Abstract
Mutations in the spin gene are characterized by an extraordinarily strong rejection behavior of female flies in response to male courtship. They are also accompanied by decreases in the viability, adult life span, and oviposition rate of the flies. In spin mutants, some oocytes and adult neural cells undergo degeneration, which is preceded by reductions in programmed cell death of nurse cells in ovaries and of neurons in the pupal nervous system, respectively. The central nervous system (CNS) of spin mutant flies accumulates autofluorescent lipopigments with characteristics similar to those of lipofuscin. The spin locus generates at least five different transcripts, with only two of these being able to rescue the spin behavioral phenotype; each encodes a protein with multiple membrane-spanning domains that are expressed in both the surface glial cells in the CNS and the follicle cells in the ovaries. Orthologs of the spin gene have also been identified in a number of species from nematodes to humans. Analysis of the spin mutant will give us new insights into neurodegenerative diseases and aging.
The central nervous system (CNS) is comprised of various types of neurons and glial cells. Neurons obviously play a central role in neural integration, and the data showing the importance of glial cells in the CNS are now increasing. Glial cells provide growth factors, nutrition, and insulation and are responsible for the maintenance of ionic homeostasis and guiding the migration of neuronal cells and axons. Furthermore, glial cells also promote the formation and functioning of synapses (29). Abnormal glial cell function has been implicated in various neurodegenerative diseases in vertebrates (34, 35). In Drosophila melanogaster, several mutations that cause neurodegeneration have now been isolated (26) and some are shown to be associated with glial cell function. In the drop-dead mutant, glial cells have stunted processes and, as a result, neurons lack the glial sheath (4). In reverse polarity (repo) mutant flies, glial cells in the optic lobe degenerate and this leads to the degeneration of neurons (46). The swiss cheese mutation results in glial hyperwrapping and brain degeneration (21). All of these mutations result in a shortened life span and specific behavioral aberrations.
During the development of the Drosophila CNS, a large number of glial cells and neurons are eliminated by programmed cell death (PCD) (41). PCD is an evolutionarily conserved cell death process that plays a major role in normal development and homeostasis (44). In flies, the reaper (rpr), grim, and head involution defective (hid) genes are crucial for the regulation of PCD (45). Although the homologs of these genes have not yet been identified in other animals, these genes have been shown to function in vertebrates (8, 11, 14). Many of the doomed neurons in the Drosophila CNS express high levels of the A isoform of the ecdysteroid receptor (EcR-A) (31) prior to expression of rpr or grim and/or hid genes (10). However, the mechanism of cell death, particularly postembryonic PCD, is still poorly understood.
This paper describes the molecular characterization of the spin mutation. The Drosophila spin mutation was originally isolated as a mutation in which the females exhibited a strong rejection behavior toward courting males (37, 49). The spin gene, to which the mutagenic P-element impinges at 52E on the second chromosome, encodes an evolutionarily conserved novel protein containing multiple transmembrane domains. The spin gene transcripts are expressed mainly in a subset of surface glial cells in the nervous system and the follicle cells of the ovaries. Loss of the spin gene product from these two types of cells interferes with PCD of neurons and nurse cells in their respective systems. Persistence of unnecessary cells results in the degeneration of oocytes in the ovaries and of neurons in the CNS. The Spin protein is therefore postulated to function in mediating apoptotic signals conveyed from follicle cells to nurse cells or from glia to neurons. The accumulation of lipofuscin-like materials in spin mutant neurons further implies the possible function of the spin gene in regulating lysosomal turnover in nerve cells.
MATERIALS AND METHODS
Mutagenesis, mutant screening, and behavior analysis.
Mutagenesis and mutant screening were carried out as described previously (18, 37). For a detailed characterization of the behavioral phenotype, paired flies were placed in circular mating chambers and their behaviors were videotaped. The behavioral actions undertaken by the females during the 10-min period following the first attempt at courtship by the partner males were then analyzed. If the female exhibited fending, flicking, kicking, punching, curling, spreading, extrusion, or decamping at least once during the 10-min period, the female was classified as positive for the respective behavioral action. The number of positive females was counted for each behavioral action, and the proportion of the total number of observed females was calculated to quantify the intensity of the rejection behavior displayed by the females of the different genotypes. The mating success rate and the sex appeal parameter index (SAPI) were estimated as described previously (18). The locomotive activity of spin flies was examined according to the method of Nilsson et al. (28). The spinR4 revertant line was obtained by introducing the P(ry+Δ2–3) chromosome into the spinP1 line. Excision of the BmΔ-w insertion in spinR4 was confirmed by Southern blot analysis (37).
Rescue experiment.
For the generation of spin transgenic flies, the full-length sequences of the type I, type II, type IV, and type V cDNAs were inserted into the pUAST vector (3). Type III cDNA was inserted into the CaSpeR-hs vector. Plasmids were injected into w1118 embryos together with the phsπ helper plasmid (32); independent lines harboring the transgene on the third chromosome were then isolated. As a driver, a 2.5-kb fragment of the spin 5′ region was inserted into the pGaTB vector (3) and used to generate several transgenic lines having a spin-Gal4 insertion on the third chromosome. Fly lines carrying the transgene and either the spinP1 or the spinP2 mutation were generated and then, crossed. In the case of the type III transgene-carrying flies, heat shock treatments were carried out at 37°C for 1 h at various developmental stages between the embryo and the adult stages.
Molecular analysis.
Genomic DNA adjacent to the P-element insertion site in the spinP1 mutant was isolated by plasmid rescue and subsequently used to screen a Drosophila genomic library in λEMBL3 (Clontech Laboratories, Inc.) and cDNA libraries in λgt11 (Clontech Laboratories, Inc.). Recombinant-DNA techniques were used as previously described (33). The nucleotide sequences of the cDNAs for both strands were determined by dideoxy sequencing using Sequenase (United States Biochemical Corporation) and a 377 DNA sequencer (Applied Biosystems, Inc.). The spin genomic DNA was partially sequenced to confirm the cDNA sequences and to map the exon-intron boundaries. Different DNA and protein databases were searched for homologous sequences using the BLAST program (1), and the protein sequences were then aligned using MacDNAsis (Hitachi Tech).
RNA analysis and in situ hybridization.
RNA preparation and Northern blot analysis were performed as previously described by Miyamoto et al. (25). In Northern blotting, a digoxigenin-labeled 3-kb fragment from the full-length type I cDNA was used to detect spin mRNA, and the same blot was also probed with the ribosomal protein gene rp49 as a control. To measure the relative amount of each type of transcript, reverse transcription-PCR (RT-PCR) was performed using a set of primers, A (5′-TTTGGAGGCCACTACCTACAAGCAGGACAT-3′) and B (5′-CTCTGAGTGCGCAGCATAACCTCCAGAATC-3′). This set of primers could amplify all five transcripts. After size fractionation, the distinction between the type I and II or type III and IV transcripts was carried out by digestion using a unique BamHI site of exon 5. For in situ hybridization, spin antisense RNA or cDNA probes were synthesized and used to detect the spin transcripts expressed in Drosophila tissues using the method previously described (39) with some minor modifications (18).
Isolation and analysis of vertebrate orthologs.
Two degenerate PCR primers, P1 [5′-GTNGGNATNGGNGA(A/G)GC-3′] and P2 [5′-AT(C/T)TG(A/G)AANGC(C/T)TCNGCNGT-3′] were designed based on comparisons of the amino acid sequences of the fly Spin and Caenorhabditis elegans C39E9.10 gene products. Fourteen-day-old mouse embryonic cDNA (Clontech Laboratories, Inc.) was amplified with the P1 and P2 primers for 35 cycles (94°C for 15 s, 50°C for 30 s, and 72°C for 90 s). The PCR products at the expected sizes were subcloned into the T7Blue vector (Novagene) and then sequenced. An 800-bp fragment corresponding to amino acids 161 to 261 in Fig. 7A was first obtained by PCR; the 5′ region was then cloned by the 5′ rapid amplification of cDNA ends method. Human and mouse cDNA clones were obtained by screening the database of Expressed Sequence Tags at the National Center for Biotechnology Information using the BLAST network service (1). The following human and mouse cDNA clones were obtained and sequenced: cDNA clones 426031 and 645298.
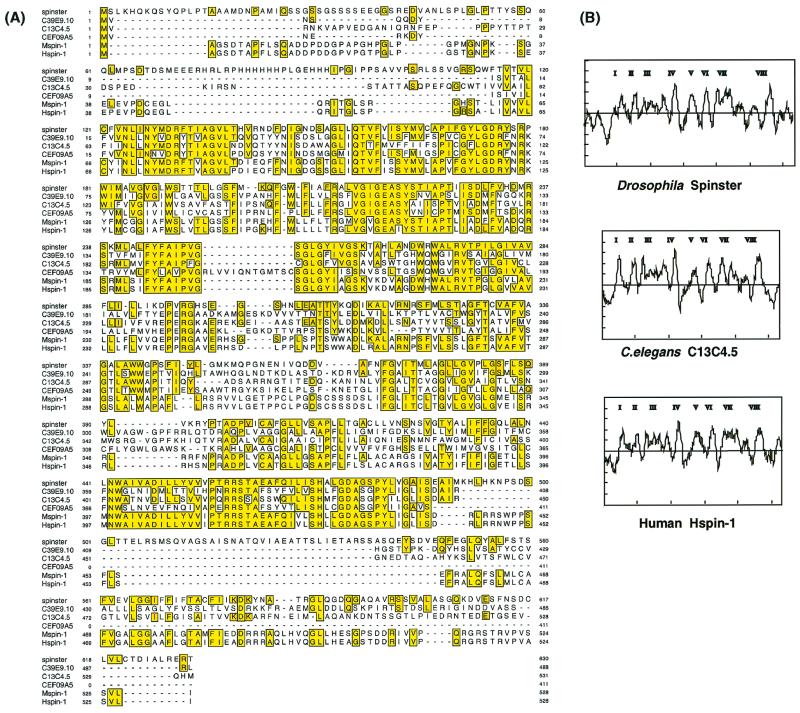
FIG. 7.
Comparison of the spin gene products in various species. (A) Alignment of the nematode (C39E9.10, C13C4.5, and CEF09A5), fly (type I), mouse (Mspin1), and human (Hspin1) spin gene products. Sequences are aligned using MacDNAsis, and residues that are identical with fly Spin are highlighted. The predicted cyclic-AMP- and cyclic-GMP-dependent protein kinase phosphorylation sites are underlined. (B) Hydrophobicity profiles of fly (type I), nematode (C13C4.5), and human Spin proteins. Eight putative transmembrane domains are present.
Immunohistochemistry.
The primary antibodies used were mouse anti β-galactosidase (β-Gal) antibody, J1E7 (Developmental Studies Hybridoma Bank), rabbit polyclonal antibody (Promega), rabbit anti-Repo antibody (gift of K. Ito), and the anti-EcR-A and anti-Elav monoclonal antibodies (gift of S. Robinow). The secondary antibodies used were an Oregon Green-conjugated goat anti-rabbit antibody (Molecular Probes) diluted 1:400 and Cy2-, Cy3-, or Cy5-conjugated goat anti-mouse or -rabbit antibody (Jackson Laboratories) diluted 1:250. Actin was visualized by staining with Texas Red-X phalloidin, and DNA was visualized with SPIF DNA stain. Apoptosis was detected using an in situ cell death detection kit, Fluorescin (Boehringer Mannheim), and the confocal images were collected by a Bio-Rad MRC 1024 confocal microscope.
TdT-mediated dUTP-biotin nick end labeling (TUNEL).
Tissue was fixed for 30 min at room temperature in 4% paraformaldehyde–0.1 M sodium phosphate buffer (pH 7.3), rinsed briefly in PBS-TX (0.5% Triton X-100 in phosphate-buffered saline[PBS]), incubated in 100 mM SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% Triton X-100 at 65°C for 30 min, and rinsed twice in PBS-TX. Tissue was washed for 10 min in 1 × terminal deoxynucleotidyl transferase (TdT) buffer (Roche) and then incubated in TdT reaction mix (Roche) at 37°C for 3 h. Tissue was washed in PBS-TX and incubated overnight with converter-POD (horseradish peroxidase-labeled anti-fluorescein antibody [Roche]) at 4°C, followed by three washes in PBS-TX. After horseradish peroxidase was reacted with diaminobenzidine-H2O2, tissue was observed in a whole-mount preparation. Digital camera lucida images of whole-mount tissues were composed by using Adobe Photoshop and a Macintosh computer with a Kontron Progress 3012 digital scanning camera mounted on a Zeiss Axiophoto microscope.
Electron microscopy (EM).
For transmission electron microscopy, pupal and adult specimens were fixed by 4% glutaraldehyde in 0.1 M sodium cacodylate buffer, (pH 7.4 to 7.6) for 1.5 to 2 h, washed in 0.1 M cacodylate for 20 to 30 min, and then postfixed with 1% OsO4 in 0.1 M cacodylate for 1 h, with all stages being performed at room temperature. The tissue was then dehydrated in a graded ethanol series, the ethanol was replaced with propylene oxide, and the tissue was embedded in epoxy resin. Ultra-thin (60- to 80-nm-thick) sections were obtained on a Reichert Ultracut E ultramicrotome, double stained with uranyl acetate and lead citrate, and then viewed on a Zeiss 10/A or LEO 912AB transmission electron microscope at 80 or 100 kV.
Lipid analysis.
Lipids from the heads from spin and wild-type flies were extracted with chloroform-methanol (2:1, vol/vol), and the analysis of the fluorescence spectra of the lipid extracts was performed according to the method of Fletcher et al. (13) using a Hitachi F-2000 fluorescence spectrophotometer. Thiobarbituric acid-reactive substances were measured by the method of Buege and Aust (5) with the modification of Yagi (48).
Nucleotide sequence accession number.
The GenBank accession numbers for the sequences described in this paper are as follows: spinster (type I), AF212366; spinster (type II), AF212367; spinster (type III), AF212368; spinster (type IV), AF212369; spinster (type V), AF212370; Hspin1, AF212371; and Mspin1, AF212372.
RESULTS
Behavioral phenotypes of spin mutants.
In addition to the spinP1 line isolated by ourselves, two fly lines with a P-element insertion in the spin locus identified from the Barkeley Drosophila Genome Project gene disruption project (www.fruitfly.org) were used in this study. The lethal line l(2)10403, designated the spinP2 line, was used in the rescue of the spin lethal phenotype. The EP822 line exhibited a low viability similar to that of the spinP1 line; therefore, we used this line to investigate the adult spin phenotypes.
Single male and female pairs were placed in a plastic syringe for 1 h. During this time the mating success was measured (50); it was found that, while 70% of the wild-type pairs copulated, only 4% of the spinP1 mutant females that were paired with wild-type males copulated under the same conditions (Fig. 1C). Females of a revertant line, obtained by P-element excision, exhibited essentially the same level of mating success as the wild-type females (Fig. 1C). This result demonstrates that this phenotype is caused by the P-element insertion, as excision was able to restore normal receptivity in females. The intensity of the male courtship can be quantified by the SAPI (19); this index represents the percentage of time spent by the male performing unilateral wing vibration during a 10-min observation period. The SAPI was found to be almost the same for wild-type, spinP1, and revertant pairs, thus indicating that the females of these strains were able to elicit similar levels of courtship from the males (Fig. 1C). This means that the low mating success observed in spinP1 females cannot be accounted for by reduced attractiveness but rather that the low mating success may reflect the unwillingness of the spinP1 females to copulate.
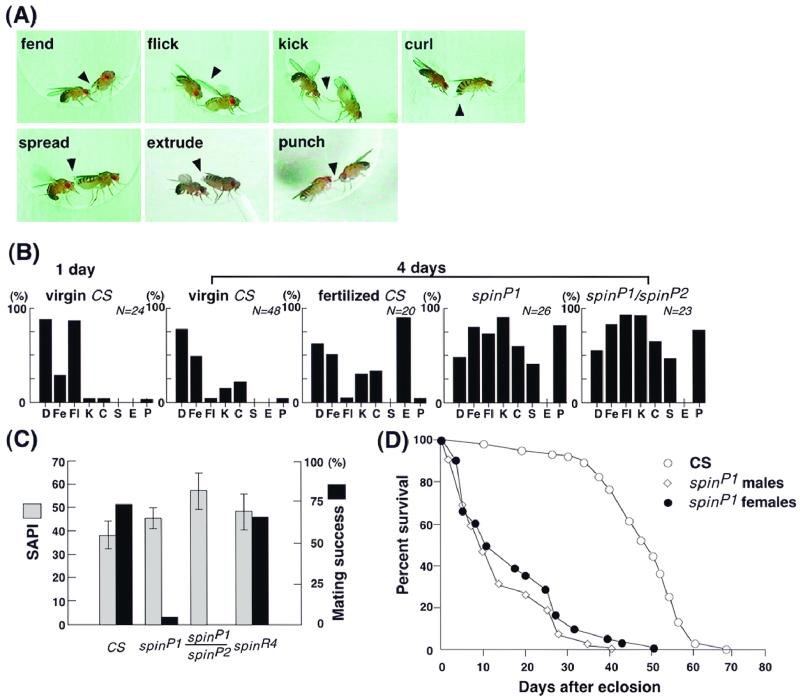
FIG. 1.
Rejection behavior displayed by spin mutant females. (A) Typical repelling postures taken by spinP1 female flies. The pictures were selected from continuous videotape recordings. The picture illustrating extrusion was obtained using a wild-type fertilized female, as spinP1 females do not exhibit this behavior. (B) Relative contributions of different types of repelling actions compared among wild-type, spinP1, and spinP1/spinP2 females. The wild-type females were divided into three categories, 1-day-old virgin, 4-day-old virgin, and 4-day-old fertilized females. The females of spinP1, spinP1/spinP2, and spinR4 lines were 4 days old, and the males were all wild type. The behavior of single females was recorded on a video tape for 10 min after pairing, and the numbers of females exhibiting decamping (D), fending (Fe), flicking (F1), kicking (K), curling (C), spreading (S), extrusion (E), and punching (P) actions were counted and are shown as percentages with the respect to the total number of females observed. A value of 100% means that all of the females observed exhibited that action at least once during the recorded period, while 0% means that none of the females did so. The counts obtained from at least 20 females were summed and are illustrated as frequency histograms. CS, CS strain. (C) The percentage of mating success (dark columns) was decreased in spinP1 and spinP1/spinP2 females compared to that of CS wild-type and spinR4 females. The differences in levels of mating success between the wild-type and mutant (spinP1 and spinP1/spinP2) females were found to be statistically significant at a P of <0.001 using Student's t-test. No difference was detected between the four genotypes in the females' ability to elicit male courtship songs (SAPI, paler columns). The bars on the columns represent the standard errors of the mean values for SAPI. The numbers of flies observed in order to estimate the mating success were 114 (wild type), 62 (spinP1), 44 (spinP1/spinP2), and 44 (spinR4). The number of flies observed to calculate the SAPI was 20 for each of the three genotypes. Males were from the CS wild-type strain, and all flies were aged for 3 days following eclosion. (D) Longevity of adult flies. The percentage of survival after eclosion is plotted for wild-type (CS) females spinP1 females, and spinP1 males.
Indeed, the spinP1 females consistently displayed a number of rejection responses (9) against the courting males; these included fending, kicking, flicking, curling, punching, and decamping (Fig. 1A and B); extrusion was rarely seen (Fig. 1B). The pattern of rejection displayed by spinP1 females resembled that of immature wild-type virgin females rather than that of fertilized females, in that extrusion did not occur (9, 49). However, the spinP1 females did exhibit kicking and curling behavior much more frequently than the wild-type females (Fig. 1B). In response to approaching males, the spinP1 females tended to raise their abdomens while spreading their vaginal plates (Fig. 1A and B). This spreading was unique to the spin mutant females and was also distinctly different from extrusion, in which the ovipositor protrudes from the female terminalia. Furthermore, the spinP1 female often rushed toward the courting male, pushing the male's head with her forelegs; this aggressive behavior is termed punching and is rare among wild-type females (Fig. 1B). A similarly pronounced refusal of suitors was observed in heteroallelic spinP1/spinP2 females (Fig. 1B and C) (see below). spinP1 male flies exhibited no obvious abnormality in their courtship behavior, while general locomotive activity was reduced in both sexes (12% reduction in females and 38% reduction in males 3 days old).
Apart from the regulation of female sexual behavior, the spin gene plays an additional vital role, as the partial loss-of-function mutation (spinP1) reduces viability and life span (Fig. 1D) and the spinP2 mutation is lethal, yielding no adult flies. This reduction in viability caused by the spinP1 mutation was actually more extreme in males than in females (the viability of females was 22% and that of males was 11%), resulting in an uneven sex ratio (females/males, 2:1) of the emerged homozygous adults.
spin mutant flies have a long abdominal ganglion.
In an attempt at determining the cellular basis for the behavioral phenotypes of the spin mutant, we have analyzed the morphology of the CNS and have found that the abdominal ganglia in spinP1 mutant flies of both sexes are longer than those of the wild type (Fig. 2 and 3). Figure 3A shows the ventral nerve cords (VNCs) of the wild type and spinP1 mutant stained with SPIF DNA stain 24 h after eclosion. The thoracic segment of the VNC appears normal, while the abdominal segment is abnormally long in the spinP1 mutant. The ratio of the length of the posterior part over the total length of the VNC (Fig. 3A) was estimated as follows: for wild-type males, it was 0.260 ± 0.006 (mean ± standard error of the mean) (n = 27); for wild-type females, it was 0.265 ± 0.004 (n = 25); for spin males, it was 0.343 ± 0.016 (n = 15), and for spin females, it was 0.338 ± 0.010 (n = 23). The unshortened morphology of the VNC was still evident 2 weeks after eclosion in spinP1 flies. The same phenotype was also observed in the EP822 line. Apart from the VNC, we were unable to find any gross morphological abnormalities in spin mutant flies.
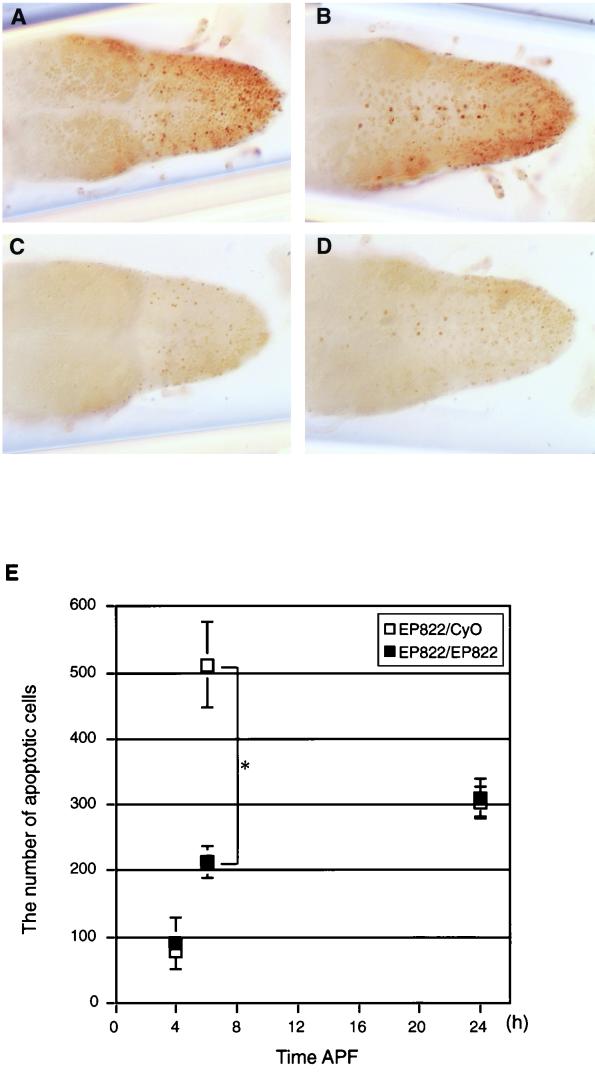
FIG. 2.
Reduced number of cells undergoing apoptosis in the spin mutant VNC. (A to D) VNCs at 6 h APF stained by the TUNEL method as viewed from the dorsal (A and C) or ventral (B and D) side. (A and B) EP822/CyO; (C and D) EP822/EP822. (E) Numbers of cells undergoing apoptosis at three different time points APF (4, 6, and 24 h) in spin heterozygotes (open squares) or homozygotes (filled squares). The mean and standard deviation of the mean are shown by a symbol and bar, respectively. The values for heterozygotes and homozygotes are significantly different at 6 h APF (P < 0.01 by Student's t test). At other time points, the difference between the values for heterozygotes and homozygotes is statistically insignificant (P > 0.05).
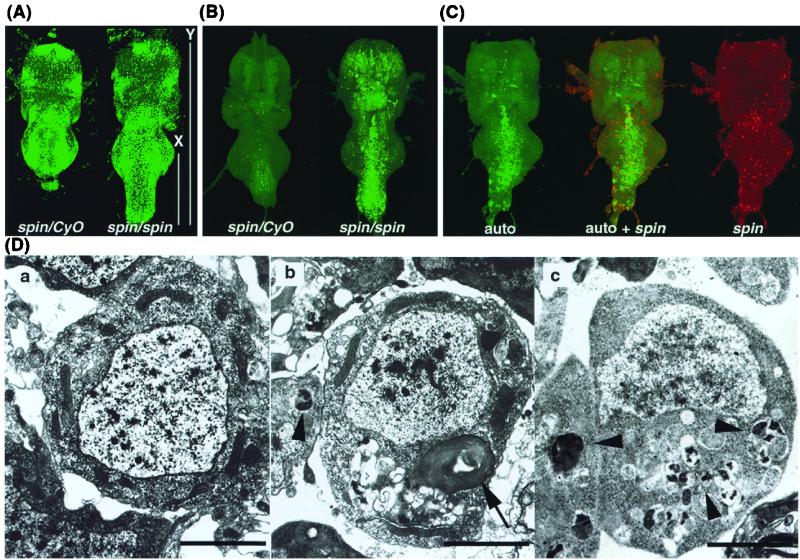
FIG. 3.
CNS abnormalities in spin mutant flies. (A) VNCs of the spinP1/spinP1 and spinP1/CyO flies 24 h after eclosion and staining with SPIF DNA stain. The spinP1 homozygous fly has a long abdominal ganglion. X, the length from the center of the third thoracic ganglion to the posterior edge of the abdominal ganglion; Y, the total length of the thoracic and abdominal ganglion. (B) Autofluorescence in the spinP1 mutant VNC. Tissues were fixed by 4% paraformaldehyde for 1 h, and the images were obtained by confocal microscopy using the blue excited light channel. The spin mutant CNS exhibits autofluorescence. (C) Expression of the spin gene and the distribution of the autofluorescent material. spin gene expression was examined in spinP1/spinP2 transheterozygotes. Only a portion of the autofluorescence overlaps with the expression of the spin gene. EM analysis of spinP1/CyO (Da) and spinP1/spinP1 (Db) cells in the abdominal ganglia 24 h after eclosion and spinP1/spinP1 cells at an early pupal stage (Dc). Nerve cells in the spinP1 homozygotes have lipofuscin-like materials inside them. Arrows indicate multilamellate bodies. Arrowheads indicate electron-dense lobulated granules. Scale bars, 2 μm.
PCD of the neurons in the VNC occurs shortly after pupariation and after the emergence of the adult; typically, the ventral abdominal ganglion cells die during metamorphosis and after eclosion (20, 31). The abdominal portion of the VNC shortens after the first wave of PCD in the mid-pupal stage.
The absence of condensation of the abdominal ganglia in the spin mutants implies that some cells in the VNC are prevented from undergoing PCD. To evaluate this possibility, we compared the numbers of cells undergoing apoptosis between the spin heterozygous and homozygous VNCs by selectively staining fragmented DNA using TUNEL (10). Consistent with the previous observations, PCD in the VNC occurred in two phases, one during the first several hours after pupation and the other immediately after adult eclosion (20, 31, 40). No difference was found in the numbers of apoptotic cells between the spin heterozygote and homozygote after adult emergence (Y. Nakano, unpublished observation). In contrast, the profile of PCD in the VNC of the spin homozygote was distinctly different from that in the spin heterozygote during the pupal stage (Fig. 2): the number of cells undergoing PCD in heterozygotes was maximum at 6 h after puparium formation (APF) as reported for the wild type (Fig. 2E, open squares), whereas the number of apoptotic cells remained quite low at this developmental stage in homozygotes (Fig. 2E, filled squares). The difference in the numbers of the cells undergoing apoptosis at 6 h APF between heterozygotes and homozygotes was statistically significant (P < 0.01 by Student's t test). No significant diference was found in the number of apoptotic cells between heterozygotes and homozygotes at 4 and 24 h APF (Fig. 2E). These observations suggest that many cells that will be eliminated at 6 h APF remain alive through the pupal stage in the spin mutant.
spin mutant cells contain lipofuscin-like materials in the CNS.
During the tissue staining of spinP1 flies, we observed the existence of autofluorescent material in the blue channel. In the VNC, the autofluorescent material was observed mainly in the central region of the VNC, particularly in the abdominal ganglia (Fig. 3B) of pupae and adults. The lethal allele, spinP2, showed an earlier onset of the accumulation of autofluorescent materials in the CNS (from larval stages). In the brain, this material was observed in the central brain and in the optic lobe. Figure 3C shows the distribution of the autofluorescent material (green) and spin gene expression (red), as marked by β-Gal expression in the VNC of spinP1/spinP2 transheterozygotes. It was found that part of the autofluorescence overlapped with spin gene expression; however, most did not. Figure 3D shows the EM analysis of the VNC cells in spinP1 homozygotes and heterozygotes at the early pupal stage and 24 h after eclosion. The spin mutant samples exhibited cellular disorganization in that most of the spin mutant cells, including both the neurons and glial cells, contained multilamellate bodies and electron-dense lobulated granules (Fig. 3Db and -c), these structures were never observed in the spin/CyO cells. These aberrant structures were observed in the CNS from the early pupal stage (Fig. 3Dc) and increased thereafter in spinP1 homozygotes. The early pupal VNC contained electron-dense lobulated granules (Fig. 3Dc), which appeared to be precursors of the multilamellate bodies seen in the adult VNC. These structures were observed in both sexes, and no differences were observed between the sexes at the cellular level. Neither structure was found in gut or muscle cells from the spin mutant; in addition, the nucleus, mitochondria, and endoplasmic reticulum all appeared to be normal in the spin mutant. The aberrant structures contained within spin mutant nerve cells were found to be very similar to lipofuscin, which is known to be induced by oxidative stress, some proteinase inhibitors, inherited lysosomal storage diseases, and the normal aging process (17).
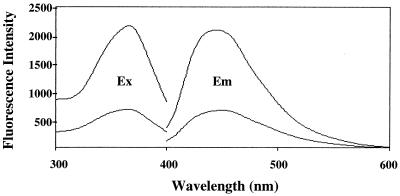
In order to identify the nature of these materials, biochemical analyses were performed. Fletcher et al. (13) have reported that a large proportion of the fluorescent pigments in the tissues can be extracted by a chloroform-methanol solution and offer a sensitive fluorometric assay for the measurement of fluorescent lipid peroxidation products that have accumulated in the various tissues. According to their method, the lipids were extracted from the heads of spin and wild-type flies and fluorescence spectra of the lipid extracts were then measured. The lipid extracts from the heads of spin flies had an excitation maximum at 368 nm and an emission maximum at 450 nm; these are characteristic of those observed with the lipofuscin pigments (13, 38, 42) (Fig. 4). The fluorescence intensity of the spin flies was 3.1 times higher than that of wild-type flies, suggesting that the accumulation of lipid-soluble lipofuscin-like substances was significant in spin flies. The accumulation of lipid peroxides was also examined using a thiobarbituric acid assay. The amounts of thiobarbituric acid-reactive substances in the homogenates and the lipid extracts from the heads of spin flies were found to be 30.2 ± 0.17 pmol/mg of head (mean ± standard deviation of results of three independent experiments) and 104.3 ± 43.6 pmol/mg of head (mean ± standard deviation of results of four independent experiments), respectively. These values were significantly higher than those observed with wild-type flies, namely, 21.5 ± 0.24 and 51.0 ± 6.3 pmol/mg of head, respectively. These results clearly indicate that the chemical nature of the lipofuscin-like pigments that accumulated in spin flies is quite similar to that reported for lipofuscin pigments in various mammalian tissues (38).
FIG. 4.
Fluorescence spectra of spin mutant brain extract. Fluorescence excitation and emission spectra of the lipid extracts from the heads of spin flies (upper curves). The lower curves were obtained with the lipid extracts from the heads of wild-type flies. Ex, excitation spectrum; Em, emission spectrum.
The spin gene function is required for proper ovarian development.
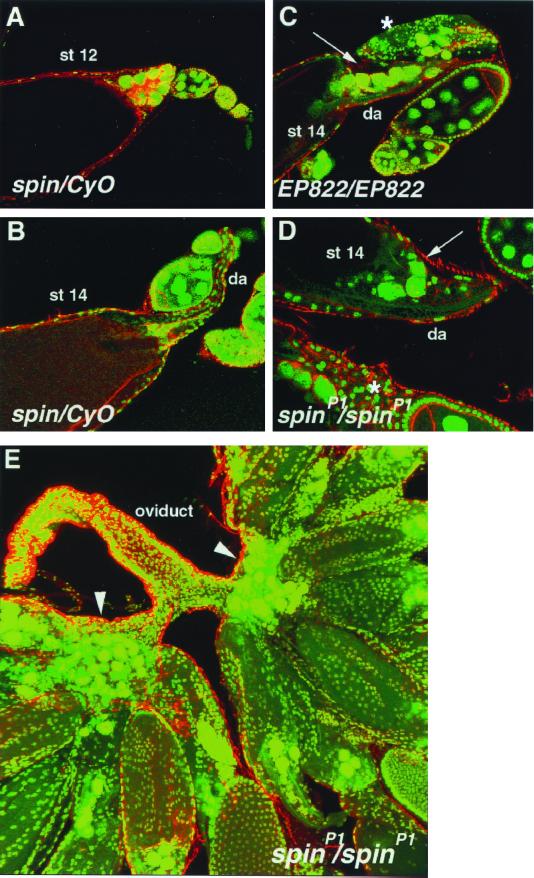
spinP1 females not only exhibit very strong rejection behavior toward courting males, they also rarely lay eggs. However, the distribution of motor nerve endings along the uterine muscles was found to be normal (see reference 17). In order to evaluate the possibility that spinP1 mutants are defective in egg production, ovarian development in spin flies was studied. Figure 5A and B show normal ovarian development by staining with SPIF DNA stain (green) (36) and Texas Red-X phalloidine (red), denoting actin staining. At stage 12, nurse cells were found to dump cytoplasmic components into the oocyte, their nuclei accumulated at the anterior of the oocyte, and the actin bundles were well formed. At stage 14, the dorsal appendages were well formed and nurse cell nuclei had disappeared due to PCD in spinP1/CyO flies. In spinP1 flies, the dorsal appendages were again well formed at stage 14, but the nurse cell nuclei were still present and some oocytes were found to be degenerated (Fig. 5D); spinP1 mutant mature ovaries exhibited an accumulation of hundreds of nurse cell nuclei near the basal stalk (Fig. 5E). The same phenotype was also observed in the EP822 line (Fig. 5C). In contrast, spin mutant males produce normal offspring; therefore, male germ cells seem to develop normally in these flies.
FIG. 5.
The spin gene is required for proper ovarian development. Confocal images of control (A and B) and spin (C to E) mutant egg chambers labeled with Texas Red-X phalloidin (red) and SPIF DNA stain (green). (A) Stage 12 (st 12) control egg chamber showing nurse cell nuclear accumulation at anterior and cytoplasmic actin bundles. (B) Stage 14 control egg chamber showing dorsal appendage (da) formation and no nurse cell nuclei. (C and D) Stage 14 EP822/EP822 and spinP1 mutants egg chambers. Nurse cell nuclei still remain (arrows), cytoplasmic actin bundles are now absent, but the dorsal appendages (da) are well formed. Some earlier-stage egg chambers are degraded (asterisks). (E) Mature ovaries in the spinP1 mutant. Nurse cell nuclei are accumulated at the basal stalk region (arrowheads).
Molecular cloning of the spin gene.
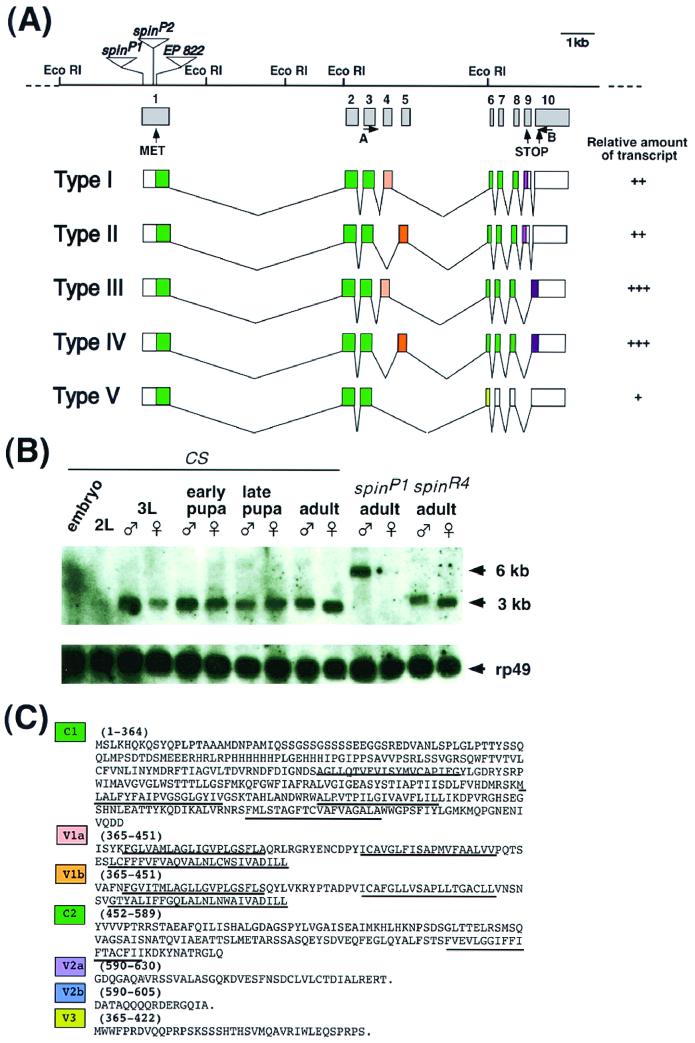
Genomic DNA flanking the P element in the spinP1 mutant was cloned by plasmid rescue (16) and subsequently used to screen genomic and cDNA libraries. Sequence analysis of the identified clones revealed that there were five different transcript forms of approximately 3 kb in length (Fig. 6); these were produced by the alternative processing of a primary transcript (Fig. 6A). The P-element was found to be inserted 8 bp downstream of the transcription initiation site of this transcription unit in the spinP1 mutant.
FIG. 6.
Molecular characterization of the spin gene and its products. (A) Genomic structure of the spin gene and the structures of five transcripts produced by alternative splicing. The restriction sites are shown for the EcoRI restriction enzyme. The P-element insertion site in the spinP1 mutant is located 8 bp downstream of the transcription initiation site and is indicated by a triangle. The enhancer trap vectors in the spinP2 and EP822 genes are inserted in the middle of exon 1. The relative amount of each type of transcript is represented as follows: +++, abundant; ++; modest; or +, rare. No obvious differences were found during development between the sexes or between wild-type and spinP1 mutant flies. The primers used for RT-PCR were primers A and B. (B) Developmental Northern blots probed by spin cDNA. Twenty microgram samples of poly(A)+ RNAs extracted from embryos, second-instar larvae, third-instar larvae, early pupae, late pupae, adults of the wild type, adults with the spinP1 mutation, and adults with the revertant spinR4 mutation were loaded onto the gel. The blots were then probed with the spin type I cDNA as well as the ribosomal protein gene rp49 cDNA in order to assess the amount of RNA loaded in each lane. (C) Amino acid sequences of each of the domains of Spin. C1 and C2 represent common domains. The V1, V2, and V3 domains are variable (see panel A). The predicted transmembrane domains are underlined.
The P-element insertion in the spinP1 allele was found to reduce the amount of the 3-kb transcript and also produced an additional mRNA (∼6 kb) from this transcription unit (Fig. 6B). This 6-kb transcript was found to be a read-through product from the neomycin gene of the P-element vector. These observations suggest that this transcription unit corresponds to the spin gene. Two out of five alternative splicing events resulted in the exclusion of exon 9, which contains an in-frame termination codon, thereby producing proteins containing different C-terminal amino acids (type III and type IV in Fig. 6A). A different type of splicing variation led to the mutually exclusive utilization of exon 4 in the case of type I and type III and of exon 5 in type II and type IV transcripts (Fig. 6A). Exons 4 and 5 were found to be 262 bp long and encode amino acid sequences that are 53% identical to each other (Fig. 6C). The predicted lengths of the polypeptides encoded by the cDNAs described are 630 amino acids for type I and type II, 605 amino acids for type III and type IV, and 422 amino acids for type V proteins (Fig. 6C).
A hydropathy plot analysis (22) indicates that Spin proteins have multiple membrane-spanning domains with no cleavable signal sequence (Fig. 7B). Spin exhibits no significant homology to any of the membrane proteins of known function, such as transporters, ion channels, and receptors. A search of the sequence database has allowed us to identify three C. elegans genes of unknown function, C39E9.10, C13C4, and CEF09A5 as orthologs of the Drosophila spin gene (Fig. 7A). However, we were unable to find homologs in Saccharomyces cerevisiae.
The full-length coding sequence of the mouse and human spin genes were determined by sequencing cDNA clones obtained from the Expressed Sequence Tags data bank. Both mouse and human cDNAs (designated Mspin1 and Hspin1, respectively) were found to encode proteins of 528 amino acids (Fig. 7A). The identities between Spin (type I) and Mspin1, Spin and Hspin1, and Mspin1 and Hspin1 were 41, 42, and 94%, respectively. Hydropathy analyses (22) for the human, mouse, fly, and nematode proteins show that these proteins are remarkably similar to each other (Fig. 7B), thus suggesting that they share a common topological structure. The predicted cyclic-AMP- and cyclic-GMP-dependent protein kinase phosphorylation site is conserved from C. elegans to humans; therefore, this indicates that phosphorylation and/or dephosphorylation events may play an important role in Spin function.
The spin gene is expressed in the surface glial cells in the nervous system and follicle cells in the ovary.
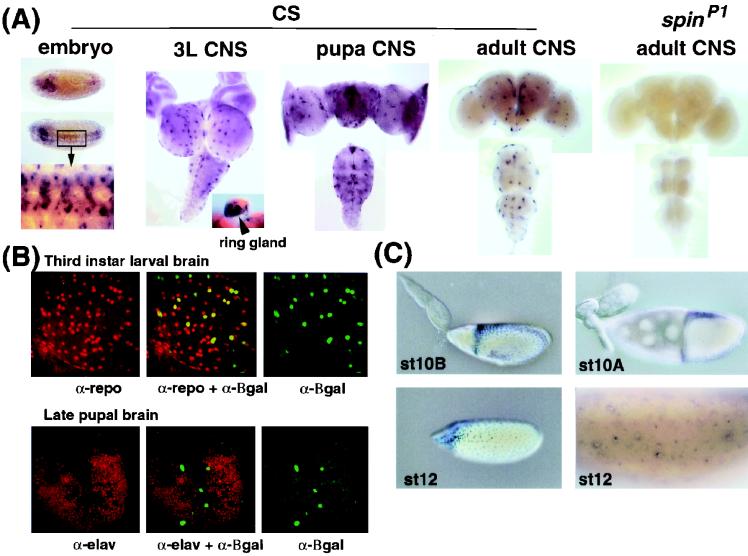
The expression pattern of the spin gene was analyzed by Northern blotting. A 3-kb transcript was observed throughout development, although the level of expression was very low in the embryonic and second-instar larval stages (Fig. 8B). The relative amount of each type of transcript was examined by RT-PCR methods. The type III and type IV transcripts were found to be abundant, while types I and II were expressed at a moderate level and type V was found to be very rare (Fig. 6A). No major differences were found during development between the sexes or between wild-type and spinP1 flies in this experiment. The spatial expression of the spin transcripts was examined by whole-mount in situ hybridization using an antisense RNA or cDNA probe that was able to detect all five types of transcript (Fig. 8A). The spin transcripts were detectable at the beginning of the germ band retraction (stage 12) in a subset of cells in the VNC and the brain, and this expression pattern continued throughout development (Fig. 8A).
FIG. 8.
Expression of the spin gene during development. (A) In situ hybridization of the spin mRNA probe to whole-mount wild-type embryos at stage 12 is shown in the first set of images. A dorsal view (top) and a dorsolateral view (middle) of an embryo are shown. The boxed region is illustrated at a higher magnification in the bottom image. Expression is evident in the developing VNC and brain. The three middle panels illustrate mRNA localization in the CNS of a third-instar larva (3L) (left), pupa (middle), and adult (right) of the CS wild type. The eye-antennal disks attached to the third-instar brain can also be seen. Expression in the larval ring gland is shown in the inset. The far-right images show mRNA expression in the spinP1 adult. The four CNS images show the dorsal side up. (B) The top three images show the double staining of the spinP2/CyO larval brains for the spin reporter β-Gal (green) and the Repo protein (red). Colocalization of β-Gal and the Repo protein results in a yellow signal. The bottom three images show the double staining of the spinP2/CyO pupal brains for spin reporter β-Gal (green) and the Elav protein (red). The spin gene is not expressed in the neurons. α, antibody. (C) The left two images show β-Gal expression in spinP2/CyO ovaries. The right two images show the in situ hybridization analysis of spin gene expression in ovaries at stage 10A (st10A) and st12. The spin gene is expressed in the follicle cells at st10B and st12.
In order to establish the identity of the Spin-expressing cells, we performed double staining using an anti-Repo antibody and an antisense probe to spin mRNA. Repo is a glia-specific homeobox protein expressed in all glial cells except for the midline glial and two segmental nerve root glial cells (6, 15, 47). We observed that more than 95% of Spin-expressing cells overlapped with Repo-expressing cells in the VNC and the brain. This expression pattern was confirmed by using spinP2, which carries a P-element with an enhancer trap reporter inserted in the middle of the first exon of the spin gene (Fig. 6A). The β-Gal expression pattern observed in the embryos and the third-instar larval brain of the spinP2 heterozygotes was found to correspond well with spin expression as detected by in situ hybridization. Figure 8B shows the double staining of larval and pupal brains by anti-Repo or anti-Elav and anti-β-Gal antibodies in spinP2/CyO flies. The spin gene is expressed in the surface glial cells, which include the peripheral exit glia, the subperineural glia, and the channel glia of the nervous system. In addition, β-Gal expression in spinP2/CyO flies in the larval and pupal stages was also observed in the trachea, gut, salivary glands, and ring gland.
The expression of the spin gene was also observed in adult ovaries; expression was observed in the follicle cells in the manner of dorsal-ventral and anterior-posterior gradients (Fig. 8C) but not in the nurse cells or the oocyte. This expression pattern was also confirmed by in situ hybridization (Fig. 8C).
Rescue of the spin phenotype by specific transcripts.
To confirm that the cloned transcriptional unit is the spin gene and whether these different transcripts have different functions, the cDNA corresponding to each transcript was expressed in vivo using the Gal4-upstream activation sequence (UAS) system (3). A 2.5-kb upstream region of the spin gene was used to drive the Gal4 vector. Each type of cDNA (types I, II, IV, and V) was also inserted in the UAS vector. Type III cDNA was examined under the regulation of the heat shock promoter. In the behavioral-rescue experiment, the transgene was examined in spinP1/spinP1 flies, while in the lethality rescue experiment, the transgene was introduced into the spinP2/spinP2 flies. Table 1 shows the results of these rescue experiments. Type I cDNA was found to rescue both the behavioral and lethal phenotypes. Type V cDNA, which encodes half the protein of type I, was also found to rescue the behavioral phenotype; however, it was not able to rescue the lethality phenotype. Type II, III, and IV cDNAs were unable to rescue the behavioral and lethal phenotypes.
TABLE 1.
Rescue of the spin behavior and lethal phenotype
| cDNA typea | % of females receptiveb (n) | Lethalityc |
|---|---|---|
| I | 49.1 (120) | + |
| II | 0 (17) | − |
| III | 0 (120) | − |
| IV | 0 (24) | − |
| V | 46.1 (13) | − |
cDNA expression was induced by the spin-GAL4–UAS-cDNA system except for type III cDNA, which was driven by the hs70 promoter and thus induced by heat shock application throughout development. spin-GAL4 simulates authentic spin gene expression in the nervous system and ovaries.
Female receptivity was examined in spinP1/spinP1 flies.
Lethality was examined in spinP2/spinP2 flies.
Given that the sole difference between the type I and type II transcripts is an alternative usage of exon 4 in type I and of exon 5 in type II, the exon 4 sequence is necessary for rescuing the behavioral and lethal phenotypes. These two exons both encode the same number of amino acids and share 56% homology, and although their sequences are different, the predicted topology does not change. Since the difference between type I and type III is located in the C-terminal region, the C terminus also seems to play an important role in Spin function. In spinP1 mutants, a 6-kb transcript was produced by read-through from the inserted P-element vector and the amount of the authentic 3-kb transcripts was dramatically reduced. However, the relative amount of the each transcript was not changed in spinP1 mutants; thus, the effect of mutations on spin transcription was not isoform specific. These data suggest that reduced amounts of Spin proteins, particularly the type I and type V products, induce spinP1 phenotypes.
DISCUSSION
We demonstrated that spin mutations interfere with PCD in both ovaries and the nervous system. These cellular defects should contribute at least in part to behavioral phenotypes of spin mutant female flies, i.e., the reduction in oviposition and sexual receptivity. It is evident that some immature oocytes degenerate in spin mutant ovaries (Fig. 5); thus, the total number of eggs available for oviposition decreases. However, this finding does not necessarily exclude the possibility that the spin mutant female flies are accompanied by some other deficits that hamper normal oviposition. It is important to note that the spin gene product is expressed in the follicle cells of ovaries and not in nurse cells or oocytes. This observation suggests that the Spin protein is required by follicle cells for inducing the PCD of nurse cells, a group of support cells which are crucial for the normal development of oocytes (36). The degeneration of immature oocytes in spin mutant female flies may thus result from the disturbance of the process of PCD of nurse cells as a consequence of the loss of Spin from follicle cells.
The link between the PCD defect in the nervous system and enhanced mate refusal in spin mutant female flies is less obvious. By analogy with the effect on ovaries, it is conceivable that the spin mutations likely affect female sexual behavior through degeneration of cells in the nervous system. Indeed, neurodegeneration is widespread in the spinP1 mutant nervous system, which accumulates lipofuscin-like materials. In the spin mutant, the neurodegenerative phenotype was observed in both sexes, but germ cell abnormality and abnormal sexual behavior were found to be female specific. It may be that changes in female sexual receptivity are the easiest parameter by which to detect the malfunctioning of the nervous system due to the accumulation of lipofuscin-like materials in neurons. Alternatively, the reduced receptivity to copulation in spinP1 mutant females and the neurodegeneration observed in both sexes of the spin mutant flies might be unrelated. This issue remains to be resolved by future analysis of spin mutant flies.
There are interesting parallels in the effect of spin mutations on ovaries and the nervous system. Like those in the ovaries, PCD defects in the nervous system are most pronounced in cells not expressing the spin gene product, i.e., neurons. In the nervous system, the spin gene is expressed exclusively in glial cells that play a number of different roles in the development and maintenance of the nervous system. The distribution of the observed autofluorescent material in the nervous system does not correlate with the distribution of the Spin-expressing cells (Fig. 3B); in addition, the time-dependent accumulation of lipofuscin-like materials is observed in most of the neuronal and glial cells in spinP1 mutant flies. Thus, the spin gene product is expressed in follicle cells in ovaries and glial cells, particularly surface glial cells, in the CNS, which enwrap nurse cells and neurons, respectively. Loss of spin function of follicle cells and glial cells leads to suppression of PCD in nurse cells and neurons. These observations provide us with the idea that the Spin protein functions in glial cells in the regulation of PCD of neurons, and the failure of PCD at the proper developmental timing leads to widespread neurodegeneration accompanied by accumulation of lipofuscin-like materials at a later stage.
The most striking effect of spin mutations on neuronal PCD is found in the VNC at 6 h APF, at which time the number of cells undergoing PCD reaches a maximum in the wild type (Fig. 2). In spin mutant pupae, the number of cells undergoing apoptosis is 60% less than that in wild-type pupae. Since no later or earlier peaks of PCD are observed in the spin mutant, the cells to be eliminated must remain there throughout the pupal and adult stages. The presence of supernumerary neurons would perturb the formation of adult neural circuits that are established during the pupal stage.
In fact, the structure of the nervous system of spin mutants is aberrant even when it is observed at the level of gross anatomy. The abdominal segment of the VNC is remarkably longer in the mutant than in the wild type (Fig. 3). This part of the VNC is shortened at the mid-pupal stage. The extent of PCD of neurons influences the shortening of the VNC. For example, the VNC does not shorten in the flies hemizygous for the H99 deficiency (K. Usui-Aoki, unpublished observation), in which three PCD genes, hid, rpr, and grim, are deleted (7, 45), and thus PCD in the VNC is prevented (10). Furthermore, the VNCs of H99 hemizygous pupae contain autofluorescent material (K. Usui-Aoki, unpublished observation). These results support the idea that the long-abdominal-ganglion phenotype of spin mutants results from the reduced extent of PCD in the pupal VNC.
It has been known that, in the moth pupa, damage to glial cells interferes with the shortening of the VNC (43). Another study has shown that the metamorphic shortening of the moth VNC cultured in vitro is accelerated by the insect steroid hormone ecdysteroid (30). Taking all these observations together into consideration, it is postulated that the Spin protein functions in glial cells to induce PCD of neurons during metamorphosis, which is under the control of the humoral hormone ecdysteroid.
Widespread neurodegeneration and accumulation of lipofuscin-like materials are important features of the nervous systems of spin mutant adults. Although such neurodegeneration might be a secondary result of PCD defects, it provides us with a unique model to study the basis for some human diseases with similar phenotypes. Both histological and biochemical analyses indicate that accumulated materials in spin flies are very similar to lipofuscin. Lipofuscin was discovered as an aging pigment more than a century ago. However, the mechanism of lipofuscinogenesis has not yet been fully elucidated. In humans, the abnormal accumulation of lipofuscin has been reported for several neurodegenerative disorders such as the neural ceroid lipofuscinoses and Tay-Sachs disease. Both are categorized as lysosomal storage diseases (2). Although we have cloned two human spin orthologs and mapped them to regions on chromosomes 16 and 17 (Y. Nakano, unpublished data), these two loci do not match the mapped loci of the known disease holders. However, there are still many unidentified diseases that will probably be classified as lysosomal storage diseases; therefore, spin orthologs will be good candidate genes in these diseases.
Interestingly, spin mutant flies exhibit a shortened life span (Fig. 1D). One Drosophila mutant, eggroll, has a very similar phenotype to that produced by the spin mutation; such mutants exhibit a shortened life span and have multilamellar structures in the CNS (24). We point out the similarity to some human neurodegenerative diseases such as Tay-Sachs disease and Niemann-Pick sphingomyelin storage diseases (27). Therefore, the study of the spin and eggroll mutants will provide us with a good tool for understanding the relationship between aging and lipofuscinogenesis.
ACKNOWLEDGMENTS
We thank Steve Robinow, Koji Owada, Eduardo A. Porta, Jun Motoyama, Eiki Kominami, Hidenobu Tsujimura, Andy Furley, Dave N. Palmer, and the members of the Yamamoto Behavior Genes Project for their valuable discussions and excellent assistance; Tina M. Weatherby for sectioning the EM samples; and Keiko Shukuya and Yuka Kai for their secretarial assistance.
This study was supported in part by Special Cooperation Funds for Promoting Science and Technology from the Ministry of Education, Sports, Science and Technology Agency of Japan to D.Y. and M.U. and by Waseda University grant no. 2000B-029 to D.Y.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Becker L E, Prior T W, Yates A J. Metabolic disease. In: Davis R L, Robertson D M, editors. Textbook of neuropathology. Baltimore, Md: The Williams & Willkins Co.; 1997. pp. 407–509. [Google Scholar]
- 3.Brand A H, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan R L, Benzer S. Defective glia in the Drosophila brain degeneration mutant drop-dead. Neuron. 1993;10:839–850. doi: 10.1016/0896-6273(93)90200-b. [DOI] [PubMed] [Google Scholar]
- 5.Buege J A, Aust S D. Microsomal lipid peroxidation. Methods Enzymol. 1978;52:302–310. doi: 10.1016/s0076-6879(78)52032-6. [DOI] [PubMed] [Google Scholar]
- 6.Campbell G, Goring H, Lin T, Spana E, Andersson S, Doe C Q, Tomlinson A. RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development. 1994;120:2957–2966. doi: 10.1242/dev.120.10.2957. [DOI] [PubMed] [Google Scholar]
- 7.Chen P, Rodriguez A, Erskine R, Thach T, Abrams J M. Dredd, a novel effector of the apoptosis activators reaper, grim, and hid in Drosophila. Dev Biol. 1998;201:202–216. doi: 10.1006/dbio.1998.9000. [DOI] [PubMed] [Google Scholar]
- 8.Claveria C, Albar J P, Serrano A, Buesa J M, Barbero J L, Martinez-A C, Torres M. Drosophila grim induces apoptosis in mammalian cells. EMBO J. 1998;17:7199–7208. doi: 10.1093/emboj/17.24.7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly K, Cook R. Rejection responses by female Drosophila melanogaster: their ontogeny, causality and effects upon the behavior of the courting male. Behaviour. 1973;52:142–166. [Google Scholar]
- 10.Draizen T A, Ewer J, Robinow S. Genetic and hormonal regulation of the death of peptidergic neurons in the Drosophila central nervous system. J Neurobiol. 1999;38:455–465. doi: 10.1002/(sici)1097-4695(199903)38:4<455::aid-neu2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 11.Evans E K, Kuwana T, Strum S L, Smith J J, Newmeyer D D, Kornbluth S. Reaper-induced apoptosis in a vertebrate system. EMBO J. 1997;16:7372–7381. doi: 10.1093/emboj/16.24.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finley K D, Taylor B J, Milstein M, McKeown M. dissatisfaction, a gene involved in sex-specific behavior and neural development of Drosophila melanogaster. Proc Natl Acad Sci USA. 1997;94:913–918. doi: 10.1073/pnas.94.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher B L, Dillard C J, Tappel A L. Measurement of fluorescent lipid peroxidation products in biological systems and tissues. Anal Biochem. 1973;52:1–9. doi: 10.1016/0003-2697(73)90327-8. [DOI] [PubMed] [Google Scholar]
- 14.Haining W N, Carboy-Newcomb C, Wei C L, Steller H. The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc Natl Acad Sci USA. 1999;96:4936–4941. doi: 10.1073/pnas.96.9.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halter D A, Urban J, Rickert C, Ner S S, Ito K, Travers A A, Technau G M. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development. 1995;121:317–332. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D, Lane D, Lipsich L, Wigler M, Botchan M. Characteristics of an SV-40 plasmid recombinant and its movement into and out of the genome of a murine cell. Cell. 1980;21:127–139. doi: 10.1016/0092-8674(80)90120-8. [DOI] [PubMed] [Google Scholar]
- 17.Harman D. Lipofuscin and ceroid formation: the cellular recycling system. In: Porta E A, editor. Lipofuscin and ceroid pigments. New York, N.Y: Plenum Press; 1989. pp. 3–15. [DOI] [PubMed] [Google Scholar]
- 18.Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D. Sexual orientation in Drosophila is altered by the satori mutation in the sex determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci USA. 1996;93:9687–9692. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jallon J-M, Hotta Y. Genetic and behavioral studies of female sex appeal in Drosophila. Behav Genet. 1979;9:257–275. doi: 10.1007/BF01068205. [DOI] [PubMed] [Google Scholar]
- 20.Kimura K, Truman J W. Postmetamorphic cell death in the nervous and muscular systems of Drosophila melanogaster. J Neurosci. 1990;10:403–411. doi: 10.1523/JNEUROSCI.10-02-00403.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kretzschmar D, Hasen G, Sharma S, Heisenberg M, Benzer S. The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J Neurosci. 1997;17:7425–7432. doi: 10.1523/JNEUROSCI.17-19-07425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyte J, Doolittle T G. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 23.Lundell M J, Hirsh J. A new visible light DNA fluorochrome for confocal microscopy. BioTechniques. 1994;16:434–440. [PubMed] [Google Scholar]
- 24.Min K-T, Benzer S. Spongecake and eggroll: two hereditary diseases in Drosophila resemble patterns of human brain degeneration. Curr Biol. 1997;7:885–888. doi: 10.1016/s0960-9822(06)00378-2. [DOI] [PubMed] [Google Scholar]
- 25.Miyamoto H, Nihonmatsu I, Kondo S, Ueda R, Togashi S, Hirata K, Ikegami Y, Yamamoto D. canoe encodes a novel protein containing a GLGF/DHR motif and functions with Notch and scabrous in common developmental pathways in Drosophila. Genes Dev. 1995;9:612–625. doi: 10.1101/gad.9.5.612. [DOI] [PubMed] [Google Scholar]
- 26.Mutsuddi M, Nambu J R. Neural disease: Drosophila degenerates for a good cause. Curr Biol. 1998;8:809–811. doi: 10.1016/s0960-9822(07)00506-4. [DOI] [PubMed] [Google Scholar]
- 27.Naureckiene S, Sleat D E, Lackland H, Fensom A, Vanier M T, Wattiaux R, Jadot M, Lobel P. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 28.Nilsson E E, Asztalos Z, Lukacsovich T, Awano W, Usui-Aoki K, Yamamoto D. fruitless is in the regulatory pathway by which ectopic mini-white and transformer induce bisexual courtship in Drosophila. J Neurogenet. 2000;13:213–232. doi: 10.3109/01677060009084495. [DOI] [PubMed] [Google Scholar]
- 29.Pfrieger F W, Barres B A. Synaptic efficacy enhanced by glial cells in vitro. Science. 1997;277:1684–1687. doi: 10.1126/science.277.5332.1684. [DOI] [PubMed] [Google Scholar]
- 30.Robertson J, Pipa R. Metamorphic shortening of interganglionic connectives of Galleria mellonella (Lepidoptera) in vitro: stimulation by ecdysone analogues. J Insect Physiol. 1973;19:673–679. doi: 10.1016/0022-1910(73)90076-0. [DOI] [PubMed] [Google Scholar]
- 31.Robinow S, Talbot W S, Hogness D S, Truman J W. Programmed cell death in the Drosophila CNS is ecdysone-regulated and coupled with a specific ecdysone receptor isoform. Development. 1993;119:1251–1259. doi: 10.1242/dev.119.4.1251. [DOI] [PubMed] [Google Scholar]
- 32.Rubin G M, Spradling A. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Scherer S S. Molecular genetics of demyelination: new wrinkles on an old membrane. Neuron. 1997;18:13–16. doi: 10.1016/s0896-6273(01)80042-8. [DOI] [PubMed] [Google Scholar]
- 35.Siman R, Card J P, Nelson R B, Davis L G. Expression of β-amyloid precursor protein in reactive astrocytes following neuronal damage. Neuron. 1989;3:275–285. doi: 10.1016/0896-6273(89)90252-3. [DOI] [PubMed] [Google Scholar]
- 36.Spradling A. Developmental genetics of oogenesis. In: Bate M, Martinez-Arias A, editors. The development of Drosophila. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- 37.Suzuki K, Juni N, Yamamoto D. Enhanced mate refusal in female Drosophila induced by a mutation in the spinster locus. Appl Entomol Zool. 1997;32:235–243. [Google Scholar]
- 38.Tappel A L, Fletcher B L, Deamer D. Effect of antioxidants and nutrients on lipid peroxidation fluorescent products and aging parameters in the mouse. J Gerontol. 1973;28:415–424. doi: 10.1093/geronj/28.4.415. [DOI] [PubMed] [Google Scholar]
- 39.Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 40.Tissot M, Stocker R F. Metamorphosis in Dorosophila and other insects: the fate of neurons throughout the stages. Prog Neurobiol. 2000;62:89–111. doi: 10.1016/s0301-0082(99)00069-6. [DOI] [PubMed] [Google Scholar]
- 41.Truman J W, Thorn R S, Robinow S. Programmed neuronal death in insect development. J Neurobiol. 1992;23:1295–1311. doi: 10.1002/neu.480230917. [DOI] [PubMed] [Google Scholar]
- 42.Tsuchida M, Miura T, Aibara K. Lipofuscin and lipofuscin-like substances. Chem Phys Lipids. 1987;44:297–325. doi: 10.1016/0009-3084(87)90055-7. [DOI] [PubMed] [Google Scholar]
- 43.Tung A S, Pipa R L. Insect neurometamorphosis. V. Fine structure of axons and neuroglia in the transforming interganglionic connectives of Galleria mellonella (L.) (Lepidoptera) J Ultrastruct Res. 1972;39:556–567. doi: 10.1016/s0022-5320(72)90121-9. [DOI] [PubMed] [Google Scholar]
- 44.Vaux D L, Korsmeyer S J. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 45.White K, Grether M E, Abrams J M, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 46.Xiong W-C, Montell C. Defective glia induce neuronal apoptosis in the repo visual system of Drosophila. Neuron. 1995;14:581–590. doi: 10.1016/0896-6273(95)90314-3. [DOI] [PubMed] [Google Scholar]
- 47.Xiong W-C, Okano H, Patel N H, Blendy J A, Montell C. repo encodes a glia-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- 48.Yagi K. A simple fluorometric assay for lipoperoxide in blood plasma. Biochem Med. 1976;15:212–216. doi: 10.1016/0006-2944(76)90049-1. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto D, Jallon J M, Komatsu A. Genetic dissection of sexual behavior in Drosophila melanogaster. Annu Rev Entomol. 1997;42:551–585. doi: 10.1146/annurev.ento.42.1.551. [DOI] [PubMed] [Google Scholar]
- 50.Yokokura T, Ueda R, Yamamoto D. Phenotypic and molecular characterization of croaker, a new mating behavior mutant of Drosophila melanogaster. Jpn J Genet. 1995;70:103–117. doi: 10.1266/jjg.70.103. [DOI] [PubMed] [Google Scholar]