Abstract
Background & Aims
Colonic motor patterns have been described by a number of different groups, but the neural connectivity and ganglion architecture supporting patterned motor activity have not been elucidated. Our goals were to describe quantitatively, by region, the structural architecture of the mouse enteric nervous system and use functional calcium imaging, pharmacology, and electrical stimulation to show regional underpinnings of different motor patterns.
Methods
Excised colon segments from mice expressing the calcium indicator GCaMP6f or GCaMP6s were used to examine spontaneous and evoked (pharmacologic or electrical) changes in GCaMP-mediated fluorescence and coupled with assessment of colonic motor activity, immunohistochemistry, and confocal imaging. Three-dimensional image reconstruction and statistical methods were used to describe quantitatively mouse colon myenteric ganglion structure, neural and vascular network patterning, and neural connectivity.
Results
In intact colon, regionally specific myenteric ganglion size, architecture, and neural circuit connectivity patterns along with neurotransmitter-receptor expression underlie colonic motor patterns that define functional differences along the colon. Region-specific effects on spontaneous, evoked, and chemically induced neural activity contribute to regional motor patterns, as does intraganglionic functional connectivity. We provide direct evidence of neural circuit structural and functional regional differences that have only been inferred in previous investigations. We include regional comparisons between quantitative measures in mouse and human colon that represent an important advance in showing the usefulness and relevance of the mouse system for translation to the human colon.
Conclusions
There are several neural mechanisms dependent on myenteric ganglion architecture and functional connectivity that underlie neurogenic control of patterned motor function in the mouse colon.
Keywords: Gastrointestinal, Colonic Enteric Nervous System, Functional Neural Circuitry, Quantitative Morphology
Abbreviations used in this paper: 3D, 3-dimensional; 5-HT, serotonin; ANOVA, analysis of variance; ChAT, choline acetyltransferase; CMC, colonic motor complex; CGRP, calcitonin gene–related peptide; DMPP, dimethylphenyl-piperazinium; ENS, enteric nervous system; FOV, field of view; GABA, γ-aminobutyric acid; HS, horse serum; IPAN, intrinsic primary afferent neuron; MP, myenteric plexus; nAChR, nicotinic acetylcholine receptor; NOS, nitric oxide synthase; RNA-seq, RNA sequencing; VIP, vasoactive intestinal polypeptide
Graphical abstract
Summary.
Colonic motor complexes are neurogenic in nature but underlying mechanisms linking neural activity, neural network connectivity, and muscle activity have not been defined. We show that regional differences in functional connectivity in the enteric nervous system mechanistically underpin different colonic motor patterns.
See editorial on page 346.
The enteric nervous system (ENS) lies within the gastrointestinal tract and consists of 2 ganglionated neural plexuses, the myenteric plexus (MP) lying between the inner circular and outer longitudinal muscle layers, and the submucosal plexus located in the submucosal layer of connective tissue. The ENS controls a multitude of functions including muscle motor control (peristalsis), secretion, blood flow, nutrient transport, and immune responses.1, 2, 3, 4 Neural regulation of gastrointestinal function relies on integration of several intrinsic reflex and motor pattern–generating circuits that intersect with sympathetic, parasympathetic, and central nervous systems,2,5 as well as signals arising from mucosal epithelial cells.3,5 MP and submucosal plexus neurons are heterogeneous and have been classified based on morphologic, projection, neurochemical, and functional criteria, into at least 20 different neuron types.2,6, 7, 8 The current view of functional architecture of ENS neural circuitry is based on results primarily from guinea pig small intestine.2,9, 10, 11 Importantly, recent evidence has indicated significant differences between small intestine and colon, as well as between species.12, 13, 14, 15 Recent studies in mouse colon16,17 have suggested that colonic architecture defines mechanistically region-specific motor patterns,10,16,17 but the defining characteristics of ganglion architecture and functional connectivity have not been elucidated.
With the advent of molecular genetics, use of mouse models to investigate ENS function, architecture, and identification of disease-related genes, has become possible but has been largely confined to embryonic tissue samples. Our recent single-cell RNA sequencing18 identified genes differentially expressed in small intestine and colon in developing and adult mice that will aid in analysis of functional and quantitative anatomic data. This and other recently published RNA sequencing (RNA-seq) data sets18, 19, 20, 21 highlights the importance of in-depth quantitative morphologic, anatomic, and functional assessment if murine data are to be used to understand better motility disorders in human beings. Lacking this detailed knowledge diminishes the utility of increasingly large data sets. Thus, the goal of the present study was to define quantitatively functional connectivity, myenteric ganglion morphology, and architecture needed to understand how regionally distinct motor patterns are generated in the colon. One aim of the studies was to provide comparative data of the human colon22 to establish whether the results of morphologic, transcriptomics, and functional studies in the mouse could be used to identify disease-related genes relevant to human colonic health.
To this end, we took advantage of mice genetically engineered to express reporters in a large number of cell types to aid in identification of neurochemical code. Tiled confocal images enabled assessment of cell–cell connectivity patterning from proximal to distal colon and 3-dimensional (3D) reconstruction of innervation architecture. Functional studies combining GCaMP6 imaging with pharmacology and electrical stimulation were used to confirm functional neural circuits affecting region-specific colonic motor patterns. We identified a new intrinsic primary afferent neuron (IPAN), and show that myenteric neurons express macrophage markers. We also show how the patterning, size, and distribution of myenteric ganglia changes from proximal to distal colon, and provide evidence of intraganglionic connectivity. In total, linking our quantitative morphology with functional studies has allowed us to show that patterns of ganglion-to-ganglion connectivity are reflected in regional functional differences in the colonic neural connectome that underlies different motor patterns prevalent in proximal compared with distal colon.
Results
Myenteric Neuron Cross-Sectional Area and Volume
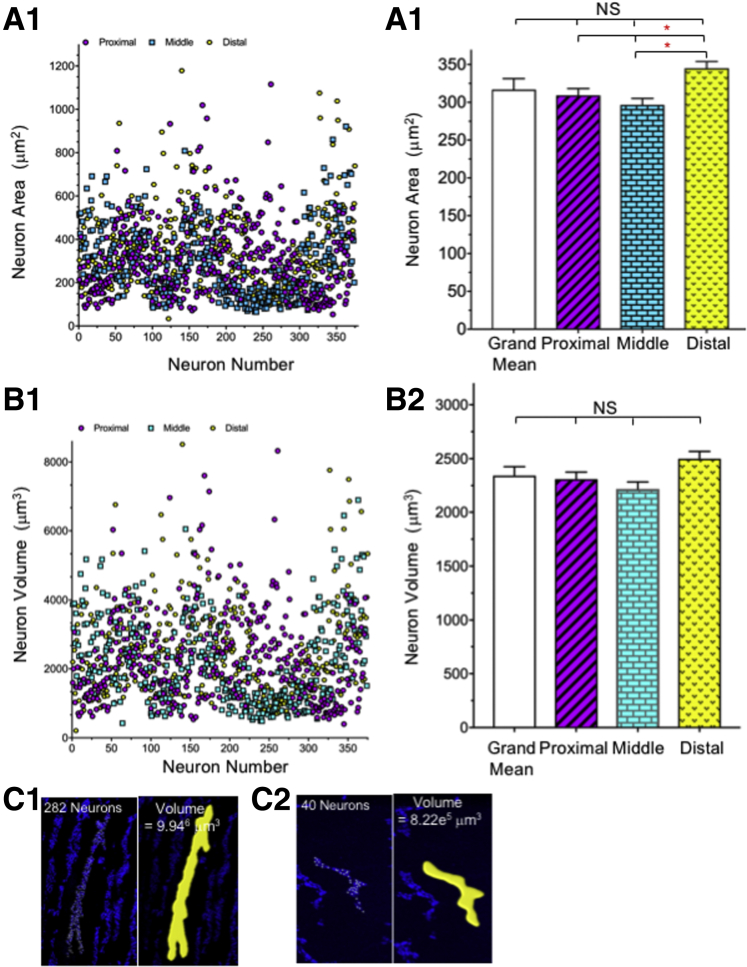
Our understanding of ENS neuron morphology and connectivity is based largely on post hoc tracings of neurons filled with dyes delivered from intracellular microelectrodes. A major limitation of this approach is that small or inaccessible neurons are difficult to impale and will go unidentified, while large surface neurons will present easier targets and therefore be over-represented. To overcome this limitation, we measured the cross-sectional area (Figure 1A) and volume (Figure 1B) of myenteric neurons and ganglia (Figure 1C) as the first step in completing a comprehensive quantitative map of the mouse colonic MP. Neuron soma sizes were measured in 2 ways: the diameters in 3 dimensions of 1121 neurons were measured in randomly chosen ganglia using the Leica (Bannockburn, IL) Application Suite of tools, and neurons isolated from MP, fluorescence-activated cell sorter–sorted. and compared with sorted size-specific beads. The 2 methods yielded comparable results, showing a broad range of small and large neuron soma sizes (Figure 1A and B). Individual neuron soma volume ranged from 243 to 8511 μm3, with a mean value of 2342 ± 83 μm3; neuron areas ranged from 34 to 1179 μm2 (mean, 317 ± 12 μm2). There was a small, but significant, difference in cross-sectional area (Figure 1A2) in the distal colon; if one considers volume (Figure 1B2), neurons are distributed randomly by size in ganglia but the number of neurons per ganglion and ganglion distribution significantly changes from proximal to distal colon.
Figure 1.
Myenteric neuron area, volume, and ganglionic volume. (A1 and A2) Cross-sectional area was measured manually in 1121 neurons (372 proximal, 374 middle, and 375 distal) from 3 wild-type and 9 ChAT reporter mice immunostained for the pan-neuronal marker HuC/D and antibody combinations recognizing neurofilaments, calbindin, calretinin, GABA, and NOS. Neuron volume (B1 and B2) was measured in the same neurons as panel A; there was no difference in neuron volume in 3 (Figure 2B1–B3) colonic regions, but a significant difference in area comparing the proximal and middle colon with the distal colon (A2). The statistics (1-way ANOVA) indicate that there is a small but significant difference in cross-sectional area between the proximal and middle colon compared with the distal colon, which is likely owing to the number of neurons included in the data set. Because there is no difference when comparing neuron volume and there is no difference in any region based on mean neuron area, we conclude that there is no regional difference in cell size. (C) Ganglion volume, in ChAT or NOS reporter mice immunolabeled with the neuronal marker, HuC/D, was measured using Imaris software (Bitplane, Belfast, United Kingdom) according to the manufacturer’s protocol; (C1) proximal colon (Figure 2D2), and (C2) distal colon (Figure 2D7).
Myenteric Ganglion Size and Distribution
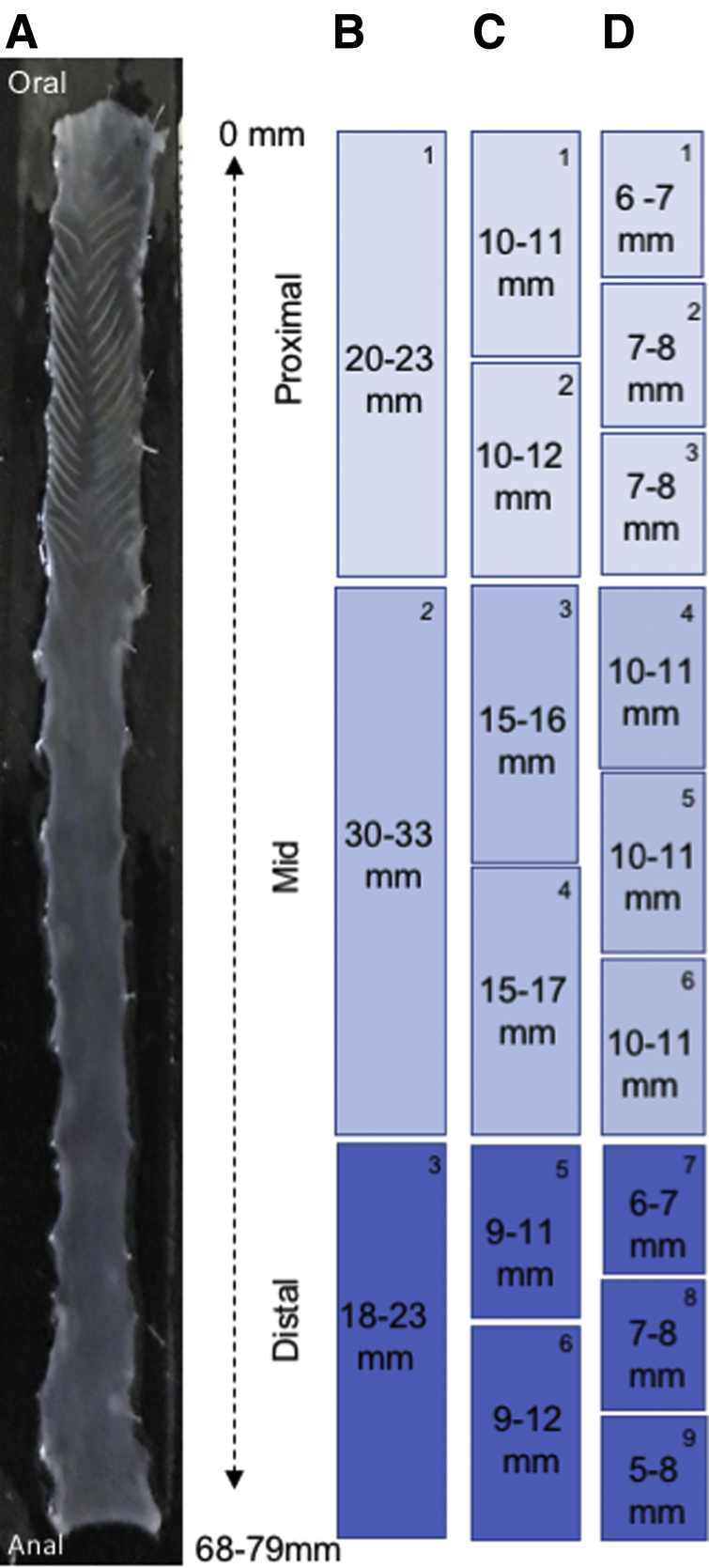
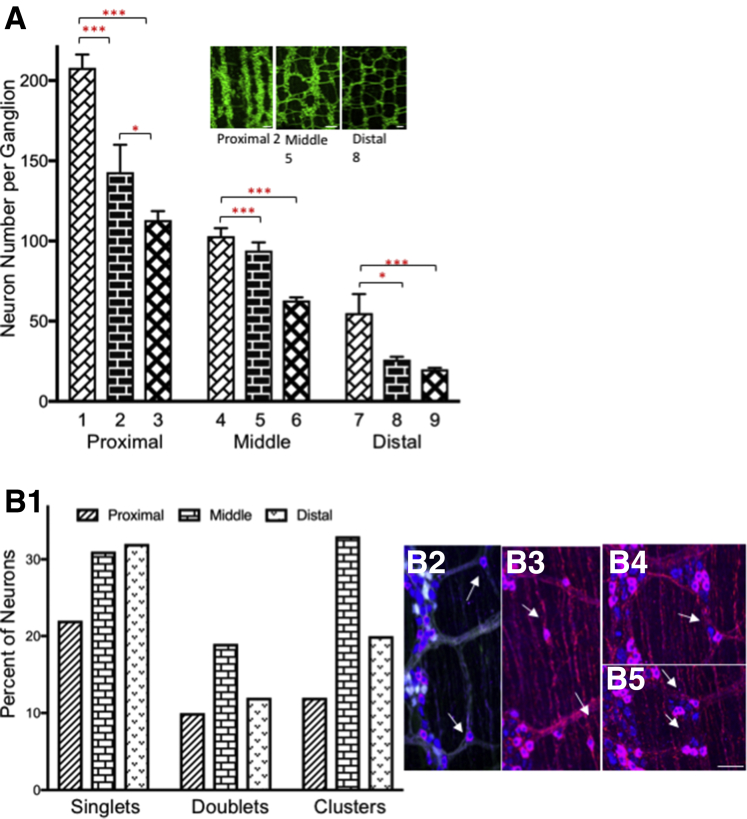
The number of neurons per ganglion was counted manually in 9 colon regions (Figure 2D). There was a significant change in neuron number per ganglion, progressing from proximal to distal colon (Figure 3A). The number of neurons decreased from 303 neurons per ganglion, the maximum counted in proximal colon area D1 (Fig. 2) (mean range in proximal colon D1 to proximal colon D3, 208 ± 8 to 113 ± 6) to 3 neurons per ganglion in distal colon (mean range in distal colon area D7 to distal area D9, 55 ± 12 to 20 ± 1). The number of neurons per ganglion in the transition zones from proximal area D3 to midcolon area D4 (113 ± 6 to 103 ± 5) and from midcolon area D6 to distal colon area D7 (63 ± 2 to 55 ± 12), showed a consistent but gradual decrease in ganglion size. The incidence of isolated neurons occurring as single-cell increases from proximal to middle and distal colon (Figure 3B1–B3). The percentage of doublet sets of neurons was highest in the midcolon (Figure 3B1 and B4), while groups of 3–5 neurons (clusters) was highest in the middle and distal colon (Figure 3B1 and B5). Individual neurons were found within intermodal strands (Figure 3B2) and circular muscle (Figure 3B3).
Figure 2.
Location and size of dissections for colon mapping. (A) Cleaned and pinned colon showing the entire colonic length. Colon length varied from 68 to 79 mm. (B–D) The colon was dissected into the sizes and locations shown to maintain uniformity for quantitative measurements. Each colon piece is numbered in the upper right corner of each box. These numbers are used to identify where samples came from for each subsequent Figure where appropriate. Each scheme is numbered starting at the same proximal colon oral site. Each set starts with the number 1 because it indicates that all 3 sectioning schemes start from the same place and show how many sections were taken in each scheme. The long section in scheme B comprises numbers 1 and 2 from scheme C and numbers 1, 2, and 3 from scheme D. The subsections in schemes C and D are smaller portions of the total region shown in scheme B. In scheme C, section 1 from scheme B is divided into 2 section, and in scheme D, it is divided into thirds.
Figure 3.
Myenteric neuron numbers. (A) Neuron number per ganglion was counted in 9 regions of colon (Figure 2D1–D9) of ChAT reporter mice immunostained for HuC/D and calbindin, calretinin, or NOS. Minimally, 50 ganglia were counted for each region. (A) Data were analyzed using 1-way ANOVA and the Tukey multiple comparison test. ∗P < .05, ∗∗∗P < .001. There was no significant difference between regions at the transition zone. Inset: ganglion size for proximal (Figure 2D2), middle (Figure 2D5), and distal colon (Figure 2D8) in wild-type reporter mice. (B) The number of neurons appearing as single isolated cells (B1 and B2), doublets (B1 and B4), or groups of 3–5 (B1 and B5), was counted in the proximal (Figure 2B1) middle (Figure 2B2), and distal colon (Figure 2B3) of samples immunolabeled with antibodies specific for enhanced green flourescent protein or TdTomato, calretinin, or calbindin and HuC/D. (B1) The percentage of neurons is shown for the proximal, middle, and distal colon; the bar symbols are defined on the graph. Scale bars: 100 μm (A, insets), and 50 μm (B2–B5).
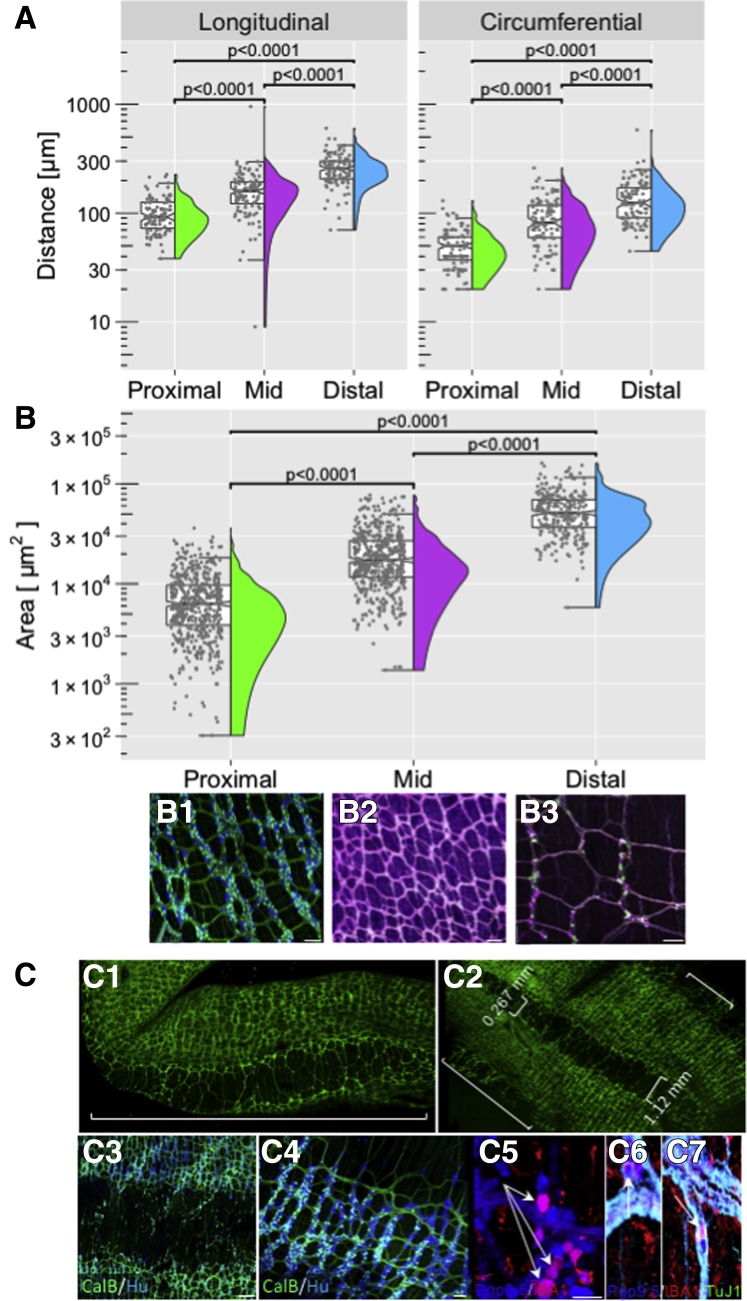
The mean longitudinal distance between ganglia increases from 95 μm in proximal to 250 μm in distal colon (Figure 4A). There was a significant increase in longitudinal distance from the middle (149 μm) to distal colon (250 μm). The average circumferential distances were lower, but they increased as well from 46 to 127 μm from proximal to distal colon (Supplementary Tables 1 and 2).
Figure 4.
Colon morphology: quantitative and descriptive. (A) Longitudinal and circumferential distance between myenteric ganglia was measured by hand on confocal projections using the Leica suite of tools, on 3 samples from each region (Figure 2D, all regions) and 123 measurements in each direction. (B) The interganglionic area was measured by hand in 4 samples of proximal (Figure 2B1), middle (Figure 2B2), and distal (Figure 2B3) colon on confocal projections with a minimum of 609 measurements for each area. (B1–B3) Examples of images that were quantified. (C) Whole-mount (C1) confocal image of proximal (Figure 2B1) colon from a Wnt-YFP reporter mouse showing taenia and intertaenia region (white bracket). (C2) Proximal colon from C1, cut along the mesenteric border and the width of the intertaenia region was measured as shown (white brackets). (C3 and C4) Myenteric ganglia get smaller toward the intertaenaie region. (C5 and C6) Proximal colon (Figure 2C2) immunolabeled for Protein Gene Product9.5 (PgP9.5) and Ionized calcium-binding molecule 1 (IBA1) show immune markers in myenteric neurons (arrows). (C6) co-labeled neurons (PgP9.5, TuJ1, IBA1) in the intertaenia region (arrows). Scale bars: 100 μm (B1), 150 μm (B2), 100 μm (B3), 200 μm (C3), 100 μm (C4), and 50 μm (C5).
Similarly, the mean interganglionic area increased from 5876 to 49,802 μm2 from proximal to distal colon (Figure 4B, Supplementary Table 3). Some of these quantitative changes may be owing to the presence of taenia in proximal colon (Figure 4C1–C3). There were no taenia in the middle or distal colon, but in the proximal colon the intertaenia distance increased (Figure 4C1 and C2) as the mucosal folds dissipated toward the midcolon on average from 0.267 ± 0.08 mm (12 measurements in 3 animals) to 1.12 ± .07 mm (23 measurements in 6 animals). There were few ganglia in the intertaenia domain but there were scattered neurons (Figure 4C3, C6, C7); the number of neurons per ganglion decreased as one approached the intertaenia region in the circumferential direction (Figure 4C4). The intertaenia region was heavily invested with macrophages. Data from RNA-seq (Marthe J. Howard, unpublished data) indicated that some neurons also express macrophage markers, which we confirmed (Figure 4C5–C7). Using the macrophage-specific antibody IBA1 (ionized calcium-binding adaptor molecule 1), which labels enteric macrophages,23 we identified myenteric neurons co-expressing the neuronal marker PgP9.5 and ionized calcium-binding adaptor molecule 1 (IBA1) (Figure 4C5–C7).
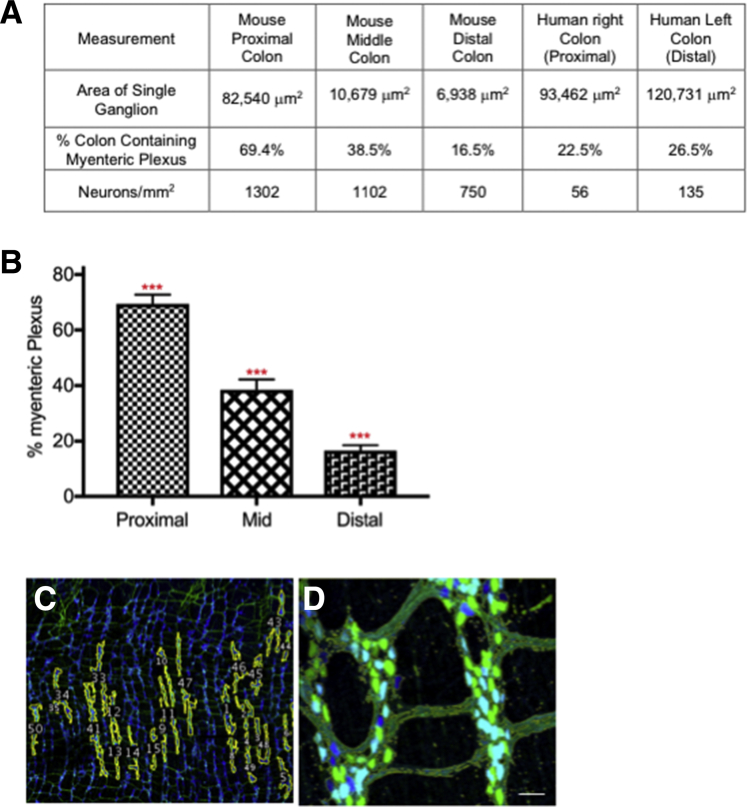
From proximal to distal colon, the average ganglionic area decreased from 82,640 μm2 in the proximal colon to 6.938 μm2 in the distal colon (Figure 5A and Supplementary Table 4). The percentage of the colon occupied by the myenteric plexus (ganglia plus internodal strands) was 69.4% ± 3.3% in the proximal, 38.5% ± 3.7% in the middle, and 16.5% ± 2% in the distal colon (Figure 5B and Supplementary Table 5. The number of neurons per square millimeter was calculated to be 1302 in the proximal colon, 1102 in the middle colon, and 750 in the distal colon (Figure 5A); these values were used to calculate the total number of colonic neurons: 361,416 (proximal), 331,250 (middle), and 145,746 (distal), yielding a total of 838,412 neurons. For comparison, data from the human22 right (ascending [proximal] and left (descending [distal]) colon (Figure 5A and Supplementary Table 6), are provided.
Figure 5.
Myenteric plexus quantitative measures: mice compared with human beings. Comparison of the colon measurements from the proximal, middle, and distal colon obtained in mice with those from human beings.22 (A) Myenteric ganglion area and volume were measured on confocal image stacks in ImageJ using the thresholding method. Data from the human samples were published previously.22 The right colon, also referred to as the ascending colon, is the beginning part of the colon and is equivalent to the mouse proximal colon. The left, or descending, colon begins at the splenic flexure and ends at the sigmoid colon and is equivalent to the mouse distal colon. The terms used in the table (right and left colon) are the same as those in our original article22 and are maintained for clarity. Ganglion area in the mouse proximal colon most approximates that of the human right colon. Overall ganglion packing in the human is best modeled by the mouse middle colon. There are significantly more neurons per square millimeter in the mouse compared with human beings. (A and B) To measure ganglion area and volume in mice, (B–D) ganglia were outlined by hand and then area and volume measures were calculated automatically. (C) Example showing the myenteric plexus outlined in yellow in thresholded sample. (D) Example of regions of interest around myenteric ganglia used for ganglion area measurements. All measurements were performed on tissue derived from the colon as described in Figure 2B1–B3. (C and D) Scale bars: 50 μm.
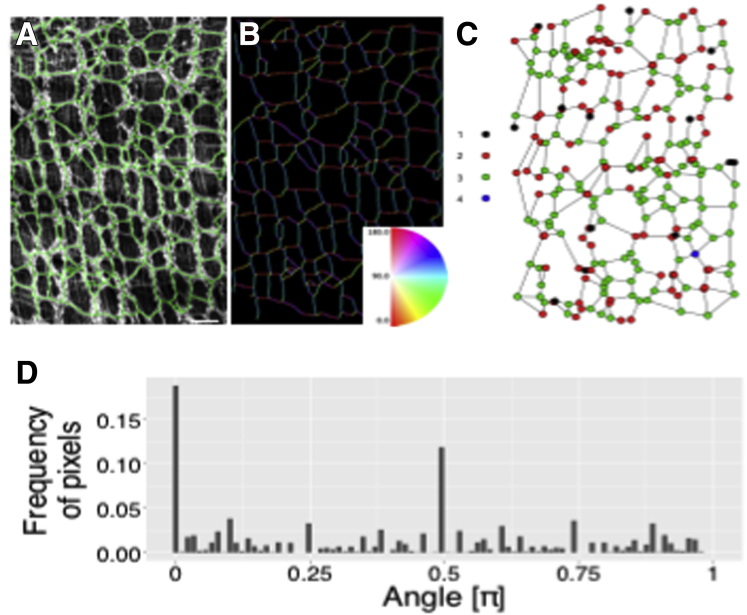
Image processing and graph analysis were used to quantify the organization of observed ENS network architecture24,25 (Figure 6). A random subset of segmented images was subjected to directionality analysis using Fiji software (National Institutes of Health, Bethesda MD) and Directionality plug-in (ImageJ, National Institues of Health, Bethesada MD) to find the dominant directions of the interganglionic connections (Figure 6A–C). Subsequently, the segmented networks were analyzed as planar graphs, with ganglion centers considered vertices and the simplified representations of interganglionic connections serving as the edges (Figure 6C). The secondary directionality analysis was performed using the edges (Figure 6B, inset, and C). This graph-based approach run in a scale-independent manner was complementary to the statistics describing ganglia and interganglionic connections. The analysis showed that the neural networks were not randomly structured but highly organized in terms of their vertex positions (Figure 6B and C, inset), the orientation of the interganglionic connections, and the number of formed connections between vertices. Both approaches to directionality analysis (bitmap-based and graph-based) repeatedly showed 2 dominant orthogonal directionalities (0/180º [π] and 90º [π/2]) (Figure 6D), rather than a uniform distribution of angles, confirming that sampled networks were arranged in a lattice-like fashion. The agreement between the results found using the intervertex edges and the directionalities computed using image-processing methods using bitmaps and gradient operators indicated that most interganglionic connections do not deviate from straight lines. These results show that the observed networks’ structure resembles a semiregular grid.26,27
Figure 6.
The organization of observed ENS network architecture quantified. A representative example of a (A and B) binarized network, (B) directional analysis image, and (C) an extracted graph that shows the valences of the vertices. (D) The computed distribution of orientations shows 2 dominating directions (0 and π/2). The colon region was as shown in Figure 2B2 and 2B3. (A) Scale bar: 100 μm.
Neurochemical Code of Mouse Colonic Neurons
We analyzed the colonic neurochemical code using identified neurotransmitters or neuromodulators previously used to define neuronal cell type in guinea pig6, 7, 8,15,28 and mouse intestine29,30 combined with additional markers and counted relative to the total number of neurons per ganglion from proximal to distal colon (Table 1). Excitatory motor neurons were classified and counted based on co-expression of choline acetyltransferase (ChAT), calretinin, γ-aminobutyric acid (GABA), and substance P. Although we identified neurons with projection patterns indicating innervation of both circular and longitudinal muscle, we did not quantify these as separate populations. There was no significant difference in the percentage of excitatory motor neurons or IPANs in proximal, middle, and distal colon. From proximal to distal colon there was an increasing number of ganglia with few to no ChAT-expressing neurons. Ganglia not expressing ChAT or nitric oxide synthase (NOS) were excluded in counts so the percentages of cells classified based on neurochemical code do not include ganglia that express neither NOS nor ChAT. We estimate that in the distal colon, approximately 10% of ganglia do not contain neurons expressing ChAT or NOS; this coincides with what we have reported in human colon22 and what has been reported previously for mouse intestine.31 We did not observe ganglia lacking ChAT or NOS in the middle or proximal colon.
Table 1.
Neurochemical Code of Myenteric Neurons in Mouse and Human Colon
| Neuron type chemical code | Total neurons, % |
Colon, % | ||
|---|---|---|---|---|
| Proximal | Middle | Distal | ||
| Excitatory Motor Neuron ACh/GABA/CalR/SubP |
18 ± 3 (N = 26) | 14 ± 2 (N = 23) | 17 ± 1 (N = 71) | 16 ± 2 |
| Inhibitory Motor Neuron NOS/GABA/VIP NOS/VIP |
15 ± 3 (N = 21) | 24 ± 2 (N = 22) | 27 ± 2 (N = 58) | 22 ± 2 |
| Intrinsic Primary Afferent Neuron ACh/CalB/CalR/SubP |
25 ± 3 (N = 50) | 17 ± 2 (N =39) | 20 ± 2 (N = 72) | 21 ± 2 |
| Descending Interneuron type I ACh/NOS/VIP |
11 ± 3 (N = 32 | 6 ± 1 (N = 32) | 10 ± 2 (N = 36) | 9 ± 2 |
| Descending Interneuron type III ACh/GABA/Som |
4 ± 1 (N = 10) | 10 ± 3 (N = 10) | 10 ± 1 (N = 27) | 8 ± 2 |
| Ascending Interneuron ACh/CalR/SubP/Enk |
5 ± 1 (N = 18) | 8 ± 1 (N = 31) | 9 ± 1 (N = 57) | 7 ± 1 |
| Comparison of mouse with human | ||
|---|---|---|
| Chemical code | Mouse | Human22 |
| NOS+/ChAT- | 22% | ∼50% |
| ChAT+/NOS- | 47% | 36%–56% |
| ChAT+/NOS+ | 8% | 4%–6% |
| Neither ChAT nor NOS | 10%–20% | 14%–19% |
NOTE. The neurochemical code was determined in 3 regions (Figure 2B1–B3) of the mouse (top) and total human colon (bottom) based on manual cell counts of neurons immunolabeled (as shown); HuC/D was used as a pan-neuronal marker. N equals the number of ganglia counted. Human data were extracted from Graham et al.22 The nomenclature matches that used by Qu et al.30
CalB, calbindin; CalR, calretinin; DIN, descending interneuron; SubP, substance P.
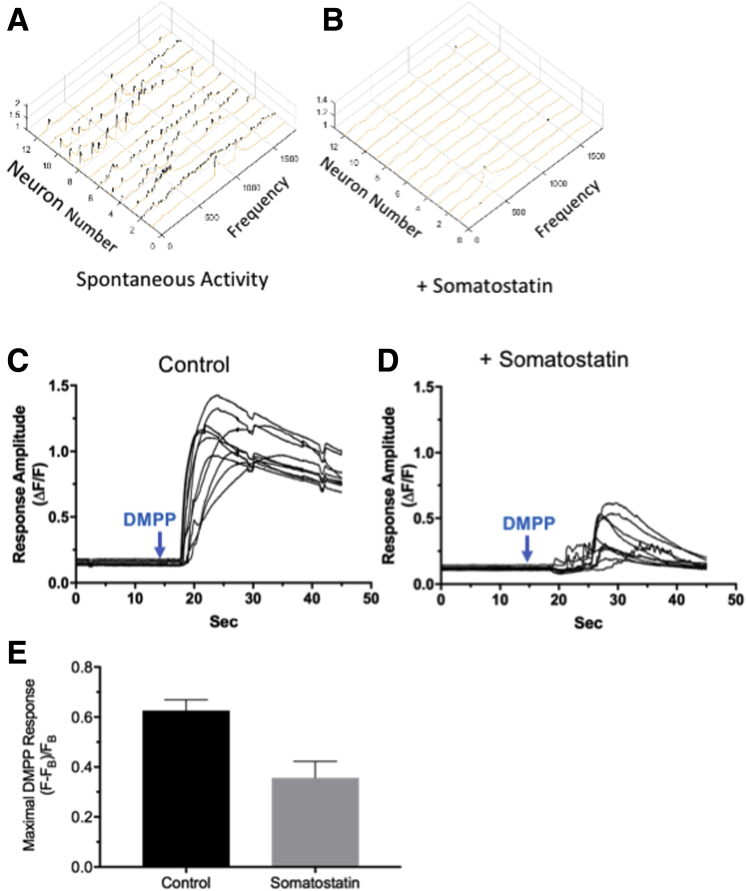
There is a significant increase in the percentage of inhibitory motor neurons from proximal (15% ± 3%) to distal (27% ± 2%) colon (P < .01) (Table 1). We distinguished 2 types of descending interneurons. The first type co-expresses ChAT, GABA, and somatostatin (descending interneuron type III30) and is expressed in a higher percentage (P < .01) of ganglia from the proximal to distal colon (Table 1). Because the localization and function of somatostatin is not well established in the murine colon, we assessed somatostatin function in mice we engineered to express the sensitive fast Ca2+ indicator, GCaMP6f, in neurons expressing the acetylcholine biosynthetic enzyme ChAT. This allowed rapid changes in Ca2+ fluorescence to be detected without disrupting intrinsic inputs exclusively in cholinergic myenteric ganglion neurons (Figure 7). Somatostatin (0.65–1.00 μmol/L) strongly depressed both spontaneous Ca2+ transients (Figure 7A and B) and responses to the nicotinic acetylcholine receptor (nAChR) agonist, dimethylphenyl-piperazinium (DMPP) (10 μmol/L), in ChAT+/GCaMP6f+ colonic myenteric neurons (Figure 7C–E). Somatostatin depressed the frequency (f) of Ca2+ transients (Figure 7A compared with 7B) per neuron without an appreciable effect on amplitude or kinetics (rise and decay times), but depressed the amplitudes of DMPP-evoked responses (Figure 7C compared with 7D) per neuron by approximately 50% (Figure 7E).
Figure 7.
Somatostatin affects spontaneous and induced myenteric neuron activity. Somatostatin (0.65–1.00 μmol/L) strongly depressed both spontaneous Ca2+ transients and responses to DMPP in Chat+/GCaMP6f+ colonic myenteric neurons. (A and B) A 3D plot showing that somatostatin strongly depresses the frequency of spontaneous Ca2+ transients recorded from ChAT+/GCaMP6f+ colonic myenteric neurons without an appreciable effect on amplitude or kinetics (rise and decay time). Ca2+ transients from 1800 simultaneous recordings from 12 neurons are shown (A) before and (B) 20 minutes after application of somatostatin. (C and D) Somatostatin depresses DMPP-evoked Ca2+ responses recorded from Chat+/GCaMP6f+ colonic myenteric neurons. Simultaneous responses evoked by rapid puffer perfusion of DMPP from 10 neurons are shown (C) before and (B) 20 minutes after bath application of somatostatin. (E) Somatostatin depresses the response to DMPP by 50%. Ca2+ transients were measured in 50 control neurons and 45 neurons treated with somatostatin (N = 3). Induced activity was measured in 30 control (+DMPP) and 30 treated (DMPP + somatostatin) neurons (N = 2) in the proximal colon (Figure 2B1).
The second type of descending interneuron co-expressed ChAT, NOS, and vasoactive intestinal polypeptide (VIP); the percentage of these neurons is lower in the midcolon compared with either the proximal or distal colon. Ascending interneurons, co-expressing ChAT, substance P, enkephalin, and/or calretinin, were distributed fairly evenly along the entire colon, albeit with a lower, but not statistically significant, percentage in the proximal colon. A comparison of mouse with human colon22 highlights the similarity in the percentage of neurons classified on the basis of ChAT and/or NOS expression (Table 1).
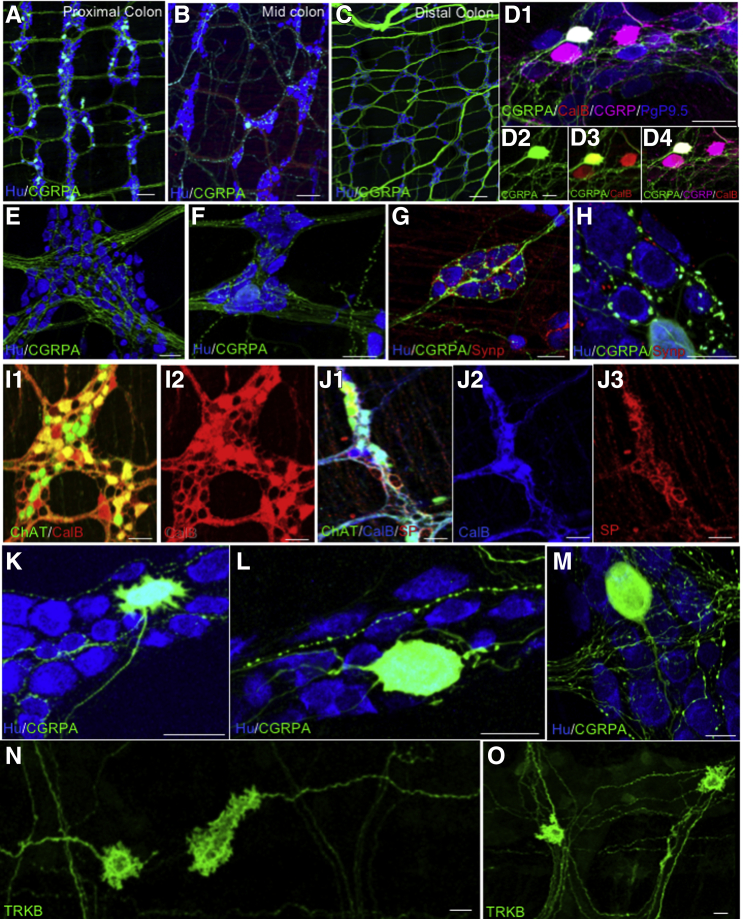
In addition to determining the percentage of neuron types based on neurochemical code, we identified a number of marker combinations and individual markers not detected previously, to our knowledge, in the colon of mice or other species (Table 2). Importantly, neurons expressing calcitonin gene-related peptide A (CGRPA) were identified in proximal and midcolon albeit in low numbers32,33 (Figure 8A–H); many ganglia have none of this cell type although we rarely found more than 4–10 per ganglion. With one exception, this cell type was absent from the distal colon. CGRPA+ neural processes run in the intrinsic neural network in the proximal and middle colon, but appear randomly organized and mostly do not run in the intrinsic neural network in the distal colon (Figure 8A–C). Using commercial anti-CGRP antibodies, 78% ± 6% of CGRPA-reporter neurons (Figure 8D1–D3) co-label, and, depending on the antibody used, 45%–100% also co-label with anticalbindin antibodies commonly used as a marker for IPANs (Figure 8D1–D4). Approximately 20% of CGRPA neurons express substance P and 100% express calretinin; in addition to IPANS, substance P and calretinin are co-expressed in excitatory motor neurons and ascending interneurons. This cell type tentatively is identified as an IPAN(A). The connectivity patterns (Figure 8E–J) coincide with basket structures described previously in distal colon,34, 35, 36 but connectivity is more complicated and diverse (Figure 8E–H). Comparison of IPANs with IPANAs shows that basket structures contain both calbindin and substance P (Figure 8I1 and I2); substance P forms more dense structures around neurons that are ChAT+, but do not express calbindin (Figure 8J1–J3). Co-labeling to localize the synaptic marker protein synaptophysin showed a large range of synaptic sites on the IPANA neurons (Figure 8G and H). We identified CGRPA Dogiel type II (Figure 8L) neurons as expected11,29,37,38 of intrinsic sensory neurons, but also several unexpected morphologies39 as well (Figure 8K–M). Many of these CGRPA neurons have only 1 apparent axon but smooth somas and are adendritic.37, 38, 39 There are neurons with short lamellar dendrites and more than 1 axon37, 38, 39 (Figure 8K). All of these characteristics have been reported for DG II neurons, but neurons with large smooth round or oval cell bodies and 1–2 long tapering axons are most common.
Table 2.
New Chemical Code Combinations in Mouse Colon
| Neurochemical code |
|---|
| ChAT+NOS+VIP- |
| VIP+ChAT-NOS- |
| ChAT+CalB+CalR-SubP- |
| CGRPA+ |
| ChAT-CalR+ |
| ChAT-NOS-SubP+ |
| NOS+CalB+ |
| ChAT-GABA+CalR+ |
| ChAT+CalB-ENK+ |
NOTE. We identified 11 new neurochemical marker combinations in mouse colonic myenteric neurons. Some neurons were identified that expressed neither ChAT or NOS, and that had 1 bona fide marker, as shown. These neurons were identified in all regions of the colon.
CalB, calbindin; CalR, calretinin; ENK, enkephalin; SubP, substance P.
Figure 8.
Myenteric neuron identity and morphology. (A–C) Neurons expressing CGRPA in the (Figure 2B1) proximal and (Figure 2B2) middle colon, (Figure 2B3) with few to none in the distal colon. CGRPA fibers in the distal colon likely arise from extrinsic dorsal root ganglion afferents. (D1) CGRPA neurons express (D3) calbindin as do (D1–D4) IPANs and (D1 and D4) CGRPA and CGRPB neurons are labeled with CGRP-specific antibody. (E–H) CGRPA axons are (E and F) mostly smooth, but those that ramify within ganglia and circumferentially (F) are varicose. (G) Clusters of neurons surrounded by networks of CGRPA fibers associated with synaptophysin-labeled (localizes synapses) terminals. (H) Some CGRPA terminals surrounding myenteric neurons appear as modified calyxes. (I1 and I2) Calbindin forms basket-like structures around neurons expressing ChAT (likely IPANS) and (J1–J3) some of which do not express ChAT but that do express substance P; the identity of this neuron is unknown. Substance P also forms dense basket-like structures around ChAT + calbindin-expressing neurons, which could be IPANS or excitatory motor neurons, and around neurons that do not express ChAT. The identity of this neuron is unknown but the basket structure likely arises from an IPAN. (K–O) CGRPA neuron morphologies; (L) Dogiel Type II, (M) adendritic with a single process, and (K) neurons with lamellar dendrites but with more than 1 process. (N and O) Filamentous tyrosine receptor kinase B (TrkB) reporter neuron fibers run (N) longitudinally and (O) circumferentially. (D–O) Samples from colon areas B1–B4 (Figure 2). Scale bars: 100 μm (A–C), 50 μm (D1–K), and 25 μm (L–O).
In tyrosine receptor kinase B reporter mice, we identified filamentous neurons (Figure 8N and O) that, based on morphologic criteria, appear to be intestinofugal neurons31,40 akin to those identified in the guinea pig. The majority are uni-axonal and have somewhat modified Dogiel type I morphology11,39 (Figure 8N and O); these neurons have both short and long branched lamellar dendrites and do not express either calbindin or NOS.
Ganglion Organization, Neurotransmitter-Receptor Expression, and Connectivity Arrangements Underlie Function of Mouse Myenteric Ganglia
Because of the diversity in myenteric ganglion organization and colonic motor patterns, we analyzed connectivity configurations (Figures 9 and 10), functional neurotransmitter-receptor expression (Figure 11), and functional connectivity (Figure 12) in the myenteric plexus.3,7,8,10,41, 42, 43
Figure 9.
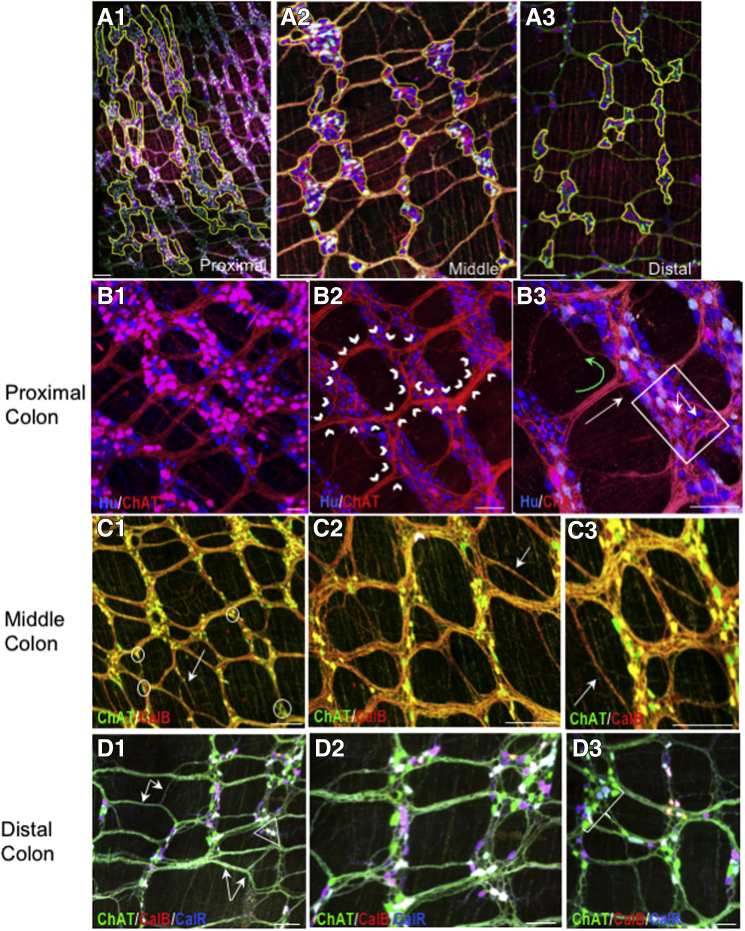
Myenteric ganglia connectivity patterns change from proximal to distal colon. (A1–A3) Examples showing syncytial patterning of myenteric ganglia in proximal (Figure 2C1 and C2) vs patterning of the middle (Figure 2C3 and C4) and distal (Figure 2C5 and C6) colon. The yellow regions of interest outline several ganglia in each region. Pictures are snapshots of confocal image stack projections. (B1–B3) In the proximal colon a ramifying pattern of interganglionic and intraganglionic connectivity (outlined by white chevrons) appears specific to this region. (B3) In the proximal colon there is significant longitudinal (white arrow) and circumferential (green curved arrow) connectivity. (B3) Basket structures (white box) are evident. (C1-C3) In the midcolon, (D1) there is increased connectivity between internodal strands (white arrows and double arrow), often bypassing ganglia (white arrow in C1). (D1–D3) The connectivity pattern in the distal colon has increasing complexity with multiple processes from 1 ganglion converging on single ganglia (bracket in D3). Small triangular ganglia often form the interstices of connection points (circles in C1 and triangles in D1 and D3). Note that cells appearing white co-express ChAT, CalB, and CalR. (B2) The relative lack of ChAT cell bodies visible is owing to unevenness of the sample and not a lack of ChAT-expressing neurons. Scale bars: 100 μm (A1–C3), 50 μm (D1–D3). CalB, calbindin; CalR, calretinin.
Figure 10.
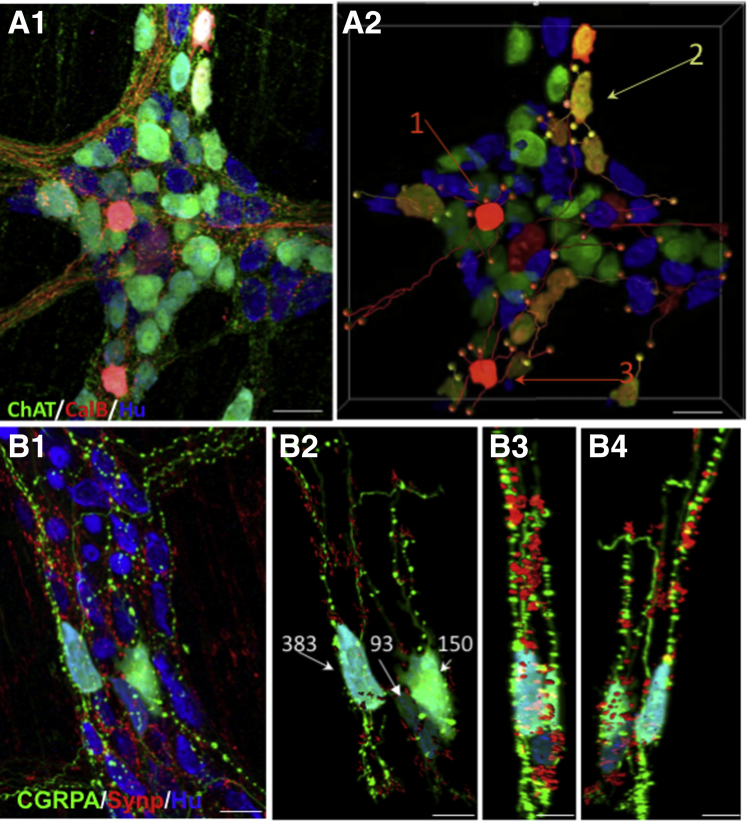
3D reconstruction of intraganglionic connectivity and synaptic sites. (A1) Intraganglionic connectivity was confirmed based on (A1) confocal imaging and (A2) 3D reconstruction using Imaris (Bitplane, Belfast, United Kingdom). The ganglion shown is from colon area in Figure 2D4. Local cell–cell contact is shown for 3 neurons (arrows). Cell #1 has close contacts (orange balls) with 4 cells. Cell #2 has 4 close contacts (yellow balls), and cell #3 has close contacts on 3 neurons with multiple sites on at least 1 (orange balls). Each of these cells expresses calbindin and ChAT, suggesting that they are IPANs. Immunolabeling with the presynaptic marker synaptophysin, confirms the presence of en passant synapses (B3 and B4). (B1) CGRPA neurons have prominent varicosities and (B2) variable numbers of synaptophysin contacts (B3 and B4) on both their somas and axons. (B1–B4) Images are from the proximal colon area D4 (Figure 2). Scale bars: 50 μm (A1 and A2), and 25 μm (B1–B4).
Figure 11.
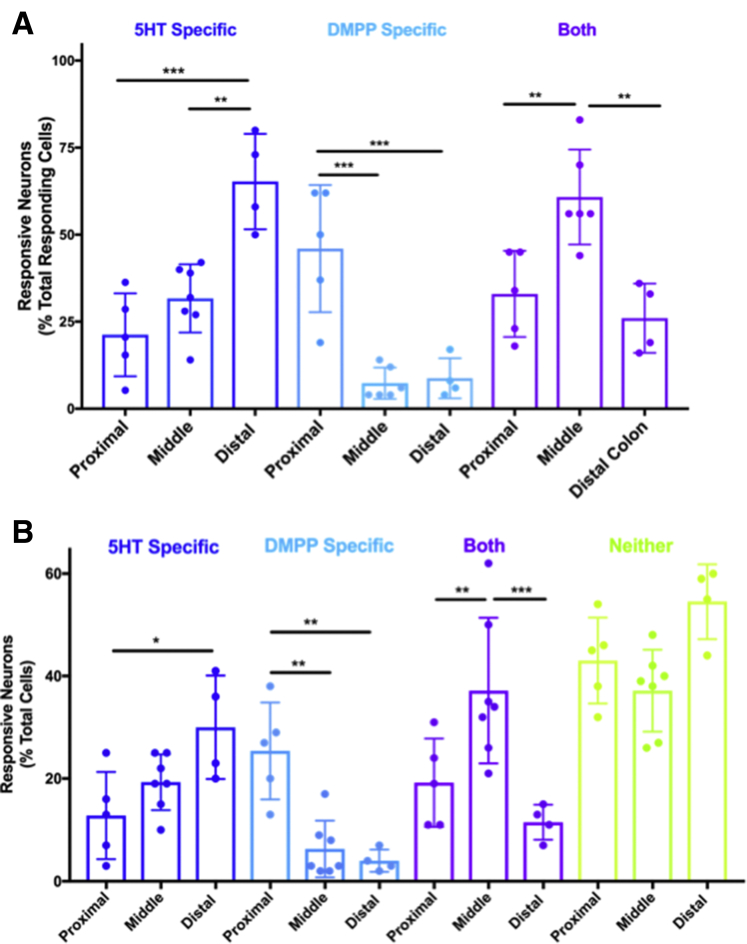
Differential distribution of myenteric neurons expressing nACh receptors and/or 5-HT-3 receptors. (A) Graphic summary of response profiles in the proximal (Figure 2B1), middle (Figure 2B2), and distal (Figure 2B3) colon of the total responding neurons. Graph shows the percentage of neurons that only responded to 5-HT (5-HT-specific, dark blue), only to DMPP (DMPP-specific, light blue), or to both agonists (Both, purple). (B) Graphic summary of response profiles in the proximal, middle, and distal colon of total neurons in the field of view. Graph shows the percentage of neurons that only responded to 5-HT (5-HT-specific, dark blue), only to DMPP (DMPP-specific, light blue), to both agonists (Both, purple), or to neither agonist (Neither, green). For each field of view, three 20-s movies were collected; spontaneous activity was imaged first and then during application of 5-HT (10 μmol/L, H9523; Sigma) and DMPP (10 μmol/L, D5891; Sigma) delivered via 2 glass pipettes placed directly above and adjacent to the imaging field on either side of the objective. Pilot studies using dye were used to confirm that the contents reached the entire field of view. The order of agonist application was randomized, as well as the region of colon tested, and the first agonist was washed-out for 10 minutes before application of the second. Amplitudes of GCaMP6s signals were analyzed by calculating dF/F0 (% = [(F-F0)/F0] × 100), where F is peak fluorescence and F0 is the average fluorescence before agonist application; dF/F0 of four standard deviations greater than baseline was considered a response. Only neurons identified in all 3 imaging fields were included for analyses. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 using repeated-measure 2-way ANOVA. n = 4–6 fields from N = 3 mice. (A) Data for the proximal colon have appeared elsewhere in different form (see Margiotta et al44).
Figure 12.
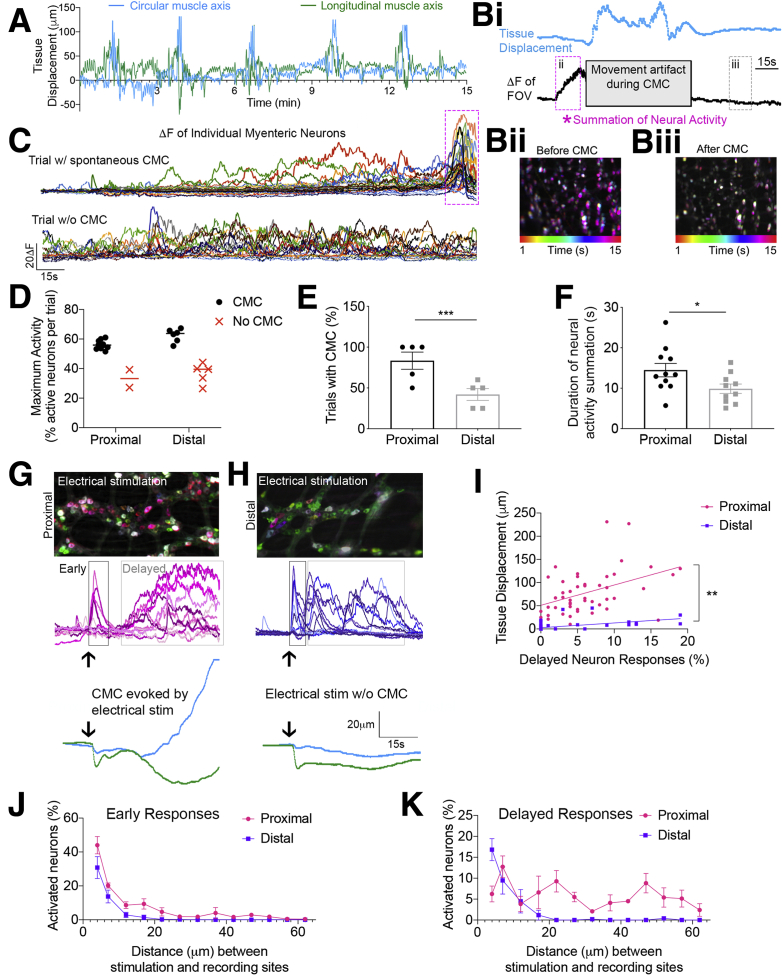
Neural circuitry differentially supports CMC activity in the proximal compared with distal colon. (A) Example traces of tissue displacement in the circular (x, blue) and longitudinal (y, green) muscle axes, measured using the ImageJ plugin Template Matching, show regularly occurring CMCs. (Bi) Traces of tissue displacement (blue) and average GCaMP6s+ signal of the entire FOV (black) show a coordinated increase or summation of myenteric neuron activity (pink asterisk and box) that precedes a CMC. (Bii and Biii) Time-lapse color-coded images of 15 s of neural activity (ii, left) before and (iii, right) after CMC. Note the numerous neurons that show maximal activity at the same time just before the CMC (indicated by magenta). (C) GCaMP6s+ traces of 3-minute recordings of spontaneous activity in the proximal colon (top) leading up to a CMC and in the distal colon (bottom) where CMCs did not occur as regularly. (D) Regardless of colon location, the maximum percentage of neurons with spontaneous activity was higher in trials with CMCs (black circles) compared with trials without CMCs (red X). n = 12 trials in the proximal colon and n = 11 trials in the distal colon from N = 5 mice. (E) The percentage of trials with neurogenic activity and corresponding CMCs was higher in the proximal colon compared with distal (P < .001, N = 5 mice), suggesting that the proximal colon myenteric neurons show coordinated increases in activity more frequently than the distal colon. (F) The average duration (in seconds) of coordinated, summated neural activity (as represented by the pink boxes in panels B and C) was longer in the proximal colon compared with the distal colon. (G) Time-lapse color-coded image (top) and GCaMP6s traces (middle, magenta) of responses from the proximal colon myenteric neurons to electrical stimulation; traces of displacement of the imaging field (bottom, blue; circular axis, green; longitudinal axis) show that the summation of neural activity produced a CMC-like event at the end of the recording. (H) Time-lapse color-coded image (top) and GCaMP6s+ traces (middle, dark blue) of responses from distal colon myenteric neurons to electrical stimulation; traces of displacement of the imaging field (bottom) show that the pattern of evoked neural activity does not lead to a CMC. Note the differences in summation between the proximal and distal colon. (I) Scatterplot showing the relationship of delayed responses and evoked tissue displacement for the proximal and distal colon. (J) Graphic summary of the percentage of neurons with early responses in the proximal and distal colon. (K) Graphic summary of the percentage of neurons with delayed responses in the proximal and distal colon. The entire length of colon was used in these experiments.
We observed differences in connectivity arrangements in proximal compared with middle and distal colon. Of interest is the overall patterning of ganglia, which are branched in the proximal colon; this structure was not seen in the midcolon or distal colon (Figure 9A1–A3). The large ganglia tend to cover 2 rows in the taenia that extend proximal to distal, but can cover 3 rows in the smaller ganglia found at the intertaenia transition. Of note is the distinct pattern of interganglionic fiber track connectivity in the proximal vs distal colon (Figure 9B1–B3 compared with Figure 9D1–D3). In the proximal colon, interganglionic fiber tracts project in both the longitudinal (Figure 9B3, white arrow) and circumferential directions (Figure 9B3, green arrow), but that connect one row of ganglia or portion of a single ganglion to the adjacent portion (Figure 9B3, white box) and that take fibers away from ganglia and mostly run in a longitudinal direction (Figure 9B1–B3); there is significant circumferential fiber distribution within large ganglia and from ganglia to ganglia residing in a single row (Figure 9B3, white box). There are several kinds of interganglionic strand structures; in the proximal colon there is a continuous convergence and divergence of fibers that run primarily longitudinally from ganglion to ganglion; fibers converge as they leave the aboral side of a ganglion, often to several points from one large ganglion and diverge as they enter the next ganglion (Figure 9B2, white chevrons, and B3). Upon entering the next ganglion, a subset of fibers spread within the ganglion making many close contacts with myenteric neurons; some of these fibers make a circumferential trajectory and then converge again onto ganglia located at some distance (Figure 9B3). Fibers can pass across or through ganglia with no apparent contribution to interganglionic or intraganglionic connectivity or to basket-like structures that are visible in many large proximal myenteric ganglia; the basket-like structures (Figure 9B3, arrows within white box) contain fibers from cells expressing calretinin, calbindin, CGRP, ChAT, and substance P (likely IPANS). It is common to find that groups of fibers upon exiting a more proximal ganglion project laterally to join a fiber tract that enters a ganglion in the more aboral row of ganglia (Figure 9B3). In the distal part of the proximal and midcolon, there are many occurrences of interstrand connectivity or internodal connectivity that bypasses a level of myenteric ganglia completely (Figure 9C1, white arrow). Upon transitioning into the midcolon, the connectivity pattern changes; the apparent divergence and convergence of fibers decreases, and the number and type of interganglionic connections become more diverse; this is partly owing to the increase in small neuron clusters (Figure 9C1, white circles) that contribute to the complicated patterns of interganglion connectivity (Figure 9C2 and C3). In the middle and distal colon, fiber tracts send collaterals to other fiber tracts independent of intersecting with neurons (Figure 9C1 and D1, white arrows). It is not uncommon to see multiple inputs from a group of neurons to single neurons in the next row of more aboral ganglia. This pattern is carried into the distal colon (Figure 9D3, white bracket). There are small triangular-shaped ganglia that form the vertices of internodal strands that then form a ganglion-to-ganglion connection. The major change in distal colon is an apparent increase in ganglion to ganglion and internodal strand to internodal strand connectivity in a circumferential direction (Figure 9D1, white triangle).
The density of fibers in the internodal strands makes it difficult to follow individual axons but we have used 3D imaging tools to assess intraganglionic connectivity (Figure 10). Within individual myenteric ganglia there were apparent close contacts between axons and somas (Figure 10A1–B2) and axo-axonal or axon to fiber bundle contacts (Figure 10B1–B4). The CGRPA neurons (Figure 10B1–B4) have both varicose (Figure 10B1) and nonvaricose axons; a subfraction of the varicosities co-labeled with the synaptic marker protein synaptophysin (Figure 10B3 and B4). Synaptic (synaptophysin+) sites were evident in varying numbers on both cell bodies and fibers (Figure 10B2) and were well distributed throughout ganglia (Figure 10B1 and B4).
Because of the differences in ganglion organization, neurochemical code, and synaptic and neural circuit connectivity observed in the proximal, middle, and distal colon, we predicted that the expression and co-expression of major ENS neurotransmitter receptors on individual myenteric neurons also was regionally organized. We tested this assumption using GCaMP6s responses to serotonin (5-hydroxytryptamine (5-HT)) and the nicotinic cholinergic receptor agonist DMPP, which were measured in individual myenteric neurons in the proximal, middle, and distal colon (Figure 11). We then compared the percentage of each response type (5-HT–specific, DMPP-specific, both) of the total number of responding neurons (Figure 11A) and of the total number of neurons (Figure 11B) per field of view. The percentage of myenteric neurons that only responded to serotonin was significantly higher in the distal colon, the percentage that only responded to DMPP was significantly higher in the proximal colon, and the percentage of myenteric neurons that responded to both 5-HT and DMPP was significantly higher in the midcolon. The regional differences in the expression and co-expression of receptors for 2 major ENS neurotransmitters likely contribute to unique functions of the proximal and distal colon, suggesting that we could expect differences in the neural response to electrical stimulation/activation of colonic myenteric ganglia.
We tested for regional differences in ENS network activity and motor output by assessing spontaneous and electrically evoked colonic motor complexes (CMCs) using tissue displacement (Figure 12A), while simultaneously recording neural activity in myenteric ganglia expressing the calcium indicator GCaMP6s. Before spontaneous CMCs, many myenteric neurons developed activity that increased in a coordinated manner, causing the entire field of view (FOV) to increase fluorescence intensity (Figure 12Bi and Bii, Before CMC). By contrast, fewer neurons have spontaneous activity after the CMC, and because the activity is not coordinated, the average GCaMP signal of the FOV does not deviate from baseline (Figure 12Bi and 12Biii, After CMC). Traces from individual myenteric neurons illustrate that summation of activity precedes CMCs, and CMCs do not occur in the absence of summation (Figure 12C). Comparing the maximum percentage of active neurons in trials with and without spontaneous CMCs showed an apparent activation threshold of approximately 48% of neurons per FOV regardless of colon region. Significantly more neurons were active in trials with CMCs compared with trials without CMCs for both regions (Figure 12D), but spontaneous CMCs were more frequent in the proximal colon compared with the distal colon (Figure 12E), suggesting that this activation threshold was not met as often in the distal colon. Further supporting neural circuit differences, the duration of coordinated (or summated) activity was greater in the proximal colon (Figure 12F). As shown earlier, proximal colon regions have more neurons tightly packed within closely arranged, large myenteric ganglia (Figure 3, Figure 4, Figure 5), and extensive intraganglionic synapses (Figure 10). Our combined anatomic and functional data suggest that ENS circuits in the proximal colon are better wired to produce the coordinated patterns of activity required to generate spontaneous CMCs.
We used electrical stimulation to evoke neurogenic activity that produces contractions and measured neural response patterns in the proximal and distal colon. Stimulation at close distances to the imaging field (<15 mm) produced an early response that was comparable in both colon regions, but the delayed response differed between the 2 regions. Proximal colon neurons showed gradual increases in GCaMP signal with a large degree of summation that often led to CMCs (Figure 12G). However, distal colon neurons showed GCaMP signals that returned to baseline relatively quickly, which is indicative of bursting activity too short to have significant summation (Figure 12H). As predicted, the percentage of neurons with delayed responses to stimulation was correlated linearly to tissue displacement in the proximal colon (Figure 12I).
As the distance between imaging and stimulation sites increased, the early responses decreased in both the proximal and distal colon (Figure 12J). The delayed responses in the proximal colon were produced by stimulation at short and far distances, but delayed responses in the distal colon were negligible when stimulating at distances greater than 20 mm (Figure 12K). These results highlight the differences in the extent of signal integration and long-range input to proximal vs distal ENS networks and further suggests that morphology and circuit connectivity underlie regionally diverse function.
Mouse Colonic Vasculature
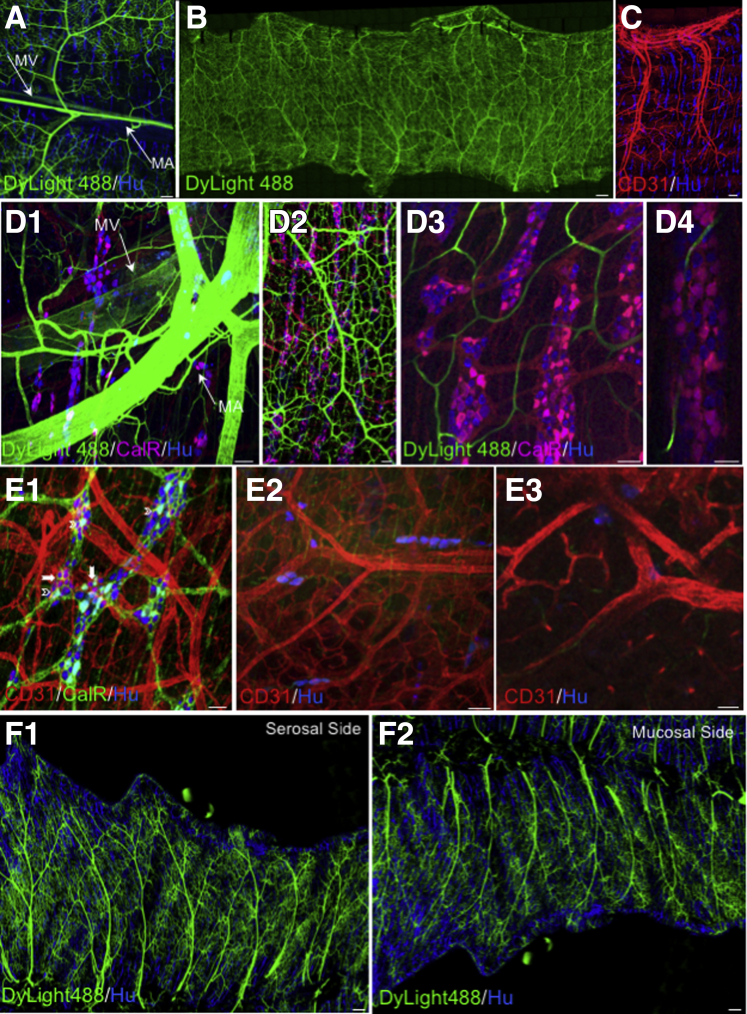
The fundamental role of the vasculature in absorption of nutrients, water balance, metabolism, and damage caused by ischemia prompted scrutiny of vessel location in relation to the colonic myenteric and submucosal ganglia. We visualized the colonic neurovascular complex using CD31 to label endothelial cells or DyLight 488-tomato Lectin to paint the vessels.45,46 The colonic vascular network derives from the superior and inferior mesenteric artery and vein (Figure 13A), with branches that do not appear to be region-specific (Figure 13B and C). The colonic vasculature parallels the overall tissue architecture, with the primary branches running circumferentially, and arterioles, venules, and capillaries running longitudinally and circumferentially (Figure 13D1, D2, F1, and F2). The vessels generally travel on the serosal side of the MP with a few on the mucosal side (Figure 13E1, F1, and F2). We did not observe ganglia completely surrounded by vasculature of any caliber. Large mesenteric artery branches occur in a nonrandom pattern with capillary beds sending fine branches toward the ganglia (Figure 13D1–D4). The deep vasculature, associated with submucosal neurons and the epithelium, has a denser, more random pattern than that closer to the myenteric plexus (Figure 13E2 and E3). These vessels are deep to the submucosal neurons closest to the epithelium but are found around ganglia scattered throughout the submucosa. Submucosal neurons were evident both close to the myenteric plexus and to the epithelium (Figure 14). There is a capillary network that sends blind-end vessels near to the plexi, but that occasionally do go around and/or through a ganglion (Figure 13D2–D4). Similar to what has been reported for nitrergic neurons,46 we did identify vascular nets or baskets around some calretinin expressing myenteric neurons (Figure 13E1, arrowheads) as well as unidentified neurons (Figure 13E1, arrows).
Figure 13.
The colonic vasculature parallels the overall colonic tissue architecture. (A–C) Vasculature was painted using Tomato lectin–Dylight 488, and confocal tile scans showed repeating patterns of vessels originating from the (A) mesenteric artery (MA) and vein (MV). (C) Patterning of vasculature was confirmed using the endothelial marker CD31. (D1–D4) Large- and small-caliber branches leave the (D1) mesenteric artery, (D2) forming a vascular bed primarily on the (F1 and F2) serosal side of the myenteric plexus but with a (D3) capillary network that wends its way around and sometimes through (D4) myenteric ganglia. (E1) Vascular nets were observed around neurons expressing calretinin (arrows) or unidentified neurons (chevrons). Toward the epithelium, the vasculature is dense and mostly below (E2) submucosal ganglia that are (E3) deep. The entire colon was assessed. Scale bars: 200 μm (A and C), 400 μm (B), 50 μm (D1–E3), 200 μm (F1 and F2).
Figure 14.
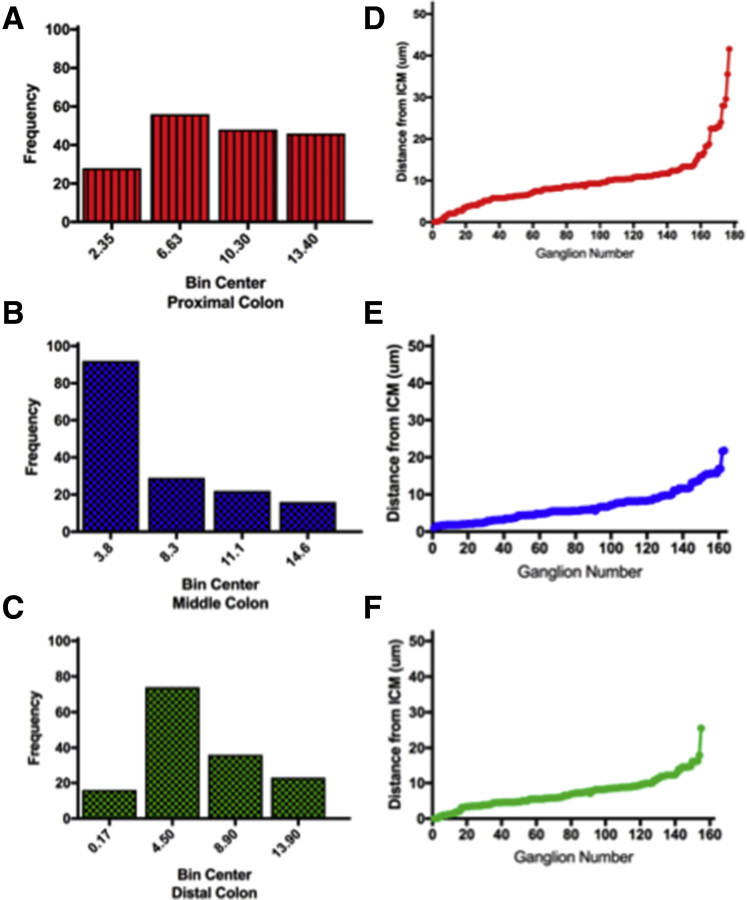
Distribution of colonic submucosal ganglia. The distance from the inner circular muscle to the epithelium was measured in a minimum of 3 samples from the proximal (N = 177 ganglia), middle (N = 163 ganglia), and distal (N = 155 ganglia) colon. For each whole-mount colon sample, the inner circular layer of muscle was located, and the micron scale was set to 0, then moved in micron steps through the submucosa measuring the distance from the circular muscle for each ganglion encountered. At least 4 different areas were measured in each sample. (A–C) The data were binned according to distance, and the number of ganglia (frequency) per bin is presented. (B) In the midcolon, there are more submucosal ganglia closer to inner circular muscle compared with the proximal or distal colon. (D–F) For each region measured, the distance between ganglia increased approaching the epithelium. The distance from the inner circular muscle was plotted for each region (proximal, middle, distal) for each ganglion measured. The data show, as we have published for human beings,22 that ganglia are distributed throughout the submucosa. The proximal colon sampled was from area B1, the midcolon was from area B2, and the distal colon was from area B3 (Figure 2B).
Discussion
Dissecting ENS circuitry is challenging because of technical limitations and the complexity of the neural network. Several groups, including ours,5,18,43,44,47 have used GCaMP6-based calcium imaging approaches to try to address these issues. This approach has not proven robust enough to show direct cell-to-cell connectivity but we do provide data showing direct vs indirect neural activity. The quantitative analyses presented here provide detailed information that will improve our ability to interpret functional and transcriptomic data and develop better treatments for gastrointestinal motility disorders. Our comparisons between human22 and mouse ENS show that mouse data are a valuable surrogate for better understanding function and diseases of the human ENS.
The complexity in ganglion size, distribution, and neuron numbers combined with connectivity and neuron distribution patterns based on neurochemical code provide insight into how ENS structural defects contribute to a multitude of bowel motility disorders. ENS connectivity patterns underlie functional differences between the proximal and distal colon that we47 and others10,43,48,49 have reported previously; the connectivity pattern both within and between myenteric ganglia is more complex in the proximal colon compared with the distal colon, but connectivity between ganglia and internodal strands increases in complexity in the middle and distal colon. The branching structure of myenteric ganglia in the proximal colon combined with the connectivity pattern suggest that the mixing movements characteristic of the proximal colon10,16,17,49,50 are underpinned by the connectome. Our data show that neural activity summation is required to initiate CMCs, and that the proximal colon is better at achieving this task, likely owing to a high density of neurons that have multiple contacts with one another, forming recurrent synapses that recruit and coordinate activity across many neurons. Our data suggest that the proximal colon, in particular, is responsible for integrating input from the entire length of colon, has the ability to spontaneously generate robust and coordinated neural activity, and ultimately decides, based on summation, when CMCs should be generated for mixing and propulsion of gut luminal contents. By contrast, the distal colon performs commands that it receives from more proximal, adjacent segments to propagate CMCs and expel gut luminal contents.
It may seem obvious that structure underlies function, but our detailed description supports the assumptions made from previous studies. The ganglion structure and connectivity pattern could explain the reported disparity between the numbers of responding neurons in the proximal vs distal colon to focal electrode stimulation.10 Although neither neuron number nor fiber density could account for the increased number of activated neurons in this previous investigation,10 our identification of intraganglionic connectivity, the extensive number of synaptic sites on individual neurons, and our assumption that varicosities are en passant synapses (close contact not at an axon terminal), suggests that simple models consisting of monoaxonal neurons making only 1 synaptic contact needs to be reconsidered; 3D reconstruction shows that some neurons make multiple contacts with neighboring neurons in addition to contacts in other ganglia. Specifically, in the proximal colon, based on neuron-to-neuron connectivity and functional studies, our data suggest that there is an increased number of short distance and polysynaptic connectivity resulting in reverberating neural activation. Our results support the idea that at least 2 types of neuron-to-neuron connectivity account for regionally identifiable motor patterns in the colon. Distal colon peristaltic activity is underpinned by monosynaptic connectivity, and the more complicated patterns in the proximal colon responsible for mixing are the result of coordinated monosynaptic, polysynaptic, and reverberating intraganglionic and interganglionic synaptic connectivity. This type of connectivity could account for the increased numbers of neurons firing in response to connective stimulation in the proximal colon. We have shown that more than 50% of proximal colon neurons show synaptic delays between 15 and 65 ms, indicating polysynaptic activation.44 The data presented in this study suggest that a unique function of proximal ENS circuits is to integrate and process information from all over the colon to make decisions regarding motor output; this likely is accomplished by parallel neural integration as opposed to neurons only integrating signals in series. There may even be specific areas in the proximal colon that receive more concentrated input, as evidenced by the bigger responses at distances of 10, 20, and 50 mm from stimulation sites. Our data confirm that differences in morphology and circuit connectivity within the proximal, middle, and distal colon have significant consequences on the overall function of these distinct colon regions, making species comparisons very important.
The ganglion morphology, neuron number, and connectivity patterns in the middle and distal colon approximate that of the mouse small intestine and human distal (left) colon22 more than the mouse proximal colon, which could account for the propulsive movements characteristic of the small intestine and more distal bowel. The percentage of colon occupied by myenteric ganglia is significantly different in mice and human beings. The mouse middle and distal colon is most similar to human colon where ganglia distribution is relatively similar throughout with large distances between ganglia; this suggests that mouse proximal colon is not a good surrogate when ganglion distribution affects function. The comparative data show that simple neural network models designed to explain functional connectivity need to take the details of the colon structure into account.
Comparison of mouse with human colon22 highlights important considerations relevant to human disease.1,19,51 Changes in the ENS or loss of neurons is associated with a plethora of pathologic conditions.1,15,22,51, 52, 53, 54 In the mouse, neurons are more densely packed in the proximal colon and the branching pattern has not been observed in the human samples examined thus far. The area of single ganglia in the human left colon (distal) more approximates the mouse proximal colon, indicating that modeling of the neural circuits must take these differences into account to fit function and circuitry to human pathophysiology. The distribution of neurons in the submucosal plexus is more similar to that of the human colon22 than other small rodents; we found that submucosal neurons were distributed throughout the submucosa from just under the inner circular muscle to the epithelium. Comparison of mouse with human22 data will influence interpretation of mouse data, both genetic and morphologic, as it relates to assessment of the salient features in mouse models of Hirschsprung’s disease. One important therapeutic goal is to improve the results of pull-through surgery, which requires a much better description of the transition zone in both mice and human beings. There is evidence that the ganglionated portions of Hirschsprung’s disease bowel are abnormal, contributing to postoperative issues.55 There is an increase in the number of NOS neurons and a decrease in ChAT neurons at the transition zone, which could result in a change in motility patterns from the mixing normal to proximal colon to the more propulsive pattern observed in the middle and distal colon53,56; this conclusion is based on a change in normal distribution that we observed in the mouse with a significant increase in NOS neurons and a decrease in ChAT neurons in the distal colon. We show that the increased reliance on cholinergic input in the distal colon reflects long axons originating oral to the recording site and not an increased number of cholinergic neurons. This increased reliance on cholinergic input also could reflect an increase in the number and/or class of neuronal nicotinic acetylcholine receptors as suggested by RNA-seq data and the patterns of functional distribution that we show. Dysmotility often is related to changes in the distribution or loss of nitrergic neuron,56 with no apparent change in cholinergic neurons as observed in the diabetic colon. These observed changes in nitrergic and cholinergic neuron numbers and distribution are significant because we have shown that under normal conditions the percentage of these neurons is comparable in mice and human beings, supporting the use of mice as a surrogate for human beings under many experimental conditions.
Our data showing investment of myenteric ganglia by small capillaries and that some myenteric neurons express mucosal macrophage markers suggests interplay between the neurons and immune system underpinning colon inflammation; choline acetyltransferase and both neuronal and muscarinic acetylcholine receptors are expressed by some immune cells.51 The significant changes in the vasculature associated with colon inflammation52,57 combined with evidence of a gut–vascular barrier58,59 suggest that a detailed investigation of interactions between capillaries and nitrergic neurons would be of particular interest owing to the increased susceptibility of these neurons to oxidative damage. Our observations suggest that further characterization of neuronally expressed immune genes could fill several holes in our understanding of neuroimmune interactions and the role that neurons could play in stimulating or inhibiting inflammatory responses in the gut.
The pattern of neural subtype marker proteins we report coincides well with that reported for human,22,60 guinea pig, and other species.2,6,7,11,12,39 The differential distribution of neural subtypes could contribute to diverse motility patterns from the proximal to distal colon. The significant increase in inhibitory motor neurons from the proximal to distal colon10 likely impacts motor patterns. The increase in interneurons expressing ChAT, GABA, and somatostatin34 in the middle and distal colon is functionally significant because stimulation enhances release of NO and VIP from inhibitory motor neurons,60 as well as a decrease in acetylcholine release,34 both of which result in decreased intestinal motility. Recent RNA-seq data21 indicate that somatostatin receptors are expressed exclusively on IPANs; this is functionally significant because IPANS initiate neurogenic signaling in the ENS. Interestingly, our data suggest more than 1 mechanism of synaptic regulation because somatostatin affects significant inhibition of both spontaneous activity and that stimulated by the nicotinic agonist DMPP. The almost-complete block of spontaneous activity suggests presynaptic inhibition of acetylcholine release, while reduced response to DMPP could be the result of changes in cyclic adenosine monophosphate mediated by somatostatin34; regulation of nAChR sensitivity in response to nicotinic agonists by cyclic adenosine monophosphate is established in autonomic neurons.61 We recently showed that ENS spontaneous activity is dependent on presynaptic activity, resulting in postsynaptic activation via nicotinic acetylcholine receptors and/or 5-HT-3 receptors.44 A change in the number or distribution of one or both of these receptor types could drastically affect motility. This conclusion is supported by the differential distribution of myenteric neurons expressing nACh receptors and/or 5-HT receptors in the proximal, middle, and distal colon. It is relevant to note that based on RNA-seq data,18,19 the 5-HT-3 receptor is most highly, if not exclusively, expressed on IPANs. The fact that we found significant differences in distribution and function along the colon coupled with the identification of a new type of IPAN(A) most highly expressed in the proximal colon shows that quantitative aspects of morphology, connectivity, and distribution based on neurochemical code must be considered in assessing function in health and disease.
The central role as initiator of enteric motor activity suggests functional significance to identification of IPANs-expressing CGRPA, which are located primarily in the proximal and middle colon; it was accepted that enteric IPANS express CGRPB and not CGRPA.32,33 The neurochemical code (IPANB) and at least 3 different morphologies (IPANA) are reminiscent of that reported in the guinea pig distal colon.3,35, 36, 37, 38, 39,62 Recent RNA-seq profiling identified 1 IPAN class expressing CGRP(A) that also express VIP and cholecystokinin (CCK),19,21 classic markers of intestinofugal neurons; this neuron is similar to that reported by Morarach et al,21 with a smooth cell body and 1 axon that we also identified as IPANA. One IPAN, with this morphology, was shown to express the mechanoreceptor Piezo II,21 raising the intriguing possibility that the neurons that we identified as IPAN(A) are a special class of intestinofugal neuron and/or mechanoreceptive sensory neuron.
We discerned calbindin baskets in ganglia associated with IPANs, similar to guinea pig distal colon,36 but IPANA fibers have a unique web-like structure that can surround many neurons. The majority of their processes ramify within 1 or several ganglia before contributing to internodal strands running both longitudinally and circumferentially. The large number of varicose axon close contacts suggests extensive synaptic connectivity. The differences between these morphologies and those identified in guinea pig suggest distinct neural circuits of a specialized IPANA that is specific to the proximal and middle colon and that contribute to driving proximal vs distal colon motor patterns. Because of their proximal location and extensive synaptic connectivity, IPANA neurons likely contribute to the coordinated ENS network activity required to generate CMCs.
With increasing appreciation of the ENS in the etiology of dysmotility disorders, availability of single-cell RNA-seq data and optogenetic approaches for investigation of neural circuit function, our detailed quantitative and descriptive data underpin the ability to make maximal use of multispecies and gut region–specific data. Comparison of mouse and human data will enhance our ability to determine which mouse data and molecular genetics can be best used as a surrogate for the human ENS. By taking a multidisciplinary approach as we have done, we have shown how morphology, connectivity, and cell-type–specific chemistry and receptor expression combine to control differential patterns of coordinated nerve and muscle function.
Material and Methods
Animals
Animal care, breeding procedures, and experimental protocols were approved by the University of Toledo Health Science Campus animal care and use committee. Animals were housed in an Association for Acreditation of Laboratory Animal Care-approved facility with a 12-hour light cycle with food (standard chow) and water ad libitum. Male and female mice aged 3–9 months were used in the reported studies. All reporter mice, except where indicated, were generated by crossing either R26R-EYFP/EGFP mice or RCL-tdTomato mice with 1 of the Cre lines (Table 3), which express the reporter in a promoter-specific manner after Cre-mediated recombination. All listed mice, except where noted (Table 3), were purchased from the Jackson Laboratories (Bar Harbor, ME). All genotyping was performed using primers recommended by the Jackson Laboratories according to their protocols.
Table 3.
Mouse Manifest
| Formal strain name | Commonly used name | Mouse genome informatics ID | Jax Lab stock number | Strain background |
|---|---|---|---|---|
| Tg(Calca-EGFP)FG104Gsat/Mmucd | Calca-EGFP | 3842528 | Gift from Dr David Ginty (Harvard Medical School) | C57BL/6J |
| B6(Cg)-Calb2<tm1)cre)Zjh>/J | Calb2-IRES-Cre | 4365736 | 010774 | C57BL/6 |
| B6;129S6-Chattm2(cre)Lowl/J | ChAT-IRES-Cre | 5475195 | 006410 | C57BL/6 |
| B6.129-NOS-TM1(CRE)MGM/J | NOS1-Cre | 5301174 | 017526 | C57BL/6J |
| B6.Cg-Gt(ROSA)26Sor<tm14(CAG-tdTomato)Hze>/J (RCL-tdTomato mice) | Ai14, Ai14D, or Ai14(RCL-tdT)-D | 3809524 | 007914 | 129S6/SvEvTac × C57BL/6NCrl)F1 Maintained on C57BL/6J |
| Vip<tm1(cre)Zjh>J | Vip-IRES-cre | 4431361 | 031628 | C57BL/6J |
| B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | R26R-EYFP | 2449038 | 006148 | C57BL/6J |
| B6.Cg;Gt)ROSA)26Sor<tm6(CAG-ZsGreen1) Hze/J | AI6 (R26R-EGFP) | 3809522 | 007906 | 129S6/SvEvTac × C57BL/6NCrl)F1 Maintained on C57BL/6 |
| B6.FVB-Tg(EIIa-cre)C5379Lmgd/J | E2a-Cre | 2137691 | 003724 | C57BL/6 FVB/N |
| B6J.Cg-Gt(ROSA)26Sortm95.1(CAG-GCaMP6f)Hze/MwarJ | Ai95(RCL-GCaMP6f)-D (C57BL/6J) or Ai95D (C57BL/6J) | 5558090 | 028865 | C57BL/6J |
| Ai96(RCL-GCaMP6s) (C57BL/6J) or Ai96 (C57BL/6J) | Ai96 | 5558094 | 028866 | C57BL/6J |
NOTE. All data needed to identify mice used in the reported studies is provided. All mice, with the exception of Calca mice (a generous gift from Dr David Ginty, Harvard Medical School), were purchased from the Jackson Laboratory, Bar Harbor, ME).
EGFP, enhanced green fluorescent protein; EYFP, enhanced yellow fluorescent protein.
Immunohistochemistry
Whole-mount immunostaining methods were reported previously.63,64 Briefly, cleaned whole colons were fixed in fresh 4% paraformaldehyde prepared in Dulbecco’s modified phosphate-buffered saline for 1–2 hours at room temperature or overnight at 4ºC. After washing in Dulbecco’s modified phosphate-buffered saline, tissue was incubated in 0.1 mol/L Tris, 1.5% NaCl, 0.5% TX-100 (Sigma-Millipore, St. Louis, MO) 3 × 30 minutes, blocked in the same buffer containing 20% horse serum (HS) 2 × 1 hour with shaking, followed by a quick rinse in the same buffer without HS. Primary antibody was applied in the same buffer containing 10%–20% HS for 48 hours at 4ºC with rocking. After washing in the same buffer for 3 × 1 hour, and 3 × 10-minute washes in buffer without HS, a secondary antibody was applied in the same buffer with 10%–20% HS for 48 hours at 4ºC with rocking. After washing in the same buffer without HS for 6 × 10 minutes, samples were mounted in Fluoromount-G (Thermo Fisher Scientific, Waltham, MA) and visualized. All antibodies used are listed in Table 4. Species-specific secondary antibodies were chosen to minimize spectral overlap. Specifically, Alexa Fluor 488 (Thermo Fisher Scientific), yellow fluorescent protein, and =green fluorescent protein (Aves Labs, Inc, Tigard, OR) were excited at 488 nm with an emission peak at 519 nm. Alexa Fluor 555 excitation and emission peaks were 488 nm and 568 nm, respectively. The Alexa Fluor 594 excitation and emission peaks were 561 nm and 620 nm, respectively. The Alexa Fluor 647 excitation and emission peaks were 633 nm and 670 nm, respectively. Images were stitched and 2-dimensional projections were made from the XYZ stacks.
Table 4.
Primary and Secondary Antibodies
| Antigen detected by primary antibody | Host | Dilution | Source | Research Resource Identifier (RRID) | |
|---|---|---|---|---|---|
| Calbindin D-28 | Rabbit | 1:400 | Swant (CB38A); Berne, Switzerland | RRID:AB_10000340 | |
| Calretinin | Rabbit | 1:400 | Swant (7697) | RRID:AB_2721226 | |
| Calretinin | Goat | 1:400 | Swant (CG1) | RRID:AB_10000342 | |
| CD31 | Rat | 1:300 | BD Pharmingen (557355); San Diego, CA | RRID:AB_396660 | |
| CGRP | Mouse | 1:200 | Abcam (ab81887); Boston, MA | RRID:AB_1658411 | |
| αCGRP | Rabbit | 1:500 | Peninsula Labs (T4032), San Carlos, CA | RRID:AB_518147 | |
| ChAT | Goat | 1:100 | Millipore (AB 144P), Burlington, MA | RRID:AB_262156 | |
| Enkephalin | Mouse | 1:200 | Millipore (MAB350) | RRID:AB_2088149 | |
| Enkephalin | Mouse | 1:150 | Abcam (ab150346) | Not Found | |
| GABA | Rabbit | 1:300 | Sigma (A2052), St. Louis, MO | RRID:AB_477652 | |
| Green Fluorescent Protein | Chicken | 1:300 | AVES (GFP1020), Davis, CA | RRID:AB_10000240 | |
| Green Fluorescent Protein | Goat | 1:400 | Abcam (ab5450) | RRID:AB_304897 | |
| Green Fluorescent Protien | Rabbit | 1:200 | Abcam (ab6556) | RRID:AB_305564 | |
| HuC/D (ANNA-1) | Human | 1:20,000 | Gift from Dr V. A. Lennon | RRID:AB_2314657 | |
| Iba1 | Rabbit | 1:500 | Wako Chemicals (019-19741), Richmond, VA | RRID:AB_839504 | |
| Neurofilaments | Mouse | 1:500 | ThermoFisher (13-1300), Waltham, MA | RRID:AB_2532999 | |
| neuronal nitric oxide synthase | Rabbit | 1:400 | Millipore (AB5380) | RRID:AB_91824 | |
| neuronal nitric oxide synthase | Rabbit | 1:300 | Abcam (ab76067) | RRID:AB_2152469 | |
| neuronal nitric oxid synthase | Goat | 1:200 | Invitrogen (OSN00004G), Waltham, MA | RRID:AB_10709561 | |
| PGP9.5 | Guinea pig | 1:500 | Neuromics (GP14104), Edina, MN | RRID:AB_2210625 | |
| red fluorescent protein | Chicken | 1:600 | Rockland (600-901-379), Philadelphia, PA | RRID:AB_10704808 | |
| red fluorescent protien | Rabbit | 1:600 | Rockland (600-401-379) | RRID:AB_2209751 | |
| S-100 | Rabbit | 1:350 | Abcam (ab52642) | RRID:AB_882426 | |
| Serotonin (5-HT) | Rabbit | 1:1000 | Immunostar 20080, Hudson, WI | RRID:AB_572263 | |
| Serotonin (5-HT) | Goat | 1:1000 | Immunostar 20079 | RRID:AB_572262 | |
| Somatostatin | Rat | 1:200 | Abcam (ab30788) | RRID:AB_778010 | |
| Somatostatin | Rat | 1:200 | Millipore (MAB354) | RRID:AB_2255365 | |
| Somatostatin | Rabbit | 1:200 | Abcam (ab108456) | RRID:AB_11158517 | |
| Substance P | Rabbit | 1:200 | Abcam (ab67006) | RRID:AB_1143173 | |
| Substance P | Rat | 1:150 | Abcam (ab7340) | RRID:AB_305866 | |
| Substance P | Guinea pig | 1:200 | Abcam (ab06291) | No longer available | |
| Synaptophysin | Rabbit | 1:150 | Abcam (ab32127) | RRID:AB_2286949 | |
| Synaptotagmin 1 | Rabbit | 1:100 | Abcam (ab131551) | RRID:AB_11157546 | |
| Synaptotagmin 1 | Chicken | 1:100 | Abcam (ab133856) | RRID:AB_2885088 | |
| TUJ1 (β III tubulin) | Rabbit | 1:1000 | Covance (AB 291637) discontinued; now BioLegend (PRB-435P-100), Saan Diego, CA | RRID:AB_291637 | |
| TUJ1 (β III tubulin) | Chicken | 1:500 | Abcam (ab107216) discontinued | RRID:AB_10899689 | |
| VIP | Rabbit | 1:300 | Immunostar (20077) | RRID:AB_572270 | |
| VIP | Rabbit | 1:100 | Abcam (ab22736) (used in initial experiments, new lots do not work) | RRID:AB_447294 | |
| VIP | Rabbit | 1:500 | Sigma (MPA017324) | Not found | |
| VIP | Guinea pig | 1:300 | Invitrogen (PA1-36030) | RRID:AB_1088264 | |
| Secondary antigen | Host | Conjugate | Dilution | Source | RRID |
| Chicken (IgY, H+L) | Goat | Alexa Fluor 488 | 1:300 | Invitrogen (A11039) | RRID:AB_142924 |
| Chicken (IgY, H+L) | Goat | Alexa Fluor 568 | 1:500 | Invitrogen (A11041) | RRID:AB_2534098 |
| Chicken (IgY, H+L) | Goat | Alexa Fluor 594 | 1:500 | Invitrogen (A11042) | RRID:AB_2534099 |
| Goat (IgG, H+L) | Donkey | Alexa Fluor 488 | 1:500 | Invitrogen (A11055) | RRID:AB_2534099 |
| Goat (IgG, H+L) | Donkey | Alexa Fluor 555 | 1:500 | Invitrogen (A21423) | Not Found |
| Goat (IgG, H+L) | Donkey | Alexa Fluor 647 | 1:500 | Invitrogen (A21447) | RRID:AB_141844 |
| Guinea pig (IgG, H+L) | Goat | Alexa Fluor 555 | 1:500 | ThermoFisher A-21435 | RRID:AB_2535856 |
| Guinea pig (IgG, H+L) | Goat | Alexa Fluor 594 | 1:500 | Invitrogen (A11076) | RRID:AB_141930 |
| Human (IgG, H+L) | Goat | Alexa Fluor 555 | 1:500 | Invitrogen (A21433) | RRID:AB_2535854 |
| Human (IgG, H+L) | Goat | Alexa Fluor 647 | 1:500 | Invitrogen (A 21445) | RRID:AB_2535862 |
| Human (IgG, H+L) | Goat | DyLight 405 | 1:500 | Novus Biologicals (NBP1-72871) lLittleton, CO | RRID:AB_11008096 |
| Human (IgG, H+L) | Alpaca | DyLight 405 | 1:100 | Jackson IR (609-475-213), West Grove, PA | RRID:AB_2721858 |
| Mouse (IgG, H+L) | Goat | Alexa Fluor 594 | 1:500 | Invitrogen (A11005) | RRID:AB_141372 |
| Rabbit (IgG, H+L) | Donkey | Alexa Fluor 488 | 1:500 | Invitrogen (A21206) | RRID:AB_2535792 |
| Rabbit (IgG, H+L) | Donkey | Alexa Fluor 555 | 1:500 | Invitrogen (A31572) | RRID:AB_162543 |
| Rabbit (IgG, H+L) | Donkey | Alexa Fluor 647 | 1:500 | Invitrogen (A31573) | RRID:AB_2536183 |
| Rabbit (IgG, H+L) | Goat | Alexa Fluor 488 | 1:500 | Invitrogen (A11034) | RRID:AB_2576217 |
| Rabbit (IgG, H+L) | Goat | Alexa Fluor 546 | 1:500 | Invitrogen (A11010) | RRID:AB_2534077 |
| Rabbit (IgG, H+L) | Goat | Alexa Fluor 568 | 1:500 | Invitrogen (A11011) | RRID:AB_143157 |
| Rabbit (IgG, H+L) | Goat | Alexa Fluor 594 | 1:500 | Invitrogen (A21207) | RRID:AB_141637 |
| Rabbit (IgG, H+L) | Goat | Alexa Fluor 647 | 1:500 | Invitrogen (A21245) | RRID:AB_2535813 |
| Rat (IgG, H+L) | Donkey | Alexa Fluor 594 | 1:400 | Invitrogen (A21209) | RRID:AB_2535795 |
| Rat (IgG, H+L) | Goat | Alexa Fluor 647 | 1:500 | Invitrogen (A21247) | RRID:AB_141778 |
| Sheep (IgG, H+L) | Donkey | Alexa Fluor 488 | 1:300 | Invitrogen (A11015) | RRID:AB_141362 |
| Sheep (IgG, H+L) | Donkey | Alexa Fluor 594 | 1:500 | Invitrogen (A11016) | RRID:AB_253408 |
| Sheep (IgG, H+L) | Donkey | Alexa Fluor 647 | 1:500 | Invitrogen (A21488) | Not found |
NOTE. All primary and secondary antibodies used in the reported studies are listed. The list includes the antigen recognized, the host that the antibody was raised in, the dilution used, the source, and RRID when available.
RRID, Research Resource Identifier; H+L, high and low.
The vasculature was visualized in wild-type and calretinin reporter mice after intravascular perfusion (in anesthetized mice) of Lycopersicon esculentum (Tomato) lectin (LEL,TL), Dylight 488 (DL-1174-1; Vector Laboratories, Burlingame, CA) through the heart. The Tomato lectin Dylight 488 was diluted in phosphate-buffered saline (20 mg/mL). The gut was removed after a 30-minute rest in situ after perfusion and fixed in fresh 4% paraformaldehyde at 4ºC overnight.
The protocol followed for colonic dissections (Figure 2) ensured that the same regions were compared for quantitative and qualitative measurements.
Confocal Imaging
Intact or flat-mount colon samples were imaged using a Leica TCS SP5 laser scanning confocal microscope (Leica Microsystems, Bannockburn, IL) equipped with continuous-wave solid-state lasers (458, 488, 514, 561, and 633 nm) and a titanium-sapphire tunable (705–980 nm) multiphoton laser (Coherent, Santa Clara, CA). Images were acquired at 512 × 512 in the XYZ planes as a single field of view or as tile scans (XYZS) in 1-μm steps with 20× (NA, 0.70), 40× (NA, 1.25), or 63× (NA, 1.4) objectives using an automated scanning stage. Images were acquired with the LAS AF (Leica Microsystems, Bannockburn, IL) software in sequential scan mode to minimize spectral overlap in the emission spectrum.
Image Analysis
For the purpose of directionality and graph analysis, the images were preprocessed and analyzed using the ImageJ/Fiji (National Institutes of Health, Bethesda, MD) software package (Figure 6). The Directionality analysis plugin was used with the local gradient orientation method, using a 5 × 5 Sobel filter, and subsequently the resultant output was used to compute local gradient orientation. The graph analyses and simulation of random planar graphs was performed using R-language (R Foundation, Vienna, Austria) for statistical computing and the igraph (The igraph Core Team) library for R.
Imaging Ca2+ Signals From Myenteric Neurons
Male and female mice aged 3–6 months were studied. To generate mice expressing the fluorescent calcium indicator GCaMP in selected neurons, mouse strains were cross-bred to express GCaMP6f selectively in cholinergic enteric neurons (Figure 7), or GCaMP6s in all neurons (Figures 11 and 12). The utility of using calcium imaging in genetically modified mice is that one is able to use changes in calcium fluorescence as a surrogate for action potentials and neural circuit dynamics. Both GCAMP6f and GCAMP6s have fast rise and decay kinetics and a dynamic range, allowing detection of action potentials as well as spontaneous synaptic activity; GCaMP6s has somewhat slower kinetics than GCaMP6f, but increased sensitivity; this is advantageous when all neurons are being examined as we have shown previously in the ENS, sympathetic, and sensory neurons.43,44,47,65 Mice that contain a floxed GCaMP6f construct within the Gt(ROSA)26 locus were bred with mice that express Cre recombinase under control of the ChAT promoter. For experiments involving electrical stimulation of myenteric ganglia connectives and nAChR/5-HT-R co-expression (Figures 11 and 12), mice were cross-bred to express GCaMP6s in cells expressing E2a, an upstream activator of neural lineage genes as well as the majority of cells in most tissue types66,67; expression of Cre recombinase initiates in preimplantation embryos. Myenteric neurons were easily distinguished from non-neuronal cells based on their location within the myenteric plexus, morphology, and calcium transient dynamics.
Excised colon segments were opened along the mesenteric border and pinned mucosal-side down onto a Sylgard surface (Dow Chemical) lining a glass coverslip attached to the bottom of a plastic imaging chamber containing Krebs solution: 10× stock was prepared as follows: 1.26 mol/L NaCL [cat. S7653; Sigma, St. Louis MO], 25 mmol/L KCL [cat. P9541; Sigma, St. Louis, MO], 250 mmol/L NaHCO3 [cat. S6014; Sigma, St. Louis, MO], 12 mmol/L NaH2PO4 [cat S8282; Sigma, St. Louis MO], 12 mmol/L MgCl2 [cat. M1028; Sigma, St. Louis, MO], 25 mmol/L CaCl2 [cat. C1016; Sigma, St. Louis, MO] diluted and oxygenated on the day of use (Figure 7) or artificial cerebrospinal fluid (CSF) (artificial cerebrospinal fluid was prepared on the day of the experiment containing: 117.9 mmol/L NaCl [cat. S9888, Sigma, St. Louis, MO], 4.7 mmol/L KCL [cat. P3911; Sigma, St. Louis, MO], 25 mmol/L NaHCO3 [catS6014; Sigma, St. Louis, MO], 1.3 mmol/L NaH2PO4, [cat S8282; Signma, St. Louis, MO] 1.2 mmol/L MgSO47H2O [cat. 230391; Sigma, St. Louis, MO], 2.5 mmol/L CaCl2 [cat. C1016; Sigma, St. Louis, MO], 11.1 mmol/L D-glucose [cat. G5767; Sigma, St. Louis, MO], 2 mmol/L sodium butyrate [cat. B5887; Sigma, St. Louis, MO], and 20 mmol/L sodium acetate [cat. S2889; Sigma, St. Louis, MO]) (Figures 11 and 12) perfused at 1–2 mL/min and bubbled with carbogen (95% O2/5% CO2) to oxygenate and achieve pH 7.4 as previously described.43,44 In experiments that involved recording neural activity leading up to spontaneous CMCs (Figure 12), artificial cerebrospinal fluid was heated to 35ºC. GCaMP-positive neurons were detected by their basal Ca2+ fluorescence using an Olympus BX-51 microscope fitted with an Olympus UMPlanFL 20×/0.5 NA water-immersion objective (Tokyo, Japan) and epifluorescence optics consisting of a 470-nm 3W LED source (BLS-LCS-0470-03-22; Mightex Systems, York, Ontario Canada) control module (BIOLed BLS-IO04-US; Mightex Systems, York, Ontario, Canada) and an appropriate 470- to 490-nm excitation and 520-nm emission filter cube set (U-MNB2; Olympus, Tokyo, Japan) (Figure 7) or an upright DM6000FS Leica fluorescence microscope (Leica, Buffalo Grove, IL) and Prime 95B Scientific CMOS camera (Photometrics, Tucson, AZ) (Figures 11 and 12). Images were collected with Metamorph software (Molecular Devices, Downington, PA) at a 40-Hz sampling rate. The following characteristics were used to distinguish myenteric neurons in E2a+/GCaMP6s+: (1) location within myenteric ganglia, (2) cell body size and shape, and (3) the latency and upstroke rate of response to electrical stimulation.43,47,68 To correlate myenteric neuron GCaMP activity with CMCs (Figure 12), which regularly occur every 3–5 minutes, several 3-minute movies were collected sequentially for 9–15 minutes in proximal and distal imaging fields. Tissue displacement in the x- (circular muscle) and y- (longitudinal) axes, shown to be correlated linearly to the force of contractions,43 was tracked and quantified with the ImageJ plugin Template Matching. Using a concentric electrode (A&M Systems, Sequim, WA) placed on interganglionic connectives at 10-mm increments, neuron responses and tissue displacement evoked by electrical stimulation (100-μs pulses, 20 Hz, for 1 sec) were measured and compared in proximal and distal colon regions. Responses that occurred after <1 second of stimulation were considered early, whereas responses that occurred later (up to 1 min poststimulation) were considered delayed. Values obtained from proximal and distal imaging fields were compared and tested for differences using the Student t test and 2-way analysis of variance (ANOVA) with the Tukey post hoc test. P < .05 was considered significant.
Spontaneous and evoked changes in of GCaMP6f-mediated Ca2+ fluorescence intensity (Figure 7) were acquired in 12-bit images using a 1.44-megapixel CMOS camera capable of capturing at up to 80 frames/sec (Prime 95B; Teledyne Photometrics, Tuscon, AZ) controlled by MetaMorph software (version 7.10.1.161; Molecular Devices, Silicon Valley, CA). Image stacks of 400–1800 frames were processed, and motion-corrected when necessary using Fiji (National Institutes of Health, Bethesda, MD) software (version 2.0.0-rc-65/1.52a).69 To segment the image stacks, regions of interest were drawn around an area of representative background fluorescence (F0), and around myenteric ganglia neuron somas based on the following criteria: (1) they showed basal levels of GCaMP-mediated Ca2+ fluorescence intensity (F), (2) they showed detectable F increase in response to applied agonist, or (3) they showed spontaneous, transient increases in F. The mean F values of regions of interest in each frame were acquired and exported to Excel (Microsoft, Redmond, WA) spreadsheets where the time-dependent, background-subtracted, and normalized changes in specific neuronal Ca2+ fluorescence ([F - F0]/F0 = ΔF/F0) were calculated for each neuron per frame (typically 15 neurons in 1800 frames) and used for subsequent analyses. The net amplitudes of peak ΔF/F0 increases induced by agonists or evoked by electrical stimulation (AA or E,Peak) were obtained from the records by subtracting basal from peak ΔF/F0 values (AA or E,Peak = peak ΔF/F0 – basal ΔF/F0). The frequency (FS) and amplitude (FA) of spontaneous Ca2+ transients were obtained from the Excel ΔF/F0 data values using a PeakCaller script70 run under Matlab (R2018b) as previously described.44
Responses to nicotinic acetylcholine neurotransmitter agonists and/or somatostatin or serotonin on myenteric ganglion neurons were assessed from AA,Peak responses evoked by fast application of receptor agonists and their inhibition by incubation with selective antagonists. Agonists were focally applied by pressure microperfusion (5 psi, 10 sec; via Picospritzer II; Parker Instrumentation Corp, Barnstaple, United Kingdon) from blunt glass micropipettes (diameter, 5–10 μm) delivered within 50 μm of an adjacent myenteric ganglion (Figures 7 and 12). DMPP was used to assay functional nAChRs because it preferentially targets and activates a ganglionic subtype containing α3 and β4 subunits.71 5-HT was applied in the same manner.
The effects of receptor agonists and antagonists on the parameters we tested (ie, AE,Peak, AS,Peak, FS, TR,1/2, TD,1/2) were assessed by comparing values obtained from cells before drug treatment (control) and then the same cells incubated with the drug (test). The values obtained for control and test conditions were considered significantly different if P < .05, with P determined using the Student paired 2-tailed t test (Prism 4; GraphPad Software, San Diego, CA). To determine if there was a relationship between colon region (proximal, middle, distal) and responses to receptor agonists, a repeated-measures 2-way ANOVA with the Tukey post hoc test was used.
Statistical Analysis
Statistical tests (t test, ANOVA, and post hoc Tukey) were performed in GraphPad Prism (La Jolla, CA) and R-language for scientific computing. Data are expressed as means ± SEM or as the mean accompanied by 95% CI, where appropriate. N represents the number of colons analyzed or the number of neurons or connections counted. Significance was defined as P < .05. Effect sizes are expressed as the standardized mean difference.
Statistical Analysis: Linear Models and Computation of Means
The statistical analysis of interganglionic distances and areas was performed using linear models, followed by a Tukey post hoc test (Figure 4). Owing to the log-normality of the data, the model incorporated log-transformation. The model was represented by the following equation:
In the equation, g is the function providing appropriate transformation (logarithm), X is an n × p matrix encoding descriptive variables (such as the measurement category and the region of the colon), β is the vector of model coefficients, ε is the error, n is the number of measurements, and p is the dimensionality required to encode the measurement (for proximal, middle, distal, and longitudinal vs circumferential, P = 6) The mean, 95th percentile CIs, corresponding confidence limits and the effect sizes were back-transformed from the log scale. The effect sizes were expressed as the standardized mean difference. The computation was performed using R-language for scientific computing and emmeans library.
Calculation of Spherical Area
The Leica application suite of tools was used to determine the long and short diameter of neurons (Figure 1A2 and B2). Z-plane thickness was determined by adding the number of 1-μm planes within each neuron and elliptical cell volume calculated using the following formula:
In the equation, A is the short radius, B is the long radius, and C is the radius of depth.
Acknowledgments
The authors thank Ms Samantha McKee, Dr Kalina Hristova-Venkova, and Dr Alexander Hristov for excellent technical support and animal husbandry. Calca-EGFP, FG104 mice Tg(Calca-EGFP)FG104Gsat, and CGRPA reporter mice were a generous gift from Dr David Ginty (Department of Neurobiology, Harvard Medical School). Tyrosine receptor kinase B CreERT2 mice, originally provided by Dr David Ginty, were crossed with mice containing a fusion protein of channelrhodopsin-2 and EYFP in the Rosa26 locus downstream of a floxed STOP cassette to generate tyrosine receptor kinase B–ChR2–YFP mice; tissues from these mice were generously provided by Dr Richard Koerber (Department of Neurobiology, School of Medicine, University of Pittsburgh). Hu ANNA-1 (HuC/D) was a generous gift from Dr Vanda A. Lennon (Neuroimmunology Laboratory, Mayo Clinic). The RNA sequencing BAM and FastQ files will be available through the National Institutes of Health Stimulating Peripheral Activity to Relieve Conditions Portal (RRID:SCR_017041) under a CC-BY 4.0 license at: https://doi.org/10.26275/sfbw-ax7q.
CRediT Authorship Contributions
Andrea Nestor-Kalinoski, PhD (Investigation: Equal; Methodology: Supporting; Resources: Equal; Visualization: Equal; Writing – review & editing: Equal)
Kristen M Smith-Edwards, PhD (Formal analysis: Supporting; Investigation: Supporting; Methodology: Equal; Resources: Equal; Writing – review & editing: Equal)
Kimberly Meerschaert, BS (Investigation: Equal; Methodology: Equal)
Joseph F Margiotta, PhD (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Resources: Equal; Software: Equal; Validation: Equal; Visualization: Equal; Writing – review & editing: Equal)
Bartek Rajwa, PhD (Investigation: Supporting; Methodology: Equal; Resources: Supporting; Writing – review & editing: Equal)
Brian M Davis, PhD (Investigation: Equal; Methodology: Equal; Resources: Supporting; Supervision: Equal; Writing – review & editing: Equal)
Marthe J Howard, Ph.D. (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Equal; Project administration: Lead; Resources: Equal; Supervision: Lead; Validation: Equal; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by the National Institutes of Health, Stimulating Peripheral Activity to Relieve Conditions (SPARC) program as an Other Transactions award, OT2OD023859 (PI: Marthe J. Howard; Co-Is Joseph F. Margiotta, Andrea Kalinoski, Brian M. Davis and NIBIB U18EB021790 (PI: Marthe J. Howard, and NIH Other Transaction award OT2OD023847 (PI: Bartek Rajwa) and The National Institutes of Health grant NIDDK122798 (PI: Brian M. Davis; CoPI: Marthe J. Howard).
Supplementary Methods
Statistical Calculations
The Supplementary tables show the measurements of interganglionic distances and areas, as well as the areas of the ganglia. The average value represents the raw arithmetic mean computed using untransformed data. The mean column represents marginal means computed from the linear model after appropriate transformation. The major differences between the arithmetic mean and the median or computed mean show substantial skew of the distributions, which appear to be log-normal. The lower/upper confidence limit columns represent the lower and upper bounds of the 95% CI around the computed mean. The maximum, minimum, and median columns are self-explanatory.
Supplementary Table 1.
Measurements of Interganglionic Distances
| Direction | Region | Average | Minimum | Maximum | Median | IQR | Mean | Lower CL | Upper CL |
|---|---|---|---|---|---|---|---|---|---|
| Longitudinal | Proximal | 101.09 | 38.20 | 226.30 | 92.50 | 53.13 | 95.04 | 88.03 | 102.62 |
| Middle | 166.74 | 9.00 | 952.10 | 159.75 | 69.95 | 149.01 | 138.02 | 160.88 | |
| Distal | 262.83 | 70.00 | 601.00 | 256.65 | 92.68 | 249.61 | 231.19 | 269.50 | |
| Circumferential | Proximal | 49.92 | 20.00 | 130.00 | 50.00 | 22.93 | 46.17 | 42.77 | 49.85 |
| Middle | 91.20 | 20.00 | 260.00 | 81.75 | 57.80 | 81.16 | 75.17 | 87.63 | |
| Distal | 138.63 | 44.60 | 580.00 | 124.25 | 78.88 | 126.86 | 117.50 | 136.96 |
NOTE: The results are depicted in Fig. 4A
CL, confidence limit; IQR, interquartile range.
Supplementary Table 2.
Comparisons of Interganglionic Areas by Region
| Measurement | Regions | Log-ratio | Difference | SMD | Lower CL | Upper CL | P value |
|---|---|---|---|---|---|---|---|
| Longitudinal | Proximal vs middle | 0.640 | 53.969 | 1.069 | 1.333 | 0.806 | <.001 |
| Proximal vs distal | 0.380 | 154.567 | 2.296 | 2.581 | 2.011 | <.001 | |
| Middle vs distal | 0.600 | 100.598 | 1.227 | 1.493 | 0.961 | <.001 | |
| Circumferential | Proximal vs middle | 0.570 | 34.989 | 1.341 | 1.609 | 1.074 | <.001 |
| Proximal vs distal | 0.360 | 80.683 | 2.404 | 2.691 | 2.116 | <.001 | |
| Middle vs distal | 0.640 | 45.695 | 1.062 | 1.326 | 0.798 | <.001 |
NOTE: The results are depicted in Figure 4B. Interganglionic areas (all values are shown in μm2).
CL, confidence limit; SMD, standardized mean difference.
Supplementary Table 3.
Measurements of Interganglionic Area
| Type | Average | Minimum | Maximum | Median | IQR | Mean | Lower CL | Upper CL |
|---|---|---|---|---|---|---|---|---|
| Proximal | 7548 | 304 | 36,108 | 6300 | 5820 | 5876 | 5551 | 6219 |
| Middle | 21,056 | 1377 | 77,034 | 17,353 | 15,372 | 16,960 | 16,015 | 17,960 |
| Distal | 55,972 | 5800 | 160,925 | 51,379 | 32,697 | 49,802 | 45,978 | 53,944 |
NOTE: Data are depicted in Fig. 4B
CL, confidence limit; IQR, interquartile range.
Supplementary Table 4.
Comparison of Mouse and Human Colon
| Regions | Log-ratio | Difference | SMD | Lower CL | Upper CL | P value |
|---|---|---|---|---|---|---|
| Proximal vs middle | 0.346 | 11,084.34 | 1.55 | 1.68 | 1.42 | <.001 |
| Proximal vs distal | 0.118 | 43,926.33 | 3.13 | 3.32 | 2.95 | <.001 |
| Middle vs distal | 0.341 | 32,841.99 | 1.58 | 1.73 | 1.42 | <.001 |
NOTE: The results are depicted from Figure 5A. Areas of the ganglia (all values are shown in μm2).
CL, confidence limit; SMD, standardized mean difference.
Supplementary Table 5.
Measurements of Mouse Colon Myenteric Ganglia Area Compared to Huamn
| Location | Average | Minimum | Maximum | Median | IQR | Mean | Lower CL | Upper CL |
|---|---|---|---|---|---|---|---|---|
| Proximal | 82,640 | 12,623 | 267,697 | 67,354 | 64,006 | 66,599 | 57,507 | 77,127 |
| Middle | 10,679 | 1336 | 46,576 | 8400 | 8706 | 8661 | 7963 | 9420 |
| Distal | 6938 | 84 | 24,427 | 6006 | 4455 | 5704 | 5152 | 6314 |
NOTE: Data are depicted in Fig. 5A.CL, confidence limit; IQR, interquartile range.
Supplementary Table 6.
Comparison by Region of Colon Occupied by Myenteric Ganglia
| Regions | Log ratio | Difference | SMD | Lower CL | Upper CL | P value |
|---|---|---|---|---|---|---|
| Proximal vs middle | 7.68 | 57,937.80 | 3.02 | 2.70 | 3.33 | <.001 |
| Proximal vs distal | 11.67 | 60,895.11 | 3.63 | 3.29 | 3.98 | <.001 |
| Middle vs distal | 1.52 | 2957.31 | 0.62 | 0.42 | 0.82 | <.001 |
NOTE: Data are depicted in Fig. 5B
CL, confidence limit; SMD, standardized mean difference.
References
- 1.Schneider S., Wright C.M., Heuckeroth R.O. Unexpected roles for the second brain: enteric nervous system as master regulator of bowel function. Annu Rev Physiol. 2019;81:235–259. doi: 10.1146/annurev-physiol-021317-121515. [DOI] [PubMed] [Google Scholar]
- 2.Furness J.B. The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol. 2012;9:286–294. doi: 10.1038/nrgastro.2012.32. [DOI] [PubMed] [Google Scholar]
- 3.Spencer N.J., Hu H. Enteric nervous system: sensory transduction, neural circuits and gastrointestinal motility. Nat Rev Gastroenterol Hepatol. 2020;17:338–351. doi: 10.1038/s41575-020-0271-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanner S., Greenwood-Van Meerveld B., Mawe G., et al. Fundamentals of neurogastroenterology: basic science. Gastroenterology. 2016;150(6):1280–1291. doi: 10.1053/j.gastro.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makadia P.A., Najjar S.A., Saloman J.L., Adelman P., Feng B., Margiotta J.F., Albers K.M., Davis B.M. Optogenetic activation of colon epithelium of the mouse produces high-frequency bursting in extrinsic colon afferents and engages visceromotor responses. J Neurosci. 2018;38:5788–5798. doi: 10.1523/JNEUROSCI.0837-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furness J.B. Types of neurons in the enteric nervous system. J Auton Nerv Syst. 2000;81:87–96. doi: 10.1016/s0165-1838(00)00127-2. [DOI] [PubMed] [Google Scholar]
- 7.Lomax A.E., Furness J.B. Neurochemical classification of enteric neurons in the guinea-pig distal colon. Cell Tissue Res. 2000;302:59–72. doi: 10.1007/s004410000260. [DOI] [PubMed] [Google Scholar]
- 8.Messenger J.P., Furness J.B. Projections of chemically-specified neurons in the guinea-pig colon. Arch Histol Cytol. 1990;53:467–495. doi: 10.1679/aohc.53.467. [DOI] [PubMed] [Google Scholar]
- 9.Costa M., Keightley L.J., Wiklendt L., Hibberd T.J., Arkwright J.W., Omari T., Wattchow D.A., Brookes S.J.H., Dinning P.G., Spencer N.J. Identification of multiple distinct neurogenic motor patterns that can occur simultaneously in the guinea pig distal colon. Am J Physiol Gastrointest Liver Physiol. 2019;316:G32–G44. doi: 10.1152/ajpgi.00256.2018. [DOI] [PubMed] [Google Scholar]
- 10.Li Z., Hao M.M., Van den Haute C., Baekelandt V., Boesmans W., Vanden Berghe P. Regional complexity in enteric neuron wiring reflects diversity of motility patterns in the mouse large intestine. Elife. 2019;8 doi: 10.7554/eLife.42914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nurgali K., Stebbing M.J., Furness J.B. Correlation of electrophysiological and morphological characteristics of enteric neurons in the mouse colon. J Comp Neurol. 2004;468:112–124. doi: 10.1002/cne.10948. [DOI] [PubMed] [Google Scholar]
- 12.Furness J.B., Stebbing M.J. The first brain: species comparisons and evolutionary implications for the enteric and central nervous systems. Neurogastroenterol Motil. 2018;30(30:13234.):1–6. doi: 10.1111/nmo.13234. [DOI] [PubMed] [Google Scholar]
- 13.Timmermans J.P., Adriaensen D., Cornelissen W., Scheuermann D.W. Structural organization and neuropeptide distribution in the mammalian enteric nervous system, with special attention to those components involved in mucosal reflexes. Comp Biochem Physiol A Physiol. 1997;118:331–340. doi: 10.1016/s0300-9629(96)00314-3. [DOI] [PubMed] [Google Scholar]
- 14.Timmermans J.P., Scheuermann D.W., Stach W., Adriaensen D., De Groodt-Lasseel M.H. Functional morphology of the enteric nervous system with special reference to large mammals. Eur J Morphol. 1992;30:113–122. [PubMed] [Google Scholar]
- 15.Zeisel A., Hochgerner H., Lonnerberg P., Johnsson A., Memic F., van der Zwan J., Haring M., Braun E., Borm L.E., La Manno G., Codeluppi S., Furlan A., Lee K., Skene N., Harris K.D., Hjerling-Leffler J., Arenas E., Ernfors P., Marklund U., Linnarsson S. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014 e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa M., Keightley L.J., Hibberd T.J., Wiklendt L., Smolilo D.J., Dinning P.G., Brookes S.J., Spencer N.J. Characterization of alternating neurogenic motor patterns in mouse colon. Neurogastroenterol Motil. 2020;33 doi: 10.1111/nmo.14047. [DOI] [PubMed] [Google Scholar]
- 17.Spencer N.J., Travis L., Wiklendt L., Hibberd T.J., Costa M., Dinning P., Hu H. Diversity of neurogenic smooth muscle electrical rhythmicity in mouse proximal colon. Am J Physiol Gastrointest Liver Physiol. 2020;318:G244–G253. doi: 10.1152/ajpgi.00317.2019. [DOI] [PubMed] [Google Scholar]
- 18.Wright C.M., Schneider S., Smith-Edwards K.M., Mafra F., Leembruggen A.J.L., Gonzalez M.V., Kothakapa D.R., Anderson J.B., Maguire B.A., Gao T., Missall T.A., Howard M.J., Bornstein J.C., Davis B.M., Heuckeroth R.O. scRNA-sequencing reveals new enteric nervous system roles for GDNF, NRTN, and TBX3. Cell Mol Gastroenterol Hepatol. 2021;11:1548–1592.e1. doi: 10.1016/j.jcmgh.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drokhlyansky E., Smillie C.S., Van Wittenberghe N., Ericsson M., Griffin G.K., Dionne D., Cuoco M.S., Goder-Reiser M.N., Sharova T., Aguirre A.J., Boland G.M., Graham D., Rozenblatt-Rosen O., Xavier R.J., Regev A. The enteric nervous system of the human and mouse colon at a single-cell resolution. bioRxiv. 2019:746743. [Google Scholar]
- 20.May-Zhang A.A., Tycksen E., Southard-Smith A.N., Deal K.K., Benthal J.T., Buehler D.P., Adam M., Simmons A.J., Monaghan J.R., Matlock B.K., Flaherty D.K., Potter S.S., Lau K.S., Southard-Smith E.M. Combinatorial transcriptional profiling of mouse and human enteric neurons identifies shared and disparate subtypes in situ. Gastroenterology. 2021;160:755–770.e26. doi: 10.1053/j.gastro.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morarach K., Mikhailova A., Knoflach V., Memic F., Kumar R., Li W., Ernfors P., Marklund U. Diversification of molecularly defined myenteric neuron classes revealed by single-cell RNA sequencing. Nat Neurosci. 2021;24:34–46. doi: 10.1038/s41593-020-00736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham K.D., Lopez S.H., Sengupta R., Shenoy A., Schneider S., Wright C.M., Feldman M., Furth E., Valdivieso F., Lemke A., Wilkins B.J., Naji A., Doolin E.J., Howard M.J., Heuckeroth R.O. Robust, 3-dimensional visualization of human colon enteric nervous system without tissue sectioning. Gastroenterology. 2020;158:2221–2235 e5. doi: 10.1053/j.gastro.2020.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avetisyan M., Rood J.E., Huerta Lopez S., Sengupta R., Wright-Jin E., Dougherty J.D., Behrens E.M., Heuckeroth R.O. Muscularis macrophage development in the absence of an enteric nervous system. Proc Natl Acad Sci U S A. 2018;115:4696–4701. doi: 10.1073/pnas.1802490115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barthelemy M. Spatial networks. Phys Rep. 2011;499:1–101. [Google Scholar]
- 25.Barthelemy M. 1st ed. Springer; Paris: 2018. Morphogenesis of spatial networks, lecture notes in morphogenesis. [Google Scholar]
- 26.Herrmann C., Barthelemy M., Provero P. Connectivity distribution of spatial networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;68 doi: 10.1103/PhysRevE.68.026128. [DOI] [PubMed] [Google Scholar]
- 27.Miftakhov R.N., Wingate D.L. Mathematic modelling of the enteric nervous network. 5. Excitation propagation in a planar neural network. Med Eng Phys. 1995;17:11–19. doi: 10.1016/1350-4533(95)90372-i. [DOI] [PubMed] [Google Scholar]
- 28.Sternini C. Structural and chemical organization of the myenteric plexus. Annu Rev Physiol. 1988;50:81–93. doi: 10.1146/annurev.ph.50.030188.000501. [DOI] [PubMed] [Google Scholar]
- 29.Furness J.B., Robbins H.L., Xiao J., Stebbing M.J., Nurgali K. Projections and chemistry of Dogiel type II neurons in the mouse colon. Cell Tissue Res. 2004;317:1–12. doi: 10.1007/s00441-004-0895-5. [DOI] [PubMed] [Google Scholar]
- 30.Qu Z.D., Thacker M., Castelucci P., Bagyanszki M., Epstein M.L., Furness J.B. Immunohistochemical analysis of neuron types in the mouse small intestine. Cell Tissue Res. 2008;334:147–161. doi: 10.1007/s00441-008-0684-7. [DOI] [PubMed] [Google Scholar]
- 31.Mazzuoli G., Schemann M. Mechanosensitive enteric neurons in the myenteric plexus of the mouse intestine. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiocchetti R., Grandis A., Bombardi C., Lucchi M.L., Dal Lago D.T., Bortolami R., Furness J.B. Extrinsic and intrinsic sources of calcitonin gene-related peptide immunoreactivity in the lamb ileum: a morphometric and neurochemical investigation. Cell Tissue Res. 2006;323:183–196. doi: 10.1007/s00441-005-0075-2. [DOI] [PubMed] [Google Scholar]
- 33.Mulderry P.K., Ghatei M.A., Spokes R.A., Jones P.M., Pierson A.M., Hamid Q.A., Kanse S., Amara S.G., Burrin J.M., Legon S., Polak J.M., Bloom S.R. Differential expression of alpha-CGRP and beta-CGRP by primary sensory neurons and enteric autonomic neurons of the rat. Neuroscience. 1988;25:195–205. doi: 10.1016/0306-4522(88)90018-8. [DOI] [PubMed] [Google Scholar]
- 34.Gonkowski S., Rytel L. Somatostatin as an active substance in the mammalian enteric nervous system. Int J Mol Sci. 2019;20:4461. doi: 10.3390/ijms20184461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smolilo D.J., Costa M., Hibberd T.J., Brookes S.J.H., Wattchow D.A., Spencer N.J. Distribution, projections, and association with calbindin baskets of motor neurons, interneurons, and sensory neurons in guinea-pig distal colon. J Comp Neurol. 2019;527:1140–1158. doi: 10.1002/cne.24594. [DOI] [PubMed] [Google Scholar]
- 36.Smolilo D.J., Costa M., Hibberd T.J., Wattchow D.A., Spencer N.J. Morphological evidence for novel enteric neuronal circuitry in guinea pig distal colon. J Comp Neurol. 2018;526:1662–1672. doi: 10.1002/cne.24436. [DOI] [PubMed] [Google Scholar]
- 37.Furness J.B., Bornstein J.C., Trussell D.C. Shapes of nerve cells in the myenteric plexus of the guinea-pig small intestine revealed by the intracellular injection of dye. Cell Tissue Res. 1988;254:561–571. doi: 10.1007/BF00226506. [DOI] [PubMed] [Google Scholar]
- 38.Lomax A.E., Sharkey K.A., Bertrand P.P., Low A.M., Bornstein J.C., Furness J.B. Correlation of morphology, electrophysiology and chemistry of neurons in the myenteric plexus of the guinea-pig distal colon. J Auton Nerv Syst. 1999;76:45–61. doi: 10.1016/s0165-1838(99)00008-9. [DOI] [PubMed] [Google Scholar]
- 39.Clerc N., Furness J.B., Bornstein J.C., Kunze W.A. Correlation of electrophysiological and morphological characteristics of myenteric neurons of the duodenum in the guinea-pig. Neuroscience. 1998;82:899–914. doi: 10.1016/s0306-4522(97)00318-7. [DOI] [PubMed] [Google Scholar]
- 40.Sharkey K.A., Lomax A.E., Bertrand P.P., Furness J.B. Electrophysiology, shape, and chemistry of neurons that project from guinea pig colon to inferior mesenteric ganglia. Gastroenterology. 1998;115:909–918. doi: 10.1016/s0016-5085(98)70263-x. [DOI] [PubMed] [Google Scholar]
- 41.Hibberd T.J., Feng J., Luo J., Yang P., Samineni V.K., RWt Gereau, Kelley N., Hu H., Spencer N.J. Optogenetic Induction of colonic motility in mice. Gastroenterology. 2018;155:514–528 e6. doi: 10.1053/j.gastro.2018.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith T.K., Park K.J., Hennig G.W. Colonic migrating motor complexes, high amplitude propagating contractions, neural reflexes and the importance of neuronal and mucosal serotonin. J Neurogastroenterol Motil. 2014;20:423–446. doi: 10.5056/jnm14092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith-Edwards K.M., Najjar S.A., Edwards B.S., Howard M.J., Albers K.M., Davis B.M. Extrinsic primary afferent neurons link visceral pain to colon motility through a spinal reflex in mice. Gastroenterology. 2019;157:522–536 e2. doi: 10.1053/j.gastro.2019.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margiotta JF, Smith-Edwards KM, Nestor-Kalinoski AL, Davis BM, Albers KM, Howard MJ. Synaptic components, function and modulation characterized by GCaMP6f Ca2+ imaging in mouse cholinergic myenteric ganglion neurons. Front Physiol.Front Physiol. 2021 Aug 2;12:652714. [DOI] [PMC free article] [PubMed]
- 45.Fu Y.Y., Peng S.J., Lin H.Y., Pasricha P.J., Tang S.C. 3-D imaging and illustration of mouse intestinal neurovascular complex. Am J Physiol Gastrointest Liver Physiol. 2013;304:G1–11. doi: 10.1152/ajpgi.00209.2012. [DOI] [PubMed] [Google Scholar]
- 46.Jabari S., Neuhuber W., Brehmer A. Neurovascular interface in porcine small intestine: specific for nitrergic rather than nonnitrergic neurons. Cells Tissues Organs. 2016;201:203–210. doi: 10.1159/000444168. [DOI] [PubMed] [Google Scholar]
- 47.Smith-Edwards K.M., Edwards B.S., Wright C.M., Schneider S., Meerschaert K.A., Ejoh L.L., Najjar S.A., Howard M.J., Albers K.M., Heuckeroth R.O., Davis B.M. Sympathetic input to multiple cell types in mouse and human colon produces region-specific responses. Gastroenterology. 2021;160:1208–1223.e4. doi: 10.1053/j.gastro.2020.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costa M., Wiklendt L., Simpson P., Spencer N.J., Brookes S.J., Dinning P.G. Neuromechanical factors involved in the formation and propulsion of fecal pellets in the guinea-pig colon. Neurogastroenterol Motil. 2015;27:1466–1477. doi: 10.1111/nmo.12646. [DOI] [PubMed] [Google Scholar]
- 49.Smith T.K., Koh S.D. A model of the enteric neural circuitry underlying the generation of rhythmic motor patterns in the colon: the role of serotonin. Am J Physiol Gastrointest Liver Physiol. 2017;312:G1–G14. doi: 10.1152/ajpgi.00337.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szurszewski J.H., Ermilov L.G., Miller S.M. Prevertebral ganglia and intestinofugal afferent neurones. Gut. 2002;51(Suppl 1):i6–10. doi: 10.1136/gut.51.suppl_1.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furness J.B. The enteric nervous system: normal functions and enteric neuropathies. Neurogastroenterol Motil. 2008;20(Suppl 1):32–38. doi: 10.1111/j.1365-2982.2008.01094.x. [DOI] [PubMed] [Google Scholar]
- 52.Bodi N., Talapka P., Poles M.Z., Hermesz E., Jancso Z., Katarova Z., Izbeki F., Wittmann T., Fekete E., Bagyanszki M. Gut region-specific diabetic damage to the capillary endothelium adjacent to the myenteric plexus. Microcirculation. 2012;19:316–326. doi: 10.1111/j.1549-8719.2012.00164.x. [DOI] [PubMed] [Google Scholar]
- 53.Cheng L.S., Schwartz D.M., Hotta R., Graham H.K., Goldstein A.M. Bowel dysfunction following pullthrough surgery is associated with an overabundance of nitrergic neurons in Hirschsprung disease. J Pediatr Surg. 2016;51:1834–1838. doi: 10.1016/j.jpedsurg.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Giorgio R., Bianco F., Latorre R., Caio G., Clavenzani P., Bonora E. Enteric neuropathies: yesterday, today and tomorrow. Adv Exp Med Biol. 2016;891:123–133. doi: 10.1007/978-3-319-27592-5_12. [DOI] [PubMed] [Google Scholar]
- 55.Musser M.A., Correa H., Southard-Smith E.M. Enteric neuron imbalance and proximal dysmotility in ganglionated intestine of the Sox10(Dom/+) Hirschsprung mouse model. Cell Mol Gastroenterol Hepatol. 2015;1:87–101. doi: 10.1016/j.jcmgh.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rivera L.R., Poole D.P., Thacker M., Furness J.B. The involvement of nitric oxide synthase neurons in enteric neuropathies. Neurogastroenterol Motil. 2011;23:980–988. doi: 10.1111/j.1365-2982.2011.01780.x. [DOI] [PubMed] [Google Scholar]
- 57.Chandrasekharan B., Nezami B.G., Srinivasan S. Emerging neuropeptide targets in inflammation: NPY and VIP. Am J Physiol Gastrointest Liver Physiol. 2013;304:G949–G957. doi: 10.1152/ajpgi.00493.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gershon M.D., Bursztajn S. Properties of the enteric nervous system: limitation of access of intravascular macromolecules to the myenteric plexus and muscularis externa. J Comp Neurol. 1978;180:467–488. doi: 10.1002/cne.901800305. [DOI] [PubMed] [Google Scholar]
- 59.Spadoni I., Zagato E., Bertocchi A., Paolinelli R., Hot E., Di Sabatino A., Caprioli F., Bottiglieri L., Oldani A., Viale G., Penna G., Dejana E., Rescigno M. A gut-vascular barrier controls the systemic dissemination of bacteria. Science. 2015;350:830–834. doi: 10.1126/science.aad0135. [DOI] [PubMed] [Google Scholar]
- 60.Krantis A. GABA in the mammalian enteric nervous system. News Physiol Sci. 2000;15:284–290. doi: 10.1152/physiologyonline.2000.15.6.284. [DOI] [PubMed] [Google Scholar]
- 61.Margiotta J.F., Pardi D. Pituitary adenylate cyclase-activating polypeptide type I receptors mediate cyclic AMP-dependent enhancement of neuronal acetylcholine sensitivity. Mol Pharmacol. 1995;48:63–71. [PubMed] [Google Scholar]
- 62.Smolilo D.J., Hibberd T.J., Costa M., Wattchow D.A., De Fontgalland D., Spencer N.J. Intrinsic sensory neurons provide direct input to motor neurons and interneurons in mouse distal colon via varicose baskets. J Comp Neurol. 2020;528:2033–2043. doi: 10.1002/cne.24872. [DOI] [PubMed] [Google Scholar]
- 63.Hendershot T.J., Liu H., Clouthier D.E., Shepherd I.T., Coppola E., Studer M., Firulli A.B., Pittman D.L., Howard M.J. Conditional deletion of Hand2 reveals critical functions in neurogenesis and cell type-specific gene expression for development of neural crest-derived noradrenergic sympathetic ganglion neurons. Dev Biol. 2008;319:179–191. doi: 10.1016/j.ydbio.2008.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hendershot T.J., Liu H., Sarkar A.A., Giovannucci D.R., Clouthier D.E., Abe M., Howard M.J. Expression of Hand2 is sufficient for neurogenesis and cell type-specific gene expression in the enteric nervous system. Dev Dyn. 2007;236:93–105. doi: 10.1002/dvdy.20989. [DOI] [PubMed] [Google Scholar]
- 65.Chen T.W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V., Looger L.L., Svoboda K., Kim D.S. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lakso M., Pichel J.G., Gorman J.R., Sauer B., Okamoto Y., Lee E., Alt F.W., Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heffner C.S., Herbert Pratt C., Babiuk R.P., Sharma Y., Rockwood S.F., Donahue L.R., Eppig J.T., Murray S.A. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat Commun. 2012;3:1218. doi: 10.1038/ncomms2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boesmans W., Martens M.A., Weltens N., Hao M.M., Tack J., Cirillo C., Vanden Berghe P. Imaging neuron-glia interactions in the enteric nervous system. Front Cell Neurosci. 2013;7:183. doi: 10.3389/fncel.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Artimovich E., Jackson R.K., Kilander M.B.C., Lin Y.C., Nestor M.W. PeakCaller: an automated graphical interface for the quantification of intracellular calcium obtained by high-content screening. BMC Neurosci. 2017;18:72. doi: 10.1186/s12868-017-0391-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou X., Ren J., Brown E., Schneider D., Caraballo-Lopez Y., Galligan J.J. Pharmacological properties of nicotinic acetylcholine receptors expressed by guinea pig small intestinal myenteric neurons. J Pharmacol Exp Ther. 2002;302:889–897. doi: 10.1124/jpet.102.033548. [DOI] [PubMed] [Google Scholar]