Abstract
CCAAT/enhancer binding protein α (C/EBPα) is an integral factor in the granulocytic developmental pathway, as myeloblasts from C/EBPα-null mice exhibit an early block in differentiation. Since mice deficient for known C/EBPα target genes do not exhibit the same block in granulocyte maturation, we sought to identify additional C/EBPα target genes essential for myeloid cell development. To identify such genes, we used both representational difference analysis and oligonucleotide array analysis with RNA derived from a C/EBPα-inducible myeloid cell line. From each of these independent screens, we identified c-Myc as a C/EBPα negatively regulated gene. We mapped an E2F binding site in the c-Myc promoter as the cis-acting element critical for C/EBPα negative regulation. The identification of c-Myc as a C/EBPα target gene is intriguing, as it has been previously shown that down-regulation of c-Myc can induce myeloid differentiation. Here we show that stable expression of c-Myc from an exogenous promoter not responsive to C/EBPα-mediated down-regulation forces myeloblasts to remain in an undifferentiated state. Therefore, C/EBPα negative regulation of c-Myc is critical for allowing early myeloid precursors to enter a differentiation pathway. This is the first report to demonstrate that C/EBPα directly affects the level of c-Myc expression and, thus, the decision of myeloid blasts to enter into the granulocytic differentiation pathway.
Hematopoiesis is the process through which mature blood cells of distinct lineages are produced from pluripotent stem cells. Like many differentiation systems, transcription factors that activate lineage-specific genes are essential to the commitment and development of specific hematopoietic lineages (57, 63). One such transcription factor essential for commitment to and development of the granulocytic lineage is CCAAT/enhancer binding protein α (C/EBPα).
C/EBPα is a basic leucine zipper protein (bZIP) that forms homodimers or heterodimers with other C/EBP proteins to activate the transcription of target genes (reviewed in references 34 and 63). In addition to granulocytes, C/EBPα is highly expressed in many differentiated cell types such as hepatocytes and adipocytes. A number of reports indicate that C/EBPα has a crucial role in regulating the balance between cell proliferation and differentiation, which is crucial for lineage commitment of any cell type. First, C/EBPα has been shown to cause growth arrest in adipocytes as well as in hepatocytes (18, 64, 67, 68, 71). C/EBPα initiates growth arrest through its ability to stabilize the expression of the cyclin-activating kinase inhibitor (CAK), p21, as well as through disruption of E2F transcriptional complexes during the G1 phase of the cell cycle (64–67). Additionally, expression of antisense C/EBPα RNA prevents both growth arrest and terminal differentiation of 3T3 L1 adipocytes (36). Finally, C/EBPα−/− mice exhibited improper development of lung and liver with increased hepatocyte proliferation, supporting the role of C/EBPα in the differentiation of these tissues (17). A striking feature of the C/EBPα−/− mice was the complete absence of any mature neutrophils (73). This result demonstrates the indispensability of C/EBPα for the granulocytic differentiation pathway.
C/EBPα−/− mice exhibit a block in granulocytic differentiation that is early in the developmental pathway. Fluorescence-activated cell sorter analysis of embryonic and newborn animals demonstrated no detectable expression of the granulocyte colony-stimulating factor (G-CSF) and interleukin-6 (IL-6) receptors, and mRNA levels for both were drastically reduced (73, 74). Consequently, C/EBPα−/− mice exhibit a reduced response to those respective cytokines. These results suggested that much of the C/EBPα−/− phenotype could be attributed to the decrease in the levels of both the G-CSF receptor and the IL-6 receptor and their respective signaling pathways. However, neither G-CSF receptor−/− mice nor IL-6−/− mice exhibit serious defects in granulocytic differentiation (39, 40). Therefore, it was hypothesized that a cross between G-CSF receptor−/− mice and IL-6−/− mice would mimic the phenotype observed with the C/EBPα−/− mice alone. However, this cross did not result in a severe defect in granulocytic differentiation, which indicates that there must be additional C/EBPα target genes in myeloid progenitor cells necessary for mature neutrophil development.
c-Myc is a basic helix-loop-helix (HLH) leucine zipper protein that dimerizes with its partner Max to activate gene transcription through consensus E-box elements located on the promoters of certain genes (6, 7). Myc was discovered to be an oncogene causing leukemia in birds and inducing in vitro transformation of avian myeloid cells (56). Dysregulated c-Myc expression has been implicated in the development of lymphoid malignancies and other tumors (13, 33), as well as in the induction of genomic instability (15). This demonstrates the importance of appropriate c-Myc regulation and the role of c-Myc for proper maintenance of the cell cycle (46). c-Myc is expressed in proliferating cells, and both c-Myc mRNA and protein levels are virtually undetectable in terminally differentiated cells (21, 32, 72). These studies indicate that down-regulation of c-Myc is a critical event for a cell to commit to a differentiation pathway (12, 25). This is particularly true in differentiation of myeloid cells (25), and treatment of myeloid cell lines with antisense oligonucleotides that inhibit c-Myc expression induces myeloid cell differentiation (26). Failure to down-regulate c-Myc in transgenic mice can lead to myeloid leukemia, a condition characterized by a block in differentiation (16).
As proliferation and differentiation are mutually exclusive, c-Myc, a proliferative factor, and C/EBPα, a differentiation factor, act in opposition to each other. First, c-Myc and C/EBPα act reciprocally during adipogenesis (18). Overexpression of c-Myc blocks the ability of adipoblasts to terminally differentiate, while the introduction of C/EBPα overcomes this c-Myc-induced differentiation block (37). Next, c-Myc can activate cyclin E complexes, which results in increased active E2F transcription complexes. This leads cells into the G1/S transition of the cell cycle. Moreover, expression of c-Myc can overcome growth arrest imposed by the p21, p27, and p16 cyclin-dependent kinase (cdk) inhibitor proteins (61). In contrast, C/EBPα achieves growth arrest through increased p21 CAK inhibitor protein, which ultimately results in decreased numbers of active E2F transcription complexes (65, 66). Most importantly for their opposing effects in cells, c-Myc and C/EBPα can reciprocally regulate the expression of their respective genes. c-Myc has previously been shown to negatively regulate C/EBPα expression and block C/EBPα transactivation function (2, 35, 43). However, the effects of C/EBPα on c-Myc regulation have not been investigated.
In order to identify C/EBPα targets in myeloid cells, we performed both representational difference analysis (RDA) and oligonucleotide array screening. From both of these independent screens, we identified c-Myc as a target gene of C/EBPα. We show that C/EBPα can directly down-regulate human c-Myc promoter activity. Moreover, we have identified a consensus E2F site located between the P1 and P2 c-Myc promoter elements as being critical for C/EBPα negative regulation. This is the first investigation to show that C/EBPα directly affects c-Myc expression levels and thus further elucidates the mechanisms through which C/EBPα induces cellular differentiation.
MATERIALS AND METHODS
Cell culture conditions.
U937 cells stably transfected with a zinc-inducible C/EBPα construct (U937α#2) or vector alone [U937(vect)#1] have been described previously (52). C/EBPα expression from the metallothionein promoter was induced by adding 100 μM ZnSO4 to the culture medium. The Tet-o-myc 1137 myeloblast cell line has been previously described (16). The addition of 20 ng of tetracycline/ml to the culture medium turns off the expression of the human c-Myc transgene which induces the cells to differentiate. 1137 cells stably transfected with a metallothionein-driven C/EBPα cDNA (1137/C/EBPα) or metallothionein vector alone (1137/vector) were generated using previously described methods (52). In these 1137 stably transfected cells, the level of human c-Myc was titrated using the indicated dilutions of tetracycline. C/EBPα expression was induced by addition of ZnSO4 as previously described (52). Differentiated 1137 cells were quantified by Wright-Giemsa staining and differential cell counts. Monkey kidney lines, CV-1 and COS7, as well as the Rb-Saos osteosarcoma cell line, were maintained in Dulbecco modified Eagle medium (BioWhittaker, Walkersville, Md.) supplemented with 10% fetal bovine serum.
Plasmids and transient transfection.
A series of 5′ deletions were generated from an EcoRI/NaeI genomic fragment of the human c-Myc gene (3) using internal restriction sites EcoRI (−6.5 kb to +49 bp), XmnI (−2,451 to +49 bp), PvuII (−511 to +49 bp), XbaI (−263 to +49 bp), and XhoI (−92 to +49 bp) and subcloned into the pXP2 firefly luciferase reporter (45). The 0.14-kb mutant E2F reporter construct was generated by PCR to produce a mutation in the consensus E2F binding site sequence from GCGGGAAA to GTTTCAAA. The 2.5-kb mutant E2F reporter construct was made by linearizing the pXP2 0.14-kb mutant E2F construct with HindIII and XhoI. A HindIII/XhoI fragment from the pXP2 2.5-kb reporter construct was subsequently cloned into corresponding sites to create the larger construct. The pcDNA3 C/EBPα construct was generated by releasing a BamHI/EcoRI fragment of rat C/EBPα cDNA from the pUC18 vector and ligating this fragment into pcDNA3 (Invitrogen) prepared with BamHI and EcoRI. 4HEP C/EBPα was a gift of Charles Vinson (National Cancer Institute, Bethesda, Md.) and has been previously described (49). pECE PU.1 was described previously (30). The reporter construct pTK81G-CSFr contains four consensus C/EBPα binding sites from the G-CSF receptor promoter linked in tandem and cloned into pTK81 luciferase (45, 60). Approximately 2 × 104 CV-1 cells (or Saos cells) were transfected by Lipofectamine according to the manufacturer's instructions (Promega, Madison, Wis.) with 200 ng of reporter gene, 20 ng of expression plasmid DNA, and 20 pg of promoterless Renilla luciferase as an internal control. Twenty-four hours later, firefly luciferase activities were determined and normalized to Renilla luciferase (4). Results are presented as the percentage of luciferase activity with pcDNA3 vector alone set to 100% activity, except for transfections with c-Myc reporter constructs containing a mutated E2F site, which are given in actual relative light units. Results are given as the averages of at least three independent experiments, and error bars represent the standard errors of the means.
Identification of C/EBPα-regulated genes.
RDA was performed as described previously (29) but with the substitution of poly(A)+ mRNA derived from U937α#2 cells stimulated with ZnSO4 for 8 and 12 h to derive the “tester” cDNA and unstimulated cells to derive the “driver” cDNA. Nucleotide array analysis was performed as described previously (62) but with the substitution of RNA isolated from U937α#2 cells stimulated with ZnSO4 for 8 and 24 h. A detailed protocol is available at http://waldo.wi.mit.edu/MPR or http://www.genome.wi.mit.edu/MPR.
Northern analysis.
U937α#2 and U937(vect)#1 cells were stimulated with ZnSO4, and total RNA was isolated at the indicated time points as described previously (29). Fifteen micrograms of each RNA sample was analyzed by Northern blotting as described previously (29). Blots were hybridized to an [α-32P]dCTP-labeled human c-Myc probe (a 305-bp XbaI/EcoRI cDNA fragment isolated from the cDNA clone obtained from the RDA screen above), an [α-32P]dCTP-labeled rat C/EBPα probe (a 300-bp HincII-BamHI cDNA fragment from the pcDNA3 C/EBPα plasmid described above), and an [α-32P]dCTP-labeled glyceraldehyde-3-phosphate dehydrogenase probe to control for RNA loading and integrity. Northern blots were stripped between hybridizations by incubation in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate (SDS) at 100°C for 20 min.
Western analysis.
At the indicated time points following treatment with ZnSO4 for U937 stable lines or tetracycline for the 1137 cell line, cells were harvested for total cell lysates with modified RIPA buffer (1% Triton X, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl2, 5 mM EDTA, 50 mM Tris, pH 8.0). Cell lysates were subsequently resolved on SDS–10% polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad). Western blots were incubated with c-Myc antisera (sc-764; Santa Cruz; 1:200 dilution), C/EBPα antisera (sc-61; Santa Cruz; 1:250 dilution), or β-tubulin monoclonal antibody (catalog no. 1111 876; Boehringer Mannheim; 1:500 dilution) followed by a 1:5,000 dilution of an appropriate anti-mouse or anti-rabbit immunoglobulin G antibody conjugated with horseradish peroxidase (Santa Cruz). Detection of immune complexes was achieved by enhanced chemiluminescence (NEN DuPont) and autoradiography. Western blots were stripped between hybridizations by incubating blots at 65°C for 5 min in buffer containing 62.5 mM Tris (pH 6.8), 0.02% SDS, and 10 mM β-mercaptoethanol.
In vitro protein-protein binding assays.
Glutathione S-transferase–DP1 (GST-DP1) and GST-E2F1 were a gift of S. Chellappan and are described in reference 70. Other GST fusion proteins have previously been described (51). GST fusion proteins were bound to a 1:1 slurry of glutathione-X-linked beads (Sigma) in GST binding buffer (phosphate-buffered saline containing 20% glycerol, 0.1% NP-40, 1 mM dithiothreitol [DTT], 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). All proteins were quantitated by SDS-polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining. [35S] methionine-labeled rat C/EBPα and E2F1 proteins were prepared using 2 μg of pcDNA3 C/EBPα and pPS75 E2F1 (gift of William Kaelin), respectively, as template for coupled in vitro transcription-translation (TNT kit; Promega). For the in vitro binding assays, equal amounts of all GST proteins were incubated with 5 μl of 35S-labeled proteins. The bead volume of all samples was adjusted to 50 μl with GST beads alone. The binding reaction mixtures were then resuspended in a total volume of 250 μl of protein binding buffer (10 mM Tris [pH 7.5], 150 mM NaCl2, 1 mM DTT, and 1 mM PMSF). Bound proteins were released by heating at 95°C in 2× SDS gel loading buffer and resolved on SDS–10% polyacrylamide gels followed by exposure to X-ray film for 24 h. The percentages of in vitro-translated protein complexed with GST fusion proteins on beads were calculated with a phosphorimager.
Coimmunoprecipitation conditions.
Cytomegalovirus-E2F1 was a gift from Jacqueline Lees (69). COS7 cells (106) for each immunoprecipitation group were either transfected with 20 μg of cytomegalovirus promoter-E2F1 and 5 μg of pcDNA3 C/EBPα (to yield equal protein expression) using Lipofectamine (Gibco-BRL) according to the manufacturer's directions or mock transfected (untransfected) by treatment with the same reagents minus plasmid DNA. Cell lysates were harvested 24 h following transfection by lysing cells in 200 μl of lysis buffer (50 mM NaCl2, 150 mM Tris [pH 7.6], 0.1% NP-40, 1 mM PMSF, and 10 μM aprotinin and leupeptin). One-thirtieth the amounts of lysate from both untransfected and transfected cells were used in Western analysis without immunoprecipitation as a control for protein expression. For immunoprecipitations performed using the U937α#2 cell line, approximately 107 cells per experimental group were treated with ZnSO4 to induce C/EBPα expression or left untreated. Cell lysates were harvested at 12 h following ZnSO4 treatment by lysing cells in 200 ml of lysis buffer. Supernatants were precleared with 50 ml of a 1:1 slurry of protein A-agarose (Santa Cruz) in lysis buffer with 6 μg of normal rabbit serum (NRS). The precleared supernatants were recovered and incubated with 12 μg of either C/EBPα antiserum or NRS antiserum (as a control) and 50 μl of a 1:1 slurry of protein A-agarose. The bound protein-protein A complexes were washed once with lysis buffer and once with wash buffer (50 mM NaCl2, 150 mM Tris [pH 7.6], 1 mM PMSF, and 10 μg of aprotinin and leupeptin/ml). Bound complexes were released by being heated to 100°C for 5 min, resolved on SDS–10% polyacrylamide gels, and analyzed by Western analysis as described above. To detect immunoprecipitated E2F1, E2F2, or E2F4 protein, membranes were hybridized with a 1:2,000 dilution of E2F1 antibody (sc-251x), E2F2 antiserum (sc-633x), or E2F4 antiserum (sc-1082x; Santa Cruz).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (41) using an E2F binding site oligonucleotide (CTCAGAGGCTTGGCGGGAAAAAGAACGGAGGG) from the human c-Myc promoter sequences located at bp −82 to −46 (GenBank accession no. J00120) or the C/EBPα binding site oligonucleotide from the G-CSF receptor promoter sequence extending from bp −57 to −38 (60). E2F binding reactions were performed in 20 mM Tris (pH 7.5)–100 mM KCl–5 mM DTT–2 mM MgCl2–10% glycerol–0.5 μg of double-stranded salmon sperm DNA with nuclear extracts from COS7 cells transfected with E2F1 or C/EBPα plasmid. C/EBPα binding was performed with in vitro-translated protein and COS7 cells transfected with C/EBPα plasmid in 10 mM HEPES (pH 7.9)–50 mM KCl–5 mM MgCl2–1 mM DTT–1 mM EDTA–1 μg of bovine serum albumin per μl–10% glycerol with 2 μg of poly(dI-dC). For specific competition, unlabeled competitor oligonucleotide was added to the binding reaction mixtures at a 200-fold molar excess. In some competition reactions, a shorter E2F double-stranded oligonucleotide (GCTTGGCGGGAAAAAG) was used based on sequences located at bp −70 to −51 in the human c-Myc promoter. For supershift experiments, 3 μl of specific C/EBPα or E2F1 antiserum (200 μg/0.1 ml) (Santa Cruz) or NRS was added to the reactions. For competition experiments with E2F1 and C/EBPα proteins, 16 μg of nuclear extract from COS7 cells transfected with E2F1 was incubated with increasing amounts of in vitro-translated C/EBPα. All binding reactions were adjusted with control unprogrammed lysate to contain a total of 25 μg of rabbit reticulocyte lysate. In competition reactions using nuclear extract from COS7 cells cotransfected with E2F1 and C/EBPα expression plasmids, 10 mg of E2F1 was cotransfected along with increasing amounts of C/EBPα or PU.1 for a control. Vector DNA (pcDNA3) was added to all transfections to ensure that equal amounts of total DNA were transfected.
RESULTS
Identification of c-Myc as a potential C/EBPα target.
Because mice devoid of the G-CSF and IL-6 signaling pathways did not duplicate the dramatic phenotype demonstrated by the C/EBPα-targeted mice (39), we hypothesized that there are additional C/EBPα-targeted genes required for appropriate granulocytic differentiation. In order to identify these additional C/EBPα-regulated genes, we performed RDA (27, 38), a PCR-based subtractive hybridization technique using mRNA derived from a U937 cell line stably transfected with a rat C/EBPα gene under the control of the human metallothionein promoter, U937α#2 (52). From this RDA screen, we identified several novel cDNAs, as well as previously identified cDNAs such as inhibitor of differentiation 2H (Id-2H), ornithine decarboxylase, and thyroid hormone binding protein. In addition, we identified the c-Myc gene as a target for regulation by C/EBPα (Table 1). The discovery of c-Myc as a C/EBPα-regulated gene is intriguing because it has been previously shown that down-regulation of the c-Myc gene can induce myeloid differentiation (16, 26). Additionally, c-Myc has been shown to negatively regulate C/EBPα expression (2, 35, 43).
TABLE 1.
C/EBPα-regulated genes identified by RDAa
| Type | Genes |
|---|---|
| Down-regulated | c-Myc,bc Id-2H (inhibitor of differentiation), Noggin, ubiquitin–52-amino-acid fusion protein,b LL Rep3, ornithine decarboxylase,c β-tubulin, thyroid hormone binding protein |
| Up-regulated | Haptoglobin,b gp 91-phox, (2′–5′) oligo (A) synthetase,bc fibrillin, expressed sequence tag (H19982), unknown 1, unknown 2, unknown 3, unknown 4 |
DNA fragments generated by RDA, U937/MT-C/EBPα zinc-stimulated cells minus unstimulated cells.
Gene identified also by the nucleotide array screen.
Known c-Myc target gene.
Although RDA is an effective technique to identify differentially regulated genes, we have found that it has some limitations. For example, some differentially expressed genes can be lost during repeated subtractive hybridization after increasing the stringency. In addition, RDA preferentially amplifies genes with significant differences in expression and thus is not effective at identifying genes with small differences in regulated expression (29). In order to overcome these limitations, we performed an additional screen for C/EBPα target genes using nucleotide array analysis (11, 20, 62). Recently developed array technologies allow for the analysis of expression patterns of thousands of genes during cellular differentiation or in response to a particular cellular signal. For this screen, we again isolated mRNA from the U937α#2 line 8 and 24 h following induction of C/EBPα expression by treatment with zinc. The c-Myc gene again was identified as a gene regulated by C/EBPα, confirming our RDA results (Table 2).
TABLE 2.
C/EBPα-regulated genes identified by nucleotide arraya
| Type of gene and sequence accession no. | Xb | Yc | Y/20/Xd | Xe | Ye |
|---|---|---|---|---|---|
| Down-regulated | |||||
| No cluster in current Unigene (T50334_f) | 191 | 74 | 0.387 | P | P |
| Platelet-endothelial tetraspan antigen 3f(R74349) | 162 | 58 | 0.358 | P | P |
| Heterogeneous nuclear ribonucleoproteinf (X16135) | 158 | 56 | 0.354 | P | P |
| Tetracycline transporter-like proteinf (H28711) | 159 | 56 | 0.352 | P | P |
| Myb proto-oncogene (M13665) | 325 | 108 | 0.332 | P | P |
| Natural killer cell protein 4 precursor (M59807) | 180 | 58 | 0.322 | P | P |
| Cathepsin G precursor (J04990) | 867 | 268 | 0.309 | P | P |
| Nonmetastatic cell 1 (NME1) (T86473) | 133 | 41 | 0.308 | P | P |
| HLA-DRB1, major histocompatibility complex class II (T62633) | 332 | 86 | 0.259 | P | P |
| MacMarcks (D44497) | 370 | 93 | 0.251 | P | P |
| c-Myc (X00364) | 82 | 10 | 0.243 | P | P |
| Calmodulin (U10117) | 105 | 25 | 0.238 | P | A |
| ARHG, Ras homolog family, rho Gg (X61587) | 140 | 33 | 0.235 | P | P |
| E1F4A (eukaryotic translation initiation factor 4A)h (T69446) | 111 | 26 | 0.234 | P | P |
| PTMA (α-prothymosin)h (R98842) | 142 | 33 | 0.232 | P | A |
| PRTN3 (proteinase 3) (M96839) | 167 | 36 | 0.215 | P | P |
| No cluster in current Unigene (H43328_i) | 95 | 7 | 0.210 | P | A |
| FLN1 (filamin 1; actin-binding protein) (R78934) | 115 | 12 | 0.173 | P | A |
| CALR (autoantigen calreticulin) (M84739) | 568 | 89 | 0.156 | P | P |
| E1F5A (eukaryotic translation initiation factor 5A)h (R72300) | 182 | −62 | 0.109 | P | P |
| Up-regulated | |||||
| Arginase (ARG1) (M14502) | 3 | 351 | 17.55 | A | P |
| Antithrombin III precursor (D29832) | 37 | 622 | 16.81 | P | P |
| Haptoglobin (HP) (T67511) | 12 | 300 | 15.00 | A | P |
| Annexin 1 (ANX1; lipocortin 1) (X05908) | 23 | 345 | 15.00 | P | P |
| HMOX1 (heme oxygenase 1) (X06985) | 11 | 278 | 13.9 | A | P |
| HLA-G, major histocompatibility complex class proteinh (M32800) | 23 | 261 | 11.35 | A | P |
| DDH1 (dihydrodiol dehydrogenase) (T64167) | −2 | 204 | 10.20 | A | P |
| dUTP pyrophosphatase (T50797) | 11 | 162 | 8.10 | P | P |
| No cluster in current Unigene (T55731) | 95 | 750 | 7.89 | P | P |
| No cluster in current Unigene (T71025) | 151 | 1,081 | 7.16 | P | P |
| NMB (neuromedin B) (X76534) | 9 | 143 | 7.15 | A | P |
| Small nuclear ribonucleoprotein polypeptides B and B1 (R8411) | 74 | 517 | 6.99 | P | P |
| Protease inhibitor 12 (H09572) | −3 | 138 | 6.90 | A | P |
| Prolactin (H51034) | 3 | 117 | 5.85 | A | P |
| Calgranulin A (T99219) | 109 | 632 | 5.80 | P | P |
| Ubiquitin A-52i | 155 | 888 | 5.73 | P | P |
| Oligo(A) synthetase (2′–5′)hi (X02875) | 12 | 113 | 5.65 | A | P |
| Ribosomal protein S4 (R05923) | 14 | 111 | 5.55 | P | P |
| No cluster in current Unigene (H29761) | 5 | 111 | 5.55 | A | P |
| Protein tyrosine phosphatase receptor type c (Y00062) | 22 | 122 | 5.55 | P | P |
| PSMB5, proteasome subunit (H87473) | 16 | 109 | 5.45 | P | P |
| Bcl-2 related (Bfl-1) (U27467) | 15 | 104 | 5.20 | P | P |
| Glycogenin (U31525) | 42 | 218 | 5.19 | P | P |
| Vimentin (T51852) | 98 | 504 | 5.14 | P | P |
| No cluster in current Unigene (R59583) | 27 | 131 | 4.85 | P | P |
| Autotaxin (L35594) | −7 | 96 | 4.80 | A | P |
| Expressed sequence tag (T94579) | 19 | 93 | 4.65 | A | P |
| No cluster in current Unigene (M26311) | 52 | 234 | 4.50 | P | P |
| H2AZ histone (M37583) | 18 | 90 | 4.50 | P | P |
| Pre-P-cell enhancing factor (U02020) | 15 | 89 | 4.45 | A | P |
| RNA polymerase II subunit (U37690) | 22 | 94 | 4.27 | P | P |
| Ferritin heavy chain (T63508) | 33 | 139 | 4.21 | P | P |
| Heat shock 10-kDa protein 1 (R08183) | 132 | 536 | 4.06 | P | P |
| Glyoxalase 1 (D13315) | 73 | 289 | 3.96 | P | P |
| Expressed sequence tag (T59954) | 98 | 383 | 3.91 | P | P |
| Cystatin A (X05978) | 114 | 403 | 3.54 | P | P |
| No cluster in current Unigene (T61661) | 149 | 509 | 3.42 | P | P |
| KIAA0108f (D14696) | 78 | 258 | 3.31 | P | P |
| Expressed sequence tag (H06970) | 82 | 268 | 3.27 | P | P |
| PABPL [poly(A)-binding protein-like 1] (T64148) | 32 | 102 | 3.19 | P | P |
| Heat shock 27-kDa protein 1 (T48904) | 760 | 2,396 | 3.15 | P | P |
| Myosin regulatory light chain (T78489) | 40 | 126 | 3.15 | P | P |
| Nucleobindin precursor (R52271) | 48 | 150 | 3.13 | P | P |
| Ribosomal protein S11 (H89983) | 41 | 128 | 3.13 | P | P |
| SSB (Sjögren syndrome antigen B)f (H29485) | 51 | 148 | 2.90 | P | P |
| Ubiquitin carrier protein (E2-EPF) (M91670) | 248 | 704 | 2.84 | P | P |
| Surface antigen (M60922) | −18 | 56 | 2.80 | A | P |
| LRPAP1 (low-density lipoprotein-related protein) (M63959) | 104 | 274 | 2.63 | P | P |
| PSMA5 (proteasome component C5) (D00761) | 60 | 157 | 2.62 | P | P |
| Eletron-transfer flavoprotein (J04058) | 51 | 134 | 2.63 | P | P |
| Adenosine receptor A1 (L22214) | 56 | 145 | 2.59 | M | P |
| No cluster in current Unigene (H05222) | 57 | 147 | 2.58 | A | A |
| Putative protein kinase C inhibitor (H46728) | 67 | 171 | 2.55 | P | P |
| ATP6B2, ATPase, H+ transporter (L35249) | 74 | 188 | 2.54 | P | P |
DNA fragments identified by nucleotide array analysis, U937/MT-C/EBPα zinc-stimulated cells compared with unstimulated cells. Results are listed by decreasing Y/20/X values.
Fluorescence intensity with RNA from cells before zinc stimulation.
Fluorescence intensity with RNA from cells 8 or 24 h following stimulation.
All intensities below 20 are adjusted to 20, which is within background. For down-regulated genes, data were sorted for expression differences of <0.4-fold using data more than 70-fold above the lower value. For up-regulated genes, data were sorted for expression differences of >2.5-fold using data more than 70-fold above the lower value.
P, present; A, absent; M, undetermined.
Gene potentially regulated by c-Myc, as indicated by Transfac transcription factor binding site search.
Gene potentially regulated by E2F, as indicated by Transfac transcription factor binding site search.
Known c-Myc target gene.
Gene identified also by RDA screen.
The endogenous c-Myc gene is negatively regulated by C/EBPα.
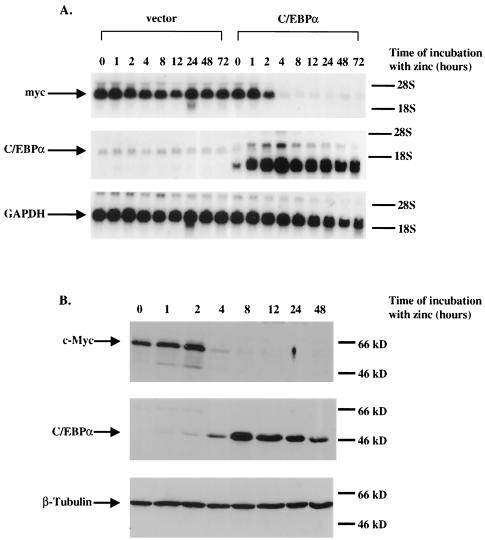
In order to confirm our screening results, we isolated RNA from both the U937α#2 stable line and U937(vect)#1 (a U937 line stably transfected with the metallothionein vector lacking the rat C/EBPα cDNA) (52) at various time points following treatment with zinc to induce metallothionein promoter-C/EBPα gene expression and used this RNA in Northern analysis (Fig. 1A). Previously, we found the induced level of C/EBPα protein to be threefold above the level of endogenous C/EBPα in these cells. This is sufficient C/EBPα expression to fully differentiate precursor cells along the granulocytic pathway (52). Following induction of C/EBPα expression, the level of endogenous c-Myc RNA dramatically decreased by 94% at 4 h following zinc treatment, corresponding to the threefold induction of C/EBPα RNA. In contrast, the level of c-Myc mRNA remained the same in the U937 vector cell line in which C/EBPα was not induced. These results indicate that the level of endogenous c-Myc RNA is substantially affected by the level of C/EBPα gene expression.
FIG. 1.
(A) The level of c-Myc RNA decreases following the induction of C/EBPα gene expression. U937 stable cell lines that contain either a rat C/EBPα cDNA under the control of the human metallothionein promoter (U937α#2) or the empty metallothionein expression vector alone were harvested for total RNA at the indicated time points following the addition of ZnSO4 to the culture medium. (Top) Northern hybridization with a c-Myc cDNA probe. (Middle) Northern hybridization with a C/EBPα cDNA probe. (Bottom) The Northern blot was stripped and rehybridized with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probe to control for RNA loading and integrity. (B) The expression of c-Myc protein decreases as the level of C/EBPα protein increases. The U937α#2 stable line was harvested for cell lysates for Western analysis at the indicated time points following incubation with ZnSO4. (Top) Western blot hybridized with c-Myc antiserum. (Middle) The same Western blot hybridized with C/EBPα antiserum. (Bottom) The same Western blot hybridized with β-tubulin antibody to control for protein loading and integrity.
In order to determine if the decrease in c-Myc mRNA corresponds to a similar decrease in the level of c-Myc protein, we again treated the U937α#2 line with zinc and harvested cell lysates for use in Western blot analysis at the indicated time points (Fig. 1B). Probing Western blots with c-Myc antiserum demonstrated that the level of c-Myc protein dramatically decreased by 80% at 4 h following treatment with zinc. This decrease corresponds to a 20-fold induction of C/EBPα protein expression at 4 h following zinc treatment. The level of c-Myc protein did not change in cell lysates isolated from the U937(vect)#1 cell line following treatment with zinc (data not shown).
C/EBPα negatively regulates the c-Myc promoter through an E2F binding site.
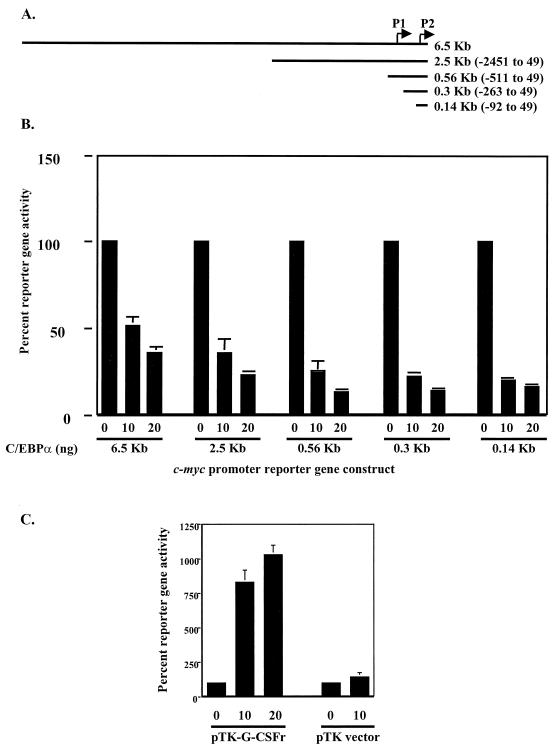
In order to determine if C/EBPα could negatively regulate the human c-Myc promoter itself, we utilized a human c-Myc promoter construct cloned into the pXP2 reporter vector and performed transfection assays to analyze c-Myc promoter activity. We cotransfected CV-1 cells with the 6.5-kb c-Myc promoter luciferase reporter gene along with increasing amounts of a C/EBPα expression plasmid. The results show that C/EBPα was able to inhibit c-Myc promoter reporter gene activity (Fig. 2) in a dose-dependent manner, as increasing amounts of C/EBPα resulted in a linear decrease in reporter gene activity (Fig. 2).
FIG. 2.
C/EBPα down-regulates c-Myc promoter activity. (A) The 6.5-kb c-Myc promoter and indicated 5′ deletions were cloned into the pXP2 luciferase reporter vector. (B) CV-1 cells were cotransfected with 200 ng of the indicated reporter gene and increasing amounts of C/EBPα expression plasmid (nanograms). Control transfection experiments indicated that C/EBPα had no effect on the pXP2 luciferase reporter vector (data not shown). (C) As a positive control for C/EBPα transactivation, CV-1 cells were cotransfected with the G-CSF receptor reporter gene containing four C/EBPα binding sites (pTK-G-CSFr). All transfection groups were normalized with a Renilla luciferase vector as an internal control. Results represent the percentages of luciferase activity with 0 ng of C/EBPα (vector alone) set to 100% activity. Results are given as the averages of at least three independent experiments, and error bars represent the standard errors of the means.
To identify the cis-acting elements on the c-Myc promoter that respond to C/EBPα, we generated 5′ deletions of the 6.5-kb promoter and cloned these deletions into the pXP2 luciferase reporter gene (Fig. 2A). Most c-Myc promoter activity is derived from two transcriptional start sites, P1 and P2, with 95% of transcription initiated from the P2 promoter site (3). We, accordingly, based our c-Myc promoter deletions relative to the P2 promoter. Cotransfection of CV-1 cells with increasing amounts of C/EBPα expression plasmid and a series of truncated c-Myc P2 promoter reporter genes resulted in a linear decrease in reporter gene activity (Fig. 2B), localizing the cis-acting element within the smallest c-Myc promoter construct (see below).
C/EBPα has previously been shown to up-regulate G-CSF receptor gene expression as well as a construct consisting of four C/EBPα binding sites derived from the G-CSF receptor upstream of a minimal reporter (pT81G-CSFr) (60). Therefore, as a positive control for C/EBPα transactivation, we cotransfected CV-1 cells with the C/EBPα expression vector and the G-CSF receptor reporter gene. Luciferase assays demonstrated that C/EBPα was able to transactivate the G-CSF receptor reporter gene, indicating that negative regulation by C/EBPα is specific to the c-Myc promoter (Fig. 2C).
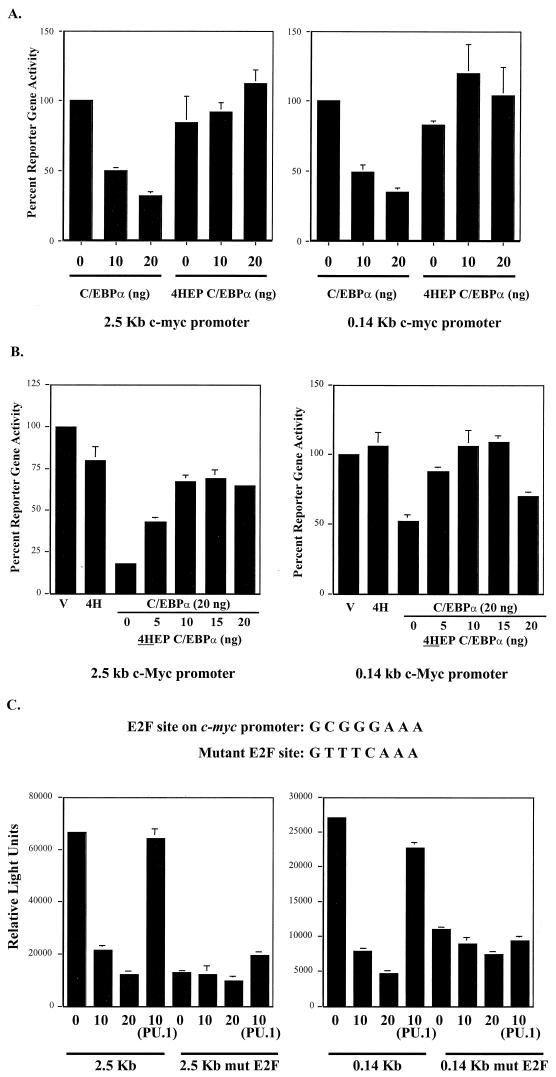
To show that C/EBPα is responsible for the negative regulation of c-Myc, we utilized a dominant-negative C/EBPα expression construct, 4HEP-C/EBPα (31, 44, 49). 4HEP-C/EBPα contains an acidic extension that extends the coiled-coiled dimerization interface from the C/EBPα leucine zipper, allowing 4HEP-C/EBPα to form stable dimers with C/EBPα without the presence of DNA. Thus, 4HEP-C/EBPα functions as a dominant negative by preventing the basic region of C/EBPα from binding to DNA. 4HEP-C/EBPα was not able to transactivate the pT81G-CSFr reporter gene, while wild-type C/EBPα can transactivate eightfold over vector alone (data not shown). Cotransfection of CV-1 cells with 4HEP-C/EBPα and c-Myc promoter reporter genes resulted in no inhibition in c-Myc reporter gene activity (Fig. 3A). We cotransfected wild-type C/EBPα with increasing amounts of 4HEP-C/EBPα. Results showed that 4HEP-C/EBPα was able to abolish the ability of wild-type C/EBPα to negatively regulate c-Myc (Fig. 3B). This indicates that a functional C/EBPα protein is required for negative regulation of the c-Myc promoter. Additionally, these results indicate that the DNA binding and dimerization regions of C/EBPα are necessary for the negative c-Myc regulation. C/EBPα negative regulation of the c-Myc promoter is specific, as the level of c-Myc reporter activity was not affected by cotransfection with the Ets transcription factor, PU.1 (Fig. 3C).
FIG. 3.
(A) Dominant-negative C/EBPα does not repress c-Myc reporter activity. CV-1 cells were cotransfected with 200 ng of the indicated c-Myc reporter construct along with wild-type C/EBPα or dominant-negative C/EBPα (4HEP C/EBPα). (B) Dominant-negative C/EBPα interferes with wild-type C/EBPα repression of c-Myc reporter activity. CV-1 cells were cotransfected with wild-type C/EBPα along with increasing amounts of dominant-negative C/EBPα and either the 2.5-kb or 0.14-kb c-Myc reporter gene. All transfection groups were cotransfected with a Renilla luciferase vector as an internal control. Results represent the percentages of luciferase activity with 0 ng of C/EBPα (vector alone) set to 100% activity. (C) Mutation of the E2F DNA binding site on c-Myc reporter constructs abolishes C/EBPα negative regulation. Wild-type and mutant sequences of the E2F DNA binding site in the c-Myc promoter located at residues −58 to −51 relative to the P2 promoter are shown at the top. CV-1 cells were cotransfected with either the 2.5-kb or 0.14-kb c-Myc reporter gene containing the wild-type or mutated E2F site along with the C/EBPα expression construct. As a control, CV-1 cells were cotransfected with a PU.1 expression construct to demonstrate that c-Myc promoter repression is specific to C/EBPα. All transfection groups were normalized with a Renilla luciferase vector as an internal control. Results are presented as the averages of at least three independent experiments, and error bars represent the standard errors of the means.
All of our 5′ c-Myc promoter deletions responded to C/EBPα regulation, and sequence analysis indicates that all c-Myc promoter constructs contain a consensus E2F binding site located between the P1 and P2 promoter elements (−65 to −58). Previous investigations have shown that c-Myc is positively regulated by E2F proteins at this site (24, 28, 53). In order to determine if C/EBPα negative regulation could act through this E2F binding site, we further deleted this site from the c-Myc promoter and subsequently cloned this truncated c-Myc promoter fragment (−57 to 49) into a luciferase reporter gene. Cotransfection of this c-Myc promoter reporter gene with a C/EBPα expression plasmid resulted in no decrease in c-Myc reporter gene activity, suggesting that down-regulation was mediated through this E2F site (data not shown).
Because the minimal −57 to 49 region of the c-Myc promoter does not possess as high a level of luciferase activity when transfected into cells, in order to demonstrate the importance of the E2F site for C/EBPα negative regulation, we mutated this E2F site in the context of our larger c-Myc promoter reporter genes (Fig. 3C). Mutation of the E2F site in these c-Myc promoter constructs abolished C/EBPα negative regulation. Cotransfection of CV-1 cells with either the 2.5- or the 0.14-kb c-Myc reporter gene containing the mutated E2F binding site along with a C/EBPα expression plasmid resulted in no decrease in reporter gene activity compared to wild-type c-Myc promoter constructs (Fig. 3C). These results demonstrate that C/EBPα negative regulation of the c-Myc promoter is mediated through this E2F binding site.
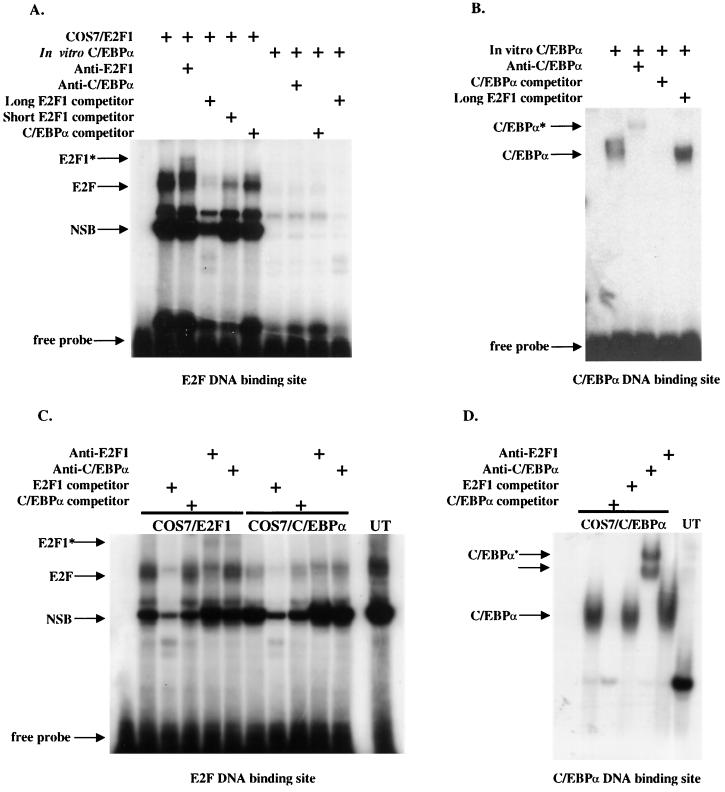
There are two possible mechanisms for how C/EBPα regulates c-Myc through this E2F binding site. First, C/EBPα may regulate c-Myc through direct binding of C/EBPα to the E2F site. However, this is unlikely, as the E2F binding site nucleotide sequence is distinct from a consensus C/EBPα DNA binding site sequence (50). In addition, we found that in vitro-translated C/EBPα protein did not bind to an E2F consensus binding site in EMSAs (Fig. 4A), while nuclear extracts from COS7 cells transfected with an E2F1 expression vector demonstrated strong binding to this E2F site. In vitro-translated C/EBPα binds strongly to a consensus C/EBP binding site. The addition of E2F oligonucleotides did not compete C/EBPα protein away from consensus C/EBPα binding site oligonucleotides in EMSAs (Fig. 4B). In order to rule out the possibility that additional cellular factors may be required for C/EBPα binding to this E2F site, we transfected COS7 cells with a C/EBPα expression plasmid. Again, EMSA was performed using nuclear extracts harvested from these cells. Results showed that no detectable C/EBPα protein was complexed on this site (Fig. 4C) In contrast, EMSA performed with a C/EBPα binding site detected C/EBPα protein complexes (Fig. 4D).
FIG. 4.
C/EBPα does not bind to the E2F DNA site in the c-Myc promoter. EMSAs using 32P-labeled, double-stranded oligonucleotides containing either an E2F site from the human c-Myc promoter (A and C) or a C/EBPα binding site from the G-CSF receptor promoter (B and D) were performed with in vitro-translated C/EBPα protein (in vitro C/EBPα) or nuclear extracts prepared from COS7 cells overexpressing C/EBPα (COS7/C/EBPα), COS7 cells overexpressing E2F1 (COS7/E2F1) as a positive control for E2F binding, or untransfected COS7 cells (UT). The migration of the free probe is indicated along with the positions of E2F1 protein complexes binding to the E2F DNA site and C/EBPα binding to the C/EBPα binding site. The asterisks indicate the positions of supershifted bands following the addition of either an E2F1 antibody or C/EBPα antisera. “competitor” refers to the 100× addition of unlabeled double-stranded E2F or C/EBPα DNA binding sites as indicated. “NSB” refers to migration of nonspecific protein complexes binding to the E2F DNA binding site. “Long” or “Short” E2F competitor refers to a double-stranded E2F oligonucleotide that contains more or less DNA sequence, respectively, flanking the E2F1 consensus site.
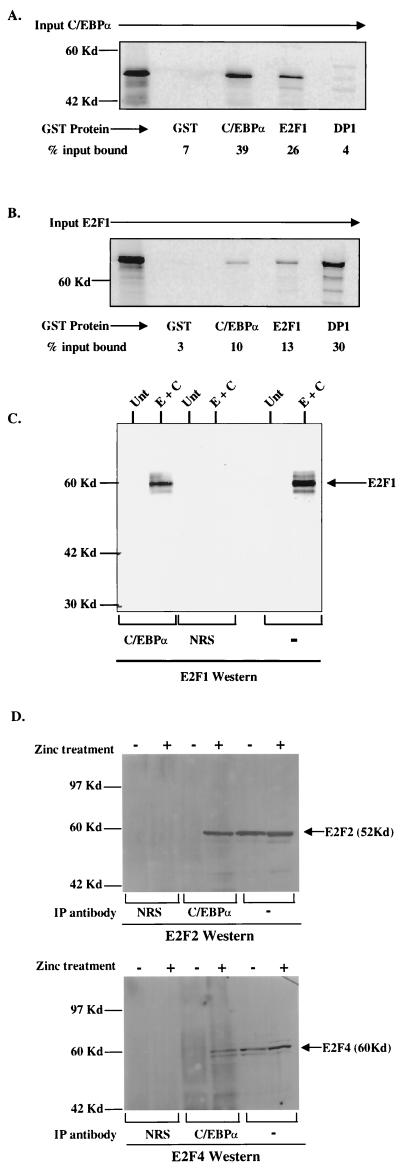
Since C/EBPα does not bind to the c-Myc promoter E2F site, C/EBPα may indirectly regulate c-Myc by disrupting E2F protein complexes at the E2F binding site. Recent reports have demonstrated that C/EBPα can disrupt E2F protein complexes in hepatocyte and adipocyte lines as well as in NIH 3T3 cells (59, 65, 66). Initial investigations of this c-Myc E2F site utilized the founding member of the E2F family, E2F1, since this was initially shown to regulate c-Myc (24). To explore the possibility that C/EBPα directly interacts with E2F1 to negatively affect its function, we used in vitro GST pull-down assays. In vitro-translated [35S]methionine-labeled C/EBPα protein was incubated with various bacterially expressed GST fusion proteins. Results of pull-down assays demonstrated that in vitro C/EBPα interacted with GST-C/EBPα and E2F1 (Fig. 5A). In vitro-translated C/EBPα did not interact with GST-DP1. In a complementary experiment using in vitro-translated [35S]methionine-labeled E2F1 incubated with the same set of GST fusion proteins, we again detected an interaction between GST-C/EBPα and E2F1 (Fig. 5B). In vitro-translated E2F1 also interacted strongly with its dimerization partner, GST-DP1, but not with GST alone. In order to demonstrate that the interaction between E2F1 and C/EBPα occurs in vivo, coimmunoprecipitation assays were performed. Because E2F1 is expressed at low levels in U937 cells (data not shown), COS7 cells were cotransfected with expression constructs for both E2F1 and C/EBPα. Whole-cell extracts were immunoprecipitated with antiserum for either C/EBPα or NRS, followed by Western analysis with an antibody to E2F1. Results demonstrated that complexes immunoprecipitated with C/EBPα antisera contained E2F1 protein (Fig. 5C). The interaction between E2F1 and C/EBPα is strong, as quantitation with a phosphorimager indicated that 86% of the transfected E2F1 protein is complexed with immunoprecipitated C/EBPα protein.
FIG. 5.
C/EBPα and E2F1 physically interact in vitro and in vivo. (A) Binding of 35S-labeled in vitro-translated C/EBPα (input C/EBPα) to GST (negative control for binding), GST-C/EBPα (positive control for binding), GST-E2F1, and GST-DP1. (B) Binding of 35S-labeled in vitro-translated E2F1 (input E2F1) to the same GST fusion proteins. Percent input bound represents the amount of in vitro-translated protein complexed with GST fusion proteins as calculated using a phosphorimager (Molecular Dynamics). (C) COS7 cells either untransfected (Unt) or transfected with E2F1 and C/EBPα expression vectors (E + C) were immunoprecipitated with C/EBPα antisera or control NRS followed by Western analysis with E2F1 antibody. As a control for E2F1 expression and migration, 1/30 of the COS7 lysate used for immunoprecipitation was resolved by SDS-PAGE (marked “−” for immunoprecipitation antibody). The position of E2F1 is indicated. (D) C/EBPα interacts with endogenous E2F proteins in myeloid cells. Uninduced (−) or induced (+) U937α#2 cells were immunoprecipitated (IP antibody) with C/EBPα antisera or control NRS followed by Western analysis with either E2F2 or E2F4 antibody. As a control for E2F2 and E2F4 expression and migration, 1/30 of the lysate used for immunoprecipitation was resolved by SDS-PAGE (marked “−” for immunoprecipitation antibody). The positions of E2F2 and E2F4 are indicated.
There are five additional E2F family members (E2F2 to E2F6). As evidenced by our EMSA, untransfected cells show significant binding to the c-Myc E2F site (Fig. 4C). We detected only a slight supershift when E2F1 antibody was added to our EMSA reactions using nuclear extracts from E2F1-transfected cells (Fig. 4A and C), suggesting that other E2F family members might bind to this E2F site. In order to demonstrate that C/EBPα interacts with other endogenously expressed E2F proteins in myeloid cells, we utilized our U937α#2 cell line. Cells were untreated or treated with ZnSO4 to induce C/EBPα expression. Whole-cell extracts were immunoprecipitated with antiserum against C/EBPα followed by Western analysis with antiserum to either E2F2 or E2F4. Results showed that, in lysates in which C/EBPα was induced, complexes containing E2F2 and E2F4 proteins were immunoprecipitated (Fig. 5D). The results demonstrate the ability of C/EBPα and E2F proteins to form complexes in mammalian cells. Taken together, our binding assays support a model in which C/EBPα may disrupt E2F protein function by directly interacting with E2F family members.
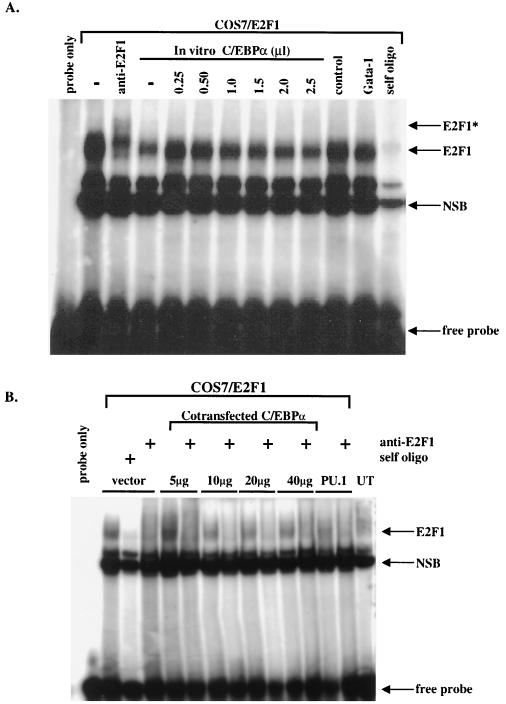
If the interaction with C/EBPα disrupts E2F protein binding, this interaction could result in negative regulation of E2F-controlled genes such as c-Myc. In order to examine the ability of C/EBPα to disrupt E2F1 DNA binding, we performed an EMSA using an E2F oligonucleotide and nuclear extracts prepared from COS7 cells overexpressing E2F1. We then mixed increasing amounts of in vitro-translated C/EBPα protein into the reactions. Results showed that increasing amounts of C/EBPα protein had no effect on E2F1 binding to a consensus E2F DNA binding site (Fig. 6A). To evaluate the influence of other cellular factors on the interaction between E2F1 and C/EBPα, COS7 cells were cotransfected with 10 mg of E2F1 plasmid and increasing amounts of C/EBPα plasmid. Again, results showed that C/EBPα did not interfere with E2F complex binding (Fig. 6B). Even though C/EBPα may not affect E2F1 binding directly, our results indicate that C/EBPα strongly interacts with E2F proteins both in vitro and in vivo. It is possible that C/EBPα interacts with other proteins that complete the active E2F transcriptional complex, or alternatively, C/EBPα may mask the E2F1 transcriptional activation domain, which ultimately results in a loss of c-Myc expression without the direct loss of E2F1 DNA binding function.
FIG. 6.
C/EBPα protein cannot disrupt the binding of E2F1 protein to DNA. (A) EMSAs using a 32P-labeled, double-stranded oligonucleotide containing the E2F site from the human c-Myc promoter were performed with nuclear extracts prepared from COS7 cells overexpressing E2F1 (COS7/E2F1). The addition of increasing amounts of in vitro-translated C/EBPα did not alter the amount of E2F1 protein binding to the E2F DNA site. The migration of the free probe is indicated along with the positions of E2F1 protein complexes. The asterisk indicates the position of a supershifted band following the addition of an E2F1 antibody. “self oligo” indicates the 100× addition of unlabeled double-stranded E2F DNA binding site. “NSB” indicates the migration of nonspecific protein complexes binding to the E2F DNA binding site. “control” indicates binding reactions performed with unprogrammed rabbit reticulocyte lysate, and “Gata-1” indicates control binding reactions performed with in vitro-translated GATA-1 protein. (B) EMSAs performed with the E2F site from the c-Myc promoter and nuclear extracts from COS7 cells cotransfected with 10 mg of E2F1 plasmid and indicated amounts of C/EBPα plasmid. For control reactions, COS7 cells were cotransfected with PU.1 or left untransfected (UT). The migration of the free probe is indicated along with the positions of the E2F1 protein complexes. “self oligo” indicates the 100× addition of unlabeled double-stranded E2F DNA binding site. “NSB” indicates the migration of nonspecific protein binding complexes.
In order to investigate these hypotheses further, we cotransfected CV-1 cells with E2F1 to activate the 0.14-kb c-Myc reporter (Fig. 7A). Cotransfection of increasing amounts of C/EBPα led to a progressive inhibition of the ability of E2F1 to transactivate c-Myc promoter activity (Fig. 7A). It has previously been shown that C/EBPα can interact with Rb (8). Since E2F proteins are regulated through their association with Rb and this association results in repression of E2F-regulated genes, we investigated the possibility that C/EBPα inhibition of E2F transactivation activity is dependent on Rb. Therefore, we utilized the Saos osteosarcoma cell line, which does not express Rb. We cotransfected E2F1 with increasing amounts of C/EBPα, along with the 0.14-kb c-Myc reporter construct. Results showed that C/EBPα also was able to block E2F1 transcriptional activation domain activity in these Rb-minus cells (Fig. 7B). Thus, an increase in C/EBPα expression interferes with E2F1 transcription activation activity in an Rb-independent fashion, and this results in repression of the c-Myc promoter.
FIG. 7.
C/EBPα interferes with E2F1 transactivation of the c-Myc promoter. CV-1 cells or Saos (Rb−) cells were cotransfected with 200 ng of the 0.14-kb c-Myc reporter gene (V), 20 ng of C/EBPα plasmid alone (α), 10 ng of E2F1 plasmid alone, or 10 ng of E2F1 plasmid along with the indicated amounts of C/EBPα plasmid. All transfection groups were normalized with a Renilla luciferase vector as an internal control. Results represent the percentages of reporter gene or luciferase activity with vector alone (V) set to 100% activity. Results are given as the averages of at least three independent experiments, and error bars represent the standard errors of the means.
c-Myc must be negatively regulated in order for myeloblasts to differentiate into neutrophils.
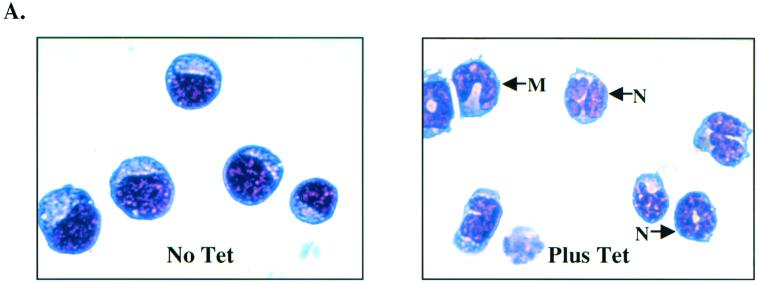
To determine if c-Myc is an important C/EBPα target gene, we investigated how Myc gene expression affected myeloblast differentiation. Tet-o-myc 1137 is a myeloblast cell line derived from a tumor from transgenic mice that express the human c-Myc cDNA under the control of a tetracycline-responsive promoter (16). Treatment with doxycycline or tetracycline turns off c-Myc expression and drives the cells to differentiate into neutrophils. We found that, in the absence of tetracycline, 85% of the cells were myeloblasts, 10% were promyelocytes, and 5% were metamyelocytes and neutrophils (Fig. 8A and B). Following treatment with tetracycline for 24 h, which turns off c-Myc expression, the culture underwent marked differentiation, with 28% myeloblasts, 38% promyelocytes, and 34% metamyelocytes and mature neutrophils (Fig. 8A and B).
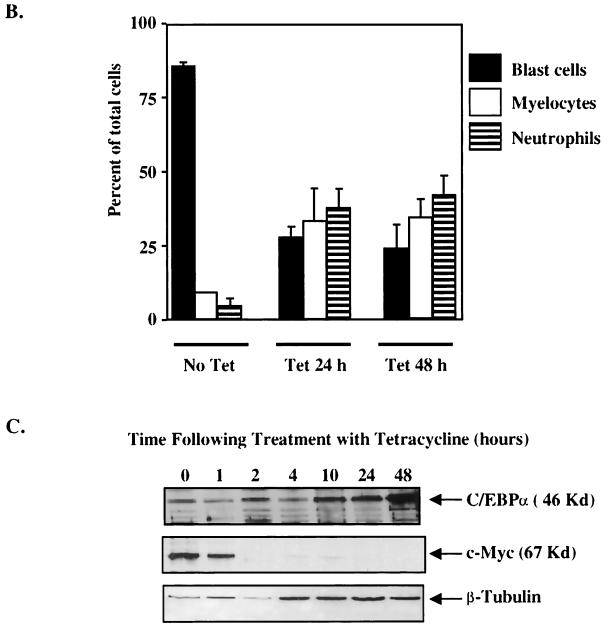
FIG. 8.
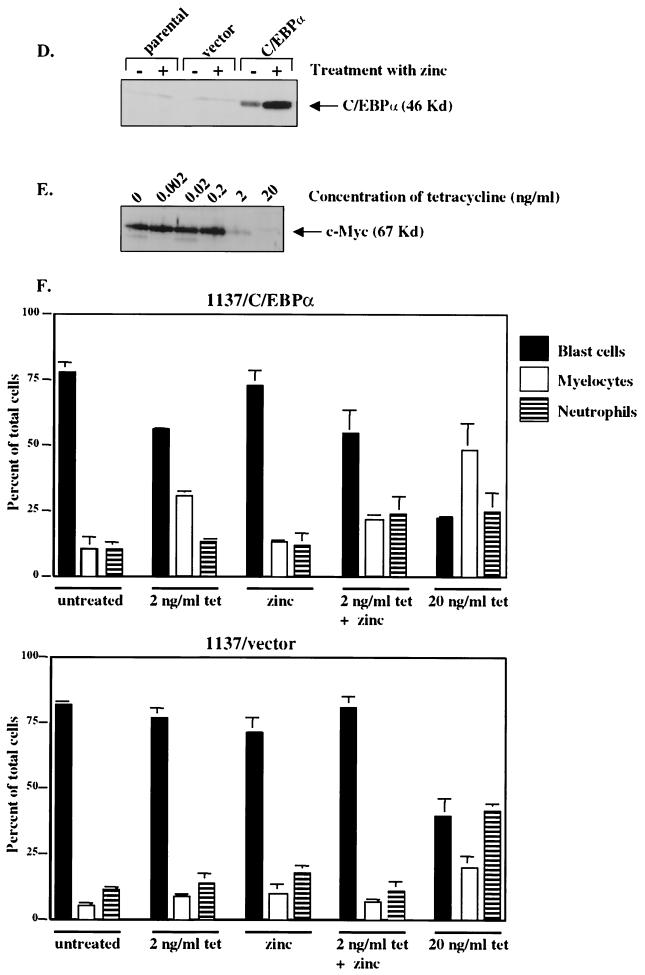
Down-regulation of c-Myc is crucial to the granulocytic differentiation pathway. The 1137 cell line was derived from murine bone marrow of a transgenic line with a human c-Myc cDNA under the control of a tetracycline-responsive promoter. The addition of tetracycline to the culture medium turns off human c-Myc expression, resulting in the differentiation of these myeloblasts to neutrophils. (A) Wright-Giemsa-stained cells without (No Tet) or with (Plus Tet) treatment with tetracycline. Cells treated with tetracycline differentiated into myelocytes (M) and neutrophils (N). (B) Differential analysis of Wright-Giemsa-stained slides following treatment with tetracycline for 0, 24, and 48 h, respectively. (C) 1137 cells were harvested for cell lysates to use in Western blotting at indicated time points following treatment with tetracycline. (Top) Western blot hybridized with C/EBPα antiserum shows that the level of endogenous C/EBPα protein increased 24 h following treatment with tetracycline. This corresponds to the shift to mature cells seen in panels A and B. (Middle) The same blot hybridized with c-Myc antiserum, showing that c-Myc protein levels dramatically decreased 2 h following treatment with tetracycline. (Bottom) The same blot hybridized with a β-tubulin antibody to control for protein loading and integrity. (D) Western analysis of 1137 stable lines with (+) and without (−) treatment with zinc. “parental” indicates the 1137 parental line, “vector” indicates the 1137 stable line with metallothionein vector, and “C/EBPα ” indicates the 1137 line with metallothionein-driven C/EBPα. (E) Western analysis of cell lysates prepared from the 1137/C/EBPα stable line with the indicated treatment with tetracycline. The level of human c-Myc protein was titrated by 100-fold dilutions of tetracycline. A 20-ng/ml concentration turns off c-Myc expression, while a 2-ng/ml concentration results in a low level of c-Myc expression. Lower concentrations of tetracycline result in no decrease in c-Myc protein expression. (F) Differential analysis of Wright-Giemsa-stained slides following treatment of 1137 stable lines with tetracycline, zinc, or the combination of tetracycline and zinc for 48 h.
When the 1137 cells were treated with tetracycline and harvested for cell lysates for Western blot analysis at distinct time points, we determined that c-Myc protein expression was not detectable by 2 h following treatment with tetracycline (Fig. 8C). Moreover, the level of c-Myc protein remains depressed throughout the entire experiment to the 48-h point. As the level of c-Myc protein decreased, we observed an increase in the level of endogenous C/EBPα protein, especially at the later time points (24 and 48 h), which correlated with the shift in 1137 cells from myeloblasts to more mature neutrophils (Fig. 8C). In order to further examine the role of negative regulation of c-Myc by C/EBPα in the differentiation process, we engineered the 1137 myeloid cells with either a metallothionein-driven C/EBPα cDNA or vector alone (Fig. 8D). In this system, the levels of C/EBPα and c-Myc could be altered independently by adjusting levels of zinc and tetracycline, respectively. Because the level of c-Myc protein in 1137 cells is highly elevated without tetracycline, we titrated the level of tetracycline such that we would observe a decrease in the level of c-Myc protein but leave a level of c-Myc high enough that cells would not differentiate (Fig. 8E). Results of Western analysis indicate that a concentration of 2 ng of tetracycline/ml dramatically lowers the level of c-Myc while the majority of cells remain undifferentiated. Treatment of 1137/C/EBPα cells with 2 ng of tetracycline/ml alone (to lower c-Myc expression), zinc alone (to turn on C/EBPα expression), or both tetracycline and zinc showed only a very slight increase in the number of differentiated cells compared to untreated cells (Fig. 8F). Again, the maximum amount of differentiation is observed with a higher concentration of tetracycline that essentially turns off c-Myc protein expression in the 1137 stable lines (Fig. 8F). The 1137/vector cells remain highly undifferentiated without the addition of the higher level of tetracycline. Therefore, increased expression of C/EBPα in 1137 cells was not able to down-regulate c-Myc expression from the tetracycline-regulatable promoter and could not overcome the block to differentiation imposed by continued expression of exogenous human c-Myc. Taken together, these data demonstrate that c-Myc protein expression must be negatively regulated in order for myeloblasts to differentiate.
DISCUSSION
C/EBPα down-regulates c-Myc through an E2F site, suggesting that other c-Myc and E2F target genes may lie downstream of C/EBPα.
In order to identify critical C/EBPα target genes involved in the differentiation of granulocytic cells, we performed both RDA and oligonucleotide array analysis (Tables 1 and 2). Both of these screens independently identified the c-Myc gene as a target for regulation by C/EBPα. This is the first report to demonstrate that C/EBPα is a negative regulator of c-Myc gene expression. Using a stable C/EBPα-inducible U937 cell line, we have shown that C/EBPα expression results in a significant decrease in the levels of endogenous c-Myc mRNA and corresponding protein (Fig. 1). Quantitation by phosphorimager indicates that the level of c-Myc RNA decreased only 30% by 2 h, compared with a dramatic decrease of 94% by 4 h (Fig. 1). The level of C/EBPα protein was induced 5-fold by 2 h and 20-fold by 4 h (Fig. 1). Therefore, the 20-fold increase in C/EBPα protein at 4 h preceded the decrease seen in c-Myc RNA and corresponding c-Myc protein levels. We have shown by luciferase reporter assays that C/EBPα protein itself negatively regulates the human c-Myc promoter (Fig. 2). Therefore, C/EBPα expression results in a linear decrease in c-Myc reporter activity.
In addition to c-Myc, both our RDA and oligonucleotide array screens identified several interesting C/EBPα candidate genes. For example, another down-regulated gene identified by the RDA screen was Id-2H, a basic HLH protein that antagonizes other basic HLH proteins to inhibit cellular differentiation and enhance cell proliferation (22, 42) (Table 1). Of the candidate genes identified by the oligonucleotide array screen (Table 2), c-Myb is a transcription factor expressed in hematopoietic cells whose expression parallels that of c-Myc, with high levels in immature hematopoietic cells and with expression decreasing during terminal differentiation (1, 5, 21, 25). As several studies have shown that c-Myb can regulate c-Myc gene expression (9, 54, 75), the down-regulation of c-Myb may contribute to the dramatic decrease that we observed for c-Myc expression in the presence of C/EBPα. Whether down-regulation of c-Myb is a direct effect or a secondary effect of C/EBPα expression remains to be determined.
Of the C/EBPα target genes identified to date, c-Myc may be the most critical target of C/EBPα, allowing myeloblasts to exit from a proliferative state and enter into a differentiation pathway. Upon further examination of our nucleotide array screen, we identified several c-Myc target genes as being regulated by C/EBPα (Table 2). Among the c-Myc target genes identified by our C/EBPα array screen are α-prothymosin, E1F4A, E1F5A, and (2′-5′) oligo(A) synthetase E, some of which were identified in Myc microarray screens (11, 47). c-Myc has been shown previously to up-regulate α-prothymosin, E1F4A, and E1F5A (10, 11, 14). In contrast, our C/EBPα screen demonstrated these c-Myc-dependent genes to be down-regulated (data not shown). As C/EBPα negatively regulates c-Myc expression, a secondary consequence is that c-Myc target genes normally activated are now down-regulated and vice versa. Additionally, following a search of the nucleotide database for consensus E-box promoter elements, we identified several additional genes potentially regulated by c-Myc. Hence, C/EBPα disruption of c-Myc expression results in a global effect on the expression of c-Myc-regulated genes required for cells to continue in a proliferative state.
The human c-Myc promoter contains no consensus C/EBPα DNA binding sites. Instead, C/EBPα regulates c-Myc promoter activity through an E2F binding site (−57 to 49) relative to the P2 promoter element (Fig. 3C). C/EBPα can regulate the expression of other genes through E2F DNA binding sites (59). However, upon searching the target genes identified through our nucleotide array screen (Table 2), we identified only one additional candidate gene for regulation by E2F, ARHG, which, like c-Myc, was negatively regulated (data not shown). To determine whether C/EBPα can indeed regulate transcription of many genes through E2F DNA elements, future studies will be needed to address the ability of C/EBPα to negatively regulate the promoter activity of other known E2F-regulated genes such as thymidine kinase; dihydrofolate reductase; or cyclin E, b-Myb, or E2F2 (19, 23, 48, 55).
Mechanism of c-Myc down-regulation through the E2F site.
As noted above, C/EBPα represses c-Myc gene expression through the consensus E2F binding site located between the P1 and P2 promoter elements (Fig. 3). However, C/EBPα protein does not bind to this site directly (Fig. 4). Whether the mechanism for C/EBPα negative regulation through this E2F site is direct or indirect through protein-protein interactions remains to be determined. We have shown that C/EBPα can physically interact with E2F1 and other E2F family members (Fig. 5), and so we hypothesized that C/EBPα might disrupt E2F protein complexes binding to this DNA site. EMSA results demonstrated that C/EBPα protein could not directly displace E2F1 binding to the c-Myc promoter E2F site (Fig. 6).
Alternatively, C/EBPα may down-regulate the c-Myc gene by masking the E2F transcriptional activation domain or through protein-protein interactions which stabilize a repressive complex at the E2F site. The E2F family of transcription factors consists of six E2F members (E2F1 to E2F6) that form heterodimers with two DP family members (DP1 and DP2). E2F transcription factors are regulated by association with the Rb protein and related p107 and p130 proteins. The interaction between Rb and E2F factors is controlled by cdk's that hyperphosphorylate Rb during the transition from G1 to the S phase of the cell cycle. Phosphorylation of Rb hinders its interaction with E2F, which allows E2F protein complexes to activate transcription of E2F-regulated genes (58). We explored the possibility that C/EBPα, through its interaction with E2F proteins, masks the E2F transcriptional activation domain, thus resulting in down-regulation of the c-Myc promoter. CV-1 cells were cotransfected with E2F1, which activates c-Myc reporter activity, and C/EBPα. Our results indicate that C/EBPα can interfere with the E2F1 transactivation of c-Myc (Fig. 7). It has previously been shown that C/EBPα can interact with Rb (8). As Rb and other pocket proteins (p107 and p130) form a repressive complex with DP-E2F dimers, it is possible that C/EBPα acts as a type of adapter molecule, linking DP-E2F complexes to Rb to form a repressive transcriptional complex. In order to investigate this hypothesis, we replicated the above cotransfection experiment in a cell line negative for Rb expression. We obtained the same result, indicating that the block to E2F1 transactivation by C/EBPα is independent of Rb and that C/EBPα is not an adapter between E2F proteins and Rb. However, we have not ruled out the possibility that C/EBPα interferes with the E2F complex formation of other proteins, such as DP1 or pocket proteins p107 and p130.
Recently, Timchenko et al. showed that C/EBPα can cause growth arrest in fetal liver cells as well as adipocytes through disruption of E2F protein complexes (65, 66). These results indicate that C/EBPα interacts with p107 in fetal liver cells. DP-E2F-p107 complexes prevail in dividing cells, and thus, C/EBPα disruption of these complexes has a negative effect on proliferation. Moreover, overexpression of C/EBPα in a preadipocyte cell line caused an increase in repressive DP-E2F-p130 complexes via an increase in the p21 protein which interferes with cdk activity (66). It is possible that C/EBPα disrupts DP-E2F-p107 and DP-E2F-p130 complexes during myeloid differentiation. In contrast to our results, Timchenko et al. did not detect any interaction between E2F proteins and C/EBPα in adipocytes. Further support of our findings that C/EBPα interacts with the E2F1 transcription complex comes from the findings of a second group, which also detected an interaction between C/EBPα and E2F in NIH 3T3 cells (59). Moreover, these investigators show that this interaction interferes with the S-phase transcription of E2F-regulated genes E2F1 and dihydrofolate reductase. Therefore, the studies of both these groups along with our own results support a role for C/EBPα gene regulation through E2F consensus binding sites. We will further explore the significance of this interaction between E2F proteins and C/EBPα and disruption of interactions with pocket proteins p107 and p130 during myeloid differentiation in future studies.
C/EBPα interacts with E2F proteins but does not directly bind to the E2F DNA site itself. When cells were cotransfected with a dominant-negative C/EBPα construct that forms stable dimers with C/EBP proteins, the ability of C/EBPα to inhibit c-Myc promoter activity was abolished (Fig. 3B). This dominant negative contains an acidic extension after the zipper region of the protein and can form strong dimers with C/EBPα without stabilizing DNA. In fact, this dominant negative was designed to stoichiometrically displace C/EBPα from DNA (31). Since the dominant-negative C/EBPα dimerizes with wild-type C/EBPα through the bZIP region, this implies that the bZIP region of C/EBPα is necessary for the interference with E2F transcriptional activation domain activity. Since we have shown that C/EBPα cannot bind to the c-Myc E2F DNA site (Fig. 4), the dominant-negative C/EBPα does not act by inhibiting C/EBPα from binding to the c-Myc promoter. Instead, the bZIP region of C/EBPα is required for the interaction between C/EBPα and E2F proteins. Future investigations will map the interaction domains of C/EBPα and E2F1.
Biological consequences of c-Myc down-regulation: role in normal myelopoiesis and leukemia.
Our previously published studies (16) and the results shown in Fig. 8 indicate that down-regulation of c-Myc expression allows myeloid cells to differentiate into mature granulocytes. Here, we provide a possible mechanism mediated through C/EBPα. The exogenous expression of c-Myc in 1137 myeloblast cells under the control of a promoter which is not responsive to C/EBPα-mediated down-regulation forces these cells to remain undifferentiated. Induced expression of C/EBPα in these 1137 cells was unable to overcome this c-Myc-mediated block to differentiation. Maintenance of c-Myc expression forces cells to remain in a proliferative state, preventing cell cycle arrest. To counteract c-Myc, C/EBPα must negatively regulate c-Myc gene expression to impose cellular growth arrest and allow cells to differentiate.
In addition to playing an important role in normal granulopoiesis, the reciprocal regulation of C/EBPα and c-Myc expression is likely to be an important factor in acute myeloid leukemia, a condition resulting from a block in myeloid maturation. In mice, maintenance of c-Myc expression using a regulatable promoter can induce myeloid leukemia (16). In humans, certain subtypes of myeloid leukemias in which C/EBPα expression is specifically down-regulated demonstrate a concomitant increase in c-Myc expression (T. L. Pabst and D. G. Tenen, unpublished results). Therefore, elucidating the mechanism of how C/EBPα down-regulates c-Myc not only will be important in understanding normal cell differentiation but may also lead to the development of novel and specific strategies for the treatment of malignancies such as myeloid leukemias that result from a block in myeloid maturation.
ACKNOWLEDGMENTS
We thank members of the Tenen lab and especially H. Radomska for suggestions; N. Timchenko for advice on E2F gel shifts; and J. Lees, S. Chellappan, J. Nevins, C. Vinson, and W. Kaelin for generous gifts of plasmids.
This work was supported by NIH postdoctoral fellowship F32DK09892 to L.M.J. and NIH grant HL56745 (to D.G.T.).
REFERENCES
- 1.Anfossi G, Gewirtz A M, Calabretta B. An oligomer complementary to c-myb-encoded mRNA inhibits proliferation of human myeloid leukemia cell lines. Proc Natl Acad Sci USA. 1989;86:3379–3383. doi: 10.1073/pnas.86.9.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonson P, Pray M G, Jacobsson A, Xanthopoulos K G. Myc inhibits CCAAT/enhancer-binding protein alpha-gene expression in HIB-1B hibernoma cells through interactions with the core promoter region. Eur J Biochem. 1995;232:397–403. doi: 10.1111/j.1432-1033.1995.397zz.x. [DOI] [PubMed] [Google Scholar]
- 3.Battey J, Moulding C, Taub R, Murphy W, Stewart T, Potter H, Lenoir G, Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983;34:779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- 4.Behre G, Smith L T, Tenen D G. Use of a promoterless Renilla luciferase vector as an internal control plasmid for transient co-transfection assays of Ras-mediated transcription activation. BioTechniques. 1999;26:24–28. doi: 10.2144/99261bm03. [DOI] [PubMed] [Google Scholar]
- 5.Bellon T, Perrotti D, Calabretta B. Granulocytic differentiation of normal hematopoietic precursor cells induced by transcription factor PU.1 correlates with negative regulation of the c-myb promoter. Blood. 1997;90:1828–1839. [PubMed] [Google Scholar]
- 6.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 7.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 8.Chen P L, Riley D J, Chen Y M, Lee W H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 9.Cogswell J P, Cogswell P C, Kuehl W M, Cuddihy A M, Bender T M, Engelke U, Marcu K B, Ting J P Y. Mechanism of c-myc regulation by c-Myb in different cell lineages. Mol Cell Biol. 1993;13:2858–2869. doi: 10.1128/mcb.13.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole M D, McMahon S B. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 11.Coller H A, Grandori C, Tamayo P, Colbert T, Lander E S, Eisenman R N, Golub T R. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppola J A, Cole M D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986;320:760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- 13.Dalla-Favera R, Martinotti S, Gallo R C, Erikson J, Croce C M. Translocation and rearrangements of the c-myc oncogene locus in human undifferentiated B-cell lymphomas. Science. 1983;219:963–967. doi: 10.1126/science.6401867. [DOI] [PubMed] [Google Scholar]
- 14.Dang C V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsher D W, Bishop J M. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci USA. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsher D W, Bishop J M. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 17.Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos K G. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein α. J Biol Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- 18.Freytag S O, Geddes T J. Reciprocal regulation of adipogenesis by Myc and C/EBP alpha. Science. 1992;256:379–382. doi: 10.1126/science.256.5055.379. [DOI] [PubMed] [Google Scholar]
- 19.Fry C J, Slansky J E, Farnham P J. Position-dependent transcriptional regulation of the murine dihydrofolate reductase promoter by the E2F transactivation domain. Mol Cell Biol. 1997;17:1966–1976. doi: 10.1128/mcb.17.4.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, Bloomfield C D, Lander E S. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 21.Gonda T J, Metcalf D. Expression of myb, myc and fos proto-oncogenes during the differentiation of a murine myeloid leukaemia. Nature. 1984;310:249–251. doi: 10.1038/310249a0. [DOI] [PubMed] [Google Scholar]
- 22.Hara E, Yamaguchi T, Nojima H, Ide T, Campisi J, Okayama H, Oda K. Id-related genes encoding helix-loop-helix proteins are required for G1 progression and are repressed in senescent human fibroblasts. J Biol Chem. 1994;269:2139–2145. [PubMed] [Google Scholar]
- 23.Hengstschlager M, Hengstschlager-Ottnad E, Pusch O, Wawra E. The role of p16 in the E2F-dependent thymidine kinase regulation. Oncogene. 1996;12:1635–1643. [PubMed] [Google Scholar]
- 24.Hiebert S W, Lipp M, Nevins J R. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc Natl Acad Sci USA. 1989;86:3594–3598. doi: 10.1073/pnas.86.10.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman-Liebermann B, Liebermann D A. Suppression of c-myc and c-myb is tightly linked to terminal differentiation induced by IL6 or LIF and not growth inhibition in myeloid leukemia cells. Oncogene. 1991;6:903–909. [PubMed] [Google Scholar]
- 26.Holt J T, Redner R L, Nienhuis A W. An oligomer complementary to c-myc mRNA inhibits proliferation of HL-60 promyelocytic cells and induces differentiation. Mol Cell Biol. 1988;8:963–973. doi: 10.1128/mcb.8.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubank M, Schatz D G. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishida S, Shudo K, Takada S, Koike K. A direct role of transcription factor E2F in c-myc gene expression during granulocytic and macrophage-like differentiation of HL60 cells. Cell Growth Differ. 1995;6:229–237. [PubMed] [Google Scholar]
- 29.Iwama A, Zhang P, Darlington G J, McKercher S R, Maki R A, Tenen D G. Use of RDA analysis of knockout mice to identify myeloid genes regulated in vivo by PU.1 and C/EBPα. Nucleic Acids Res. 1998;26:3034–3043. doi: 10.1093/nar/26.12.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klemsz J M, McKercher S R, Celada A, Van Beveren C, Maki R A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 31.Krylov D, Olive M, Vinson C. Extending dimerization interfaces: the bZIP basic region can form a coiled coil. EMBO J. 1995;14:5329–5337. doi: 10.1002/j.1460-2075.1995.tb00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsson L G, Pettersson M, Oberg F, Nilsson K, Luscher B. Expression of mad, mxi1, max and c-myc during induced differentiation of hematopoietic cells—opposite regulation of mad and c-myc. Oncogene. 1994;9:1247–1252. [PubMed] [Google Scholar]
- 33.Leder A, Pattengale P K, Kuo A, Stewart T A, Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986;45:485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- 34.Lekstrom-Himes J, Xanthopoulos K G. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 35.Li L H, Nerlov C, Prendergast G, MacGregor D, Ziff E B. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 1994;13:4070–4079. doi: 10.1002/j.1460-2075.1994.tb06724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin F T, Lane M D. Antisense CCAAT/enhancer-binding protein RNA suppresses coordinate gene expression and triglyceride accumulation during differentiation of 3T3–L1 preadipocytes. Genes Dev. 1992;6:533–544. doi: 10.1101/gad.6.4.533. [DOI] [PubMed] [Google Scholar]
- 37.Lin F T, Lane M D. CCAAT/enhancer binding protein α is sufficient to initiate the 3T3–L1 adipocyte differentiation program. Proc Natl Acad Sci USA. 1994;91:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 39.Liu F, Poursine-Laurent J, Wu H Y, Link D C. Interleukin-6 and the granulocyte colony stimulating factor receptor are major independent regulators of granulopoiesis in vivo but are not required for lineage commitment or terminal differentiation. Blood. 1997;90:2583–2590. [PubMed] [Google Scholar]
- 40.Liu F, Wu H Y, Wesselschmidt R L, Kornaga T, Link D C. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor-deficient mice. Immunity. 1996;5:491–501. doi: 10.1016/s1074-7613(00)80504-x. [DOI] [PubMed] [Google Scholar]
- 41.Lodie T A, Reiner M, Coniglio S, Viglianti G, Fenton M J. Both PU.1 and nuclear factor-kappa B mediate lipopolysaccharide-induced HIV-1 long terminal repeat transcription in macrophages. J Immunol. 1998;161:268–276. [PubMed] [Google Scholar]
- 42.Maruyama H, Kleeff J, Wildi S, Friess H, Buchler M W, Israel M A, Korc M. Id-1 and Id-2 are overexpressed in pancreatic cancer and in dysplastic lesions in chronic pancreatitis. Am J Pathol. 1999;155:815–822. doi: 10.1016/S0002-9440(10)65180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mink S, Mutschler B, Weiskirchen R, Bister K, Klempnauer K H. A novel function for myc: inhibition of C/EBP-dependent gene activation. Proc Natl Acad Sci USA. 1996;93:6635–6640. doi: 10.1073/pnas.93.13.6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moitra J, Mason M M, Olive M, Krylov D, Gavrilova O, Marcus-Samuels B, Feigenbaum L, Lee E, Aoyama T, Eckhaus M, Reitman M L, Vinson C. Life without white fat: a transgenic mouse. Genes Dev. 1998;12:3168–3181. doi: 10.1101/gad.12.20.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nordeen S K. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques. 1988;6:454–458. [PubMed] [Google Scholar]
- 46.Obaya A J, Mateyak M K, Sedivy J M. Mysterious liaisons: the relationship between c-Myc and the cell cycle. Oncogene. 1999;18:2934–2941. doi: 10.1038/sj.onc.1202749. [DOI] [PubMed] [Google Scholar]
- 47.O'Hagan R C, Schreiber-Agus N, Chen K, David G, Engleman J A, Schwab R, Alland L, Thomson C, Ronning D R, Sacchettini J C, Meltzer P, DePinho R A. Gene-target recognition among members of the MYC superfamily and implications for oncogenesis. Nat Genet. 2000;24:113–119. doi: 10.1038/72761. [DOI] [PubMed] [Google Scholar]
- 48.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olive M, Krylov D, Echlin D R, Gardner K, Taparowsky E, Vinson C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- 50.Osada S, Yamamoto H, Nishihara T, Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J Biol Chem. 1996;271:3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- 51.Petrovick S M, Hiebert S W, Friedman A D, Hetherington C J, Tenen D G, Zhang D-E. Multiple functional domains of AML1: PU.1 and C/EBPα cooperate with different regions of AML1. Mol Cell Biol. 1998;18:3915–3925. doi: 10.1128/mcb.18.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radomska H S, Huettner C S, Zhang P, Cheng T, Scadden D T, Tenen D G. CCAAT/enhancer binding protein α is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18:4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roussel F M, Davis J N, Cleveland J L, Ghysdael J, Hiebert S W. Dual control of myc expression through a single DNA binding site targeted by ets family proteins and E2F-1. Oncogene. 1994;9:405–415. [PubMed] [Google Scholar]
- 54.Schmidt M, Nazarov V, Stevens L, Watson R, Wolff L. Regulation of the resident chromosomal copy of c-myc by c-Myb is involved in myeloid leukemogenesis. Mol Cell Biol. 2000;20:1970–1981. doi: 10.1128/mcb.20.6.1970-1981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sears R, Ohtani K, Nevins J R. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17:5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheiness D, Fanshier L, Bishop J M. Identification of nucleotide sequences which may encode the oncogenic capacity of avian retrovirus MC29. J Virol. 1978;28:600–610. doi: 10.1128/jvi.28.2.600-610.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shivdasani R A, Orkin S H. The transcriptional control of hematopoiesis. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]
- 58.Slansky J E, Farnham P J. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 59.Slomiany B A, D'Arigo K L, Kelly M M, Kurtz D T. C/EBPα inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol Cell Biol. 2000;20:5986–5997. doi: 10.1128/mcb.20.16.5986-5997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith L T, Hohaus S, Gonzalez D A, Dziennis S E, Tenen D G. PU.1 (Spi-1) and C/EBPα regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88:1234–1247. [PubMed] [Google Scholar]
- 61.Spencer C A, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 62.Tamayo P, Slonim D, Mesirov J, Zhu Q, Dmitrovsky E, Lander E S, Golub T R. Interpreting gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tenen D G, Hromas R, Licht J D, Zhang D-E. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 64.Timchenko N A, Harris T E, Wilde M, Bilyeu T A, Burgess-Beusse B L, Finegold M J, Darlington G J. CCAAT/enhancer binding protein α regulates p21 protein and hepatocyte proliferation in newborn mice. Mol Cell Biol. 1997;17:7353–7361. doi: 10.1128/mcb.17.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Timchenko N A, Wilde M, Darlington G J. C/EBPα regulates formation of S-phase-specific E2F-p107 complexes in livers of newborn mice. Mol Cell Biol. 1999;19:2936–2945. doi: 10.1128/mcb.19.4.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Timchenko N A, Wilde M, Iakova P, Albrecht J H, Darlington G J. E2F/p107 and E2F/p130 complexes are regulated by C/EBPα in 3T3–L1 adipocytes. Nucleic Acids Res. 1999;27:3621–3630. doi: 10.1093/nar/27.17.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Timchenko N A, Wilde M, Nakanishi M, Smith J R, Darlington G J. CCAAT/enhancer-binding protein α (C/EBPα) inhibits cell proliferation though the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 68.Umek R M, Friedman A D, McKnight S L. CCAAT-enhancer binding protein: a component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- 69.Verona R, Moberg K, Estes S, Starz M, Vernon J P, Lees J A. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang S, Nath N, Minden A, Chellappan S. Regulation of Rb and E2F by signal transduction cascades: divergent effects of JNK1 and p38 kinases. EMBO J. 1999;18:1559–1570. doi: 10.1093/emboj/18.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watkins P J, Condreay J P, Huber B E, Jacobs S J, Adams D J. Impaired proliferation and tumorigenicity induced by CCAAT/enhancer-binding protein. Cancer Res. 1996;56:1063–1067. [PubMed] [Google Scholar]
- 72.Westin E H, Gallo R C, Arya S K, Eva A, Souza L M, Baluda M A, Aaronson S A, Wong-Staal F. Differential expression of the amv gene in human hematopoietic cells. Proc Natl Acad Sci USA. 1982;79:2194–2198. doi: 10.1073/pnas.79.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang D-E, Zhang P, Wang N D, Hetherington C J, Darlington G J, Tenen D G. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc Natl Acad Sci USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang P, Iwama A, Datta M W, Darlington G J, Link D C, Tenen D G. Upregulation of interleukin 6 and granulocyte colony-stimulating factor receptors by transcription factor CCAAT enhancer binding protein α (C/EBPα) is critical for granulopoiesis. J Exp Med. 1998;188:1173–1184. doi: 10.1084/jem.188.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zobel A, Kalkbrenner F, Guehmann S, Nawrath M, Vorbrueggen G, Moelling K. Interaction of the v- and c-Myb proteins with regulatory sequences of the human c-myc gene. Oncogene. 1991;6:1397–1407. [PubMed] [Google Scholar]