Abstract
STAM1, a member of the STAM (signal transducing adapter molecule) family, has a unique structure containing a Src homology 3 domain and ITAM (immunoreceptor tyrosine-based activation motif). STAM1 was previously shown to be associated with the Jak2 and Jak3 tyrosine kinases and to be involved in the regulation of intracellular signal transduction mediated by interleukin-2 (IL-2) and granulocyte-macrophage colony-stimulating factor (GM-CSF) in vitro. Here we generated mice lacking STAM1 by using homologous recombination with embryonic stem cells. STAM1−/− mice were morphologically indistinguishable from their littermates at birth. However, growth retardation in the third week after birth was observed for the STAM1−/− mice. Unexpectedly, despite the absence of STAM1, hematopoietic cells, including T- and B-lymphocyte and other hematopoietic cell populations, developed normally and responded well to several cytokines, including IL-2 and GM-CSF. However, histological analyses revealed the disappearance of hippocampal CA3 pyramidal neurons in STAM1−/− mice. Furthermore, we observed that primary hippocampal neurons derived from STAM1−/− mice are vulnerable to cell death induced by excitotoxic amino acids or an NO donor. These data suggest that STAM1 is dispensable for cytokine-mediated signaling in lymphocytes but may be involved in the survival of hippocampal CA3 pyramidal neurons.
STAM (signal transducing adapter molecule) was previously identified as a phosphotyrosine protein induced by stimulation with a variety of cytokines and growth factors, such as interleukin-2 (IL-2), IL-4, IL-7, IL-3, granulocyte-macrophage colony-stimulating factor (GM-CSF), platelet-derived growth factor (PDGF), and epidermal cell growth factor (34). It was demonstrated that STAM has unique structures containing a Src homology 3 (SH3) domain and a tyrosine cluster region including an immunoreceptor tyrosine-based activation motif (ITAM) (34). STAM was also found to be associated with Janus kinase 2 (Jak2) and Jak3 and to be involved in signaling for cell growth and c-myc induction mediated by IL-2 and GM-CSF in vitro (35). Recently, a new member of the STAM family, STAM2, was molecularly cloned (9, 27). Hence, we renamed the original STAM as STAM1. The in vitro function of STAM2 was not distinguishable from that of STAM1 (9, 27, 35). Although the STAM family proteins have been suggested to be important for the downstream signaling of the Jaks in vitro, their in vivo biological significance is still unknown.
Jak3 and Jak2 are associated with the cytoplasmic portions of the common cytokine receptor subunits, γc and βc chains, respectively (31, 32); they are activated by their respective cytokines to induce the downstream signal transduction including phosphorylation and activation of the Stat family proteins, which transmit signals from the receptors to the nucleus to induce the expression of target genes (14, 17). IL-2-induced cell proliferation is significantly impaired in peripheral T cells derived from Stat5A/B double-knockout mice, but these double-knockout mice show normal development of T cells (22), indicating that Stat5A and Stat5B are dispensable for T-cell development. Since the γc-Jak3 signaling pathway is indispensably involved in T-cell development (16, 25, 32), we speculate that signaling molecules other than Stat5 are critically involved in the signaling pathway directly downstream of Jak3 for T-cell development. To investigate this possibility, we focused on STAM1, associated with the Jaks, and generated STAM1 knockout mice by gene targeting. Unexpectedly, the development of lymphocytes was unaltered in STAM1−/− mice, and there was no obvious difference in IL-2-mediated DNA synthesis and c-myc induction between wild-type and STAM1−/− mice. However, histological analysis revealed a loss of hippocampal CA3 pyramidal neurons in STAM1−/− mice. The phenotypes of STAM1−/− mice suggest that STAM1 is dispensable for cytokine-mediated signaling in lymphocytes but may be involved in the survival of hippocampal CA3 pyramidal neurons in vivo.
MATERIALS AND METHODS
Targeted disruption of STAM1.
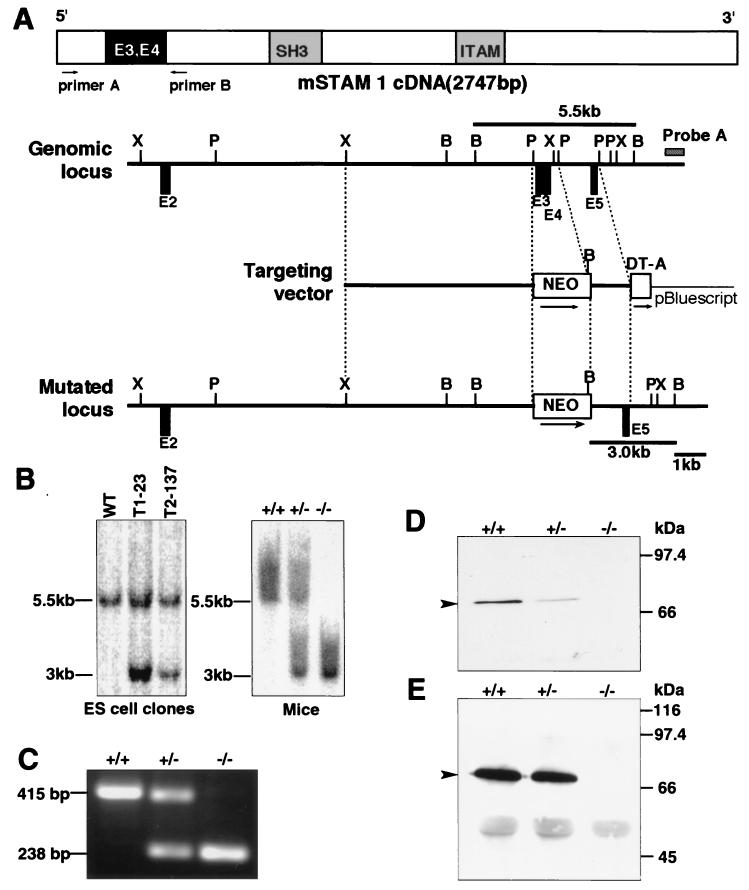
The STAM1 genomic locus was isolated from a λFixII mouse 129/Sv genomic library (Stratagene) using a 5′ region of STAM1 cDNA. The targeting vector was constructed using a pGK-neo cassette flanked by a pair of loxP sequences for positive selections and a diphtheria toxin A-chain gene cassette without a polyadenylation site for negative selection (Fig. 1A). This targeting construct replaces a 0.6-kb PstI-PstI genomic fragment encompassing exons 3 and 4, flanked by 6.5-kb (XhoI-PstI) and 1.4-kb (PstI-PstI) genomic sequences derived from the 129/Sv genomic library (Fig. 1A). The construct was linearized and electroporated into 129/Sv-derived J1 ES cells, and colonies were selected with G418 (5, 24, 30). Homologous recombination events were assessed by Southern blot hybridization (Fig. 1B). Four ES cell clones containing a mutated allele were identified. Two targeted ES clones (T1-23 and T2-137) were injected into C57BL/6 blastocysts and transferred to foster mothers to obtain chimeric mice. The chimeric male mice were mated with C57BL/6 female mice. The F1 heterozygous mice carrying the STAM1 mutation were identified by Southern blot hybridization and intercrossed to produce F2 homozygous offspring. The F2 mice were genotyped by Southern blot hybridization and by PCR with DNA from tail biopsy specimens.
FIG. 1.
Generation of STAM1-deficient mice. (A) Schematic representation of the mouse STAM1 (mSTAM1) cDNA, stam1 genomic locus, targeting vector, and stam1 mutated locus. The positions of stam1 exons are shown as boxes. The targeting vector was designed to replace exon 3 (E3) and E4, encoding amino acids 42 to 99 of STAM1. The fragments expected to be generated by BamHI digestion are 5.5 and 3.0 kb for the wild-type and the mutated alleles, respectively. B, BamHI; P, PstI; X, XbaI. (B) Southern blot analysis of the stam1 mutation in ES cell clones and mice. Lines indicate the positions of the DNA fragments corresponding to the wild-type (5.5 kb) and mutated (3.0 kb) alleles. (C) RT-PCR analysis of total RNA from purified splenocytes of STAM1+/+, STAM1+/−, and STAM1−/− mice. The primers used are primers A and B, shown in panel A. (D) Western blot analysis for STAM1. Neocortex lysates (20 μg) from STAM1+/+, STAM1+/−, and STAM1−/− mice were separated by SDS-PAGE and blotted with anti-STAM1 antibody. The position of STAM1 is marked by an arrowhead. (E) Immunoprecitipation analysis for STAM1. Lysates of activated T cells from STAM1+/+, STAM1+/−, and STAM1−/− mice were immunoprecipitated and then immunoblotted with anti-STAM1 antibody. The position of STAM1 is marked by an arrowhead.
The following oligonucleotide primers were used: STAM1FA (primer 1), CGGGACCAGAGGAAAAGCACCTGTCAC; STAM1RA (primer 2), ATCAGTGTACAAATGGGAAGGTATTAT; and PGK-2 (primer 3), TGCGAGGCCAGAGGCCACTTGTGTAGC. PCR conditions were as follows: denaturation at 94°C for 2 min, followed by 35 cycles of 1 min at 94°C, 1 min at 57°C, and 1 min at 72°C. The wild-type and mutant alleles gave rise to PCR-amplified fragments of 325 and 379 bp, respectively.
RT-PCRs.
Reverse transcription (RT)-PCRs were carried out with 5 μg of total RNA derived from splenocytes of 8-week-old STAM1+/+, STAM1+/−, and STAM1−/− mice as the template. The total RNA from each mouse was prepared by using TRIzol (Gibco-BRL). First-strand synthesis was performed using a Superscript preamplification system (Gibco-BRL). PCRs were performed with a 50-μl mixture consisting of 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 0.2 mM deoxynucleoside triphosphate mixture, 1 μM concentrations of various primers, 1.25 U of Taq DNA polymerase (Takara Shuzo), and 2 μl of the RT reaction mixture as a template. PCR conditions were as follows: denaturation at 94°C for 2 min, followed by 35 cycles of 30 s at 94°C, 30 s at 57°C, and 1 min at 72°C.
The following oligonucleotide primers were used: STAMex1F (primer A), CCCTTCGACCAGGATGTTGAGAAAGCA; and STAMex5R (primer B), CCCTTCGACCAGGATGTTGAGAAAGCA.
Immunoprecipitation and immunoblotting.
Neocortices from mice were homogenized in Nonidet P-40 lysis buffer (1% Nonidet P-40, 25 mM Tris-HCl [pH 7.5], 140 mM NaCl, 10 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 20 μg of aprotinin/ml, 1 mM Na3VO4). Supernatants of the lysates were subjected to immunoblotting. Activated T cells were lysed in Nonidet P-40 lysis buffer. Their lysates were immunoprecipitated with TUS-1 (immunoglobulin G1 [IgG1]), a monoclonal antibody (MAb) specific for STAM1 (34). The lysates or immunoprecipitates were separated by sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE) and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). After blocking with 5% nonfat milk in phosphate-buffered saline (PBS) containing 0.1% Tween 20, the filters were incubated with TUS-1, followed by incubation with anti-mouse IgG coupled with horseradish peroxidase; visualization was done by use of the enhanced chemiluminescence detection system (Amersham Pharmacia Biotech).
T-cell isolation and culturing.
Splenic cells were isolated from 4-week-old wild-type or STAM1-deficient mice. In brief, freshly isolated spleens were passed through a cell strainer to separate fibrous tissues, and red blood cells were lysed in a lysis buffer (pH 7.3) containing 150 mM NH4Cl, 1 mM KHCO3, and 0.1 mM EDTA. The splenic cells were resuspended in RPMI 1640 medium containing 10% fetal bovine serum (FBS) and 50 μM 2-mercaptomethanol at 106 cells/ml. CD4+ T cells were purified from spleens by using anti-CD4 Dynabeads (Dynal) and were further separated from the beads by using DETACHaBEAD (Dynal). The purity of the CD4+ T cells was confirmed to be greater than 98% by flow cytometry. For preparation of activated T cells, the splenic cells were stimulated with phorbol myristate acetate (PMA) (10 ng/ml; Sigma) and ionomycin (1 μg/ml; Sigma) for 24 h and then were cultured with recombinant human IL-2 (rhIL-2; 1 nM; Ajinomoto) for 7 days.
Proliferation assay.
Single-cell suspensions of spleen cells, bone marrow cells, or CD4+ T cells from spleens in RPMI 1640 medium supplemented with 10% FBS, 50 μM 2-mercaptomethanol, penicillin, and streptomycin were plated in 96-well plates at a density of 105 cells per well in 100 μl of medium. Stimuli were added and cultured for 42 h. The stimuli were rhIL-2, recombinant murine IL-7 (PeproTech), recombinant murine GM-CSF (PeproTech), an anti-CD3 MAb (145.2C11; Pharmingen), concanavalin A (ConA), PMA, ionomycin, and lipopolysaccharide (LPS; Sigma). Then the cells were pulsed with [3H]thymidine and harvested after 6 h. The incorporated [3H]thymidine was counted with a MicroBeta liquid scintillation counter (Amersham Pharmacia Biotech).
Flow cytometry.
Thymocytes and splenic cells were suspended in PBS supplemented with 3% FBS. They were preincubated in normal mouse serum to prevent labeled MAbs from nonspecific binding to the cell surface. They were then stained with MAbs conjugated with fluorescein isothiocyanate, phycoerythrin, or biotin for 30 min at 4°C. The cells were washed with PBS–3% FBS, and the biotinylated antibodies were developed with streptavidin-APC (Pharmingen). All the MAbs used were purchased from Pharmingen. The surface staining with MAbs was analyzed with a FACSCalibur flow cytometer (Becton Dickinson) in two- or three-color mode using CellQuest software.
Internalization and degradation of IL-2.
Assays of internalization and degradation of IL-2 were performed as described previously (10). In brief, cells were washed twice with PBS and incubated in RPMI 1640 medium containing 1% FBS and 25 mM HEPES (RPMI-HEPES) for 4 h. They were further incubated with 200 pM 125I-rhIL-2 (6 × 108 to 8.5 × 108 cpm/pmol) in RPMI-HEPES for 30 min at 0°C. They were then washed three times with PBS containing 3% bovine serum albumin, resuspended in 1.0 ml of RPMI-HEPES, and further incubated for indicated times at 37°C. After centrifugation, the supernatants were harvested and the cell pellets were treated with chilled 0.2 M glycine buffer (pH 2.8) for 10 min at 0°C. The radioactivities of the supernatant fractions, acid-removed glycine buffer fractions (surface ligands), and non-acid-removed cell precipitates (internalized ligands) were counted. For determination of degradation of 125I-rhIL-2, the supernatants were subjected to trichloroacetic acid precipitation. The radioactivities of the trichloroacetic acid-soluble (degraded ligands) and insoluble fractions were measured. The rate of internalization was expressed as the ratio of internalized ligands to surface ligands plus internalized ligands plus degraded ligands. The rate of degradation was expressed as the ratio of degraded ligands to surface ligands plus internalized ligands plus degraded ligands.
Histological and immunohistochemical analyses of brains.
Mice were perfused with PBS followed by 4% paraformaldehyde–PBS. Brains were removed for processing and embedding in paraffin and were sectioned at a 5- or 10-μm thickness by a microtome. For histological analyses, the 5-μm sections were stained with hematoxylin-eosin (HE). The sections were also used for detection of calbindin (23). The sections were incubated with rabbit polyclonal anti-calbindin antiserum (1:10,000) in PBS for 24 h at 4°C. They were washed in PBS and subsequently incubated with an avidin-biotin–horseradish peroxidase complex (ABC Elite; Vector) for 1 h at room temperature. The final reaction product was visualized with 3,3′-diaminobenzidine. For terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays, the 10-μm sections were deparaffinized, and terminal transferase labeling of fragmented DNA in the sections was performed with a TACS 2 TdT kit (HRP-Blue Label; Trevigen) according to the assay protocol of this kit.
For immunofluorohistochemical analyses of glial fibrillary acidic protein (GFAP) and STAM1, fresh frozen brains of mice were sectioned at a 10-μm thickness by a cryostat, fixed in 4% paraformaldehyde–PBS for 10 min at 4°C, permeabilized in 0.3% Triton X-100 for 20 min, and preincubated in PBS containing 5% horse serum for 1 h at room temperature. Subsequently, the sections were incubated overnight at 4°C with primary antibodies against GFAP (goat polyclonal, 1 μg/ml; Santa Cruz Biotechnology) or STAM1 (TUS-1; 10 μg/ml). The sections were then incubated with the corresponding fluorescein- or rhodamine-conjugated secondary antibodies (1:100; Vector) for 1 h at room temperature. Coverslips were mounted in PermaFluor (Shandon).
In situ hybridization.
In situ hybridization for mouse STAM1 was performed according to a modified method described previously (28). Fresh frozen whole bodies at embryonic day 18 (E18) and brains at postnatal week 5 of wild-type and mutant mice were sectioned at a 30-μm thickness by a cryostat. After fixation in 4% paraformaldehyde–0.1 M sodium phosphate buffer (pH 7.2), the sections were acetylated with 0.25% acetic anhydride in 0.1 M triethanolamine (pH 8.0) and prehybridized for 1 h in a buffer containing 50% deionized formamide, 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 7.4]), 0.02% polyvinylpyrrolidone, 0.02% bovine serum albumin, 0.02% Ficoll, 1% sodium N-lauroyl sarcosinate (Sarkosyl), 0.1 M sodium phosphate buffer, and 100 μg of tRNA/ml. Hybridization was performed overnight at 42°C with the prehybridization buffer supplemented with 10% dextran sulfate, 100 mM dithiothreitol, and 35S-labeled oligonucleotide probes (2 × 107 cpm/ml). The sections were washed with 0.1× SSC–0.1% Sarkosyl four times for 30 min each time at 50°C. They were exposed to Hyperfilm β-max (Amersham Pharmacia Biotech) for 2 weeks at room temperature. They were subsequently autoradiographed using NTB2 nuclear track emulsion (Kodak) for 3 weeks at 4°C.
A 35S-labeled oligonucleotide probe for mouse STAM1 was prepared as follows. An antisense 30-mer oligonucleotide was synthesized, which was complementary to nucleotide residues 173 to 202 contained in exon 3 of mouse STAM1. The oligonucleotide was labeled using terminal deoxynucleotidyltransferase (Gibco-BRL) with [α-35S]dATP (NEN Life Science Products).
Neuronal culturing.
Primary hippocampal neurons were isolated from wild-type and STAM1−/− embryos at E18.5. Fetal hippocampi were dissected and minced with scissors. Individual cells were mechanically isolated by trituration in calcium- and magnesium-free Hanks' balanced salt solution with a siliconized 9-in. Pasteur pipette with a tip barely fire polished. The cells were plated on poly-d-lysine-coated plates (Falcon) and maintained in Neurobasal medium (Gibco-BRL) plus B27 supplement (Gibco-BRL) at 37°C in a humidified atmosphere of 5% CO2 and 95% room air. Cells were seeded at a density of 600 cells/mm2.
Immunostaining of cells.
Cells were fixed in 4% paraformaldehyde in PBS for 10 min at room temperature, permeabilized in 0.3% Triton X-100 for 20 min, and rinsed in PBS. The cells were preincubated in 5% normal goat serum–PBS at room temperature for 1 h and incubated overnight at 4°C with primary antibodies against STAM1 (TUS-1; 10 μg/ml), GluR1 (rabbit polyclonal, 1 μg/ml; Upstate Biotechnology), synapsin I (rabbit polyclonal, 1:1,000; Chemicon), and SNAP-25 (rabbit polyclonal, 1:1,000; Chemicon). The sections were then incubated with the corresponding fluorescein- and/or rhodamine-conjugated secondary antibodies (1:100; Vector) for 1 h at room temperature. Coverslips were mounted in PermaFluor (Shandon). They were analyzed by confocal laser scanning microscopy (TCS NT; Leica).
Subcellular fractionation.
Subcellular fractions were prepared from adult C57BL/6 mouse brains as described previously (7, 13, 21, 26) but with minor modifications. Briefly, cerebral cortices were homogenized in ice-cold buffered sucrose (0.32 M sucrose, 10 mM Tris buffer [pH 7.4]) and centrifuged for 10 min at 4°C and 1,000 × g. The resulting pellet (P1) was discarded, while the supernatant (S1) was collected and centrifuged for 30 min at 15,000 × g. The supernatant (S2; cytosolic fraction) was removed, and the pellet (P2) was washed by resuspension in buffered sucrose and recentrifuged for 30 min at 15,000 × g to yield a supernatant (S2′) and a pellet (P2′).
The pellet (P2′) was resuspended in buffered sucrose and applied to a gradient containing 0.8 and 1.2 M sucrose solutions. The sucrose density gradients were centrifuged for 2 h at 63,000 × g. The band between 0.8 and 1.2 M containing synaptosomes was collected (Syn). To prepare the synaptosomal membrane fraction, the synaptosomal fraction was lysed by suspension in 5 mM Tris–0.1 mM EDTA (pH 8.0) and stirring of the suspension at 4°C for 1 h. The synaptosomal membranes were spun down for 1 h at 100,000 × g and resuspended in buffered sucrose. Then the suspension was applied to the gradient containing 0.85, 1.0, and 1.2 M sucrose solutions. The sucrose density gradients were centrifuged for 20 min at 48,200 × g. The band between 1.0 and 1.2 M sucrose containing synaptosomal membranes was collected (Sm). To prepare postsynaptic density (PSD) fractions, the synaptosomal fraction was incubated with ice-cold 0.5% Triton X-100 and then centrifuged for 20 min at 32,000 × g to obtain the pellet. This pellet was resuspended, incubated a second time in 0.5% Triton X-100, and centrifuged for 1 h at 201,800 × g to obtain the PSD pellet (PSD).
To prepare the synaptic vesicle fraction, the P2′ pellet was lysed by hypo-osmotic shock (suspended in ice-cold water). The suspension was then centrifuged for 20 min at 25,000 × g. The supernatant was collected and recentrifuged for 2 h at 165, 000 × g. The pellet was resuspended in 40 mM sucrose and applied to the 50 to 800 mM sucrose gradient. The sucrose density gradients were centrifuged for 5 h at 65,000 × g. The 200 to 400 mM sucrose region containing synaptic vesicles was collected (Sv).
Electrophoresis was performed with SDS-polyacrylamide gels and 10 μg of protein from each fraction and then transferred to PVDF membranes (Millipore). After blocking with 5% nonfat milk in PBS containing 0.1% Tween 20, the filters were incubated with primary antibodies against STAM1, Synapsin-I, SNAP-25, or PSD-95 (Transduction Laboratories) followed by incubation with appropriate secondary antibodies coupled with horseradish peroxidase and visualized by using the enhanced chemiluminescence detection system.
Clinical signs of drug-induced seizures.
Seizure responses to drugs were observed in C57BL/6-background 3-week-old wild-type and STAM1−/− mice, because the loss of CA3 pyramidal neurons occurred in older STAM1−/− mice. All drugs were administrated intraperitoneally. Seizures were scored as described previously (43). At least six mice in each group were observed and scored to derive the temporal response curve. Since pentetrazol (PTZ) provoked rapid and abrupt general tonic-clonic convulsions, the same criteria were used to record PTZ-induced seizures within 5 min of injection.
Cell survival assay.
Primary neurons were cultured for 12 days in Neurobasal medium plus B27 supplement. The cultured neurons were resuspended in Neurobasal medium plus B27 supplement but minus AO (Life Technologies) supplement, because B27 supplement contains some antioxidants. They were exposed to 150 μM kainic acid or 50 μM sodium nitroprusside (SNP) for 24 h and stained for 2 h with 10% Alamar blue dye (Alamar Biosciences, Sacramento, Calif.), which is a redox indicator, to assess the viability and metabolic activity of the cells (37, 42). The light absorbance of the reduced form of the dye was measured at 540 nm, whereas the oxidized form was measured at 620 nm. The viability of cells was expressed as the optical density at 540 nm (OD540) minus the OD620. The viability of primary STAM1+/+ neurons with no drug treatment was taken to be 100%.
RESULTS
Generation of mice targeted for STAM1.
To generate STAM1-deficient mice, J1 ES cells were transfected with the targeting construct shown in Fig. 1A. The gene encoding STAM1 was inactivated by replacing exons 3 and 4, which encode amino acids 44 through 99, with the targeting construct containing a neomycin resistance (neo) gene. Two independent clones of J1 ES cells which carried the disrupted STAM1 gene were used to generate heterozygous (STAM1+/−) and homozygous (STAM1−/−) mice (Fig. 1B). To assess if exons 3 and 4 were deleted, we performed RT-PCR analysis of purified splenocytes from STAM1+/+, STAM1+/−, and STAM1−/− mice. A pair of primers spanning exons 3 and 4 amplified a 415-bp fragment from STAM1+/+ cells and a 238-bp fragment from STAM 1−/− cells (Fig. 1C). Sequence analysis of the RT-PCR fragments showed that the STAM1 mRNA transcript from STAM1−/− cells contained a deletion of nucleotides 170 through 341, resulting in a frameshift and a stop codon in exon 5. Moreover, immunoblotting analyses with an anti-STAM1 MAb, TUS-1, revealed that STAM1 was undetectable in extracts of neocortices of STAM1−/− mice but was detectable in those of STAM1+/+ and STAM1+/− mice (Fig. 1D). Similarly, the expression of STAM1 was undetectable in activated T cells derived from STAM1−/− mice but was detectable in those derived from STAM1+/+ and STAM1+/− mice (Fig. 1E). Collectively, these data show that the homologous mutation of the STAM1 gene leads to a deficiency of STAM1 in mice.
Viability, behavioral phenotype, and fertility of STAM1−/− mice.
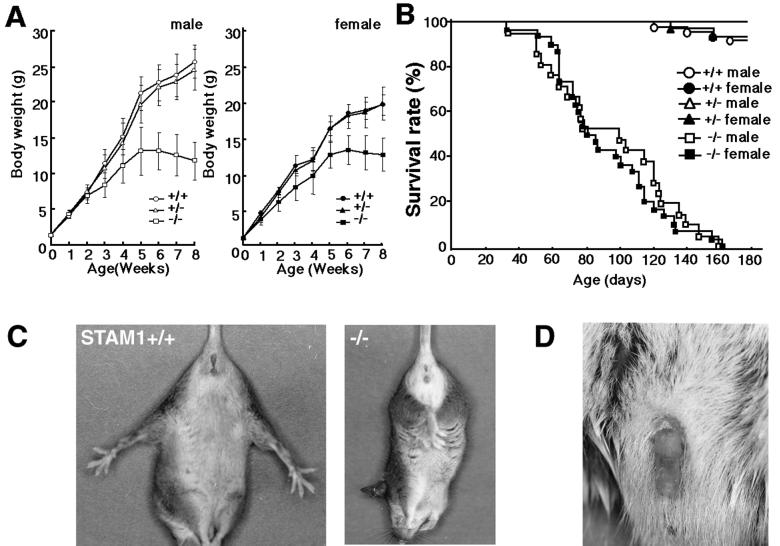
STAM1-deficient mice were morphologically indistinguishable from their littermates at birth. Genotypic analysis of neonatal offspring (n = 134) from STAM1+/− matings revealed the expected Mendelian ratios of STAM1+/+ (26%), STAM1+/− (49%), and STAM1−/− (24%) animals (data not shown), indicating that STAM1 is not essential for embryonic development. Heterozygous mice did not differ in growth, viability, or behavior from their wild-type littermates. However, growth retardation of STAM1−/− mice became detectable by 2 weeks of age and then gradually increased, and most of the STAM1−/− mice did not show any increase in their body weights after 5 weeks of age (Fig. 2A). The STAM1−/− mice began to die after 6 weeks of age and could not survive for more than 6 months (Fig. 2B).
FIG. 2.
Phenotypes of STAM1−/− mice. (A) Growth curves for STAM1+/+ (male, n = 26; female, n = 9), STAM1+/− (male, n = 36; female, n = 30), and STAM1−/− (male, n = 21; female, n = 12) mice. One male mouse died at 7 weeks, and four male mice and two female mice died at 8 weeks. Error bars indicate standard errors. (B) Survival curves for STAM1+/+ (male, n = 21; female, n = 31), STAM1+/− (male, n = 27; female, n = 31), and STAM1−/− (male, n = 21; female, n = 30) mice. (C) Abnormal limb reflex of STAM1−/− mice. STAM1+/+ mice extend their legs when lifted by the tail (left panel), whereas STAM1−/− mice lack this reflex and show clasping of the hind limbs (right panel). The mice were 4-week-old littermates. (D) Priapism in STAM1−/− mice. Priapism was observed in male STAM1−/− mice surviving for more than 8 weeks. About 70% of STAM1−/− male mice suffered from priapism during their lives.
A neurological abnormality was suspected on the basis of the observation that the STAM1−/− mice retracted their hind limbs toward the trunk when they were lifted by their legs, in contrast with their wild-type and heterozygous littermates, which invariably responded by extending their legs (Fig. 2C). This abnormal leg-clasping reflex, which became evident at 2 weeks of age, was pronounced by 3 to 4 weeks and was attenuated in surviving adults. This phenotype was observed for more than 200 mice tested, although the severity of the symptoms varied from mouse to mouse. Apart from this phenotype, coordination of movement appeared normal.
In STAM1−/− male mice surviving for more than 8 weeks, priapism, persistent penile erection, was observed (Fig. 2D). About 70% of the STAM1−/− male mice suffered from priapism during their lives. Since we could not histologically find any embolus in the veins of the penises of these affected mice, we suspect that this phenomenon may have been due to a neurological abnormality. Moreover, adult STAM1−/− males with or without priapism housed with wild-type females failed to produce litters, although histological examination of STAM1−/− mouse testes revealed that mature spermatozoa were present in the lumens of seminiferous tubules (data not shown). The same phenotype was observed for mutant strains derived from two independent clones with mixed 129/Sv × C57BL/6J and congenic C57BL/6J (≥10-generation backcross) genetic backgrounds. These observations suggest that the infertility of the STAM1−/− males is caused by impotence.
Normal development, proliferative responses, and IL-2 internalization of T cells in STAM1−/− mice.
To analyze the contribution of STAM1 in lymphoid development, a number of parameters were examined with 4-week-old mice. With regard to the anatomy of the lymphoid organs, there was no apparent defect in any of the STAM1−/− mice tested. The thymuses of the STAM1−/− and wild-type mice were equal in size, and the ratios of CD4+ or CD8+ cells in the STAM1−/− thymuses were the same as those in the wild-type thymuses (Fig. 3A). The spleens from the STAM1−/− mice were smaller than those from the wild-type mice, but the ratios of CD4+ or CD8+ cells in the spleens were not significantly different between the STAM1−/− and wild-type mice (Fig. 3A). We also analyzed markers for B-cell (IgM and B220), myeloid (Gr1 and CD11b), and erythroid (Ter119) lineages in splenocytes, but there was no significant difference between wild-type and STAM1−/− mice (data not shown).
FIG. 3.
Comparison of T-cell development, proliferative responses to cytokines, and internalization of IL-2 between STAM1+/+ and STAM1−/− mice. (A) Flow cytometric analysis for CD4 and CD8. Thymic and splenic lymphocytes derived from 5-week-old STAM1+/+ (n = 7) and STAM1−/− (n = 7) mice were doubly stained with anti-CD4 and anti-CD8 MAbs. Numbers indicate the average percentages of the gated cellular subpopulations within the lymphocyte population. (B) Proliferative responses of bone marrow and spleen cells. Total splenocytes or bone marrow cells (105 per well) were stimulated with the indicated ligands: 10 μg of ConA/ml, 3 μg of anti-CD3 MAb/ml, 10 nM IL-2, 100 ng of LPS/ml, 5 ng of IL-7/ml, and 30 ng of GM-CSF/ml. They were cultured for 42 h, pulsed with [3H]thymidine, and harvested after 6 h. (C) Proliferative responses of T cells. CD4+ T cells (105 per well) purified from spleens were stimulated with anti-CD3 MAb (145.2C11) or PMA (10 ng/ml) plus ionomycin (1 μg/ml) for 42 h (left panel). ConA-activated splenic T cells (105 per well) were preincubated with RPMI 1640 medium containing 1% FBS and 50 μM 2-mercaptoethanol for 12 h and then stimulated with rhIL-2 for 42 h (right panel). The cells were then pulsed with [3H]thymidine and harvested. (D) Internalization and degradation of IL-2 in activated T cells. Activated T cells from STAM1+/+ or STAM1−/− mice were tested for 125I-IL-2 internalization (left panel) and degradation (right panel). Data represent means and standard errors for three independent experiments performed in triplicate.
To analyze perturbations in the intracellular signaling pathways and proliferative responses of STAM1−/− mice, splenocytes or bone marrow cells were compared with those of wild-type mice. The proliferative responses to ConA, LPS, anti-CD3, IL-2, or IL-7 and to their combinations were not significantly different between the STAM1−/− and STAM1+/+ mice (Fig. 3B). The proliferative responses of purified CD4+ T cells to anti-CD3 and activated T cells to IL-2 were also compared between the STAM1−/− and STAM1+/+ mice. Similar magnitudes of proliferative responses to various doses of anti-CD3 and IL-2 were seen for the STAM1+/+ and STAM1−/− mice (Fig. 3C). We also confirmed that IL-2 induced the expression of c-myc, c-fos, and bcl-2 in activated T cells derived from STAM1+/+ and STAM1−/− mice equally well (data not shown).
EAST and HBP, respectively, are chicken and mouse molecules homologous to STAM1 and were recently reported to be involved in the receptor-mediated endocytosis of epidermal cell growth factor (18) and the vesicular transport of PDGF-PDGF receptor complexes through early endosomes (33). Hence, we examined whether or not STAM1 is involved in the internalization and degradation of IL-2. Activated T cells derived from the spleens of STAM1+/+ and STAM1−/− mice were examined for internalization and degradation of IL-2, and no significant difference in these processes was observed between these mice (Fig. 3D).
Abnormalities in hippocampal CA3 subfields in STAM1−/− mice.
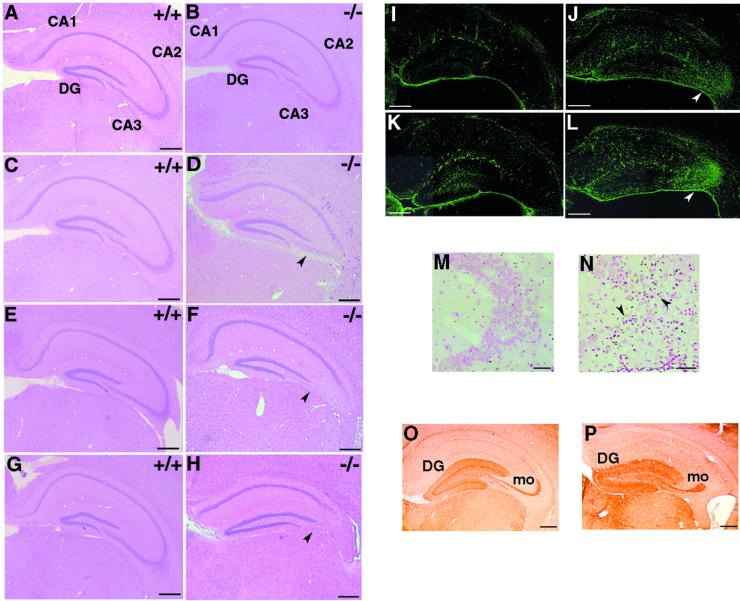
Histopathological examinations of STAM1−/− mice revealed normal morphogenesis in all tissues, including lymphoid tissues. However, one clear exception was provided by analysis of the brains of these mice. HE staining of hippocampus sections showed little difference between 3-week-old STAM1+/+ and STAM1−/− mice (Fig. 4A and B). However, the numbers of pyramidal cells in the hippocampal CA3 subfields were significantly reduced in 5- and 7-week-old STAM1−/− mice (Fig. 4C to F). Few pyramidal cells could still be observed in the hippocampal CA3 subfields of STAM1−/− mice at 9 weeks of age, whereas the numbers of these cells were unchanged in STAM1+/+ mice of a similar age (Fig. 4G and H). The same abnormality in the hippocampal CA3 subfields was observed for mutant strains derived from two independent clones with mixed 129/Sv × C57BL/6J and congenic C57BL/6J (≥10-generation backcross) genetic backgrounds. These observations indicated that the defect in CA3 pyramidal cells in STAM1−/− mice was due to their degeneration after normal hippocampi initially developed.
FIG. 4.
Abnormalities in hippocampal CA3 subfields of STAM1−/− mice. (A to H) HE staining of anterior coronal hippocampus sections of mice. Mice were STAM1+/+ (A, C, E, and G) and STAM1−/− (B, D, F, and H) and were 3 weeks old (A and B), 5 weeks old (C and D), 7 weeks old (E and F), and 9 weeks old (G and H). Note the loss of pyramidal cells in the CA3 subfields in STAM1−/− mice (arrowheads). DG, dentate gyrus. (I to L) Anti-GFAP antibody staining. Brain sections derived from STAM1+/+ and STAM1−/− mice were immunostained with anti-GFAP antibody. Gliosis (arrowheads) in hippocampal CA3 subfields was observed in STAM1−/− mice that were 4 weeks old (J) and 7 weeks old (L) but not in STAM1+/+ mice that were 4 weeks old (I) and 7 weeks old (K). (M and N) TUNEL staining of hippocampal CA3 subfields. Hippocampus sections of 4-week-old STAM1+/+ and STAM1−/− mice were stained for TUNEL. Arrowheads indicate positive staining for TUNEL. (O and P) Calbindin immunostaining. The sections were prepared from the brains of 9-week-old STAM1+/+ and STAM1−/− mice. DG and mo, dentate gyrus and mossy fibers, respectively. Bars, 0.4 mm.
To assess the gliosis caused by neural cell destruction in the STAM1−/− hippocampal CA3 subfield, we performed immunostaining assays for GFAP. The numbers of GFAP-positive astrocytes in the hippocampal CA3 subfields were significantly higher in the STAM1−/− mice and increased as the mice aged from 4 to 7 weeks old relative to the results for the STAM1+/+ mice (Fig. 4I to K). Moreover, moderate gliosis also occurred in amygdaloid nuclei and thalamic nuclei in 7-week-old STAM1−/− mice (data not shown). Next we performed TUNEL staining, which detects DNA fragmentation in dying cells. Numerous TUNEL-positive cells were detected in the hippocampal CA3 subfields of STAM1−/− mice, but few were detected in those of STAM1+/+ mice (Fig. 4M and N). These results suggest that neuronal cell death occurs in a specific region, especially the hippocampal CA3 subfields, of the STAM1−/− mouse brain.
To examine the mossy fiber pathway that connects granule cells to CA3 pyramidal cells, we performed immunostaining assays for calbindin, which selectively stains neurons in the dentate gyrus containing the mossy fiber pathway. Despite the profound reduction of pyramidal cells in the hippocampal CA3 subfields of STAM1−/− mice, the staining patterns of calbindin were not significantly different between STAM1+/+ and STAM1−/− mice (Fig. 4O and P). This result suggested that the mossy fiber pathway was normal in its overall projections in the hippocampi of STAM1−/− mice.
Expression of STAM1 in mouse brains and embryos.
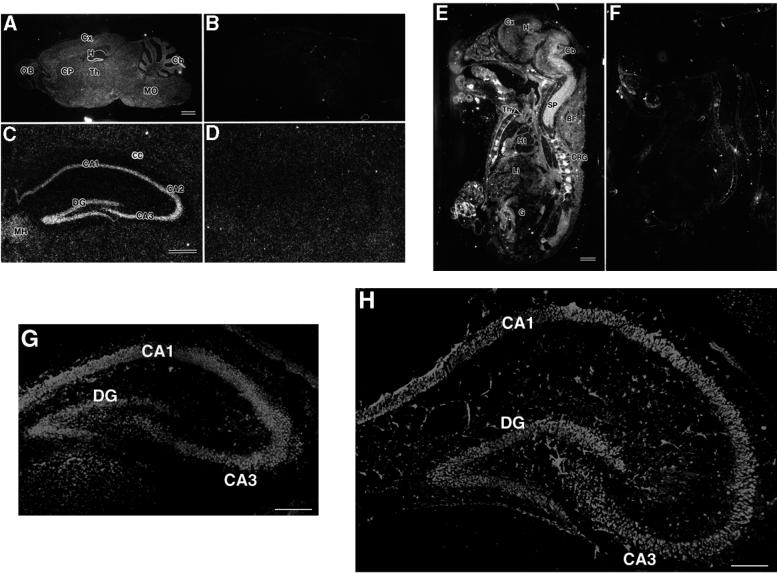
Histopathological examinations revealed abnormalities in the hippocampal CA3 subfields of STAM1−/− brain tissues. Hence, in situ hybridization was performed in order to examine the expression of the STAM1 gene in the central nervous system. In the brains of 4-week-old STAM1+/+ mice, STAM1 mRNA was expressed throughout the neuroaxis, including the olfactory bulb, cerebral cortex, caudate putamen, thalamus, cerebellar cortex, medulla oblongata, and spinal cord (Fig. 5A). An apparently higher level of STAM1 mRNA expression was observed for the hippocampal pyramidal and dentate granule cell layers without any obvious differences among sectors of Ammon's horns (CA1 to CA3) (Fig. 5C). Weak hybridization signals were also detected in the white matter, such as the corpus callosum, hippocampal fimbria, and cerebellar medulla, suggesting that both neuronal and glial cells expressed STAM1 mRNA. At E18, STAM1 mRNA was ubiquitously expressed in organs and tissues, including the brain, spinal cord, dorsal root ganglia, thymus, heart, lungs, liver, kidneys, gut, and brown adipose tissues (Fig. 5E). In STAM1−/− mice, the hybridization signals described above were completely abolished, suggesting the specificity of the oligonucleotide probe and further confirming the disruption of the STAM1 gene (Fig. 5B, D, and F). Furthermore, we performed immunohistochemical analysis to examine the expression of the STAM1 protein in the mouse hippocampus at postnatal days 5 and 14 (Fig. 5G and H). Hippocampal neuronal labeling was conspicuous throughout the CA1, CA3, and dentate gyrus areas. Staining patterns of STAM1 were not altered during brain development.
FIG. 5.
Expression of STAM1 in adult mouse brains and embryos. (A and B) Expression of STAM1 mRNA in brains. Brain sections prepared from 4-week-old STAM1+/+ and STAM1−/− mice were used for in situ hybridization for STAM1 mRNA. Cb, cerebellar cortex; CP, caudate putamen; Cx, cerebral cortex; H, hippocampal formation; MO, medulla oblongata; OB, olfactory bulb; Th, thalamus. Bar, 1 mm. (C and D) Expression of STAM1 in hippocampal formation. Hippocampal sections prepared from 4-week-old STAM1+/+ and STAM1−/− mice were used for in situ hybridization for STAM1 mRNA. Note STAM1 mRNA in a hippocampal pyramidal cell layer from CA1 to CA3 and the dentate gyrus (DG). MH, medial habenular nucleus; CC, corpus callosum. Bar, 0.5 mm. (E and F) Expression of STAM1 mRNA in embryos. STAM1+/+ and STAM1−/− embryos at E18.5 were tested for STAM1 mRNA expression by in situ hybridization. BF, brown fatty tissue; Cb, cerebellar cortex; Cx, cerebral cortex; DRG, dorsal root ganglia; G, gut; H, hippocampal formation; Ht, heart; Li, liver; SP, spinal cord; Thy, thymus. Bar, 1 mm. (G and H) STAM1 immunostaining of mouse hippocampal formation at postnatal days 5 (G) and 14 (H). DG, dentate gyrus. Bars, 0.2 mm.
Subcellular localization of STAM1 in primary cultured neurons.
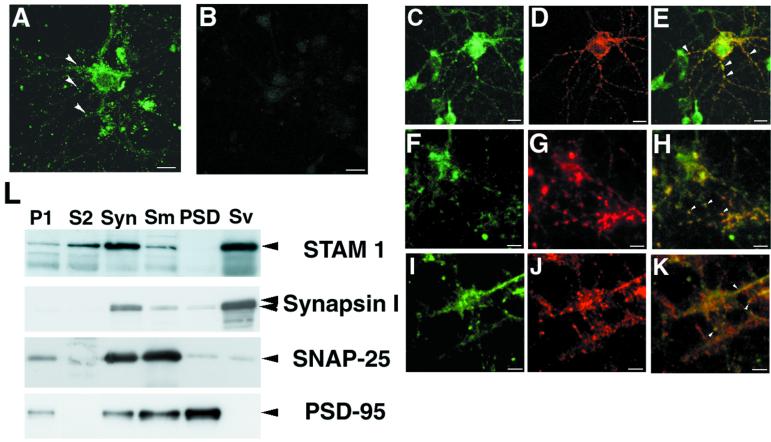
To investigate the function of STAM1 in neurons, we performed indirect immunofluorescence staining of STAM1 in primary hippocampal neurons derived from embryos. STAM1 staining was detected in the cytoplasm of dendrites and somata but not in the nuclei of wild-type neurons (Fig. 6A), while it was undetectable in STAM1−/− neurons, confirming the specificity of the anti-STAM1 antibody (Fig. 6B). In dendrites, a spot-like staining pattern was observed, suggesting that STAM1 might be present in synaptic regions. To test this idea, wild-type primary neocortical neurons were doubly stained for STAM1 and the synaptic markers GluR1 (8), Synapsin-I (13), and SNAP-25 (26). Overlapping staining between STAM1 and the synaptic markers was observed (Fig. 6E, H, and K). Furthermore, to examine the subcellular localization of STAM1, we prepared immunoblots of subcellular fractions of synaptosomal components prepared from mouse cerebral cortices. STAM1 was enriched in the synaptosomal fraction and more enriched in the synaptic vesicle fraction (Fig. 6L). These results suggest that STAM1 is present in synaptic regions.
FIG. 6.
Subcellular localization of STAM1. Primary hippocampal neurons from STAM1+/+ (A) and STAM1−/− (B) embryos at E18.5 were immunostained with anti-STAM1 antibody after 14 days of culturing. A spot-like staining pattern (arrowheads) was observed in STAM1+/+ neurons. Bars, 20 μm. (C to K) Coimmunostaining for STAM1 and synaptic markers. Primary hippocampal neurons from a STAM1+/+ embryo were stained with anti-STAM1 antibody (green, C, F, and I), anti-GluR1 antibody (red, D), anti-Synapsin-I antibody (red, G), or anti-SNAP-25 antibody (red, J) or doubly stained with anti-STAM1 and each marker (E, H, and K). Considerable overlap (arrowheads) was observed between the spots of STAM1 and synaptic markers. Bars, 20 μm. (L) Subcellular fractionation of synaptosomal components prepared from mouse cerebral cortices as described in Materials and Methods. Ten micrograms of each fraction was analyzed by SDS-PAGE, electrophoretically transferred to PVDF membranes, and incubated with anti-STAM1, anti-Synapsin-I, anti-SNAP-25, and anti-PSD-95 antibodies. Syn, synaptosome.
Susceptibility of STAM1−/− mice to kainic acid-induced seizures.
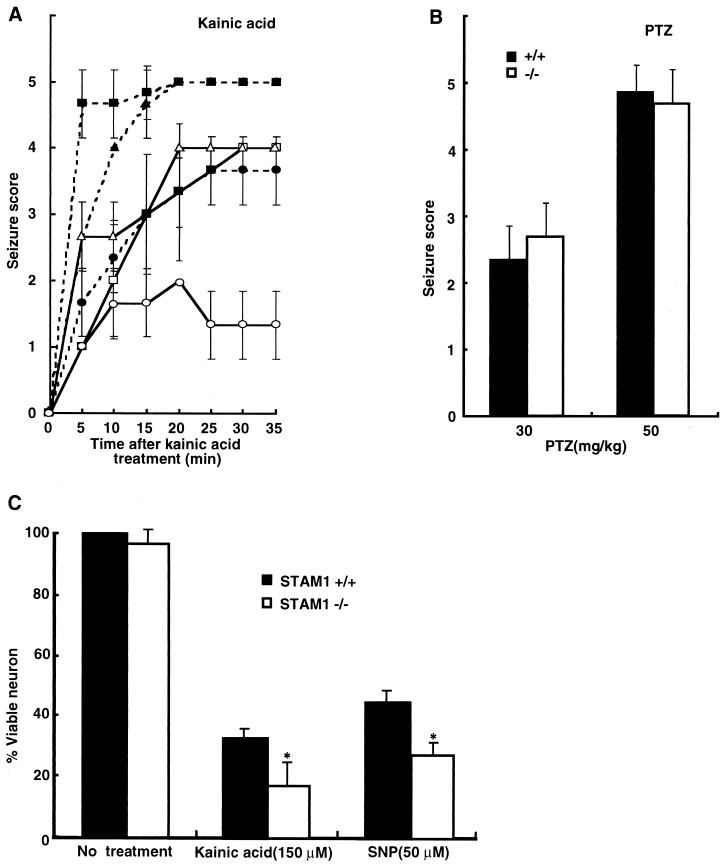
STAM1 might contribute to signal transduction and vesicle transport at synaptic sites. Moreover, STAM1−/− mice had abnormalities in hippocampi and amygdaloid nuclei, which are concerned with seizure attacks. Kainic acids elicit seizures directly by stimulation of glutamate receptors and indirectly by increasing the release of excitatory amino acids from nerve terminals (4, 41). The region most vulnerable to kainic acid-induced neuronal damage is the hippocampus (4). On the other hand, PTZ is known to induce seizures by blocking the γ-aminobutyric acid inhibitory postsynaptic potential (43). Hence, we tested the induction of seizures in C57BL/6-STAM1−/− mice initially upon stimulation with kainic acid and demonstrated that the seizure susceptibility of C57BL/6-STAM1−/− mice (≥10-generation backcross with congenic C57BL/6J) was greater than that of wild-type C57BL/6 mice (Fig. 7A). However, no difference in seizure susceptibility was observed between C57BL/6-STAM1−/− and wild-type C57BL/6 mice following injection with PTZ at both low (30-mg/kg) and high (50-mg/kg) doses (Fig. 7B). These results suggest that the greater susceptibility of STAM1−/− mice to kainic acid-induced seizures is not due to alterations of drug delivery to the brain or to blockage of the inhibitory postsynaptic potential.
FIG. 7.
Susceptibility to kainic acid-induced seizures of STAM1−/− mice. (A and B) Scores for seizures induced by kainic acid and PTZ. Four-week-old C57BL/6-STAM1+/+ (open symbols in A) and C57BL/6-STAM1−/− (filled symbols in A) mice were injected intraperitoneally with kainic acid (A) at doses of 10 (circles), 20 (triangles), and 30 (squares) mg per kg of body weight and with PTZ (B) at 30 and 50 mg per kg. Seizures were scored as follows: 1, arrest of motion; 2, myoclonic jerks of the head and neck with brief twitching movements; 3, unilateral clonic activity; 4, bilateral forelimb tonic and clonic activities; and 5, generalized tonic and clonic activities with loss of posture and death from continuous convulsions. At least six mice in each group were observed and scored to derive the temporal response curve. PTZ provoked rapid and abrupt general tonic-clonic convulsions, so the same criteria were used to record the PTZ-induced seizures within 5 min of injection. (C) Vulnerability of STAM1−/− primary hippocampal neurons to cell death induced by kainic acid and an NO donor. Hippocampal neurons were isolated from C57BL/6-STAM1+/+ and C57BL/6-STAM1−/− embryos at E18.5. Cultured neurons were treated with 150 μM kainic acid or 50 μM SNP. After 24 h, they were examined for cell viability by the Alamar blue assay. Data represent means and standard errors for three independent experiments performed in triplicate. An asterisk indicates a P value of <0.01 for a comparison with the wild type.
Since our histological observations indicated that the loss of neurons in STAM1−/− mice was due to a degenerative process after normal hippocampi initially developed, we thought that this degenerative process might be an accelerated vulnerability of STAM1−/− neurons to death induced by various stresses. To examine this possibility, we purified primary hippocampal neurons from wild-type and STAM1−/− embryos and tested them for sensitivity to kainic acid and an NO donor, SNP. The neurons from STAM1−/− mice showed greater sensitivity to death induced by kainic acid and SNP than did those from STAM1+/+ mice (Fig. 7C).
DISCUSSION
STAM1, in association with Jak2 and Jak3, was previously shown to be involved in signaling for cell growth and c-myc induction mediated by IL-2 and GM-CSF (35). In spite of this revelation of the in vitro functional role of STAM1, the present study demonstrated that targeted disruption of STAM1 had little effect on the development of hematopoietic cells, including T, B, myeloid, and erythroid cells, and on the proliferative responses of bone marrow cells and splenocytes to IL-2 and GM-CSF. These data suggest that STAM1 is not critically required for the development of lymphocytes and cytokine-mediated signaling in lymphocytes. However, using STAM1−/− mice, we documented here that STAM1 is essential for the survival of CA3 pyramidal cells in vivo. Mutant mice having a specific abnormality in the CA3 pyramidal cells of the hippocampus have not been reported thus far. Since the hippocampal CA3 subfields are known to be implicated in learning and memory (12, 20), our STAM1−/− mice may provide a useful tool for revealing the neurological significance of CA3 subfields as well as the regulatory mechanism of CA3 pyramidal cell survival.
STAM1 is involved in the survival of CA3 pyramidal neurons.
STAM1−/− mice showed neurological abnormalities, including defects in the hippocampal CA3 region and a leg-clasping reflex. The abnormal leg reflex has been observed for several mutant mice as an early symptom of neurological defects (15, 40). Histological analysis of the nervous systems of the STAM1−/− mice revealed an apparent reduction in the numbers of pyramidal cells in the CA3 subfields of the hippocampi. We thought that such a phenomenon in STAM1−/− mice was not due to their growth retardation because no reduction in the numbers of other vulnerable neurons, for example, CA1 pyramidal cells and cerebellar Purkinje cells, was seen. The fact that the loss of CA3 pyramidal cells was observed in adult STAM1−/− mice but not in young STAM1−/− mice indicated that CA3 pyramidal cells initially developed in STAM1−/− mice and were then extinguished. GFAP immunostaining and TUNEL staining also disclosed neuronal cell death, in particular, in the hippocampal CA3 subfields of the STAM1−/− mice. These data suggest that STAM1 is dispensable for the development of CA3 pyramidal cells but is involved in their survival. We confirmed the significant expression of STAM2 in the hippocampal CA3 subfields of the STAM1−/− mice (data not shown), suggesting that STAM2 is unable to compensate for STAM1 in the promotion of CA3 pyramidal cell survival in STAM1−/− mice.
In situ hybridization and immunohistochemical analyses showed that STAM1 was highly expressed in the hippocampal pyramidal and dentate granule cell layers without any obvious difference among subfields CA1 to CA3 in the wild specimen. Although this result is insufficient to explain the specific destruction of CA3 pyramidal cells in STAM1−/− hippocampi, a high level of STAM1 expression in hippocampal neurons suggests that STAM1 may play an important role in the hippocampus. On the other hand, we found that STAM1−/− mice were highly susceptible to kainic acid-induced seizures. Since kainic acid causes neuronal damage, especially in the hippocampus (4), the susceptibility to kainic acid-induced seizures may result from hippocampal vulnerability of STAM1−/− mice. In this context, we demonstrated that STAM1−/− primary hippocampal neurons were more sensitive to death induced by kainic acid and an NO donor in vitro than were wild-type neurons. Since both kainic acid (3, 29) and NO donors (29, 36, 37) are known to induce neuronal cell death, it can be assumed that STAM1 may have a role in protection against these stresses in hippocampal neurons. The NO donor-induced neuronal apoptosis could be inhibited by antiapoptotic proteins, such as Bcl-2 and Bcl-X (36); however, when we investigated the expression of Bcl-2 and Bcl-X in STAM1−/− primary hippocampal neurons, no significant difference was seen between STAM1−/− and STAM1+/+ mice (data not shown). Although we have not yet elucidated the exact role of STAM1 in sustaining the survival of CA3 pyramidal cells in hippocampi, the loss of CA3 neurons and the susceptibility to kainic acid-induced seizures in STAM1−/− mice suggest that STAM1 is involved in the survival of CA3 pyramidal cells.
STAM1 is associated with Hgs (Hrs), an FYVE finger protein (1, 6, 11, 19). A variant of Hrs, Hrs-2, has been shown to play a regulatory role in the docking and/or fusion of synaptic vesicles to plasma membranes through a calcium-regulated interaction with SNAP-25 (2, 39). The Hrs-2 expression pattern is very similar to that of STAM1 in adult mouse brains and embryos (38). Moreover, our data suggest that STAM1 may be present in synaptic sites. Hence, it can be hypothesized that STAM1 is also involved in the regulation of the docking and/or fusion of synaptic vesicles. However, we showed that STAM1 was not essential for IL-2-mediated vesicular transport in STAM1−/− lymphocytes. Although the processes of ligand-mediated vesicular transport may not be identical between synaptic sites and lymphocytes, it is possible that STAM1 does not contribute to vesicular transport in neurons.
STAM1−/− mice showed growth retardation and had a short lifespan. To investigate the mechanism of these phenotypes, we examined various metabolic markers and hormones in the blood and urine, such as levels in serum of total protein, glucose, glutamic-oxaloacetic transaminase, glutamic-pyruvic transaminase, lactic dehydrogenase, creatine phosphokinase, blood urea nitrogen, electrolytes, growth hormone, thyroid-stimulating hormone, insulin, leptin, and corticosterone. We could not detect any abnormalities in these indices, and the serum leptin level and food intake of the STAM1−/− mice were not significantly different from those of the wild-type mice (data not shown). We observed that gliosis occurred not only in the hippocampal CA3 region but also in the thalamic nuclei and amygdaloid nuclei in older STAM1−/− mice (data not shown). So, we can hypothesize that a systemic deficiency in the nervous system leads to the death of older STAM1−/− mice. However, we have no evidence suggesting the direct mechanism causing the phenotypes, including death, of the STAM1−/− mice.
Deletion of STAM1 does not compromise lymphocyte responses to cytokines.
The bone marrow cells and splenocytes derived from the STAM1−/− mice responded to various mitogens, such as anti-CD3, IL-2, and GM-CSF and their combinations, as well as did those derived from the wild-type mice. Similarly, IL-2-induced c-myc expression was not impaired in activated T cells derived from the STAM1−/− mice. These observations obtained with STAM1−/− cells suggest that STAM1 is dispensable for cytokine-mediated signaling in lymphocytes. Since STAM2, like STAM1, was involved in signaling for DNA synthesis and c-myc induction mediated by IL-2 and GM-CSF in vitro (9, 27), it is possible that STAM2 compensates for STAM1 in the intracellular signaling mediated by cytokines in STAM1−/− mice. In order to test the in vivo relevance of such compensation, the generation of STAM2-deficient mice and their crossing with STAM1-deficient mice are required.
ACKNOWLEDGMENTS
We thank T. Noda for providing the J1 ES cell line, pLoxp-neo plasmids, and pMC1 DT-A plasmids and L. C. Ndhlovu for critically reading the manuscript.
This work was supported in part by Core Research for Evolutional Science and Technology, Japan Science and Technology Corporation, and a grant-in-aid for scientific research on priority areas from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Asao H, Sasaki Y, Arita T, Tanaka N, Endo K, Kasai H, Takeshita T, Endo Y, Fujita T, Sugamura K. Hrs is associated with STAM, a signal-transducing adaptor molecule. J Biol Chem. 1997;272:32785–32791. doi: 10.1074/jbc.272.52.32785. [DOI] [PubMed] [Google Scholar]
- 2.Bean A J, Seifert R, Chen Y A, Sacks R, Scheller R H. Hrs-2 is an ATPase implicated in calcium-regulated secretion. Nature. 1997;385:826–829. doi: 10.1038/385826a0. [DOI] [PubMed] [Google Scholar]
- 3.Behrens A, Sibilia M, Wagner E F. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nat Genet. 1999;21:326–329. doi: 10.1038/6854. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Ari Y. Limbic seizure and brain damage produced by kainic acid mechanisms and relevance to human temporal lobe epilepsy. Neuroscience. 1985;14:375–403. doi: 10.1016/0306-4522(85)90299-4. [DOI] [PubMed] [Google Scholar]
- 5.Bermingham J R, Jr, Scherer S S, O'Connell S, Arroyo E, Kalla K A, Powell F L, Rosenfeld M G. Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- 6.Burd C G, Emr S D. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 7.Cohen R S, Blomberg F, Berzins K, Siekevitz P. The structure of postsynaptic densities isolated from dog cerebral cortex. I. Overall morphology and protein composition. J Cell Biol. 1977;74:181–203. doi: 10.1083/jcb.74.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig A M, Blackstone C D, Huganir R L, Banker G. The distribution of glutamate receptors in cultured rat hippocampal neurons: postsynaptic clustering of AMPA-selective subunits. Neuron. 1993;10:1055–1068. doi: 10.1016/0896-6273(93)90054-u. [DOI] [PubMed] [Google Scholar]
- 9.Endo K, Takeshita T, Kasai H, Sasaki Y, Tanaka N, Asao H, Kikuchi K, Yamada M, Chen M, O'Shea J J, Sugamura K. STAM2, a new member of the STAM family, binding to the Janus kinases. FEBS Lett. 2000;477:55–61. doi: 10.1016/s0014-5793(00)01760-9. [DOI] [PubMed] [Google Scholar]
- 10.Fujii M, Sugamura K, Sano K, Nakai M, Sugita K, Hinuma Y. High-affinity receptor-mediated internalization and degradation of interleukin 2 in human T cells. J Exp Med. 1986;163:550–562. doi: 10.1084/jem.163.3.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaullier J M, Simonsen A, D'Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P. Nature. 1998;30:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 12.Gluck M A, Myers C E. Psychobiological models of hippocampal function in learning and memory. Annu Rev Psychol. 1997;48:481–514. doi: 10.1146/annurev.psych.48.1.481. [DOI] [PubMed] [Google Scholar]
- 13.Huttner W B, Schiebler W, Greengard P, De Camilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 15.Klein R, Silos-Santiago I, Smeyne R J, Lira S A, Brambilla R, Bryant S, Zhang L, Snider W D, Barbacid M. Disruption of the neurotrophin 3 receptor gene trkC eliminates Ia muscle afferents and results in abnormal movements. Nature. 1994;368:249–251. doi: 10.1038/368249a0. [DOI] [PubMed] [Google Scholar]
- 16.Leonard W J, Noguchi M, Russell S M, McBride O W. The molecular basis of X-linked severe combined immunodeficiency: the role of the interleukin-2 receptor gamma chain as a common gamma chain, gamma c. Immunol Rev. 1994;138:61–86. doi: 10.1111/j.1600-065x.1994.tb00847.x. [DOI] [PubMed] [Google Scholar]
- 17.Leonard W J, O'Shea J J. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 18.Lohi O, Lehto V P. VHS domain marks a group of proteins involved in endocytosis and vesicular trafficking. FEBS Lett. 1998;440:255–257. doi: 10.1016/s0014-5793(98)01401-x. [DOI] [PubMed] [Google Scholar]
- 19.Mao Y, Nickitenko A, Duan X, Lloyd T E, Wu M N, Bellen H, Quiocho F A. Crystal structure of VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell. 2000;100:447–456. doi: 10.1016/s0092-8674(00)80680-7. [DOI] [PubMed] [Google Scholar]
- 20.Martin S J, Grimwood P D, Morris R G. Synaptic plasticity and memory: an evaluation of the hypothesis. Annu Rev Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- 21.Morgan I G. Synaptosome and cell separation. Neuroscience. 1976;1:159–165. doi: 10.1016/0306-4522(76)90072-5. [DOI] [PubMed] [Google Scholar]
- 22.Moriggl R, Topham D J, Teglund S, Sexl V, McKay C, Wang D, Hoffmeyer A, van Deursen J, Sangster M Y, Bunting K D, Grosveld G C, Ihle J N. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–259. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa S, Watanabe M, Isobe T, Kondo H, Inoue Y. Cytological compartmentalization in the staggerer cerebellum, as revealed by calbindin immunohistochemistry for Purkinje cells. J Comp Neurol. 1998;395:112–120. doi: 10.1002/(sici)1096-9861(19980525)395:1<112::aid-cne8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Nakai S, Kawano H, Yudate T, Nishi M, Kuno J, Nagata A, Jishage K, Hamada H, Fujii H, Kawamura K. The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 1996;9:3109–3121. doi: 10.1101/gad.9.24.3109. [DOI] [PubMed] [Google Scholar]
- 25.O'Shea J J. Jaks, STATs, cytokine signal transduction, and immunoregulation: are we there yet? Immunity. 1997;7:1–11. doi: 10.1016/s1074-7613(00)80505-1. [DOI] [PubMed] [Google Scholar]
- 26.Oyler G A, Higgins G A, Hart R A, Battenberg E, Billingsley M, Bloom F E, Wilson M C. The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J Cell Biol. 1989;109:3039–3052. doi: 10.1083/jcb.109.6.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey A, Fernandez M M, Steen H, Blagoev B, Nielsen M M, Roche S, Mann M, Lodish H F. Identification of a novel immunoreceptor tyrosine-based activation motif-containing molecule, STAM2, by mass spectrometry and its involvement in growth factor and cytokine receptor signaling pathways. J Biol Chem. 2000;275:38633–38639. doi: 10.1074/jbc.M007849200. [DOI] [PubMed] [Google Scholar]
- 28.Sakagami H, Saito S, Kitani T, Okuno S, Fujisawa H, Kondo H. Localization of the mRNAs for two isoforms of Ca2+/calmodulin-dependent protein kinase kinases in the adult rat brain. Brain Res Mol Brain Res. 1998;54:311–315. doi: 10.1016/s0169-328x(97)00362-8. [DOI] [PubMed] [Google Scholar]
- 29.Sastry P S, Rao K S. Apoptosis and the nervous system. J Neurochem. 2000;74:1–20. doi: 10.1046/j.1471-4159.2000.0740001.x. [DOI] [PubMed] [Google Scholar]
- 30.Schonemann D M, Ryan A K, McEvilly R J, O'Connell S M, Arias C A, Kalla K A, Li P, Sawchenko P E, Rosenfeld M G. Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 1996;9:3122–3155. doi: 10.1101/gad.9.24.3122. [DOI] [PubMed] [Google Scholar]
- 31.Sugamura K, Asao H, Kondo M, Tanaka N, Ishii N, Nakamura M, Takeshita T. The common gamma-chain for multiple cytokine receptors. Adv Immunol. 1995;59:225–277. doi: 10.1016/s0065-2776(08)60632-x. [DOI] [PubMed] [Google Scholar]
- 32.Sugamura K, Asao H, Kondo M, Tanaka N, Ishii N, Ohbo K, Nakamura M, Takeshita T. The interleukin-2 receptor gamma chain: its role in the multiple cytokine receptor complexes and T cell development in XSCID. Annu Rev Immunol. 1996;14:179–205. doi: 10.1146/annurev.immunol.14.1.179. [DOI] [PubMed] [Google Scholar]
- 33.Takata H, Kato M, Denda K, Kitamura N. A Hrs binding protein having a Src homology 3 domain is involved in intracellular degradation of growth factors and their receptors. Genes Cells. 2000;5:57–69. doi: 10.1046/j.1365-2443.2000.00303.x. [DOI] [PubMed] [Google Scholar]
- 34.Takeshita T, Arita T, Asao H, Tanaka N, Higuchi M, Kuroda H, Kaneko K, Munakata H, Endo Y, Fujita T, Sugamura K. Cloning of a novel signal-transducing adaptor molecule containing an SH3 domain and ITAM. Biochem Biophys Res Commun. 1996;225:1035–1039. doi: 10.1006/bbrc.1996.1290. [DOI] [PubMed] [Google Scholar]
- 35.Takeshita T, Arita T, Higuchi M, Asao H, Endo K, Kuroda H, Tanaka N, Murata K, Ishii N, Sugamura K. STAM, signal transducing adaptor molecule, is associated with Janus kinases and involved in signaling for cell growth and c-myc induction. Immunity. 1997;6:449–457. doi: 10.1016/s1074-7613(00)80288-5. [DOI] [PubMed] [Google Scholar]
- 36.Tamatani M, Che Y H, Matsuzaki H, Ogawa S, Okada H, Miyake S, Mizuno T, Tohyama M. Tumor necrosis factor induces Bcl-2 and Bcl-X expression through NFκB activation in primary hippocampal neurons. J Biol Chem. 1999;274:8531–8538. doi: 10.1074/jbc.274.13.8531. [DOI] [PubMed] [Google Scholar]
- 37.Toku K, Tanaka J, Yano H, Desaki J, Zhang B, Yang L, Ishihara K, Sakanaka M, Maeda N. Microglial cells prevent nitric oxide-induced neuronal apoptosis in vitro. J Neurosci Res. 1998;53:415–425. doi: 10.1002/(SICI)1097-4547(19980815)53:4<415::AID-JNR3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 38.Tsujimoto S, Pelto-Huikko M, Aitola M, Meister B, Vik-Mo E O, Davanger S, Scheller R H, Bean A J. The cellular and developmental expression of hrs-2 in rat. Eur J Neurosci. 1999;11:3047–3063. doi: 10.1046/j.1460-9568.1999.00716.x. [DOI] [PubMed] [Google Scholar]
- 39.Tsujimoto S, Bean A J. Distinct protein domains are responsible for the interaction of Hrs-2 with SNAP-25. J Biol Chem. 2000;274:2938–2942. doi: 10.1074/jbc.275.4.2938. [DOI] [PubMed] [Google Scholar]
- 40.Urbánek P, Wang Z-Q, Fetka I, Wanger E F, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–902. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 41.Watkins J C, Evans R H. Excitatory amino acid transmitters. Annu Rev Pharmacol Toxicol. 1981;21:165–204. doi: 10.1146/annurev.pa.21.040181.001121. [DOI] [PubMed] [Google Scholar]
- 42.White J M, Dicaprio M J, Greenberg D A. Assessment of neuronal viability with Alamar blue in cortical and granule cell cultures. J Neurosci Methods. 1996;70:195–200. doi: 10.1016/s0165-0270(96)00118-5. [DOI] [PubMed] [Google Scholar]
- 43.Yang D D, Kuan C-Y, Whitmarsh A J, Rincón M, Zheng T S, Davis R J, Raskic P, Flavell R A. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature. 1997;389:865–870. doi: 10.1038/39899. [DOI] [PubMed] [Google Scholar]