flower, the transcript of group IV CA10g20690 showed the higher level than that of other CaSAPs. Under water deficit conditions, dehydration is typically caused, which enhances biosynthesis and accumulation of the phytohormone ABA as signal molecules (Cutler et al., 2010). Cold stress also leads to dehydration in plant by reducing root water uptake (Steponkus, 1984). Based on these, we initially analyzed whether proteins in the pepper SAP family (CaSAP) play functional roles in response to water stress caused by dehydration and low temperature. First, we performed qRT-PCR using template cDNA obtained from plants that had been subjected to different stress treatments (low temperature, dehydration, ABA, and mannitol), and the primers listed in Supplementary Table 1. As a positive control for stress treatment, we used CaOSR1, which is homologous to Arabidopsis RD29B (Park et al., 2016; Lim et al., 2020). It is noteworthy that members of the aforementioned six groups tended to show differential responses to different stresses (Figure 2B). For example, after 6 and 12 h of treatment, the expression levels of group II CaSAP genes were found to increase more prominently in response to low temperature (10°C) than those of the other groups. Similarly, we detected significant increase in the expression levels of group IV, V, and VI CaSAP genes in plants subjected to the dehydration stress. In particular, compared with the other CaSAP family genes, CA01g16220, a group IV CaSAP gene, showed the highest level of expression at 12 h after dehydration treatment. In response to mannitol treatment, most of the CaSAP genes showed significantly increased expression compared with that in the control. ABA is a key phytohormone that regulates abiotic stress responses (Hasegawa et al., 2000; Shinozaki and Yamaguchi-Shinozaki, 2000; Schroeder et al., 2001); hence, we sought to determine whether expression of the CaSAP genes shows differential responses to ABA. Six hours after ABA treatment, we detected the upregulated expression of seven of the 14 CaSAP genes. Subsequently, we analyzed the promoter region cis-regulatory elements to gain an understanding of stress- and ABA-induced transcriptional regulation of the CaSAP genes (Table 2). Specifically, we obtained 3 kb upstream sequences of the CaSAP genes from the Capsicum annuum cv. CM334 (Criollo de Morelos 334) genome database accessed from SGN2 and used the New PLACE web tool3 to predict the cis-regulatory elements. We accordingly identified a number of stress- and ABA-responsive cis-elements, including stress response element (STRE), low temperature-responsive element (LTR), dehydration-responsive element (DRE), TC-rich repeats, ABA-responsive element (ABRE), MYB, MYC, and MBS (Narusaka et al., 2003; Yamaguchi-Shinozaki and Shinozaki, 2005; Table 2). A majority of the CaSAP genes were found to contain an abundance of ABRE, MYB, MBS, and MYC motifs, whereas TC-rich repeats, DRE1, and DRE core were rarely predicted in these genes. Furthermore, with the exception of group VI CaSAP genes, STRE was detected in abundance. These findings provide evidence to indicate that members of the CaSAP family are regulated by different cis-regulatory elements, resulting in transcriptional regulation in response to multiple abiotic stresses.
FIGURE 2.
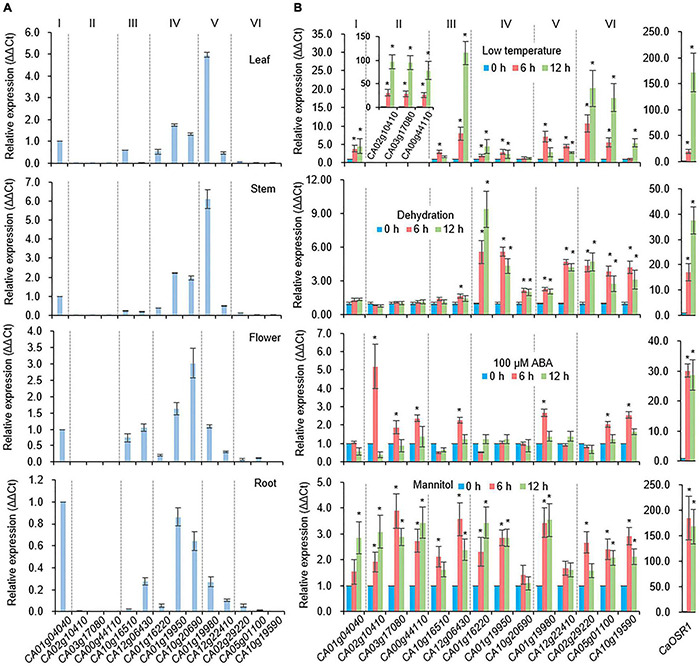
Expression patterns of CaSAP genes. (A) Tissue-specific expression of CaSAP genes. The expression levels of CaSAP genes were determined via qRT-PCR analysis using cDNA derived from the first- and second-leaves, stems, and roots harvested from the six-leaf stage pepper plants and fully opened flowers. Values for CA01g04040 were set to 1.0. (B) Expression levels of CaSAP genes in response to different abiotic stresses. Expression levels of CaSAP genes were determined via qRT-PCR analysis using cDNA derived from the first and second leaves of pepper plants (six-leaf stage) subjected to different abiotic stresses: dehydration (shoot detachment), ABA (100 μM), mannitol (600 mM), and low temperature (10°C). The relative expression (ΔΔCT) of CaSAP genes was normalized to that of CaACT1, which was used as an internal control gene, and values at 0 h were set to 1.0. Values are presented as the mean ± standard error of values from three independent experiments. Asterisks indicate significant differences compared to the value at 0 h for each gene (Student’s t-test; *P < 0.05).
TABLE 2.
Cis-regulatory elements information of CaSAP subfamily. Gene locus are from Capsicum annuum cv. ‘CM334’ genome (release 1.55).
|
Subcellular Localization of CaSAPs
In previous studies on rice, Arabidopsis, soybean, and tomato, most of the SAP family proteins have been detected in the nucleus and cytoplasm (Kothari et al., 2016; Dixit et al., 2018; Zhang et al., 2019; Zhao et al., 2020). In the present study, we similarly predicted that these proteins would show nuclear and cytoplasmic distribution in pepper (Table 1). On the basis of these predictions, we performed subcellular localization experiments for six of the identified CaSAP proteins (CA02g10410, CA03g17080, CA00g44110, CA01g16220, CA01g19950, and CA10g20690) tagged with green fluorescent protein (Figure 3). We designed A. tumefaciens strain GV3101 constructs harboring the p35S-CaSAPs-GFP vector, and transiently expressed the fusion proteins in leaf epidermal cells of N. benthamiana. Consistent with the findings obtained for other plants, we found that the CaSAP proteins were distinctly localized in the nucleus, cytoplasm, and plasma membrane (Figure 3), which implied that these six CaSAPs are functionally active in the pepper cell.
FIGURE 3.
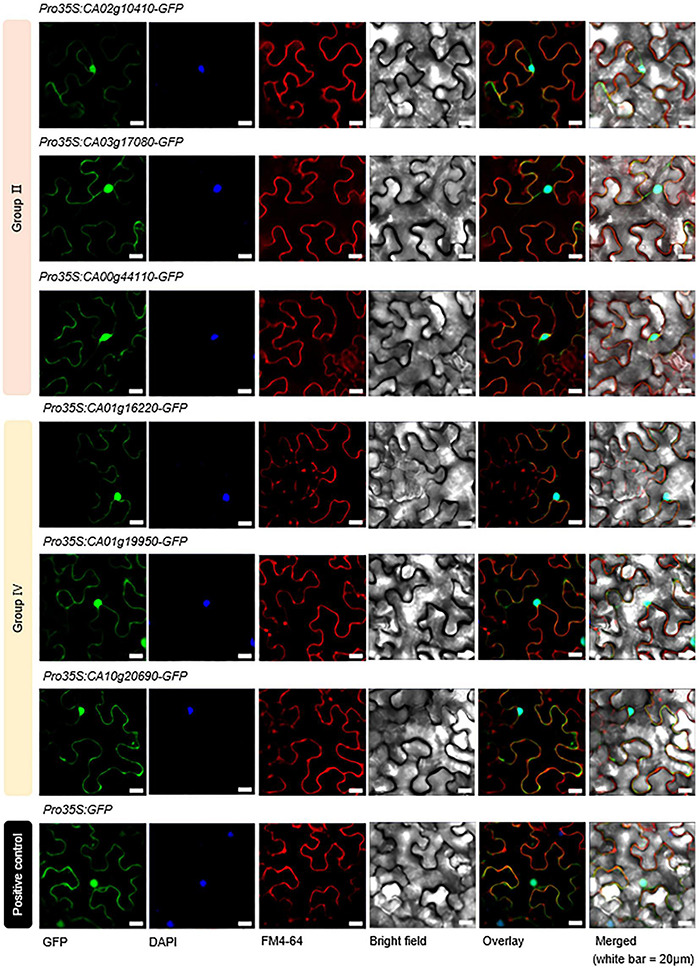
Subcellular localization of CaSAP proteins. The leaves of Nicotiana benthamiana were transformed with constructs harboring SAP–GFP fusion proteins via Agrobacterium-mediated infiltration. Confocal images show transiently transformed N. benthamiana epidermal cells expressing group II (CA02g10410, CA03g17080, and CA00g44110) and group IV (CA01g16220, CA01g19950, and CA10g20690) SAP–GFP fusion proteins. The 35S-GFP empty vector was used as a positive control. White bar = 20 μm.
Group II CaSAP-Silenced Pepper Plants Show Altered Phenotypes in Response to the Low Temperature Stress
The expression of CaSAP genes was induced by subjecting plants to water stress caused by low temperature, dehydration, and osmotic stress (Figure 2); then, we examined the stress-related functions of these genes using a tobacco rattle virus-based virus-induced gene silencing (VIGS) system (Figures 4, 5). To verify the efficiency of VIGS, we performed RT-PCR analysis (Supplementary Figure 1) and established that the levels of CaSAP gene expression were lower in CaSAP-silenced pepper plants than in control plants. Under normal growth conditions, we were unable to detect any phenotypic differences between the control and CaSAP-silenced pepper plants (Figures 4, 5). For group II CaSAP genes, we investigated the response to low temperature (Figure 4) by placing CaSAP-silenced and control (TRV2:00) pepper plants in a cold chamber at 4°C for 2 days and thereafter allowing the plants to recover at 25°C for 8 h (Figure 4A). Under low temperature conditions, we observed a shrinkage of the first and second leaves of both control and CaSAP-silenced pepper plants (Figure 4A, middle panel). During the recovery phase under normal growth conditions, we noted that recovery status of the different CaSAP-silenced pepper plants differed from that of the control plants. For example, while the recovery of CA02g10410-silenced pepper plants was more evident than that of the control plants, recovery of the CA03g17080-silenced pepper was found to be less pronounced, and CA00g44110-silenced pepper showed no appreciable differences in revival compared with the control. To quantify these observations, we calculated the survival rate and fresh weights of the first and second leaves before and after recovery (Figures 4A,B). Prior to chilling treatment, we detected no significant differences between the CaSAP-silenced and control plants. Nevertheless, in line with our expectations, we found that the fresh weights of CaSAP-silenced pepper leaves differed significantly (CA02g10410 and CA03g17080) or not (CA00g44110) from those of the controls post-recovery. Low temperature reduces the ability root water uptake and causes dehydration stress (Steponkus, 1984). This stress leads to increasing electrolyte leakage and lipid peroxidation, so we examine these physiological responses. As expected, electrolyte leakage and malondialdehyde (MDA) content showed significantly lower in CA02g10410-silenced pepper plants and significantly higher in CA03g17080-silenced pepper plants than that of control plants. CA00g44110-silenced pepper plants showed no differences (Figures 4C,D). To examine the corresponding expression patterns of stress-responsive genes, we conducted qRT-PCR analysis of control and CaSAP-silenced pepper plants (Figure 4E). The expression levels of CaOSR1 and CaRAB18 showed patterns consistent with the findings of previous phenotypic analysis. Among the CaSAP-silenced plants, expression levels of the two genes in TRV2:CA02g10410 pepper were found to be significantly higher than those in control plants at 4 h after chilling treatment, whereas the transcript levels of TRV2:CA03g17080 pepper were found to be lower than those in control plants in response to chilling treatment. Notably, however, the expression levels of CaDREB1 showed no significant correlation with drought phenotypes. Under basal conditions, with the exception in TRV2:CA00g44110 pepper plants, the expression levels of CaDREB1 did not show significant differences under basal conditions, whereas the expression of TRV2:CA02g10410 and TRV2:CA03g17080 pepper was lower and higher, respectively, than that of the control plants after chilling treatment. Moreover, we detected no significant differences between control and CaSAP-silenced lines with respect to CaNCED3 expression.
FIGURE 4.
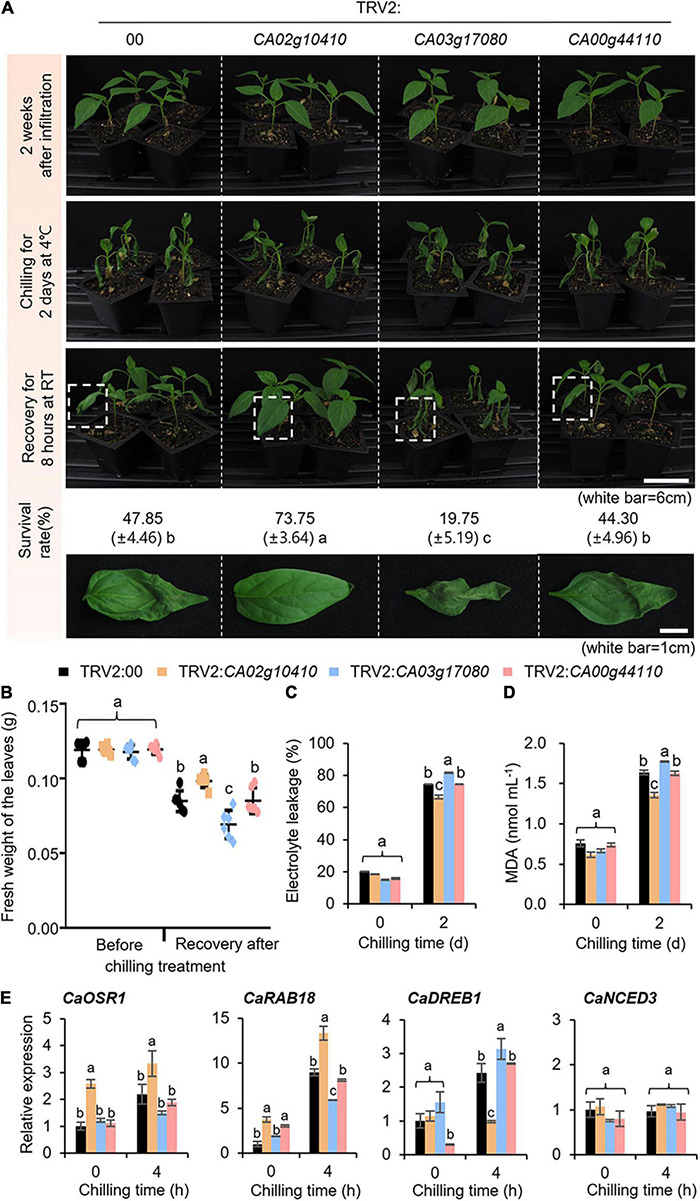
Phenotypic analysis of group II CaSAP genes to low temperature. (A) The low-temperature phenotypes of group II TRV2:CaSAP. Four-week-old plants of each line were subjected to the low temperature stress by chilling (4°C) for 2 days, followed by recovery for 8 h and the survival rates of each line were calculated. (B) The relative fresh weights of each line were calculated prior to chilling and after recovery. (C,D) Effect of low temperature stress on electrolyte leakage (C) and lipid peroxidation (D) in the leaves of TRV2:00 and group II TRV2:CaSAP pepper plants. Four-week-old plants were subjected to the low temperature (4°C) for 2 days, as shown in (A) (n = 20 plants of each line per replicate, 3 replicates). (E) Expression analysis of low temperature-inducible genes in the leaves of TRV2:00 and group II TRV2:CaSAP pepper plants. The relative expression (ΔΔCT) of CaSAP was normalized to that of CaACT1, which was used as an internal control gene. All values are presented as the mean ± standard error of values obtained from three independent experiments. Different letters indicate significant differences among the control and TRV2:CaSAP pepper plants (ANOVA; P < 0.05).
FIGURE 5.
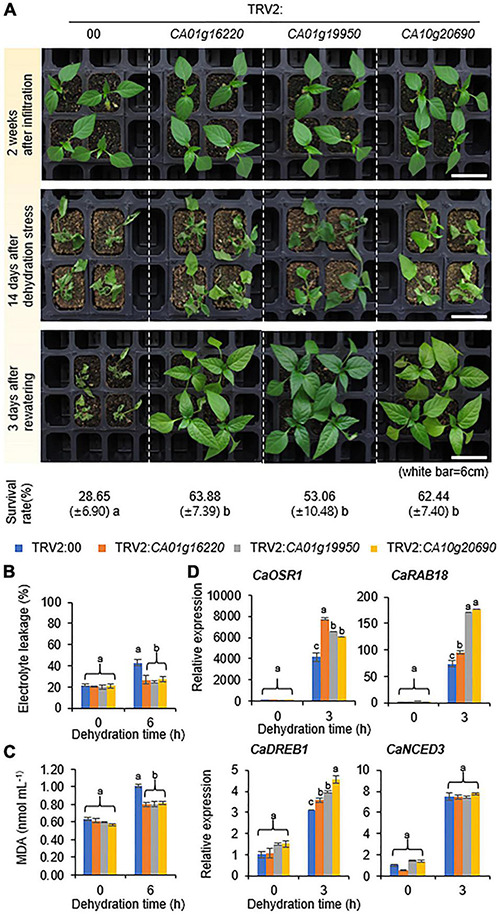
Phenotypic analysis of group IV CaSAP genes to the dehydration stress. (A) Dehydration-tolerant phenotypes of plants transformed with group IV TRV2:CaSAP. Four-week-old plants of each line were subjected to the dehydration stress by withholding watering for 14 days followed re-watering for 3 days. The percentages of surviving plants were calculated after re-watering. (B,C) Effect of dehydration stress on electrolyte leakage (B) and lipid peroxidation (C) in the leaves of TRV2:00 and group IV TRV2:CaSAP pepper plants. Four-week-old plants were subjected to the dehydration stress by detaching shoot for 6 h (n = 20 plants of each line per replicate, 3 replicates). (D) Expression analysis of dehydration-inducible genes in the leaves of TRV2:00 and group IV TRV2:CaSAP pepper plants. The relative expression (ΔΔCT) of CaSAP was normalized to that of CaACT1, which was used as an internal control gene. All values are presented as the mean ± standard error of values from three independent experiments. Different letters indicate significant differences among control and TRV2:CaSAP pepper plants (ANOVA; P < 0.05).
Group IV CaSAP-Silenced Pepper Plants Confer Tolerance to the Dehydration Stress
To investigate the response to the dehydration stress, we subjected group IV CaSAP-silenced and control (TRV2:00) pepper plants to dehydration by withholding water for 14 days and thereafter re-watering for 3 days (Figure 5A). Notably, all group IV CaSAP-silenced pepper plants showed less wilted phenotypes than the control plants. To quantify these responses, we calculated survival rates; in line with our expectations, the survival rates of the group IV CaSAP-silenced pepper (63.88% ± 7.39%, 53.06% ± 10.48%, and 62.44% ± 7.40%) were found to be significantly higher than those of control plants (28.65% ± 6.90%). To identify whether these phenotypes were related to physiological responses, we examined electrolyte leakage and lipid peroxidation. For this, the shoots of group IV CaSAP-silenced and control pepper plants were detached and dehydrated for 6h. As expected, electrolyte leakage and malondialdehyde (MDA) content showed significantly lower in group IV CaSAP-silenced pepper plants than control (Figures 5B,C). To establish whether these tolerance phenotypes were associated with the expression levels of stress-responsive genes, we performed qRT-PCR as described in the previous section (Figure 5D and Supplementary Table 1). Under normal growth conditions, we detected no significant differences between control and CaSAP-silenced pepper plants with respect to stress-responsive gene expression. However, the expression levels of CaOSR1, CaRAB18, and CaDREB1, but not CaNCED3, were found to be higher in CaSAP-silenced pepper than in the control plants after 3 h of dehydration treatment. These findings provide evidence that group IV CaSAP genes play a negative functional role in the response of pepper to dehydration.
Discussion
Stress-associated proteins zinc-finger proteins have been reported to be associated with the immune system in humans and multiple stress responses in plants (Evans et al., 2004; Chang et al., 2011; Giri et al., 2013; Zhang et al., 2019). To date, SAP genes have been identified in various monocot and dicot plant species, including Arabidopsis thaliana, Oryza sativa, Solanum lycopersicum, and Medicago truncatula (Vij and Tyagi, 2006; Solanke et al., 2009; Gimeno-Gilles et al., 2011; Gao et al., 2016; Dong et al., 2018; Saad et al., 2018, 2019; Xu et al., 2018; He et al., 2019; Li et al., 2019; Zhang et al., 2019; Lai et al., 2020; Wang et al., 2021). However, prior to the present study, there had been no similar characterization of SAP genes in Capsicum annuum.
In this study, we identified 14 SAP genes in Capsicum annuum. However, unlike our study, there is a wide variability for number of gene members in various plant such as 27 SAP genes in Glycine max and 57 genes in Brassica napus (He et al., 2019; Zhang et al., 2019). As suggested by previous studies (Gao et al., 2016; Dong et al., 2018; Lai et al., 2020), this phenomenon is possibly due to gene duplications. Consistently, we found 4 pairs of CaSAP genes have undergone dispersed and transposed gene duplication events in pepper plants (Supplementary Figure 2A).
In plants, cis-regulatory elements play an important role in the transcriptional regulation involved in growth, development, and different stress responses (Narusaka et al., 2003; Yamaguchi-Shinozaki and Shinozaki, 2005). In the present study, we found that the promoter regions of CaSAP genes are characterized by the presence of multiple types of these regulatory elements, including ABRE, MYB, MYC, STR, STRE, and MBS, which are associated with the responses to abiotic stresses (Table 2). In previous studies, it has been noted that a common feature of the SAP genes identified in different species is that a large proportion is intron-less genes. In rice, tomato, soybean, and castor bean, for example, a majority of the SAP genes lack introns, whereas a small number SAP genes have very few introns in their genomic information (Vij and Tyagi, 2006; Solanke et al., 2009; Zhang et al., 2019; Wang et al., 2021). Similarly, in pepper, type I CaSAPs, which contain A20 or AN1 domains, are lacking in introns, whereas the AN1 or C2H2 domain-contain type II CaSAPs are characterized by a single intron in their genomic region (Table 1 and Supplementary Figure 2B). Conceivably, by minimizing the number of steps necessary for post-transcriptional processing, this absence of intronic regions may confer the ability to mount a more rapid and precise response to stress stimuli (Jeffares et al., 2008; Grzybowska, 2012; Liu et al., 2021). These findings thus provide evidence indicating that CaSAP genes may play role in the rapid responses to different abiotic stresses.
In the present study, compared with the genes in other CaSAP groups, group II CaSAP genes were found to be highly induced following exposure to a low temperature (Figure 2). The CA02g10410-, CA03g17080-, and CA00g44110-silenced pepper plants showed different phenotypes under low-temperature conditions (Figure 4). Moreover, we observed that the expression of group IV CaSAPs was induced by drought stress and accordingly predicted that group IV CaSAPs would act as positive regulators of drought stress. However, contrary to our expectations, we found that pepper plants in which these genes had been knocked down displayed drought-tolerant phenotypes (Figure 5). There are two plausible explanations that could account for the observed phenotypes: (i) the induced genes may play a role in the defense response or contribute to the recovery of plants to normal growth and development; and (ii) gene transcription is regulated by multiple processes; hence, both positive and negative regulators may be induced by the same signal. In this context, the transcripts of group A PP2Cs have been shown to accumulate under drought stress conditions via a negative feedback regulatory loop (Merlot et al., 2001) and are induced by ABA and function as negative regulators of this phytohormone (Lim et al., 2015).
Abscisic acid is a major phytohormone that regulates seed germination, stomata closure, plant growth, and stress responses (Schroeder et al., 2001; Fujita et al., 2011; Nakashima and Yamaguchi-Shinozaki, 2013); previous studies have reported that several SAP genes, including AtSAP13, AtSAP9, OsSAP1, and GmSAP16, are induced by ABA and confer resistance to several abiotic stresses. In addition, GmSAP16 and AtSAP9 affect the expression levels of stress-related genes (Mukhopadhyay et al., 2004; Kang et al., 2017; Dixit et al., 2018; Zhang et al., 2019). Numerous stress-related genes associated with defense responses are induced when plants are exposed to different stress conditions (Gonzalez-Guzman et al., 2012; Park et al., 2015). For example, NCED3, induced by several sources of abiotic stress, is a key enzyme in ABA biosynthesis (Iuchi et al., 2001; Endo et al., 2008). In the present study, we found that three of the 14 identified CaSAP genes, all of which clustered in the group II category of SAPs, were significantly induced by exogenous ABA (Figure 2). Notably, in a manner similar to that reported in previous studies, we found that two of the group II CaSAP genes (CA02g10410 and CA03g17080) had considerable effects on the expression of other stress-related genes, including CaOSR1 (homologous to RD29B), CaRAB18, and CaDREB1, in response to the low temperature stress (Figure 4C). Moreover, whereas the group IV CaSAP genes were not induced by exogenous ABA, we found that they contributed to an increase in the expression levels of CaOSR1, CaRAB18, and CaDREB1 in response to the dehydration stress (Figure 5B). In contrast, however, we detected no significant differences in the expression of CaNCED3 between group II or IV CaSAP-silenced pepper and control plants (Figures 4C, 5B). These observations indicate that the group II and IV CaSAP genes may play a more weighted role in the stress response by binding to a specific site of the stress-responsive gene promoter rather than being involved in the ABA biosynthetic process.
Conclusion
In conclusion, on the basis of our findings, we suggest that CaSAPs can function either positively or negatively in mediating abiotic stress responses in Capsicum annuum. We identified 14 SAP genes in pepper plants, which were characterized genomically. Moreover, we were able to classify these genes based on domain and phylogenetic analyses and provisionally characterize the functions of the encoded SAP proteins. However, we were unable to precisely elucidate CaSAP function, nor were we able to identify the downstream genes. Thus, in further studies, we intend to focus on the mechanisms underlying SAP-mediated stress response based on molecular approaches and seek to identify the corresponding target genes.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YB and CL performed the experiments and analyzed the results. SL designed the experiments. YB and SL wrote the manuscript. All the authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This study was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Korean Government (MSIT) (No. 2021R1A2C2006338), Rural Development Administration, Republic of Korea and the Chung-Ang University Graduate Research Scholarship (Academic scholarship for College of Biotechnology and Natural Resources) in 2019.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.756068/full#supplementary-material
References
- Ben Saad R., Ben Romdhane W., Zouari N., Ben Hsouna A., Harbaoui M., Brini F., et al. (2020). Characterization of a novel LmSAP gene promoter from Lobularia maritima: tissue specificity and environmental stress responsiveness. PLoS One 15:e0236943. 10.1371/journal.pone.0236943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E. J., Ha J., Kang S. S., Lee Z. H., Kim H. H. (2011). AWP1 binds to tumor necrosis factor receptor-associated factor 2 (TRAF2) and is involved in TRAF2-mediated nuclear factor-kappaB signaling. Int. J. Biochem. Cell Biol. 43 1612–1620. 10.1016/j.biocel.2011.07.010 [DOI] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 1194–1202. 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Chen R., Guo W., Yin Y., Gong Z.-H. (2014). A novel F-box protein CaF-box is involved in responses to plant hormones and abiotic stress in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 15 2413–2430. 10.3390/ijms15022413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Jagendorf A., Zhu J.-K. (2005). Understanding and improving salt tolerance in plants. Crop Sci. 45 437–448. [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61 651–679. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- Ding Y., Shi Y., Yang S. (2020). Molecular regulation of plant responses to environmental temperatures. Mol. Plant 13 544–564. 10.1016/j.molp.2020.02.004 [DOI] [PubMed] [Google Scholar]
- Dixit A., Tomar P., Vaine E., Abdullah H., Hazen S., Dhankher O. P. (2018). A stress-associated protein, AtSAP13, from Arabidopsis thaliana provides tolerance to multiple abiotic stresses. Plant Cell Environ. 41 1171–1185. 10.1111/pce.13103 [DOI] [PubMed] [Google Scholar]
- Dixit A. R., Dhankher O. P. (2011). A novel stress-associated protein ‘AtSAP10’ from Arabidopsis thaliana confers tolerance to nickel, manganese, zinc, and high temperature stress. PLoS One 6:e20921. 10.1371/journal.pone.0020921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q., Duan D., Zhao S., Xu B., Luo J., Wang Q., et al. (2018). Genome-wide analysis and cloning of the apple stress-associated protein gene family reveals MdSAP15, which confers tolerance to drought and osmotic stresses in transgenic arabidopsis. Int. J. Mol. Sci. 19:2478. 10.3390/ijms19092478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A., Sawada Y., Takahashi H., Okamoto M., Ikegami K., Koiwai H., et al. (2008). Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 147 1984–1993. 10.1104/pp.108.116632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. C., Ovaa H., Hamon M., Kilshaw P. J., Hamm S., Bauer S., et al. (2004). Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem. J. 378 727–734. 10.1042/BJ20031377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124 509–525. 10.1007/s10265-011-0412-3 [DOI] [PubMed] [Google Scholar]
- Gao W., Long L., Tian X., Jin J., Liu H., Zhang H., et al. (2016). Genome-wide identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cotton. Mol. Genet. Genomics 291 2199–2213. 10.1007/s00438-016-1252-6 [DOI] [PubMed] [Google Scholar]
- Gimeno-Gilles C., Gervais M.-L., Planchet E., Satour P., Limami A. M., Lelievre E. (2011). A stress-associated protein containing A20/AN1 zing-finger domains expressed in Medicago truncatula seeds. Plant Physiol. Biochem. 49 303–310. 10.1016/j.plaphy.2011.01.004 [DOI] [PubMed] [Google Scholar]
- Giri J., Dansana P. K., Kothari K. S., Sharma G., Vij S., Tyagi A. K. (2013). SAPs as novel regulators of abiotic stress response in plants. BioEssays 35 639–648. 10.1002/bies.201200181 [DOI] [PubMed] [Google Scholar]
- Giri J., Vij S., Dansana P. K., Tyagi A. K. (2011). Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytologist 191 721–732. 10.1111/j.1469-8137.2011.03740.x [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guzman M., Pizzio G. A., Antoni R., Vera-Sirera F., Merilo E., Bassel G. W., et al. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24 2483–2496. 10.1105/tpc.112.098574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzybowska E. A. (2012). Human intronless genes: functional groups, associated diseases, evolution, and mRNA processing in absence of splicing. Biochem. Biophys. Res. Commun. 424 1–6. 10.1016/j.bbrc.2012.06.092 [DOI] [PubMed] [Google Scholar]
- Guo W.-L., Chen R.-G., Du X.-H., Zhang Z., Yin Y.-X., Gong Z.-H., et al. (2014). Reduced tolerance to abiotic stress in transgenic Arabidopsis overexpressing a Capsicum annuum multiprotein bridging factor 1. BMC Plant Biol. 14:138. 10.1186/1471-2229-14-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Liu D., Chong K. (2018). Cold signaling in plants: insights into mechanisms and regulation. J. Integrat. Plant Biol. 60 745–756. 10.1111/jipb.12706 [DOI] [PubMed] [Google Scholar]
- Han G., Lu C., Guo J., Qiao Z., Sui N., Qiu N., et al. (2020). C2H2 Zinc finger proteins: master regulators of abiotic stress responses in plants. Front. Plant Sci. 11:115. 10.3389/fpls.2020.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa P. M., Bressan R. A., Zhu J. K., Bohnert H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 463–499. 10.1146/annurev.arplant.51.1.463 [DOI] [PubMed] [Google Scholar]
- He M., He C.-Q., Ding N.-Z. (2018). Abiotic stresses: general defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 9:1771. 10.3389/fpls.2018.01771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Xie S., Xie P., Yao M., Liu W., Qin L., et al. (2019). Genome-wide identification of stress-associated proteins (SAP) with A20/AN1 zinc finger domains associated with abiotic stresses responses in Brassica napus. Environ. Exp. Bot. 165 108–119. [Google Scholar]
- Huang J., Wang M.-M., Jiang Y., Bao Y.-M., Huang X., Sun H., et al. (2008). Expression analysis of rice A20/AN1-type zinc finger genes and characterization of ZFP177 that contributes to temperature stress tolerance. Genes (Basel) 420 135–144. 10.1016/j.gene.2008.05.019 [DOI] [PubMed] [Google Scholar]
- Hulse-Kemp A. M., Maheshwari S., Stoffel K., Hill T. A., Jaffe D., Williams S. R., et al. (2018). Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Hortic. Res. 5:4. 10.1038/s41438-017-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27 325–333. 10.1046/j.1365-313x.2001.01096.x [DOI] [PubMed] [Google Scholar]
- Jeffares D. C., Penkett C. J., Bähler J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24 375–378. 10.1016/j.tig.2008.05.006 [DOI] [PubMed] [Google Scholar]
- Jeong S., Lim C. W., Lee S. C. (2020). The pepper MAP kinase CaAIMK1 positively regulates ABA and drought stress responses. Front. Plant Sci. 11:720. 10.3389/fpls.2020.00720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., Fokar M., Abdelmageed H., Allen R. D. (2011). Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 75 451–466. 10.1007/s11103-011-9748-2 [DOI] [PubMed] [Google Scholar]
- Kang M., Lee S., Abdelmageed H., Reichert A., Lee H. K., Fokar M., et al. (2017). Arabidopsis stress associated protein 9 mediates biotic and abiotic stress responsive ABA signaling via the proteasome pathway. Plant Cell Environ. 40 702–716. 10.1111/pce.12892 [DOI] [PubMed] [Google Scholar]
- Kang W.-H., Sim Y. M., Koo N., Nam J.-Y., Lee J., Kim N., et al. (2020). Transcriptome profiling of abiotic responses to heat, cold, salt, and osmotic stress of Capsicum annuum L. Sci. Data 7 1–7. 10.1038/s41597-020-0352-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti V., Gupta A. K. (2008). Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol. Biol. 66 445–462. 10.1007/s11103-007-9284-2 [DOI] [PubMed] [Google Scholar]
- Kim S., Park M., Yeom S.-I., Kim Y.-M., Lee J. M., Lee H.-A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46 270–278. 10.1038/ng.2877 [DOI] [PubMed] [Google Scholar]
- Kothari K. S., Dansana P. K., Giri J., Tyagi A. K. (2016). Rice stress associated protein 1 (OsSAP1) interacts with aminotransferase (OsAMTR1) and pathogenesis-related 1a protein (OsSCP) and regulates abiotic stress responses. Front. Plant Sci. 7:1057. 10.3389/fpls.2016.01057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lademann U., Kallunki T., Jäättelä M. (2001). A20 zinc finger protein inhibits TNF-induced apoptosis and stress response early in the signaling cascades and independently of binding to TRAF2 or 14-3-3 proteins. Cell Death Differ. 8 265–272. 10.1038/sj.cdd.4400805 [DOI] [PubMed] [Google Scholar]
- Lai W., Zhou Y., Pan R., Liao L., He J., Liu H., et al. (2020). Identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cucumber. Plants (Basel) 9:400. 10.3390/plants9030400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sun P., Xia Y., Zheng G., Sun J., Jia H. (2019). A stress-associated protein, PtSAP13, from populus trichocarpa provides tolerance to salt stress. Int. J. Mol. Sci. 20:5782. 10.3390/ijms20225782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. W., Baek W., Jung J., Kim J. H., Lee S. C. (2015). Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 16 15251–15270. 10.3390/ijms160715251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. W., Baek W., Lee S. C. (2018a). Roles of pepper bZIP protein CaDILZ1 and its interacting partner RING-type E3 ligase CaDSR1 in modulation of drought tolerance. Plant J. 96 452–467. 10.1111/tpj.14046 [DOI] [PubMed] [Google Scholar]
- Lim C. W., Hong E., Bae Y., Lee S. C. (2018b). The pepper dehydration-responsive homeobox 1, CaDRHB1, plays a positive role in the dehydration response. Environ. Exp. Bot. 147 104–115. [Google Scholar]
- Lim C. W., Baek W., Lim J., Hong E., Lee S. C. (2021). Pepper ubiquitin-specific protease, CaUBP12, positively modulates dehydration resistance by enhancing CaSnRK2.6 stability. Plant J. 107 1148–1165. 10.1111/tpj.15374 [DOI] [PubMed] [Google Scholar]
- Lim J., Lim C. W., Lee S. C. (2021). Pepper novel pseudo response regulator protein CaPRR2 modulates drought and high salt tolerance. Front. Plant Sci. 12:736421. 10.3389/fpls.2021.736421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. W., Jeong S., Lee S. C. (2020). Differential expression of MEKK subfamily genes in Capsicum annuum L. in response to abscisic acid and drought stress. Plant Signal Behav. 15:1822019. 10.1080/15592324.2020.1822019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. W., Lim J., Lee S. C. (2019). The pepper AP2 domain-containing transcription factor CaDRAT1 plays a negative role in response to dehydration stress. Environ. Exp. Bot. 164 170–180. [Google Scholar]
- Linnen J. M., Bailey C. P., Weeks D. L. (1993). Two related localized mRNAs from Xenopus laevis encode ubiquitin-like fusion proteins. Gene 128 181–188. 10.1016/0378-1119(93)90561-g [DOI] [PubMed] [Google Scholar]
- Liu H., Lyu H.-M., Zhu K., Van De Peer Y., Cheng Z.-M. (2021). The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 105 1072–1082. 10.1111/tpj.15088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Yuan X., Wang Y., Wang H., Wang J., Shen Z., et al. (2019c). Tomato stress-associated protein 4 contributes positively to immunity against necrotrophic fungus botrytis cinerea. Mol. Plant Microbe Interact. 32 566–582. 10.1094/MPMI-04-18-0097-R [DOI] [PubMed] [Google Scholar]
- Liu H., Yang Y., Zhang L. (2019a). Identification of upstream transcription factors and an interacting PP2C protein of dehydrin WZY2 gene in wheat. Plant Signal Behav. 14:1678370. 10.1080/15592324.2019.1678370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Wang J., Jiang S., Wang H., Gao Y., Zhang H., et al. (2019b). Tomato SlSAP3, a member of the stress-associated protein family, is a positive regulator of immunity against Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 20 815–830. 10.1111/mpp.12793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhou Y., Xiao J., Bao F. (2018). Effects of chilling on the structure, function and development of chloroplasts. Front. Plant Sci. 9:1715. 10.3389/fpls.2018.01715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret A., Conejero A., Leida C., Petri C., Gil-Muñoz F., Burgos L., et al. (2017). Dual regulation of water retention and cell growth by a stress-associated protein (SAP) gene in Prunus. Sci. Rep. 7:332. 10.1038/s41598-017-00471-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S., Gosti F., Guerrier D., Vavasseur A., Giraudat J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25 295–303. 10.1046/j.1365-313x.2001.00965.x [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A., Vij S., Tyagi A. K. (2004). Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. U.S.A. 101 6309–6314. 10.1073/pnas.0401572101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32 959–970. 10.1007/s00299-013-1418-1 [DOI] [PubMed] [Google Scholar]
- Narusaka Y., Nakashima K., Shinwari Z. K., Sakuma Y., Furihata T., Abe H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34 137–148. 10.1046/j.1365-313x.2003.01708.x [DOI] [PubMed] [Google Scholar]
- Parihar P., Singh S., Singh R., Singh V. P., Prasad S. M. (2015). Effect of salinity stress on plants and its tolerance strategies: a review. Environ. Sci. Pollut. Res. 22 4056–4075. 10.1007/s11356-014-3739-1 [DOI] [PubMed] [Google Scholar]
- Park C., Lim C. W., Baek W., Lee S. C. (2015). RING Type E3 ligase CaAIR1 in pepper acts in the regulation of ABA signaling and drought stress response. Plant Cell Physiol. 56 1808–1819. 10.1093/pcp/pcv103 [DOI] [PubMed] [Google Scholar]
- Park C., Lim C. W., Lee S. C. (2016). The pepper CaOSR1 protein regulates the osmotic stress response via abscisic acid signaling. Front. Plant Sci. 7:890. 10.3389/fpls.2016.00890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflieger S. P., Richard M. M. S., Blanchet S., Meziadi C., Geffroy V. R. (2013). VIGS technology: an attractive tool for functional genomics studies in legumes. Funct. Plant Biol. 40 1234–1248. 10.1071/FP13089 [DOI] [PubMed] [Google Scholar]
- Qin C., Yu C., Shen Y., Fang X., Chen L., Min J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111 5135–5140. 10.1073/pnas.1400975111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad R. B., Hsouna A. B., Saibi W., Hamed K. B., Brini F., Ghneim-Herrera T. (2018). A stress-associated protein, LmSAP, from the halophyte Lobularia maritima provides tolerance to heavy metals in tobacco through increased ROS scavenging and metal detoxification processes. J. Plant Physiol. 231 234–243. 10.1016/j.jplph.2018.09.019 [DOI] [PubMed] [Google Scholar]
- Saad R. B., Safi H., Hsouna A. B., Brini F., Romdhane W. B. (2019). Functional domain analysis of LmSAP protein reveals the crucial role of the zinc-finger A20 domain in abiotic stress tolerance. Protoplasma 256 1333–1344. 10.1007/s00709-019-01390-2 [DOI] [PubMed] [Google Scholar]
- Sah S. K., Reddy K. R., Li J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. 10.3389/fpls.2016.00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J. I., Kwak J. M., Allen G. J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410 327–330. 10.1038/35066500 [DOI] [PubMed] [Google Scholar]
- Shi Y., Ding Y., Yang S. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23 623–637. 10.1016/j.tplants.2018.04.002 [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3 217–223. [PubMed] [Google Scholar]
- Solanke A. U., Sharma M. K., Tyagi A. K., Sharma A. K. (2009). Characterization and phylogenetic analysis of environmental stress-responsive SAP gene family encoding A20/AN1 zinc finger proteins in tomato. Mol. Genet. Genomics 282 153–164. 10.1007/s00438-009-0455-5 [DOI] [PubMed] [Google Scholar]
- Steponkus P. L. (1984). Role of the plasma membrane in freezing injury and cold acclimation. Annu. Rev. Plant Physiol. 35 543–584. [Google Scholar]
- Ströher E., Wang X. J., Roloff N., Klein P., Husemann A., Dietz K. J. (2009). Redox-dependent regulation of the stress-induced zinc-finger protein SAP12 in Arabidopsis thaliana. Mol. Plant 2 357–367. 10.1093/mp/ssn084 [DOI] [PubMed] [Google Scholar]
- Thomashow M. F. (1999). Plant Cold Acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 571–599. 10.1146/annurev.arplant.50.1.571 [DOI] [PubMed] [Google Scholar]
- Tuteja N. (2007). Mechanisms of high salinity tolerance in plants. Methods Enzymol. 428 419–438. 10.1016/S0076-6879(07)28024-3 [DOI] [PubMed] [Google Scholar]
- Tyagi H., Jha S., Sharma M., Giri J., Tyagi A. K. (2014). Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco. Plant Sci. 225 68–76. 10.1016/j.plantsci.2014.05.016 [DOI] [PubMed] [Google Scholar]
- Vij S., Tyagi A. K. (2006). Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Genet. Genomics 276 565–575. 10.1007/s00438-006-0165-1 [DOI] [PubMed] [Google Scholar]
- Vij S., Tyagi A. K. (2008). A20/AN1 zinc-finger domain-containing proteins in plants and animals represent common elements in stress response. Funct. Integrat. Genomics 8 301–307. 10.1007/s10142-008-0078-7 [DOI] [PubMed] [Google Scholar]
- Volkov V. (2015). Salinity tolerance in plants. Quantitative approach to ion transport starting from halophytes and stepping to genetic and protein engineering for manipulating ion fluxes. Front. Plant Sci. 6:873. 10.3389/fpls.2015.00873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fu X., Xie L., Qin W., Li L., Sun X., et al. (2019). Stress associated protein 1 regulates the development of glandular trichomes in Artemisia annua. Plant Cell Tissue Organ Culture (PCTOC) 139 249–259. [Google Scholar]
- Wang Z., Kuang J., Han B., Chen S., Liu A. (2021). Genomic characterization and expression profiles of stress-associated proteins (SAPs) in castor bean (Ricinus communis). Plant Divers 43 152–162. 10.1016/j.pld.2020.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waqas M. A., Kaya C., Riaz A., Farooq M., Nawaz I., Wilkes A., et al. (2019). Potential mechanisms of abiotic stress tolerance in crop plants induced by Thiourea. Front. Plant Sci. 10:1336. 10.3389/fpls.2019.01336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Cheng J., Hu F., Qin C., Xu X., Hu K. (2020). The SnRK2 family in pepper (Capsicum annuum L.): genome-wide identification and expression analyses during fruit development and under abiotic stress. Genes Genomics 42 1117–1130. 10.1007/s13258-020-00968-y [DOI] [PubMed] [Google Scholar]
- Xu Q.-F., Mao X.-G., Wang Y.-X., Wang J.-Y., Xi Y.-J., Jing R.-L. (2018). A wheat gene TaSAP17-D encoding an AN1/AN1 zinc finger protein improves salt stress tolerance in transgenic Arabidopsis. J. Integrat. Agric. 17 507–516. [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10 88–94. 10.1016/j.tplants.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Yoon S.-K., Bae E.-K., Lee H., Choi Y.-I., Han M., Choi H., et al. (2018). Downregulation of stress-associated protein 1 (PagSAP1) increases salt stress tolerance in poplar (Populus alba × P. glandulosa). Trees 32 823–833. [Google Scholar]
- Zhang L. L., Shao Y. J., Ding L., Wang M. J., Davis S. J., Liu J. X. (2021). XBAT31 regulates thermoresponsive hypocotyl growth through mediating degradation of the thermosensor ELF3 in Arabidopsis. Sci. Adv. 7:eabf4427. 10.1126/sciadv.abf4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-Z., Zheng W.-J., Cao X.-Y., Cui X.-Y., Zhao S.-P., Yu T.-F., et al. (2019). Genomic analysis of stress associated proteins in soybean and the role of GmSAP16 in abiotic stress responses in arabidopsis and soybean. Front. Plant Sci. 10:1453. 10.3389/fpls.2019.01453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Mejias J., Quentin M., Chen Y., De Almeida-Engler J., Mao Z., et al. (2020). The root-knot nematode effector MiPDI1 targets a stress-associated protein (SAP) to establish disease in Solanaceae and Arabidopsis. New Phytol. 228 1417–1430. 10.1111/nph.16745 [DOI] [PubMed] [Google Scholar]
- Zhu J.-K. (2002). Salt anddroughtstresssignaltransduction inplants. Annu. Rev. Plant Biol. 53 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.


