Clostridium difficile is a causative agent of nosocomially acquired antibiotic-associated diarrhea and pseudomembranous colitis (3). Typing schemes include serogrouping, pyrolysis mass spectroscopy, restriction endonuclease analysis, whole-cell protein profiling, arbitrarily primed PCR, pulsed-field gel electrophoresis (PFGE), and PCR ribotyping (2). PFGE is a highly discriminatory method, but a number of strains of C. difficile are reported to be untypeable due to degradation of DNA during the procedure (4, 5). Such strains can be typed by other methods and have been assigned predominantly to PCR ribotype 1 and serogroup G (4, 5, 7). In the United Kingdom, PCR ribotype 1 accounts for 57% of isolates from hospitalized patients and is endemic in 33 of 58 hospitals surveyed (2). To date all PCR ribotype 1 isolates tested by the Anaerobe Reference Laboratory (Cardiff, United Kingdom) have been untypeable by PFGE; therefore, a method capable of enhanced discrimination of this prevalent ribotype would improve the epidemiology of C. difficile. The aim of this study was to develop a refined PFGE method capable of analyzing previously untypeable strains.
The reasons for the degradation of DNA in certain strains of C. difficile during PFGE are unclear. Traditionally, proteinase K has been used to inactivate nucleases liberated after cell lysis, and our initial, unsuccessful efforts were directed toward protecting DNA during, and after, cell wall removal. Protocols included increased EDTA concentration in suspension buffers, heat shock at 70°C, and the addition of diethylpyrocarbonate to lysozyme-induced spheroplasts prior to the addition of sodium dodecyl sulfate (1). Recently, Römling and Tümmler (10) documented the protection of degradation-sensitive DNA in Pseudomonas aeruginosa during PFGE by the addition of thiourea to Tris buffer. Thiourea is thought to neutralize a nucleolytic peracid derivative of Tris that is formed at the anode during electrophoresis (9).
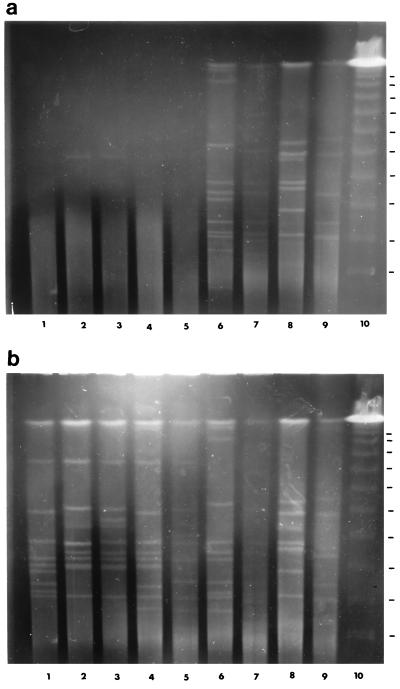
We have evaluated the effect of thiourea in PFGE on a small number of C. difficile isolates that had been analyzed previously by PCR ribotyping (8). Five PCR ribotype 1 isolates (one isolate from the United States and four isolates from disparate hospitals in the United Kingdom) and single representatives of ribotypes 2, 10, 15, and 23 were examined. Our standard preparative PFGE method was used (6), and SmaI fragments were separated in 1.0% agarose with an initial pulse of 3 s that increased linearly to 30 s after 20 h. Electrophoresis was performed with and without thiourea (50 μM) in the running buffer (Fig. 1).
FIG. 1.
PFGE patterns obtained with SmaI-digested DNA from C. difficile isolates of known PCR ribotype. Electrophoresis was performed in the absence (a) and in the presence (b) of 50 μM thiourea. Lanes 1 to 5, five degradation-sensitive PCR ribotype 1 isolates (from the United States and Newcastle, Wrexham, Tooting, and Truro, United Kingdom, respectively); lane 6, ribotype 2 isolate (Wrexham); lane 7, ribotype 10 isolate (Manchester, United Kingdom); lane 8, ribotype 15 isolate (Carshalton, United Kingdom); lane 9, ribotype 23 isolate (Jersey, United Kingdom); lane 10, λ DNA standard (48.5-kbp concatemer).
Genomic DNA was degraded in all PCR ribotype 1 isolates in the absence of thiourea (Fig. 1a), while band patterns were observed when thiourea was present (Fig. 1b). Initial results with the limited sample set seem to suggest that discrimination (subtyping) of isolates belonging to ribotype 1 may be possible since PFGE profiles differ by a significant number of bands (11). The results indicate that addition of thiourea may permit previously untypeable isolates, belonging to the most common PCR ribotype, to be examined by PFGE. Hopefully, this method will facilitate international comparison of typing results and permit genotype subdivision, leading to improved epidemiology.
REFERENCES
- 1.Blaschek H P, Klacik M A. Role of DNase in recovery of plasmid DNA from Clostridium perfringens. Appl Environ Microbiol. 1984;48:78–181. doi: 10.1128/aem.48.1.178-181.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brazier J S. The epidemiology and typing of Clostridium difficile. J Antimicrob Chemother. 1998;41(Suppl.):47–57. doi: 10.1093/jac/41.suppl_3.47. [DOI] [PubMed] [Google Scholar]
- 3.George W L, Sutter V L, Finegold S M. Antimicrobial agent-induced diarrhoea—a bacterial disease. J Infect Dis. 1977;136:822–828. doi: 10.1093/infdis/136.6.822. [DOI] [PubMed] [Google Scholar]
- 4.Hyett A P, Brazier J S, O'Neill G L. Pulsed-field gel electrophoresis as a method for typing Clostridium difficile in the routine laboratory. Rev Med Microbiol. 1997;8(Suppl. 1):S63–S64. [Google Scholar]
- 5.Kato H, Kato N, Watanabe K, Ueno K, Sakata Y, Fujita K. Relapses or reinfections: analysis of a case of Clostridium difficile-associated colitis by two typing systems. Curr Microbiol. 1996;33:220–223. doi: 10.1007/s002849900103. [DOI] [PubMed] [Google Scholar]
- 6.Ledson M J, Gallagher M J, Corkill J E, Hart C A, Walshaw M J. Cross infection between cystic fibrosis patients colonised with Burkholderia cepacia. Thorax. 1998;53:432–436. doi: 10.1136/thx.53.5.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Neill G L, Adams J E, Bowman R A, Riley T V. A molecular characterization of Clostridium difficile isolates from humans, animals and their environment. Epidemiol Infect. 1993;111:257–264. doi: 10.1017/s095026880005696x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Neill G L, Ogunsola F T, Brazier J S, Duerden B I. Modification of a PCR ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe. 1996;2:205–209. [Google Scholar]
- 9.Ray T, Mills A, Dyson P. Tris-dependent oxidative DNA strand scission during electrophoresis. Electrophoresis. 1995;16:888–894. doi: 10.1002/elps.11501601149. [DOI] [PubMed] [Google Scholar]
- 10.Römling U, Tümmler B. Achieving 100% typeability of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. J Clin Microbiol. 2000;38:464–465. doi: 10.1128/jcm.38.1.464-465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]