Highlights
-
•
Panobinostat in combination with olaparib reduces homologous recombination (HR) proficient tumor cell viability.
-
•
This combination favors anti-tumorigenic macrophages and enhances CD8+ T cell infiltration into tumors.
-
•
As a result, this combination targets HR-proficient ovarian tumors directly and through the tumor microenvironment.
Keywords: Ovarian cancer, Histone deacetylase inhibitors, PARP inhibitors, Homologous recombination repair, Immune response
Abbreviations: HDACi, histone deacetylase inhibitors; HRP, homologous recombination proficient; PARPi, polymerase ADP ribose phosphate inhibitor; Ola, olaparib; Pano, panobinostat; IP, intraperitoneal; PO, per os (oral gavage)
Abstract
Histone deacetylase inhibitors (HDACi) sensitize homologous recombination (HR)-proficient human ovarian cancer cells to PARP inhibitors (PARPi). To investigate mechanisms of anti-tumor effects of combined HDACi/PARPi treatment we performed transcriptome analysis in HR- proficient human ovarian cancer cells and tested drug effects in established immunocompetent mouse ovarian cancer models. Human SKOV-3 cells were treated with vehicle (Con), olaparib (Ola), panobinostat (Pano) or Pano+Ola and RNA-seq analysis performed. DESeq2 identified differentially expressed HR repair and immune transcripts. Luciferised syngeneic mouse ovarian cancer cells (ID8-luc) were treated with the HDACi panobinostat alone or combined with olaparib and effects on cell viability, apoptosis, DNA damage and HR efficiency determined. C57BL/6 mice with intraperitoneally injected ID8-luc cells were treated with panobinostat and/or olaparib followed by assessment of tumor burden, markers of cell proliferation, apoptosis and DNA damage, tumor-infiltrating T cells and macrophages, and other immune cell populations in ascites fluid. There was a significant reduction in expression of 20/37 HR pathway genes by Pano+Ola, with immune and inflammatory-related pathways also significantly enriched by the combination. In ID8 cells, Pano+Ola decreased cell viability, HR repair, and enhanced DNA damage. Pano+Ola also co-operatively reduced tumor burden and proliferation, increased tumor apoptosis and DNA damage, enhanced infiltration of CD8+ T cells into tumors, and decreased expression of M2-like macrophage markers. In conclusion, panobinostat in combination with olaparib targets ovarian tumors through both direct cytotoxic and indirect immune-modulating effects.
Introduction
Epithelial ovarian cancer is the deadliest gynecologic cancer [1]. Approximately 60% of women are diagnosed with advanced stage III/IV disease, characterized by peritoneal metastases and ascites. Poly ADP ribose polymerase inhibitors (PARPi) have shown remarkable promise as single agents in treating advanced stage BRCA1/2 mutant homologous recombination (HR)-deficient ovarian cancer2., 3., 4., 5., 6., 7., 8., 9.. However, effective treatment options for women with advanced stage HR-proficient ovarian cancers remains an unmet need. Approximately half of all women diagnosed with ovarian cancer are diagnosed with HR-proficient disease, including a subset with CCNE1 amplified/gain with poor clinical outcomes 10., 11., 12.. We found that combination treatment with histone deacetylase inhibitors (HDACi) synergize with PARPi in HR proficient ovarian cancer, in part through downregulation of HR repair 13., 14., 15., 16., 17., 18., 19., 20..
Peritoneal metastases and ascites associated with advanced state ovarian cancer involves complex interactions between ovarian tumor cells and host inflammatory cells, including tumor promoting peritoneal macrophages [21,22]. Our group and others have shown that pro-tumorigenic macrophages, which are abundant in ovarian cancer ascites, can be “re-educated” toward an anti-tumorigenic phenotype for therapeutic benefit 22., 23., 24.. Both PARPi and HDACi are known to alter function of specific immune cell types in the tumor microenvironment to promote anti-tumor immunity 25., 26., 27., 28., 29., 30.. However, the combined effects of HDACi and PARPi on peritoneal metastases and ascites in ovarian cancer are not known.
The purpose of this study was to elucidate the combined effects of HDACi and PARPi in ovarian cancer. We hypothesized that HDACi sensitize ovarian tumors to PARPi through downregulation of HR genes and alterations in immune gene transcripts and functions. We performed whole transcriptome sequencing in HR proficient SKOV-3 human epithelial ovarian cancer cells treated with the HDACi panobinostat and the PARPi olaparib, and confirmed that panobinostat causes significant downregulation of HR genes. We also discovered that both panobinostat and olaparib cause alterations in immune and inflammatory-related genes. The combination of panobinostat and olaparib was tested in ID8-luc cells, a syngeneic mouse model of HR-proficient epithelial ovarian cancer. We validated that the combination of panobinostat and olaparib is synergistic in reducing cell viability in cultured mouse epithelial ovarian cancer cells. As expected, panobinostat reduced HR gene expression and function. Further, panobinostat and olaparib reduced peritoneal metastases and tumor burden in a syngeneic mouse model of ovarian cancer. Peritoneal macrophages derived from ascites after treatment with panobinostat and olaparib showed a shift towards an anti-tumor phenotype. As a result, this study provides new insight into dual mechanisms of HDACi combined with PARPi that can be used to inform our ongoing clinical trial (https://clinicaltrials.gov/ct2/show/NCT03924245) and future immunotherapy combinations for women diagnosed with HR-proficient ovarian cancer.
Materials and methods
Cell culture
SKOV-3 cell line was obtained from American Type Cell Culture (ATCC). Mouse ID8 epithelial ovarian cancer cell lines expressing a constitutive luciferase reporter plasmid (ID8-Luc) were obtained from Dr. Frances Balkwill (Barts Cancer Institute, Queen Mary University of London) [24]. The cell lines were maintained in culture as previously described [19,23,31]. All cell lines used tested negative for mycoplasma.
Drugs and reagents
Panobinostat and olaparib were purchased from Selleck Chemicals (Houston, TX). Both drugs were reconstituted to prepare 100 mM stock solution using DMSO. Cell culture media was used to prepare further dilutions as described below.
Panobinostat / olaparib treatment of cell lines
For cell viability studies in ID8-luc cells, we tested effects of 1, 2.5, 5, 10 and 20 nM panobinostat (Pano) and 2, 5, 10, 20, and 40 µM olaparib (Ola) either as single drugs or as combined treatment (Pano+Ola) for 72 h. The control (Con) group was treated with vehicle only (0.01% DMSO diluted in media). For transcriptome analysis, ID8-luc cells were treated with vehicle only, 25 nM panobinostat alone, 10 µM olaparib alone or 25 nM panobinostat and 10 µM olaparib for 8 h to parallel our SKOV-3 experiments described below. For all other in vitro experiments using ID8-luc cells, the cells were treated with vehicle, 25 nM panobinostat alone, 10 µM olaparib alone, 25 nM panobinostat and 10 µM olaparib for 24 h. The concentration of DMSO added to cells was equalized among all treatment groups.
RNA-seq analysis
For transcriptome analysis, human SKOV-3 ovarian cancer cells were treated with vehicle only (0.01% DMSO diluted in media) in the control group (Con), 25 nM panobinostat alone (Pano), 10 µM olaparib alone (Ola) and combined 25 nM panobinostat and 10 µM olaparib (Pano+Ola) for 8 h. Three independent passages of cells were treated and assayed by RNA-seq in the Vanderbilt Technologies for Advanced Genomics Core (VANTAGE) core using the Illumina HiSeq2500 platform. RNA-seq reads were aligned to the human genome hg19 using TopHat2 [32] and the number of reads mapped to each gene was calculated by HTseq (http://www-huber.embl.de/users/anders/HTSeq/). Differentially expressed genes between drug treatment groups were detected by DESeq2 [33]. The P values were corrected for multiple testing using the Benjamini-Hochberg procedure. The significantly changed genes were determined based on fold change greater than 1.5 (FC>1.5) and the corrected P value less than 0.01 (FDR<0.01). Functional enrichment analysis on differentially expressed genes to infer pathways and regulatory mechanisms associated with drug treatments was performed by WebGestalt (Web-based gene set analysis toolkit; http://bioinfo.vanderbilt.edu/webgestalt/) [34]. Enrichment p-values were generated by the Fisher's exact test and adjusted by the Benjamini-Hochberg's multiple test correction. RNA-seq data files will be made publicly available at the GenBank repository (BioProject Accession PRJNA767427).
Quantitative real time RT-PCR (qRT-PCR)
Using parallel RNA samples, we performed qRT-PCR analysis to measure steady-state mRNA levels of selected HR and immune and inflammation-related genes as validation of the drug-induced changes detected in our RNA-seq experiments in SKOV-3 cells. We also used qRT-PCR analysis to validate changes in expression of a subset of genes in ID8-luc cells treated with the same drug concentrations and time points as described above. RNA isolation and cDNA synthesis was performed as previously described [23]. Levels of mRNA expression of selected HR-related and immune and inflammation-related genes were quantified using human Taqman probes (ThermoFisher Scientific, Waltham, MA) as described [35]. A list of probes used in our human and mouse ovarian cancer cell lines including GAPDH as internal control is shown in Supplemental Table 2. We also used Taqman qRT-PCR analysis to measure mRNA expression of validated markers in the cellular component of mouse ascites fluid [23].: the epithelial marker CK18 to measure tumor cell content; the macrophage marker F4/80; a mediator of cytotoxic T cell responses, CXCL9; the angiogenesis factor VEGFA; the M1 macrophage markers CCL3 and iNOS, and the M2 markers arginase-1 and mannose receptor (see Supplemental Table 3 for probe information) [23]. For both sets of analysis, we determined expression using the ΔΔCT method relative to corresponding GAPDH levels.
Cell viability assay
500 ID8-luc cells were plated in 384 well plates, in triplicates and treated as described above. Sulforhodamine B (SRB) assay was carried out as described [18] and Combination Index determined using CompuSyn software following Chou-Talay's method [36] to assess interactions between the panobinostat/olaparib combinations.
Homologous recombination functional assays
Plasmid-based DNA repair assay
For measuring drug effects on homologous recombination (HR) efficiency, the HR reporter plasmid pDRGFP and endonuclease encoding pCBASce1 (I-Sce1) (both gifts from Maria Jasin; Addgene plasmids #26475 and #26477, respectively) [37,38] were used. ID8-luc cells grown on glass coverslips were transfected with the plasmids using Lipofectamine 2000 according to manufacturer's instructions (Thermo Fisher Scientific, Waltham, MA), and then treated with vehicle, panobinostat, olaparib or panobinostat/olaparib for 24 h as described above and visualized for GFP expression using fluorescence microscopy as previously described [39].
Measurement of RAD51 and BRCA1 repair foci
ID8-luc cells grown on glass coverslips were pretreated for 6 h with the known inducer of double-strand DNA breaks, cisplatin (0.5 µM; Sigma-Aldrich). The cells were then washed with PBS, and treated with vehicle, panobinostat, olaparib or panobinostat/olaparib for 24 h as described above. Following cessation of drug treatment, the cells were fixed, stained with mouse monoclonal anti-BRCA1 (1:100 MilliporeSigma, Burlington, MA) or rabbit polyclonal anti-RAD51 (1:100 MilliporeSigma), with acquisition and analysis of fluorescent images as previously described [39].
High-content fluorescent microscopy
Following treatment of ID8-luc cells with vehicle, panobinostat, olaparib or panobinostat/olaparib for 24 h, we performed high-content IF imaging of cells using the ImageXpress Micro XL imaging platform (Molecular Devices (Sunnyvale, CA). Images of cells stained with DAPI (Sigma) to mark cell nuclei and were acquired in nine fields of view per well. Analysis of adherent cell number and the cell cycle indices %G0/G1, %S and %G2/M were performed using MetaXpress software as previously described [39].
Western blot
For ID8-luc cells treated with vehicle, panobinostat, olaparib or panobinostat/olaparib for 24 h as described above, whole cell protein isolation, hydrochloric acid extraction of histones, western blotting and signal detection were performed as previously described [15,39]. Antibodies used were: rabbit polyclonal anti-cleaved PARP (1:1000 Cell Signaling Technology, Danvers, MA) to assess apoptosis, mouse monoclonal anti-γH2AX (Ser139) (1:1000 MilliporeSigma) to assess DNA damage, mouse monoclonal anti-β-actin (1:10,000 Sigma-Aldrich) as a loading control for whole cell extracts and mouse monoclonal anti-histone H3 (1:1000 MilliporeSigma) as a loading control for histone extracts.
Animal studies
Six to eight-week-old C57BL/6 female mice were used for these studies. All mice were maintained and handled at Vanderbilt University as per respective Institutional Animal Use and Care Committee (IACUC) guidelines. Experiments were conducted in accordance with American Association for Laboratory Animal Science (AALAS) and ARRIVE guidelines. Mice were group housed (maximum 5 mice/cage) with standard bedding and ad libitum access to food and water. Animal rooms were maintained at constant temperature and humidity with a 12 h light/dark cycle.
Panobinostat and olaparib treatment of mice
All experimental C57BL/6 mice used were bred in the colony of Dr. Fiona Yull at Vanderbilt. Mice were injected intraperitoneally with 5 × 106 ID8-luc cells as previously described [23]. After 30 days, mice underwent baseline bioluminescence imaging (BLI) as previously described [40] and randomized into 4 groups (n = 5) to ensure equivalent baseline tumor burden before the start of drug treatment. Mice in the following treatment groups were treated for 4 weeks: Vehicle (1% DMSO in PBS five times weekly IP and PO); Panobinostat (2.5 mg/kg five times weekly IP, 1% DMSO in PBS five times weekly PO; Olaparib (olaparib 100 mg/kg five times weekly PO & 1% DMSO in PBS five times weekly IP); and Panobinostat±olaparib (panobinostat 2.5 mg/kg five times weekly IP, olaparib 100 mg/kg five times weekly PO). Animals were examined visually biweekly for the effects of tumor burden and tumor growth, including weekly weight measurements and body condition scoring throughout the duration of drug treatment. Pre-euthanasia BLI was measured to determine tumor burden. Mice were euthanized by carbon dioxide inhalation and secondary cervical dislocation according to IACUC-approved protocols.
Following sacrifice, omental tumor implants were harvested, weighed and snap-frozen, or fixed in 10% neutral-buffered formalin for 24 h for immunostaining as described below. Ascites was collected by withdrawing fluid from the peritoneal cavity with a hypodermic syringe, and the volume measured. If no measurable ascites was present, peritoneal lavage was performed by injecting 10 ml PBS IP and carefully extracting the fluid with a hypodermic syringe. The cellular content of ascites fluids or peritoneal lavages was isolated as previously described [23] and snap-frozen for RNA extraction for qRT-PCR analysis.
Immunostaining of tumor sections
Tissue fixation, processing and sectioning methods of harvested ID8-luc xenografts were performed as previously described [18,23]. In ID8-luc tumors, immunohistochemistry (IHC) for mib-1/Ki67 (1:1000; rabbit polyclonal anti-mib-1/Ki67, Vector Laboratories, Burlingame, CA), cleaved caspase-3 (1:300; rabbit polyclonal anti- cleaved caspase-3, Cell Signaling Technology, Danvers, MA), and γH2AX (Ser139) (1:100; mouse monoclonal MilliporeSigma) were performed as previously described [16]. Fluorescent-IHC image acquisition and analysis of co-staining for the pan-macrophage marker F4/80 (1:200 rat polyclonal anti-mouse F4/80, AbD Serotec, Oxford, UK), and the M2 macrophage marker arginase-1 (1:200; rabbit polyclonal anti-arginase-1, GeneTex, Irvine, CA), and separately for the cytotoxic T cell marker CD8 (1:100; rat monoclonal anti-mouse CD8, Novus Biologicals, Centennial, CO), was performed as previously described [23]. Cell nuclei were identified by staining with DAPI (Sigma-Aldrich). At least 1000 cells were counted in at least 5 independent fields under high power (x40) for these counts.
Statistical analysis
Statistical analyses were performed using GraphPad Prism, Version 8 (La Jolla, California, USA). Comparisons of group means in cultured cells were performed by 2-tailed Student's t test. For mouse experiments, group means were compared by 2-tailed Mann-Whitney test. A p-value <0.05 was considered statistically significant.
Results
Transcriptome sequencing reveals dual regulation of HR repair and immune gene transcripts in cells treated with panobinostat and olaparib
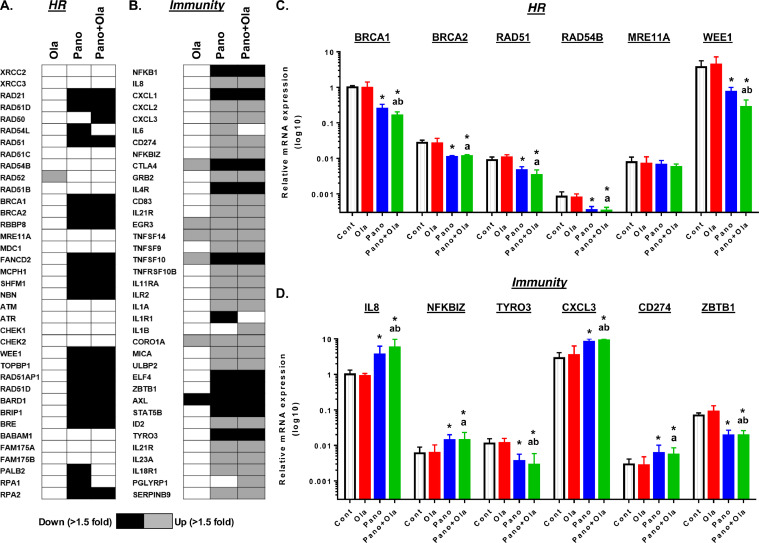
We have previously shown that HDACi such as SAHA, entinostat and panobinostat downregulate expression of selected HR genes in HR-proficient human ovarian cancer cells 18., 19., 20.. To identify genome-wide alterations associated with HDACi and PARPi combination treatment, we performed whole transcriptome sequencing of SKOV-3 cells using the Illumina HiSeq2500 platform. We extracted RNA after treatment with 10 µM olaparib, 25 nM panobinostat, or the combination of 10 µM olaparib and 25 nM panobinostat for 8 hr. Robust changes in transcript expression were found compared to control in cells treated with olaparib (2929 up, 90 down), panobinostat (6477 up, 3078 down), and the combination (5985 up, 2958 down) using a criterion of FDR<0.05, >1.5 fold increase or decrease in expression (Supplemental Fig. 1). As expected, panobinostat treatment, alone and combined with olaparib, led to coordinated downregulation of HR machinery transcripts such as RAD21, RAD51D, RAD51, FANCD2, WEE1, and RPA1/2 (black boxes) (Fig. 1A). Downregulation of HR transcripts were validated using qRT-PCR, and gene expression levels of BRCA1, BRCA2, RAD51, RAD54B and WEE1 were significantly downregulated in both the panobinostat and the combination treatment groups (Fig. 1C). There was no significant change in HR genes in cells treated with olaparib alone, with the exception of RAD52 which showed a significant upregulation (FDR<0.05, 1.7 fold increase) by RNA-seq analysis (blue box).
Fig. 1.
Coordinated downregulation of homologous recombination gene transcripts and modulation of immune gene transcripts in cells treated with panobinostat and olaparib in SKOV-3 cells. SKOV-3 cells were treated with vehicle (0.01% DMSO), panobinostat (25 nM), olaparib (10 µM) or the combination for 8 h. Heat map of drug effects on expression of (A) a panel of 37 HR-related genes and (B) immune and inflammation-related genes by RNA-seq. Differentially expressed genes were detected using DESeq2 (Fold change>1.5 and FDR<0.05). Parallel Taqman qRT-PCR validation of selected (C) HR genes and (D) immunity-related genes normalized for corresponding GAPDH levels. Values are mean+SEM expressed relative to control treatment; *p < 0.05 single drug effect relative to vehicle; ap < 0.01 combination drug effect relative to olaparib alone, bp < 0.01 combination drug effect relative to panobinostat alone; Student's t test (For interpretation of the references to color in this figure, the reader is referred to the web version of this article.).
Next, we performed functional enrichment analysis using Geneset ontology (WebGestalt). In the cells treated with the panobinostat and olaparib combination, there was enrichment in gene alterations associated with immune and inflammation response pathways (Supplementary Table 1, FDR< 0.05). Genes associated with tumor immunity were both upregulated (grey) or downregulated (black) (Fig. 1B). We validated the expression of immune regulatory transcripts by qRT-PCR, and confirmed that panobinostat alone and in combination with olaparib (green bars) showed a significant upregulation in IL8, NFKBIZ, CXCL3, and CD274 transcripts compared to vehicle or olaparib treatment (Fig. ID). Whereas, TYRO3 and ZBTB1 transcripts were both significantly downregulated in the panobinostat and combination treatment groups (Fig. 1D).
Panobinostat decreases HR gene expression and function and synergizes with olaparib in ID8-luc HR proficient mouse epithelial ovarian cancer cells
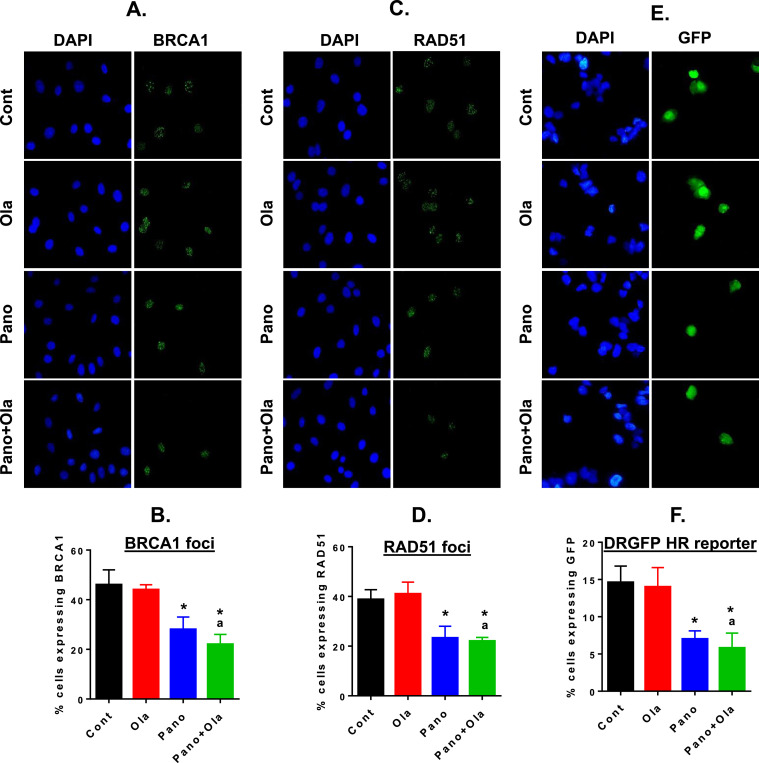
Having identified modulation of multiple HR, immune and inflammation-related genes by the panobinostat/olaparib combination in SKOV-3 cells, we then tested drug effects in a stable luciferised clone of the ID8 mouse ovarian cancer cell line (ID8-luc). We have experience with multiple intraperitoneal ovarian cancer models in immunocompetent mice, including the ID8-luc model [21,23]. We first validated that drug treatment of ID8-luc cells induced similar changes in expression of a subset of HR and immunity-related genes as those observed in SKOV-3 cells (Supplemental Fig. 2). To measure HR efficiency in ID8-luc cells, we pre-treated cells with the DNA damaging drug 0.5 µM cisplatin for 6 h to induce double strand breaks, and then treated cells with panobinostat, olaparib or the combination for an additional 24 h. After cell fixation, we performed IF staining for BRCA1 and RAD51. We counted cells with BRCA1 staining and RAD51 foci and found significant reduction in panobinostat-treated cells, with additional reduction in combination-treated cells (Fig. 2A–D). In the second approach, we co-transfected ID8 cells with the pDRGFP and I-Sce1 plasmids, and treated with cells as above (Fig. 2E&F). In panobinostat treated cells, there was a significant reduction in cells expressing DRGFP. Thus, the reduction in HR efficiency in ID8-luc cells is consistent with the coordinated decrease in HR gene transcripts in SKOV-3 and ID8-luc cells after panobinostat treatment, whether alone or combined with olaparib.
Fig. 2.
Panobinostat alone and combined with olaparib reduces HR proficiency in ID8-luc cells. HR repair efficiency of DNA double-strand breaks in drug-treated ID8-luc cells was measured in three assays. Cells were pre-treated with 0.5µM cisplatin for 6 h were then treated with panobinostat (25 nM), olaparib (10 μM) or the combination for a further 24 h, and formation of BRCA1 (A&B) and RAD51 foci (C&D) analyzed by IF. Representative BRCA1 and RAD51 images (green) are shown; DAPI-stained nuclei are in blue. (E&F) IF analysis of GFP expression in ID8-luc cells co-transfected with the pDRGFP and I-Sce1 plasmids (both 1 µg). Cells were then treated with panobinostat (25 nM), olaparib (10 μM) or the combination for 24 h. At least 100 cells were counted (x40) for each treatment in the three assays. Values are mean+SEM; *p < 0.01 compared to control; ap < 0.01 relative to olaparib alone, Student's t test (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
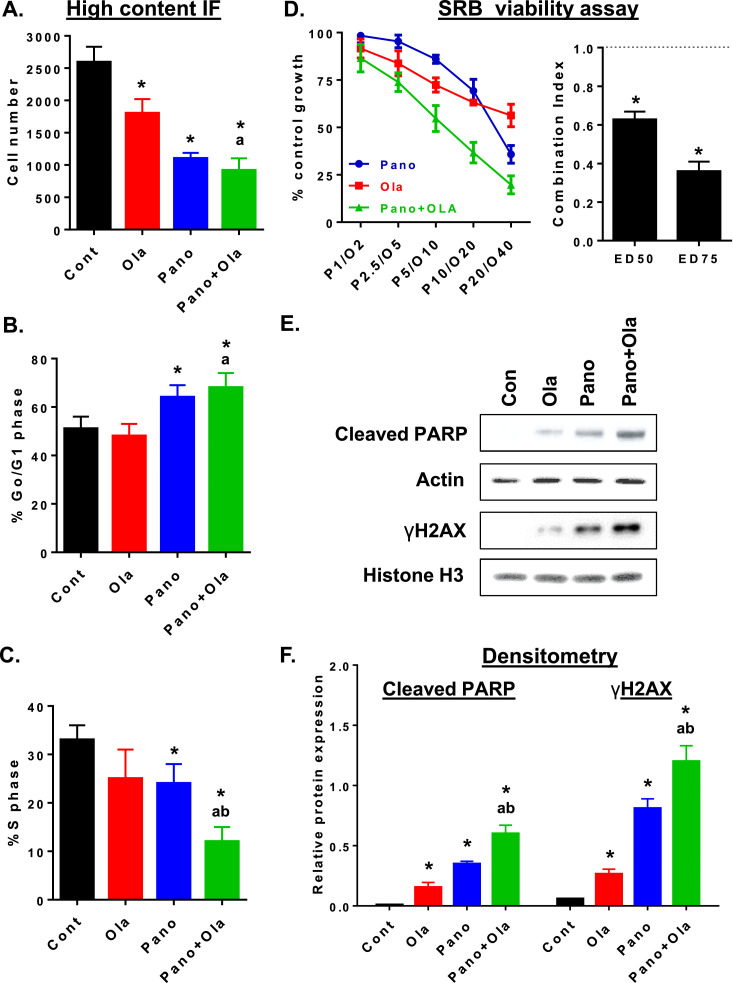
We then performed high content immunofluorescence (IF) to measure treatment effects on cell number and cell cycle indices. Panobinostat, alone and combined with olaparib, had a greater effect on reducing cell number compared to olaparib alone, which is consistent with HR-proficient status and relative resistance to olaparib (Fig. 3A). Panobinostat alone and combined with olaparib significantly increased G0/G1 and decreased the S phase fraction (Fig. 3B&C). We also assessed drug effects on viability of ID8-luc cells treated with increasing concentrations of panobinostat and/or olaparib for 72 h using SRB assays. As shown in Fig. 3D, panobinostat and olaparib had relatively minimal effects on cell viability as single agents, but were synergistic in combination. This was demonstrated by isobologram analysis which showed a Combination Index (CI) value for ED50 and ED75 that were significantly <1, indicative of synergism (p < 0.05, Student's t test) [15,16,19,36]. Finally, we showed that reduced HR repair efficiency in ID8-luc cells by panobinostat + olaparib combination treatment was associated with increased expression levels of the DNA damage marker, γH2AX, and increased expression of the apoptosis marker cleaved PARP, compared to either drug alone (Fig. 3E&F).
Fig. 3.
Panobinostat alone and combined with olaparib co-operatively reduces cell viability in ID8-luc cells. In ID8-luc cells treated with panobinostat (25 nM), olaparib (10 μM) or the combination (24 h), DAPI-stained nuclei were analyzed by IF to measure (A) adherent cell number and percentage of cells in (B) Go/G1 and (C) S phase of the cell cycle. (D) ID8-luc cells were treated with increasing concentrations of panobinostat and olaparib for 72 h, and cell viability was measured using SRB assay. Combination Index (CI) for ED50 and ED75 was calculated by isobologram analysis. CI<1 is synergistic. *p < 0.05 compared to a CI of 1, Student's t test. (E) Western blot analysis of expression of the apoptosis marker cleaved PARP in whole cell extracts and the DNA damage marker γH2AX in isolated histone extracts in ID8-luc cells treated as described in B. (F) Densitometry of western blots. Cleaved PARP and γH2AX was measured relative to corresponding actin and histone H3 expression, respectively, Values are mean+SEM; *p < 0.01 compared to control; ap < 0.01 relative to olaparib alone, bp < 0.01 relative to panobinostat alone, Student's t test.
The combination of panobinostat and olaparib reduces tumor burden in a syngeneic model of ovarian cancer
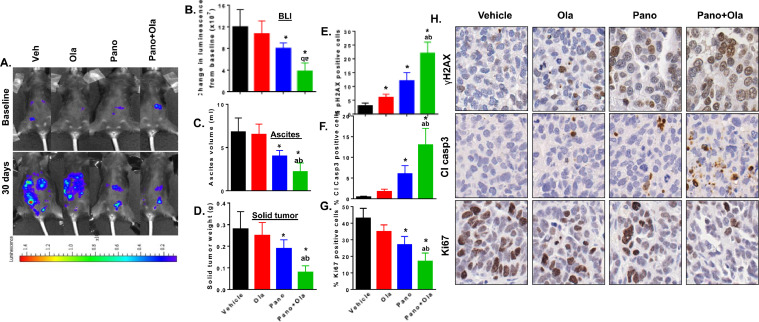
Having confirmed similar co-operative anti-tumor effects of combining HDACi and PARPi in ID8-luc cells than those observed in human ovarian cancer cell lines 18., 19., 20., we tested our drug combination in C57BL/6 mice injected intraperitoneally with 5 × 106 ID8-luc cells. Peritoneal metastases were allowed to establish for 4 weeks, and mice were subsequently treated with vehicle, panobinostat (2.5 mg/kg 5 times weekly PO), olaparib (100 mg/kg 5 times weekly PO) and the panobinostat/olaparib combination for 4 weeks after randomization. Noninvasive BLI measurements were obtained at baseline and compared to the end of treatment (Fig. 4A, B). After sacrifice, ascites solid tumors from peritoneal implants and omentum were collected and measured (Fig. 4C&D). As expected, there was no significant difference in tumor burden in olaparib-treated mice compared to controls. However, panobinostat significantly reduced tumor burden as a single agent, and these effects were potentiated by panobinostat+olaparib combination treatment. All mice tolerated the treatments well and no significant toxicity or change in weights were noted (Supplemental Fig. 3).
Fig. 4.
Panobinostat alone and combined with olaparib reduce tumor burden in ID8-luc-injected mice associated with reduced proliferation and increased apoptosis and DNA damage. C57BL/6 mice were injected with 5 × 106 ID8-luc cells. 30 days after tumor cell injection, mice underwent bioluminescence imaging (BLI) to establish baseline luminescence and then treated with vehicle, panobinostat (2.5 mg/kg 5 times weekly PO), olaparib (100 mg/kg 5 times weekly PO) and the panobinostat/olaparib combination for 4 weeks. Immediately before sacrifice, mice were re-imaged and solid tumor and ascites collected at necropsy. (A) Change in bioluminescence from baseline. Representative images from BLI analysis are shown in (B). Values are mean±SEM of n = 5 per group. (C) Ascites volume at sacrifice. (D) Harvested omental tumor weight. In harvested tumors, the percentage of cells staining for (E) γH2AX, (F) cleaved caspase-3 and (G) Ki67. Values are mean±SEM for 3 independent experiments. At least 1000 cells were counted (40X) for each drug treatment per experiment. *p < 0.02 compared to vehicle; ap < 0.02 relative to SAHA alone; bp < 0.01 relative to olaparib alone, Mann-Whitney test. (H) High power (x40) representative images of γH2AX, cleaved caspase-3 and Ki67, staining for each drug treatment.
Tumor sections were stained for molecular markers for proliferation (Ki67), apoptosis (cleaved caspase 3), and DNA damage (γH2AX) (Fig. 4E–H). Panobinostat treatment led to a significant decrease in Ki67 expression and an increased expression of cleaved caspase 3 and γH2AX compared to vehicle, effects which were potentiated by its combination with olaparib.
Panobinostat and olaparib alter the immune phenotype in tumors and ascites fluid
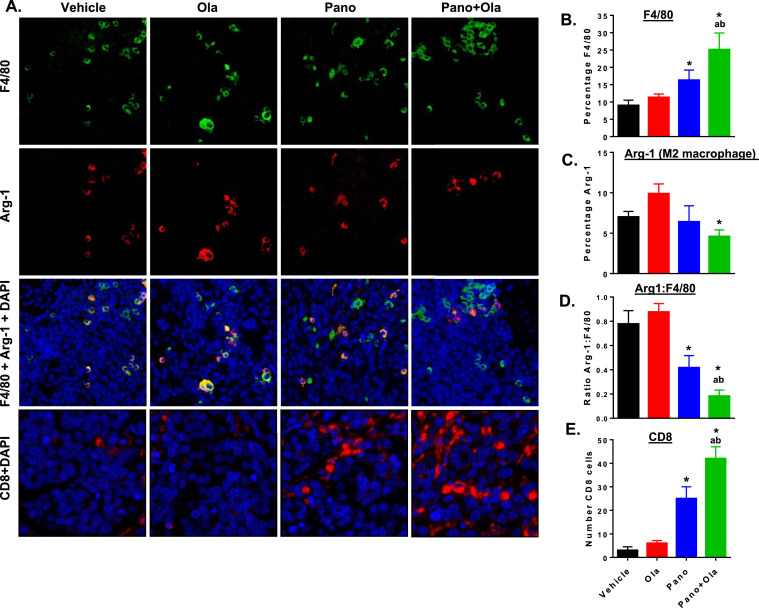
We have shown that targeting macrophages to alter functions from an overall M2-like pro-tumor to an M1-like anti-tumor phenotype can reduce tumor burden in ID8 syngeneic models of ovarian cancer accompanied by increased tumor-infiltrating cytotoxic T cells [23]. To evaluate drug effects on phenotype of tumor-infiltrating immune cells, we first performed immunofluorescence analysis of tumor sections co-stained for the pan-macrophage marker F4/80 and the M2 marker arginase-1 (Arg-1). Representative tumor sections stained for F4/80, Arg-1, CD8 and cell nuclei (DAPI) are shown in Fig. 5A. As shown in Fig. 5B–D, the panobinostat/olaparib combination results in significantly increased numbers of F4/80-positive, Arg-1-negative tumor-infiltrating macrophages compared to vehicle or either drug alone. Since M2-like tumor-associated macrophages are known to induce immunosuppressive effects, especially limiting cytotoxic T cell responses [21,41,42], we also stained tumors for CD8-positive T cells. Treatment of mice with panobinostat/olaparib combination showed higher numbers of tumor-infiltrating CD8-positive T cells above that of either drug alone (Fig. 5E).
Fig. 5.
Panobinostat alone and combined with olaparib increases macrophage recruitment, decreases M2 macrophages and increases CD8-positive T cell infiltration in the tumor microenvironment. C57BL/6 mice were injected with ID8-luc cells. 30 days after injection, mice were treated with vehicle, panobinostat (2.5 mg/kg 5 times weekly PO), olaparib (100 mg/kg 5 times weekly PO) or the panobinostat + olaparib combination or 4 weeks. (A) Representative images of tumors stained for F4/80 (green), Arg-1 (red), and DAPI (nuclei; blue). Separate staining for CD8 (red) is also shown. In harvested tumors, (B) counts of the percentage of F4/80-positive macrophages per high-powered field (HPF), (C) percentage of arginase-1 (Arg-1) positive cells, (D) the ratio of Arg-1 positive to F4/80 positive cells, and (E) the of CD8 positive cells. Counts were quantified from 5 representative HPF from each sample. *p < 0.05 single drug effect relative to vehicle; ap < 0.01 combination drug effect relative to olaparib alone, bp < 0.01 combination drug effect relative to panobinostat alone; Mann-Whitney test (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
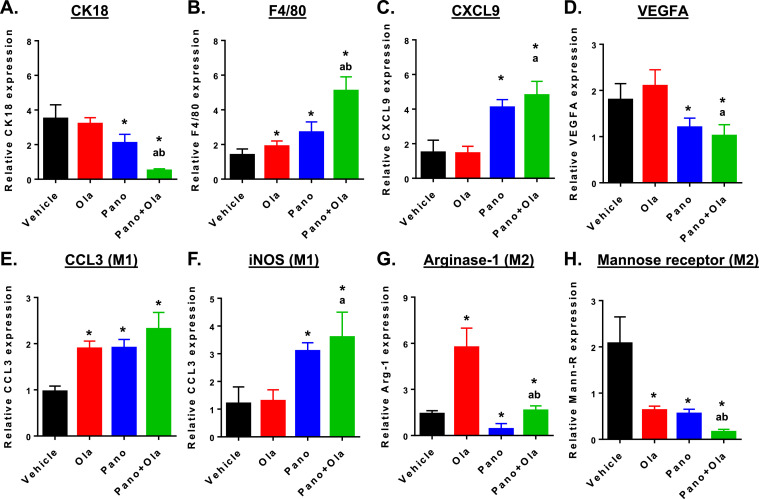
We have also shown that cell pellets from ascites predominantly contain a mix of tumor cells and macrophages (>90% of the immune cells) [21,40]. Therefore, we performed qRT-PCR after RNA extraction from ascites cells. Panobinostat decreased expression of CK18, an epithelial marker, consistent with a decrease in tumor burden (Fig. 6A). These effects were potentiated in combination with olaparib, F4/80 is a marker of macrophages, while CXCL9 promotes T cell recruitment and cytotoxic, anti-tumor responses [43]; both increased by panobinostat and in the panobinostat + olaparib combination (Fig. 6B&C). The expression levels of VEGFA, a marker of angiogenesis secreted by tumor cells and macrophages into ascites fluid [21,44,45], was also significantly reduced by panobinostat alone and in combination with olaparib (Fig. 6D).
Fig. 6.
Panobinostat reduces expression of tumor epithelial and M2 macrophage markers and increases expression of pan-macrophage and M1 markers in ascites cells from ID8-luc-injected mice. C57BL/6 mice were injected with ID8-luc cells. 30 days after injection, mice were treated with vehicle, panobinostat (2.5 mg/kg 5 times weekly PO), olaparib (100 mg/kg 5 times weekly PO) or panobinostat + olaparib combination for 4 weeks. After sacrifice, ascites was harvested and mRNA extracted from the cell pellet. Drug effects on steady state mRNA expression of (A) epithelial marker CK18, (B) macrophage marker F4/80, (C) T cell recruitment factor CXCL9, (D) angiogenic factor VEGFA, (E, F) M1 macrophage markers CCL3 and iNOS) and (G, H) M2 macrophage markers, arginase-1 and mannose receptor were measured by qRT-PCR. Relative expression values are expressed as mean±SEM, calculated using the 2−ΔΔCt method relative to corresponding GAPDH levels. *p < 0.01 single drug effect relative to control; ap < 0.01 relative to olaparib alone; bp < 0.01 relative to panobinostat alone, Mann-Whitney test.
We also assessed expression levels of macrophage M1 and M2 markers in ascites cells (Fig. 6E–H). Both panobinostat and olaparib increased the expression of CCL3 (M1) and decreased expression of the mannose receptor (M2). In contrast, panobinostat alone and combined with olaparib increased the expression of iNOS (M1) and decreased the expression of arginase-1 (M2). These results, along with the cytotoxic T cell infiltration, suggest a relative shift from a pro-tumorigenic to an anti-tumorigenic macrophage phenotype that is driven primarily by panobinostat and potentiated by olaparib.
Discussion
While PARPi are promising single agents in treating advanced stage BRCA1/2 mutant homologous recombination (HR)-deficient ovarian cancer 2., 3., 4., 5., 6., 7., 8., a persistent problem is the lack of effective treatment options for women with advanced stage HR-proficient ovarian cancers. We have shown that epigenetic drugs such as HDACi can sensitize HR-proficient ovarian cancer cells to PARPi, at least in part through downregulation of HR-related genes 18., 19., 20.. In this study, whole transcriptome sequencing in HR proficient SKOV-3 human epithelial ovarian cancer cells confirmed that there is coordinated downregulation of HR genes in cells treated with the combination of the HDACi panobinostat and the PARPi olaparib. We also discovered that both panobinostat and olaparib cause alterations in multiple genes implicated in immune and inflammatory responses in these cells. This motivated us to test the panobinostat + olaparib combination in the well-characterized ID8 syngeneic mouse ovarian cancer model to determine drug effects on tumor development in the context of the full immune repertoire [21,23,40,46].
Using luciferised ID8 cells (ID8-luc), we first established that panobinostat + olaparib combination has similar anti-tumor effects as previously observed in human ovarian cancer cell lines treated with HDACi and PARPi 18., 19., 20.. The drug combination was synergistic in reducing cell viability, and reduced efficiency of HR repair resulting in increased DNA damage-induced apoptosis. Panobinostat and olaparib also co-operatively decreased the overall tumor burden in C57BL/6 mice injected IP with ID8-luc cells, reducing both solid tumors and accumulation of ascites fluid. Consistent with the tumor burden data and our in vitro observations, the combination also co-operatively reduced tumor proliferation and increased DNA damage and apoptosis as observed in harvested tumors.
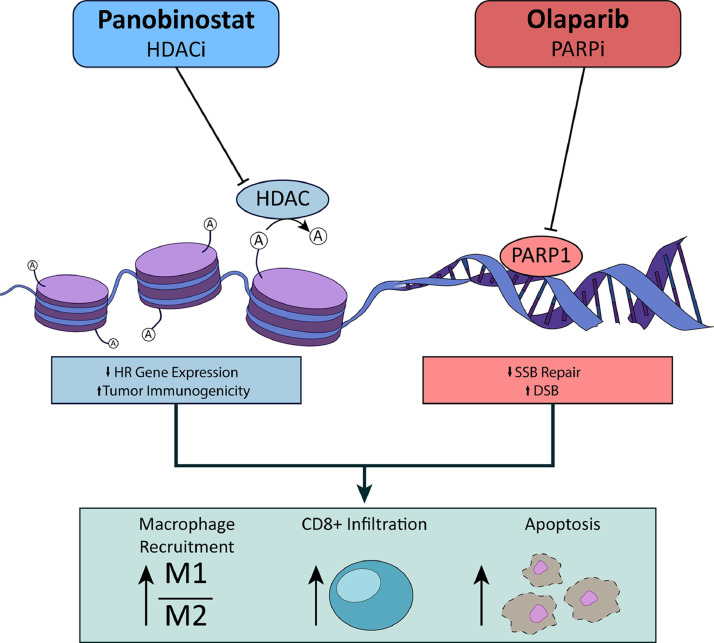
The panobinostat + olaparib combination also induced striking changes in immune cells in the tumor microenvironment, with a strong shift towards an anti-tumorigenic phenotype. Notably, panobinostat + olaparib combination induced higher overall numbers of macrophages in both tumors and in ascites fluid, based on expression of the pan macrophage marker F4/80. Importantly, these recruited macrophages were less likely to have an M2-like pro-tumorigenic phenotype, since we observed reduced overall numbers and proportion of M2-like tumor-infiltrating macrophages, and increased expression of M1 macrophages markers accompanied reduced expression of M2 markers in the cellular content of ascites fluid. We also observed a co-operative increase in tumor-infiltrating cytotoxic T cells in mice treated with panobinostat + olaparib combination, accompanied by increased levels of CXCL9 in cells in ascites fluid. CXCL9 has recently been identified as a critical mediator of anti-tumor T cell responses, and high CXCL9 levels are associated with increased numbers of CD8 positive T cells in ascites and better prognosis in ovarian cancer patients [43,47]. Graphical representation of these findings is illustrated in Fig. 7.
Fig. 7.
Graphical abstract illustrating the mechanism of action of panobinostat in combination with olaparib.
The shift towards a less immunosuppressive tumor microenvironment are consistent with studies showing that modulating the phenotype of M2-like tumor-associated macrophages towards M1-like cytotoxic, pro-inflammatory functions is a promising therapeutic strategy [22,23,48]. We recently showed that specific activation of canonical NF-kappa B in macrophages in tumor-bearing IKFM transgenic mice induces potent anti-tumorigenic effects, accompanied by increases in infiltration of M1 polarized macrophages and cytotoxic T cells into tumors, and increased levels of CXCL9 levels in ascites fluid [23]. Moreover, epigenetic drugs such as HDACi are known to have immunomodulatory functions that increase cytotoxic T cell functions 25., 26., 27.. A limitation of our current study is that SKOV-3 cells and ID8 cells do not represent high grade serous ovarian cancer (HGSOC) cells, but HR proficient ovarian cancer cells. However, we have shown that entinostat, a class 1/2 specific HDACi in combination with olaparib significantly improved survival in a patient-derived xenograft (PDX) mouse model of HGSOC [20]. Further, ovarian cancer occurs mostly in post-menopausal women, whereas mouse models used are 6–8-week-old representing the immune-landscape of younger women. To address this caveat, age-appropriate HGSOC models, in particular those correlating with post-menopausal women, will be used for detailed mechanistic explorations. In addition, post-menopausal women have compromised immune functions. To address this, studies segregating the relative effects of HR downregulation and immune environment modulation will be carried out.
Conclusions
We have observed enhanced anti-tumorigenic effects of HDACi/PARPi combinations in both immunocompromised and immunocompetent mouse models 18., 19., 20.. This suggests that the rational combination of panobinostat + olaparib targets ovarian tumors through both direct cytotoxic and indirect immune-modulating effects. This study provides new insight into mechanisms of HDACi combined with PARPi that can be used to inform our current clinical trial testing the entinostat/olaparib combination in women with HR-proficient tumors (NCT03924245) and to inform epigenetic drug combinations using immunotherapy.
CRediT authorship contribution statement
Andrew J. Wilson: Methodology, Software, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization. Vijayalaxmi G Gupta: Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization. Qi Liu: Writing – review & editing. Fiona Yull: Conceptualization, Resources, Writing – review & editing, Supervision, Funding acquisition. Marta A. Crispens: Conceptualization, Formal analysis, Investigation, Resources, Writing – review & editing, Supervision. Dineo Khabele: Conceptualization, Formal analysis, Investigation, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
Andrew J. Wilson, Vijayalaxmi G Gupta, Fiona Yull and Qi Liu declare that they do not have any competing interests. Marta A. Crispens reports grants from Astra-Zeneca during the conduct of the study. Dineo Khabele reports grant from NIH R01, Astra Zeneca, during the conduct of the study; receives personal fees from Astra Zeneca and received grants from Deciphera Pharmaceuticals, outside the submitted work.
Acknowledgments
Acknowledgments
The Translational Pathology Shared Resource is supported by NCI/NIH Cancer Center Support Grant 2P30 CA068485-14. The Vanderbilt Technologies for Advanced Genomics Core is supported by the CTSA Grant (5UL1 RR024975-03), the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Vision Center (P30 EY08126), and NIH/NCRR (G20 RR030956). Members of the Vanderbilt Ovarian Cancer Alliance (VOCAL) are gratefully acknowledged for their support of this work.
Financial support
This research was funded in part by R01CA243511 (DK), R21CA210210 (DK), and R01CA214043 (FY/DK)
Informed consent statement
Not applicable.
Data availability statement
RNA-seq data files will be made publicly available at the GenBank repository (BioProject Accession PRJNA767427).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.12.002.
Contributor Information
Vijayalaxmi G Gupta, Email: ggupta@wustl.edu.
Dineo Khabele, Email: khabeled@wustl.edu.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. PubMed PMID: 33433946. [DOI] [PubMed] [Google Scholar]
- 2.Coleman R.L., Oza A.M., Lorusso D., Aghajanian C., Oaknin A., Dean A., Colombo N., Weberpals J.I., Clamp A., Scambia G., Leary A., Holloway R.W., Gancedo M.A., Fong P.C., Goh J.C., O'Malley D.M., Armstrong D.K., Garcia-Donas J., Swisher E.M., Floquet A., Konecny G.E., McNeish I.A., Scott C.L., Cameron T., Maloney L., Isaacson J., Goble S., Grace C., Harding T.C., Raponi M., Sun J., Lin K.K., Giordano H., Ledermann J.A., on behalf of theARIEL3 investigators Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390(10106):1949–1961. doi: 10.1016/S0140-6736(17)32440-6. PubMed PMID: 28916367; PubMed Central PMCID: PMC5901715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domchek S.M., Aghajanian C., Shapira-Frommer R., Schmutzler R.K., Audeh M.W., Friedlander M., Balmana J., Mitchell G., Fried G., Stemmer S.M., Hubert A., Rosengarten O., Loman N., Robertson J.D., Mann H., Kaufman B. Efficacy and safety of olaparib monotherapy in germline BRCA1/2 mutation carriers with advanced ovarian cancer and three or more lines of prior therapy. Gynecol Oncol. 2016;140(2):199–203. doi: 10.1016/j.ygyno.2015.12.020. PubMed PMID: 26723501; PubMed Central PMCID: PMC4992984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ledermann J., Harter P., Gourley C., Friedlander M., Vergote I., Rustin G., Scott C., Meier W., Shapira-Frommer R., Safra T., Matei D., Macpherson E., Watkins C., Carmichael J., Matulonis U. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366(15):1382–1392. doi: 10.1056/NEJMoa1105535. PubMed PMID: 22452356. [DOI] [PubMed] [Google Scholar]
- 5.Ledermann J., Harter P., Gourley C., Friedlander M., Vergote I., Rustin G., Scott C.L., Meier W., Shapira-Frommer R., Safra T., Matei D., Fielding A., Spencer S., Dougherty B., Orr M., Hodgson D., Barrett J.C., Matulonis U. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15(8):852–861. doi: 10.1016/S1470-2045(14)70228-1. PubMed PMID: 24882434. [DOI] [PubMed] [Google Scholar]
- 6.Mirza M.R., Monk B.J., Herrstedt J., Oza A.M., Mahner S., Redondo A., Fabbro M., Ledermann J.A., Lorusso D., Vergote I., Ben-Baruch N.E., Marth C., Madry R., Christensen R.D., Berek J.S., Dorum A., Tinker A.V., du Bois A., Gonzalez-Martin A., Follana P., Benigno B., Rosenberg P., Gilbert L., Rimel B.J., Buscema J., Balser J.P., Agarwal S., Matulonis U.A., Investigators E-ON Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375(22):2154–2164. doi: 10.1056/NEJMoa1611310. PubMed PMID: 27717299. [DOI] [PubMed] [Google Scholar]
- 7.Pujade-Lauraine E., Ledermann J.A., Selle F., Gebski V., Penson R.T., Oza A.M., Korach J., Huzarski T., Poveda A., Pignata S., Friedlander M., Colombo N., Harter P., Fujiwara K., Ray-Coquard I., Banerjee S., Liu J., Lowe E.S., Bloomfield R., Pautier P., SOLO2/ENGOT-Ov21 investigators Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18(9):1274–1284. doi: 10.1016/S1470-2045(17)30469-2. PubMed PMID: 28754483. [DOI] [PubMed] [Google Scholar]
- 8.Swisher E.M., Lin K.K., Oza A.M., Scott C.L., Giordano H., Sun J., Konecny G.E., Coleman R.L., Tinker A.V., O'Malley D.M., Kristeleit R.S., Ma L., Bell-McGuinn K.M., Brenton J.D., Cragun J.M., Oaknin A., Ray-Coquard I., Harrell M.I., Mann E., Kaufmann S.H., Floquet A., Leary A., Harding T.C., Goble S., Maloney L., Isaacson J., Allen A.R., Rolfe L., Yelensky R., Raponi M., McNeish I.A. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18(1):75–87. doi: 10.1016/S1470-2045(16)30559-9. PubMed PMID: 27908594. [DOI] [PubMed] [Google Scholar]
- 9.Poveda A., Floquet A., Ledermann J.A., Asher R., Penson R.T., Oza A.M., Korach J., Huzarski T., Pignata S., Friedlander M., Baldoni A., Park-Simon T.W., Tamura K., Sonke G.S., Lisyanskaya A., Kim J.H., Filho E.A., Milenkova T., Lowe E.S., Rowe P., Vergote I., Pujade-Lauraine E., investigators SOE-O Olaparib tablets as maintenance therapy in patients with platinum-sensitive relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a final analysis of a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2021;22(5):620–631. doi: 10.1016/S1470-2045(21)00073-5. PubMed PMID: 33743851. [DOI] [PubMed] [Google Scholar]
- 10.Petersen S., Wilson A.J., Hirst J., Roby K.F., Fadare O., Crispens M.A., Beeghly-Fadiel A., Khabele D. CCNE1 and BRD4 co-amplification in high-grade serous ovarian cancer is associated with poor clinical outcomes. Gynecol Oncol. 2020;157(2):405–410. doi: 10.1016/j.ygyno.2020.01.038. PubMed PMID: 32044108; PubMed Central PMCID: PMC7217738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. PubMed PMID: 21720365; PubMed Central PMCID: PMC3163504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etemadmoghadam D., Weir B.A., Au-Yeung G., Alsop K., Mitchell G., George J., Davis S., D'Andrea A.D., Simpson K., Hahn W.C., Bowtell D.D., Australian Ovarian Cancer Study Group Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc Natl Acad Sci U S A. 2013;110(48):19489–19494. doi: 10.1073/pnas.1314302110. PubMed PMID: 24218601; PubMed Central PMCID: PMC3845173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khabele D., Son D.S., Parl A.K., Goldberg G.L., Augenlicht L.H., Mariadason J.M., Rice VM. Drug-induced inactivation or gene silencing of class I histone deacetylases suppresses ovarian cancer cell growth: implications for therapy. Cancer Biol Ther. 2007;6(5):795–801. doi: 10.4161/cbt.6.5.4007. PubMed PMID: 17387270. [DOI] [PubMed] [Google Scholar]
- 14.Son D.S., Wilson A.J., Parl A.K., Khabele D. The effects of the histone deacetylase inhibitor romidepsin (FK228) are enhanced by aspirin (ASA) in COX-1 positive ovarian cancer cells through augmentation of p21. Cancer Biol Ther. 2010;9(11):928–935. doi: 10.4161/cbt.9.11.11873. PubMed PMID: 20404564; PubMed Central PMCID: PMC4047717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson A.J., Holson E., Wagner F., Zhang Y.L., Fass D.M., Haggarty S.J., Bhaskara S., Hiebert S.W., Schreiber S.L., Khabele D. The DNA damage mark pH2AX differentiates the cytotoxic effects of small molecule HDAC inhibitors in ovarian cancer cells. Cancer Biol Ther. 2011;12(6):484–493. doi: 10.4161/cbt.12.6.15956. PubMed PMID: 21738006; PubMed Central PMCID: PMC3218590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson A.J., Lalani A.S., Wass E., Saskowski J., Khabele D. Romidepsin (FK228) combined with cisplatin stimulates DNA damage-induced cell death in ovarian cancer. Gynecol Oncol. 2012;127(3):579–586. doi: 10.1016/j.ygyno.2012.09.016. PubMed PMID: 23010348; PubMed Central PMCID: PMC3541411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson A.J., Cheng Y.Q., Khabele D. Thailandepsins are new small molecule class I HDAC inhibitors with potent cytotoxic activity in ovarian cancer cells: a preclinical study of epigenetic ovarian cancer therapy. J Ovar Res. 2012;5(1):12. doi: 10.1186/1757-2215-5-12. PubMed PMID: 22531354; PubMed Central PMCID: PMC3394212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konstantinopoulos P.A., Wilson A.J., Saskowski J., Wass E., Khabele D. Suberoylanilide hydroxamic acid (SAHA) enhances olaparib activity by targeting homologous recombination DNA repair in ovarian cancer. Gynecol Oncol. 2014;133(3):599–606. doi: 10.1016/j.ygyno.2014.03.007. PubMed PMID: 24631446; PubMed Central PMCID: PMC4347923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson A.J., Sarfo-Kantanka K., Barrack T., Steck A., Saskowski J., Crispens M.A., Khabele D. Panobinostat sensitizes cyclin E high, homologous recombination-proficient ovarian cancer to olaparib. Gynecol Oncol. 2016;143(1):143–151. doi: 10.1016/j.ygyno.2016.07.088. PubMed PMID: 27444036; PubMed Central PMCID: PMC5031537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta V.G., Hirst J., Petersen S., Roby K.F., Kusch M., Zhou H., Clive M.L., Jewell A., Pathak H.B., Godwin A.K., Wilson A.J., Crispens M.A., Cybulla E., Vindigni A., Fuh K.C., Khabele D. Entinostat, a selective HDAC1/2 inhibitor, potentiates the effects of olaparib in homologous recombination proficient ovarian cancer. Gynecol Oncol. 2021;162(1):163–172. doi: 10.1016/j.ygyno.2021.04.015. PubMed PMID: 33867143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson A.J., Saskowski J., Barham W., Khabele D., Yull F. Microenvironmental effects limit efficacy of thymoquinone treatment in a mouse model of ovarian cancer. Mol Cancer. 2015;14:192. doi: 10.1186/s12943-015-0463-5. PubMed PMID: 26552746; PubMed Central PMCID: PMC4640396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta V., Yull F., Khabele D. Bipolar tumor-associated macrophages in ovarian cancer as targets for therapy. Cancers. 2018;10(10) doi: 10.3390/cancers10100366. PubMed PMID: 30274280; PubMed Central PMCID: PMC6210537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoover A.A., Hufnagel D.H., Harris W., Bullock K., Glass E.B., Liu E., Barham W., Crispens M.A., Khabele D., Giorgio T.D., Wilson A.J., Yull FE. Increased canonical NF-kappaB signaling specifically in macrophages is sufficient to limit tumor progression in syngeneic murine models of ovarian cancer. BMC Cancer. 2020;20(1):970. doi: 10.1186/s12885-020-07450-8. PubMed PMID: 33028251; PubMed Central PMCID: PMC7542116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagemann T., Lawrence T., McNeish I., Charles K.A., Kulbe H., Thompson R.G., Robinson S.C., Balkwill FR. Re-educating" tumor-associated macrophages by targeting NF-kappaB. J Exp Med. 2008;205(6):1261–1268. doi: 10.1084/jem.20080108. PubMed PMID: 18490490; PubMed Central PMCID: PMC2413024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen L., Ciesielski M., Ramakrishnan S., Miles K.M., Ellis L., Sotomayor P., Shrikant P., Fenstermaker R., Pili R. Class I histone deacetylase inhibitor entinostat suppresses regulatory T cells and enhances immunotherapies in renal and prostate cancer models. PLoS One. 2012;7(1):e30815. doi: 10.1371/journal.pone.0030815. PubMed PMID: 22303460; PubMed Central PMCID: PMC3267747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCaw T.R., Goel N., Brooke D.J., Katre A.A., Londono A.I., Smith H.J., Randall T.D., Arend RC. Class I histone deacetylase inhibition promotes CD8 T cell activation in ovarian cancer. Cancer Med. 2021;10(2):709–717. doi: 10.1002/cam4.3337. PubMed PMID: 33369199; PubMed Central PMCID: PMC7877343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briere D., Sudhakar N., Woods D.M., Hallin J., Engstrom L.D., Aranda R., Chiang H., Sodre A.L., Olson P., Weber J.S., Christensen JG. The class I/IV HDAC inhibitor mocetinostat increases tumor antigen presentation, decreases immune suppressive cell types and augments checkpoint inhibitor therapy. Cancer Immunol Immunother. 2018;67(3):381–392. doi: 10.1007/s00262-017-2091-y. PubMed PMID: 29124315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun R., Luo H., Su J., Di S., Zhou M., Shi B., Sun Y., Du G., Zhang H., Jiang H., Li Z. Olaparib suppresses MDSC recruitment via SDF1alpha/CXCR4 axis to improve the anti-tumor efficacy of CAR-T cells on breast cancer in mice. Mol Ther. 2021;29(1):60–74. doi: 10.1016/j.ymthe.2020.09.034. PubMed PMID: 33010818; PubMed Central PMCID: PMC7791086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding L., Kim H.J., Wang Q., Kearns M., Jiang T., Ohlson C.E., Li B.B., Xie S., Liu J.F., Stover E.H., Howitt B.E., Bronson R.T., Lazo S., Roberts T.M., Freeman G.J., Konstantinopoulos P.A., Matulonis U.A., Zhao JJ. PARP inhibition elicits STING-dependent antitumor immunity in brca1-deficient ovarian cancer. Cell Rep. 2018;25(11):2972–2980. doi: 10.1016/j.celrep.2018.11.054. e5PubMed PMID: 30540933; PubMed Central PMCID: PMC6366450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiao S., Xia W., Yamaguchi H., Wei Y., Chen M.K., Hsu J.M., Hsu J.L., Yu W.H., Du Y., Lee H.H., Li C.W., Chou C.K., Lim S.O., Chang S.S., Litton J., Arun B., Hortobagyi G.N., Hung MC. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res. 2017;23(14):3711–3720. doi: 10.1158/1078-0432.CCR-16-3215. PubMed PMID: 28167507; PubMed Central PMCID: PMC5511572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roby K.F., Taylor C.C., Sweetwood J.P., Cheng Y., Pace J.L., Tawfik O., Persons D.L., Smith P.G., Terranova PF. Development of a syngeneic mouse model for events related to ovarian cancer. Carcinogenesis. 2000;21(4):585–591. doi: 10.1093/carcin/21.4.585. PubMed PMID: 10753190. [DOI] [PubMed] [Google Scholar]
- 32.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. PubMed PMID: 23618408; PubMed Central PMCID: PMC4053844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. PubMed PMID: 25516281; PubMed Central PMCID: PMC4302049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J., Duncan D., Shi Z., Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. (Web Server issue)PubMed PMID: 23703215; PubMed Central PMCID: PMC3692109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson A.J., Fadare O., Beeghly-Fadiel A., Son D.S., Liu Q., Zhao S., Saskowski J., Uddin M.J., Daniel C., Crews B., Lehmann B.D., Pietenpol J.A., Crispens M.A., Marnett L.J., Khabele D. Aberrant over-expression of COX-1 intersects multiple pro-tumorigenic pathways in high-grade serous ovarian cancer. Oncotarget. 2015;6(25):21353–21368. doi: 10.18632/oncotarget.3860. PubMed PMID: 25972361; PubMed Central PMCID: PMC4673270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou T.C., Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzym Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. PubMed PMID: 6382953. [DOI] [PubMed] [Google Scholar]
- 37.Pierce A.J., Johnson R.D., Thompson L.H., Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. PubMed PMID: 10541549; PubMed Central PMCID: PMC317094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson C., Moynahan M.E., Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12(24):3831–3842. doi: 10.1101/gad.12.24.3831. PubMed PMID: 9869637; PubMed Central PMCID: PMC317271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson A.J., Stubbs M., Liu P., Ruggeri B., Khabele D. The BET inhibitor INCB054329 reduces homologous recombination efficiency and augments PARP inhibitor activity in ovarian cancer. Gynecol Oncol. 2018;149(3):575–584. doi: 10.1016/j.ygyno.2018.03.049. PubMed PMID: 29567272; PubMed Central PMCID: PMC5986599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson A.J., Barham W., Saskowski J., Tikhomirov O., Chen L., Lee H.J., Yull F., Khabele D. Tracking NF-kappaB activity in tumor cells during ovarian cancer progression in a syngeneic mouse model. J Ovar Res. 2013;6(1):63. doi: 10.1186/1757-2215-6-63. PubMed PMID: 24020521; PubMed Central PMCID: PMC3846584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Worzfeld T., Pogge von Strandmann E., Huber M., Adhikary T., Wagner U., Reinartz S., Muller R. The unique molecular and cellular microenvironment of ovarian cancer. Front Oncol. 2017;7:24. doi: 10.3389/fonc.2017.00024. PubMed PMID: 28275576; PubMed Central PMCID: PMC5319992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin M., Shen J., Yu S., Fei J., Zhu X., Zhao J., Zhai L., Sadhukhan A., Zhou J. Tumor-associated macrophages (TAMs): a critical activator in ovarian cancer metastasis. OncoTargets Ther. 2019;12:8687–8699. doi: 10.2147/OTT.S216355. PubMed PMID: 31695427; PubMed Central PMCID: PMC6814357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dangaj D., Bruand M., Grimm A.J., Ronet C., Barras D., Duttagupta P.A., Lanitis E., Duraiswamy J., Tanyi J.L., Benencia F., Conejo-Garcia J., Ramay H.R., Montone K.T., Powell D.J., Jr., Gimotty P.A., Facciabene A., Jackson D.G., Weber J.S., Rodig S.J., Hodi S.F., Kandalaft L.E., Irving M., Zhang L., Foukas P., Rusakiewicz S., Delorenzi M., Coukos G. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell. 2019;35(6):885–900. doi: 10.1016/j.ccell.2019.05.004. e10PubMed PMID: 31185212; PubMed Central PMCID: PMC6961655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herr D., Sallmann A., Bekes I., Konrad R., Holzheu I., Kreienberg R., Wulff C. VEGF induces ascites in ovarian cancer patients via increasing peritoneal permeability by downregulation of Claudin 5. Gynecol Oncol. 2012;127(1):210–216. doi: 10.1016/j.ygyno.2012.05.002. PubMed PMID: 22579791. [DOI] [PubMed] [Google Scholar]
- 45.Bolat F., Gumurdulu D., Erkanli S., Kayaselcuk F., Zeren H., Ali Vardar M., Kuscu E. Maspin overexpression correlates with increased expression of vascular endothelial growth factors A, C, and D in human ovarian carcinoma. Pathol Res Pract. 2008;204(6):379–387. doi: 10.1016/j.prp.2008.01.011. PubMed PMID: 18343598. [DOI] [PubMed] [Google Scholar]
- 46.Wilson A.J., Saskowski J., Barham W., Yull F., Khabele D. Thymoquinone enhances cisplatin-response through direct tumor effects in a syngeneic mouse model of ovarian cancer. J Ovar Res. 2015;8:46. doi: 10.1186/s13048-015-0177-8. PubMed PMID: 26215403; PubMed Central PMCID: PMC4517540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lieber S., Reinartz S., Raifer H., Finkernagel F., Dreyer T., Bronger H., Jansen J.M., Wagner U., Worzfeld T., Muller R., Huber M. Prognosis of ovarian cancer is associated with effector memory CD8(+) T cell accumulation in ascites, CXCL9 levels and activation-triggered signal transduction in T cells. Oncoimmunology. 2018;7(5) doi: 10.1080/2162402X.2018.1424672. PubMed PMID: 29721385; PubMed Central PMCID: PMC5927536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F., Parayath N.N., Ene C.I., Stephan S.B., Koehne A.L., Coon M.E., Holland E.C., Stephan MT. Genetic programming of macrophages to perform anti-tumor functions using targeted mRNA nanocarriers. Nat Commun. 2019;10(1):3974. doi: 10.1038/s41467-019-11911-5. PubMed PMID: 31481662; PubMed Central PMCID: PMC6722139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data files will be made publicly available at the GenBank repository (BioProject Accession PRJNA767427).