Abstract
Human HCF-1 is a large, highly conserved, and abundant nuclear protein that plays an important but unknown role in cell proliferation. It also plays a role in activation of herpes simplex virus immediate-early gene transcription by the viral regulatory protein VP16. A single proline-to-serine substitution in the HCF-1 VP16 interaction domain causes a temperature-induced arrest of cell proliferation in hamster tsBN67 cells and prevents transcriptional activation by VP16. We show here that HCF-1 is naturally bound to chromatin in uninfected cells through its VP16 interaction domain. HCF-1 is chromatin bound in tsBN67 cells at permissive temperature but dissociates from chromatin before tsBN67 cells stop proliferating at the nonpermissive temperature, suggesting that loss of HCF-1 chromatin association is the primary cause of the temperature-induced tsBN67 cell proliferation arrest. We propose that the role of HCF-1 in cell proliferation is to regulate gene transcription by associating with a multiplicity of DNA-bound transcription factors through its VP16 interaction domain.
Regulated cell proliferation requires the coordination of many distinct cellular processes. Much of this coordination occurs through the regulation of gene expression, often at the level of transcription. Consequently, transcriptional regulation is often altered when cells become oncogenic and manipulated by viruses to promote infection. For example, the DNA-bound transcriptional activator E2F and the E2F-associated retinoblastoma (Rb) transcriptional repressor protein are important players in the control of cell proliferation and their activities are often disrupted during oncogenesis and manipulated by viral regulatory proteins during DNA tumor virus infection (see references 4 and 22 for reviews). Other less well-understood regulators of cell proliferation include the cellular protein HCF-1. HCF-1 is a highly conserved and abundant nuclear protein that plays a known role in herpes simplex virus type 1 (HSV-1) infection (for reviews, see references 10 and 23) and an important but unknown role in cell proliferation (7).
Many aspects of HCF-1 make it an unusual protein. It is initially synthesized as a large precursor molecule of over 2,000 amino acids (29). After synthesis, HCF-1 migrates to the nucleus, where it is cleaved repeatedly at a series of six 26-amino-acid repeats located near the middle of the protein (11, 29, 30). These cleavages result in a heterogeneous collection of stable amino (HCF-1N)- and carboxy (HCF-1C)-terminal subunits that remain noncovalently associated (30). Although the unusual maturation process of HCF-1 has been well documented, how HCF-1 functions normally in the cell is essentially unknown.
Much more is known about the role of HCF-1 during HSV-1 infection. When HSV-1 first infects a cell, HCF-1 forms a stable heterodimeric complex with the viral transcriptional regulator VP16 (also known as Vmw65 and α-TIF), which is delivered by the infecting virion. Together, the HCF-1–VP16 complex binds to the cellular transcription factor Oct-1 on DNA to form a multiprotein transcriptional activator complex—the VP16-induced complex—on VP16-responsive elements found in HSV-1 immediate-early promoters (for reviews, see references 10 and 23). Although HCF-1 is a large protein, only the 380 amino-terminal residues of HCF-1 are necessary to associate with VP16 and promote VP16-induced complex formation (13, 28). These 380 amino acids contain six tandem repeats called HCF-1Kel repeats because they are related to repeats found in the Drosophila protein Kelch (31); these repeats probably form a stable structure called a β propeller (3, 28) and are collectively referred to, interchangeably, as the Kelch or VP16 interaction domain.
HCF-1 is known to be involved in cell proliferation because a single proline-to-serine missense mutation at position 134 in the VP16 interaction domain (P134S) causes temperature-induced cell proliferation arrest in the hamster cell line tsBN67 (7). tsBN67 cells proliferate normally at the permissive temperature of 33.5°C, but a few cell divisions after transfer to the nonpermissive temperature of 40°C, these cells enter a stable cell proliferation arrest. HCF-1 processing and stability are nearly normal in tsBN67 cells at the nonpermissive temperature, but association with VP16 is disabled, suggesting that the arrest phenotype is caused by loss of HCF-1 association with a cellular protein(s) mimicked by VP16 (7, 28). Although sufficient for formation of the VP16-induced complex, however, the HCF-1 VP16 interaction domain is not sufficient to rescue the tsBN67 cell proliferation defect; a neighboring basic region within the HCF-1N subunit is also required for rescue (28).
Although little is known of how HCF-1 promotes cell proliferation, multiple molecular targets of HCF-1 function have been inferred from the results of Saccharomyces cerevisiae two-hybrid and in vitro protein-binding assays. The basic leucine zipper proteins LZIP (also known as Luman) and Zhangfei (ZF) associate with the HCF-1 VP16 interaction domain in a manner similar to that of VP16 (5, 15–17); all three proteins possess a short tetrapeptide HCF-1-binding motif— (D/E)HXY (where X denotes a variable position)—and their association with HCF-1 is sensitive to the tsBN67 point mutation. Two transcription factors associate with the HCF-1 basic region: GABP (26) and Sp1 (8). Last, the HCF-1C subunit can associate with protein phosphatase 1 (1). Although these protein association studies (1, 5, 8, 15–17, 26) suggest that HCF-1 plays a role in transcriptional regulation, possibly through modulation of protein phosphorylation, a natural molecular function of HCF-1 remains to be elucidated.
We show here that, in uninfected cells, HCF-1 is associated with chromatin through its VP16 interaction domain. In tsBN67 cells, HCF-1 dissociates from chromatin preceding temperature-induced cell proliferation arrest. These results suggest that HCF-1 plays an active role in cell proliferation through the regulation of chromosomal gene expression.
MATERIALS AND METHODS
Small-scale biochemical fractionation.
Small-scale biochemical fractionation was performed as described previously (20). Briefly, 1 × 107 to 2 × 107 cells were collected, washed with phosphate-buffered saline (PBS), and resuspended at 4 × 107 cells/ml in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, and protease inhibitor cocktail [Boehringer]). Triton X-100 was added (0.1% final concentration), the cells were incubated on ice for 8 min, and nuclei (fraction P1) were collected by centrifugation (5 min, 1,300 × g, 4°C). The supernatant (fraction S1) was clarified by high-speed centrifugation (5 min, 20,000 × g, 4°C), and the supernatant (fraction S2) was collected. The P1 nuclei were washed once in buffer A and lysed for 30 min in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM dithiothreitol, and protease inhibitor cocktail [Boehringer]), and insoluble chromatin (fraction P3) and soluble (fraction S3) fractions were separated by centrifugation (5 min, 1,700 × g, 4°C). The P3 fraction was washed once with buffer B and resuspended either in sodium dodecyl sulfate (SDS)-Laemmli buffer (and boiled for 10 min) or in nuclease digestion buffer.
MNase treatment.
For micrococcal nuclease (MNase) treatment, the P3 pellet was resuspended in a solution containing 10 mM Tris, 10 mM KCl, and 1 mM CaCl2 and 1 U of MNase (Sigma) was added. After 37°C incubations for the times indicated in Fig. 1C, the reaction was stopped with EGTA (1 mM final concentration). Soluble and insoluble components were then separated by centrifugation (5 min, 1,700 × g, 4°C). The pellet was resuspended in SDS-Laemmli buffer and boiled for 10 min. Half of the supernatant was used for immunoblot analysis, the remainder was extracted with phenol-chloroform and ethanol precipitated, and its DNA content was analyzed by agarose gel electrophoresis.
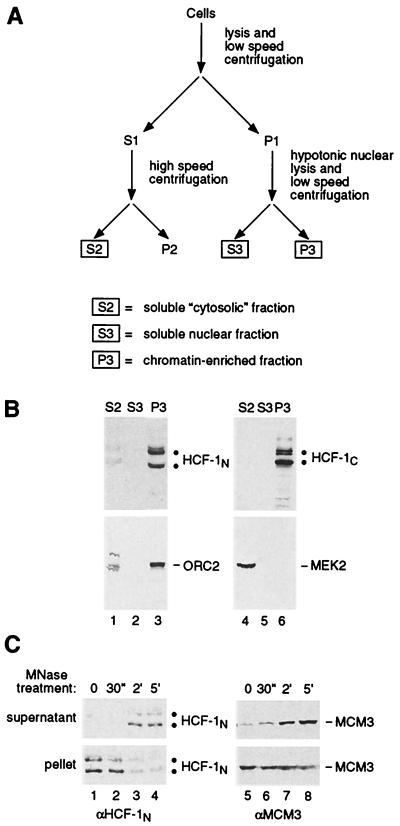
FIG. 1.
HCF-1 is chromatin associated. (A) Biochemical fractionation scheme (see Materials and Methods for details). Final fractions used for analysis—S2, S3, and P3—are boxed. (B) HCF-1 is enriched in the chromatin-containing P3 fraction. HeLa cells were subjected to biochemical fractionation, and cell-equivalent amounts of fractions S2, S3, and P3 were probed by immunoblotting with αHCF-1N (upper left panel), αHCF-1C (upper right panel), αORC2 (lower left panel), and αMEK2 (lower right panel) antisera. The positions of the heterogeneous HCF-1 subunits are indicated by double dots. (C) HCF-1 can be solubilized in the chromatin-enriched P3 fraction by treatment with MNase. The P3 fraction was resuspended in nuclease buffer and treated with MNase (see Materials and Methods) for the times indicated. The digested suspension was then centrifuged, and the supernatant (upper panel) and pellet (lower panel) were collected and analyzed by immunoblotting with αHCF-1N (lanes 1 to 4) and αMCM3 (lanes 5 to 8) antisera.
Retroviral vectors and stable HCF-1 synthesis.
A pBabeFLAG retroviral expression vector was constructed by inserting oligonucleotides encoding the FLAG epitope tag (DYKDDDDK) into the pBabe-Puro vector (21). XbaI-BamHI cDNA fragments encoding either the wild-type human HCF-1N1011 subunit (HCF-1 residues 2 to 1011), its alternative splice variant HCF-1N1011Δ382–450, its tsBN67 mutant version HCF-1N1011/P134S, or the VP16 interaction domain (HCF-1N380) alone were inserted into the pBabeFLAG vector. Populations of cells stably synthesizing FLAG-tagged forms of HCF-1 were obtained by retroviral gene transfer with the Phoenix amphotropic virus packaging line as described previously (25). Infected HeLa and IMR90 cells were selected by culturing cells in 10% fetal bovine serum (FBS) in Dulbecco's modified Eagle's medium (DMEM) containing 2 μg of puromycin per ml for 7 to 10 days. Subpopulations of infected cells were frozen for future use, and others were lysed and tested for recombinant HCF-1 synthesis by FLAG tag immunoblot analysis. Cells infected with the pBabeFLAG parent vector were used as controls.
Immunoblot analysis.
Polyclonal HCF-1N subunit (αN18 [7]) and HCF-1c subunit (αH12 [29]) antisera have been described previously. They are referred to here as αHCF-1N and αHCF-1C, respectively, and were used at a 1:3,000 dilution for immunoblot probing. Anti-FLAG (M2; Sigma) and anti-MEK2 (M24520; Transduction Laboratories) antisera were obtained commercially. ORC2 and MCM3 antisera were kind gifts of J. Méndez and B. Stillman (Cold Spring Harbor Laboratory). Immunoblots were prepared by semidry transfer and developed by chemiluminescence (SuperSignal; Pierce).
Immunofluorescence.
IMR90 cells were seeded onto sterile coverslips. After 24 h, cells were (i) washed with PBS, (ii) fixed for 20 min with 4% paraformaldehyde, (iii) permeabilized for 5 min with 0.1% Triton X-100 in PBS, (iv) washed twice with PBS, and (v) blocked for 20 min with 5% bovine serum albumin in PBS. After being rinsed with PBS, coverslips were incubated for 1 h with protein A-purified αHCF-1N antibody and excess specific (N18) or nonspecific (FLAG) peptide. The samples were then washed five times with PBS and incubated for 1 h with Texas Red-conjugated secondary antibody. After being extensively washed with PBS, the coverslips were stained with DAPI (4′,6′-diamidino-2-phenylindole) and mounted, and fluorescence was observed with a Zeiss fluorescence microscope.
HCF-1 quantitation.
A six-His tag–HCF-1N380 fusion protein was synthesized to high levels in Escherichia coli and prepared by bacterial lysis with SDS-Laemmli buffer. The levels of six-His tag–HCF-1N380 fusion protein in the lysate were determined by SDS-polyacrylamide gel electrophoresis and Coomasie staining of a twofold serial dilution of the lysate alongside a serial dilution of bovine serum albumin of known concentration. HeLa or IMR90 cells were counted and directly lysed in SDS-Laemmli buffer. The levels of HCF-1 were directly compared to those of the six-His tag–HCF-1N380 fusion protein by parallel immunoblot analysis with the αHCF-1N antiserum.
BHK21 and tsBN67 cells.
BHK21 and original tsBN67 cells were a kind gift of T. Nishimoto (Kyushu University). The tsBN67 cells were single-cell subcloned to produce the tsBN67HR1 line used here. tsBN67HR1 cells display the same temperature-sensitive proliferation defect as the original tsBN67 cells. Cells were grown in DMEM supplemented with 10% FBS. To measure tsBN67HR1 cell proliferation arrest at 40°C, 2 × 104 BHK21 or tsBN67HR1 cells were (i) seeded onto 6-cm-diameter plates, (ii) maintained at 33.5°C for 48 h, and (iii) transferred to 40°C, and portions were collected every 12 h by trypsinization and counted with a hemacytometer. To arrest tsBN67 cell proliferation by limiting serum, asynchronous populations of tsBN67HR1 cells were incubated at 33.5°C for 24 h in DMEM containing 0.25% FBS and subsequently transferred to 40°C and harvested at the times indicated in Fig. 7.
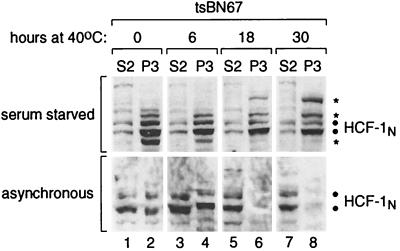
FIG. 7.
tsBN67 HCF-1P134S–chromatin association is stable at the nonpermissive temperature if tsBN67 cells are arrested before transfer to the nonpermissive temperature. tsBN67HR1 cells were arrested by growth in 0.25% serum at 33.5°C for 24 h, and the arrested cells were subsequently transferred and maintained at 40°C in 0.25% serum (upper panels). In parallel, asynchronous tsBN67HR1 cells maintained in 10% serum were analyzed (lower panels). Cells were harvested at the indicated times after transfer to 40°C and subjected to biochemical fractionation as described for Fig. 1A. The resulting S2 (odd-numbered lanes) and P3 (even-numbered lanes) fractions were analyzed by immunoblotting with the αHCF-1N antiserum. The positions of the HCF-1N subunits are indicated by the dots; extra uncharacterized bands in the serum-starved samples are indicated by asterisks.
RESULTS
HCF-1 associates with chromatin.
HCF-1 is a nuclear protein and, as part of the HSV-1 VP16-induced complex, associates with DNA. We therefore asked whether, in uninfected cells, HCF-1 is associated with DNA in the form of chromatin. We used a small-scale biochemical fractionation scheme (20) illustrated in Fig. 1A. HeLa cell lysates prepared with a nonionic detergent were divided by sequential centrifugation into three fractions: (i) a fraction largely consisting of soluble cytosolic components called S2, (ii) a fraction containing soluble nuclear components called S3, and (iii) an insoluble pellet fraction containing chromatin and nuclear matrix components called P3. The S2 fraction probably also contains soluble nuclear components owing to permeabilization of the nuclear membrane by the nonionic detergent.
To verify appropriate fractionation by this procedure, we probed the three different fractions by immunoblot analysis for the chromatin-associated DNA replication factor ORC2 and the cytoplasmic signal transduction kinase MEK2. As shown in Fig. 1B (lower panels), consistent with the desired fractionation, the majority of the ORC2 protein was in the P3 chromatin-containing pellet (compare lanes 1 to 3) whereas the MEK2 protein was in the S2 cytosolic fraction (compare lanes 4 to 6).
Using the same fractions, we analyzed the distribution of the native HCF-1N and HCF-1C subunits with HCF-1N and HCF-1C subunit-specific antisera. The large majority of the heterogeneous collection of endogenous HCF-1N (lanes 1 to 3) and HCF-1C (lanes 4 to 6) subunits were recovered in the P3 chromatin-containing pellet (Fig. 1B, upper panels). Similar results were obtained with human IMR90 and 293 cells (see below and data not shown). These results suggest that HCF-1 subunits are attached to chromatin or some other insoluble structure in the nucleus.
To distinguish between these two possibilities, we treated the P3 fraction with MNase to solubilize the chromatin. As shown in Fig. 1C, the majority of the HCF-1 in the P3 fraction becomes soluble within 2 to 5 min of MNase treatment (lanes 1 to 4, compare the pellet and supernatant panels). This pattern of HCF-1 MNase solubilization parallels that of the chromatin-bound DNA replication protein MCM3 (lanes 5 to 8). Analysis of the DNA in the MNase-treated samples showed that DNA release from the P3 pellet paralleled HCF-1 solubilization (data not shown). These results indicate that HCF-1 is associated with nuclease-sensitive chromatin in proliferating tissue culture cells.
The HCF-1C subunit is dispensable for chromatin association by the HCF-1N subunit.
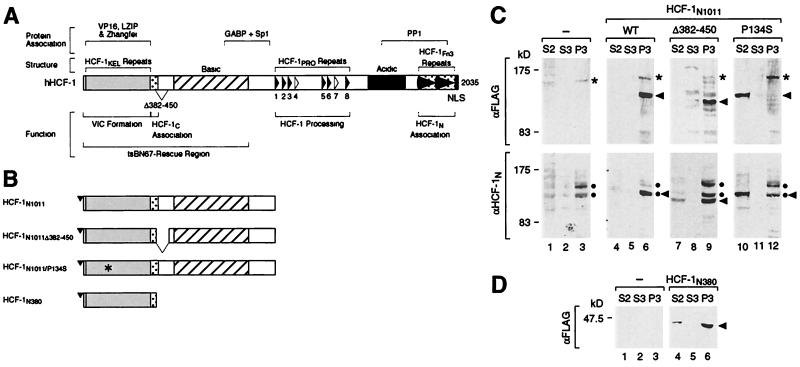
To map the regions of HCF-1 responsible for directing it to chromatin, we used a retroviral expression vector to direct synthesis of four epitope-tagged truncations of HCF-1 in HeLa cells. Figure 2A shows the overall structure of HCF-1 with the positions of (i) defined structural regions (e.g., HCF-1Kel repeats and basic region), (ii) functional regions (e.g., VP16-induced complex formation and HCF-1 subunit association), and (iii) regions involved in association with heterologous viral (VP16) and cellular (e.g., LZIP, GABP, and PP1) proteins. Figure 2B shows the series of four FLAG epitope-tagged HCF-1 proteins used to study HCF-1 recruitment to chromatin. Each of these epitope-tagged proteins is a derivative of the HCF-1N subunit: (i) HCF-1N1011 represents the wild-type HCF-1N subunit, (ii) HCF-1N1011Δ382–450 represents the product of an alternative HCF-1 pre-mRNA splice which does to associate with HCF-1C subunits (30), (iii) HCF-1N1011/P134S carries the tsBN67 P134S mutation, and (iv) HCF-1N380 represents the VP16 interaction domain alone.
FIG. 2.
The HCF-1 VP16 interaction domain is both necessary and sufficient for chromatin association. (A) Overall structure of HCF-1. Above and below a diagram of HCF-1 are shown the positions of (i) regions involved in association with the proteins indicated (top), (ii) structural or sequence elements (immediately above the diagram), and (iii) functional regions (below the diagram). Within the diagram, structural and functional features of HCF-1 are indicated from left to right as follows: the VP16 interaction or Kelch domain (gray box), the HCF-1C subunit association element (stippled box), the region deleted by the alternative pre-mRNA splice lacking residues 382 to 450 (triangle), the basic region (hatched box), the functional (filled triangles) and nonfunctional (open triangles) HCF-1PRO processing repeats, the acidic region (black box), the HCF-1N subunit association element with fibronectin type 3 repeats (stippled box with arrowheads), and the carboxy-terminal NLS (black box). VIC, VP16-induced complex. (B) Schematic of four FLAG epitope-tagged carboxy-terminal HCF-1 truncations used in this study. (C) Stable populations of HeLa cells synthesizing either FLAG-tagged HCF-1N1011 (lanes 4 to 6), HCF-1N1011Δ382–450 (lanes 7 to 9), HCF-1N1011/P134S (lanes 10 to 12), or no recombinant HCF-1 protein (lanes 1 to 3) were subjected to biochemical fractionation as described in the legend to Fig. 1A. S2 (lanes 1, 4, 7, and 10), S3 (lanes 2, 5, 8, and 11), and P3 (lanes 3, 6, 9, and 12) fractions were analyzed by immunoblotting with αFLAG (upper panel) and αHCF-1N (lower panel) antisera. The positions of endogenous HCF-1N subunits are indicated by the double dots, the positions of FLAG-tagged recombinant HCF-1N proteins are indicated by the arrowheads, and the position of an αFLAG antiserum-cross-reacting species is indicated by the asterisks. WT, wild type. (D) Stable populations of HeLa cells synthesizing FLAG-tagged HCF-1N380 (lanes 4 to 6) or no recombinant HCF-1 protein (lanes 1 to 3) were subjected to biochemical fractionation and probed with the αFLAG antiserum as described for part C. The position of the HCF-1N380 fragment is indicated by the arrowhead.
Figure 2C shows the results of biochemical fractionation of cells with each FLAG-tagged HCF-1N1011 derivative. The recombinant proteins were visualized alone with the αFLAG-tag antiserum (upper panel) or together with the endogenous proteins with the αHCF-1N subunit antiserum (lower panel). The αFLAG-tag analysis shows that the HCF-1N1011 protein is recovered in the P3 fraction (lanes 4 to 6, upper panels); this HCF-1N1011 protein could be solubilized by MNase treatment (data not shown), indicating that it is chromatin bound. The HCF-1N1011Δ382–450 protein, which does not associate with the HCF-1C subunit (30), is also present in the chromatin-containing P3 fraction (compare lanes 7 to 9, upper panel), indicating that association with the HCF-1C subunit is dispensable for chromatin association by the HCF-1N subunit.
Probing with the αHCF-1N antiserum (Fig. 2C, lower gels) shows that the recombinant HCF-1N proteins are synthesized at levels similar to those of the endogenous proteins and that the endogenous HCF-1N subunits are still largely associated with chromatin (compare lanes 1 to 12). Thus, in these cells the endogenous HCF-1N proteins are not dislodged from the chromatin to any great extent.
The HCF-1 VP16 interaction domain is necessary and sufficient for chromatin association.
The HCF-1N subunit contains two functional regions that might be involved in chromatin association: the VP16 interaction domain and the basic region. Both of these regions are required to rescue the tsBN67 HCF-1 cell proliferation defect (28) and can associate with cellular DNA-binding proteins (5, 8, 16, 26). To determine whether the VP16 interaction domain is required for HCF-1N–chromatin association, we tested whether the HCF-1N1011/P134S protein, which fails to associate with VP16 (28), LZIP (5), and ZF (15), associates with chromatin. Indeed, the HCF-1N1011/P134S subunit failed to associate with chromatin (Fig. 2C, lanes 10 and 12, upper panel) and did not cofractionate with the endogenous HCF-1N subunits (compare lanes 10 and 12, lower panel). Thus, an HCF-1 point mutation, which is known to prevent HCF-1 association with proteins that bind DNA, also prevents HCF-1N association with chromatin. Together with the lack of any evidence that HCF-1 normally binds DNA directly, these results suggest that HCF-1 is tethered to DNA by interaction with a protein receptor at least in part through its VP16 interaction domain. Because in this experiment the HCF-1N1011/P134S mutant protein is present alongside the chromatin-bound wild-type endogenous protein, the lack of chromatin-bound HCF-1N1011/P134S protein suggests that HCF-1N subunits do not recruit each other to chromatin through higher-order homomeric complex formation.
To test whether the HCF-1 VP16 interaction domain is not only necessary but also sufficient for HCF-1N–chromatin association, we asked whether the HCF-1N380 fragment (Fig. 2B) can associate with chromatin. The HCF-1N380 fragment is indeed largely present in the P3 fraction (compare lanes 4 to 6). Thus, the VP16 interaction domain is both necessary and sufficient for HCF-1N–chromatin association. These results suggest that HCF-1N subunits are tethered to DNA in chromatin through their VP16 interaction domains and that the basic region is dispensable for HCF-1–chromatin association.
Nuclear localization of HCF-1N1011 but not the HCF-1N1011/P134S mutant.
Given the results of previous studies of HCF-1 subcellular localization (12, 27), HCF-1N subunit–chromatin association is paradoxical. The previous studies have shown that HCF-1 nuclear localization is directed by a single nuclear localization signal (NLS) located at the carboxy terminus of HCF-1 (Fig. 2A). When it is synthesized independently of the HCF-1C subunit by transient overexpression, the HCF-1N subunit remains in the cytoplasm (12, 27). How then is the HCF-1N subunit present in the chromatin-containing P3 fraction after stable synthesis in HeLa cells? We hypothesized that, if not synthesized at very high levels as during transient overexpression, the majority of the HCF-1N subunits may be brought to chromatin and hence the nucleus by the VP16 interaction domain.
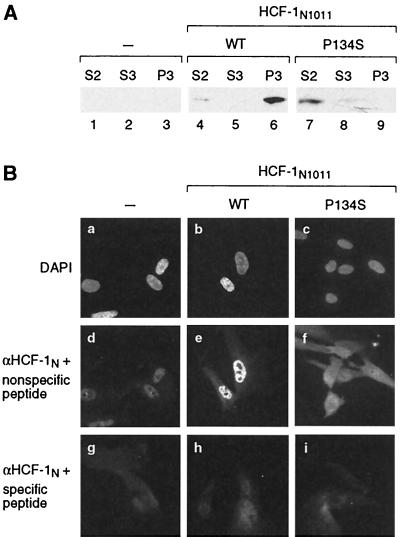
To test this hypothesis, we assayed HCF-1N1011 and HCF-1N1011/P134S localization by immunofluorescence in IMR90 cells carrying the FLAG tag retroviral expression vectors as shown in Fig. 3. We chose IMR90 cells because the suspension HeLa cells used for recombinant HCF-1 synthesis are less amenable to immunofluorescence analysis. As in HeLa cells, in these IMR90 cells, the wild-type HCF-1N1011 (Fig. 3A, lanes 4 to 6), but not the HCF-1N1011/P134S fragment (lanes 7 to 9), is associated with the chromatin-containing P3 fraction.
FIG. 3.
Nuclear localization of the HCF-1N1011 but not the HCF-1N1011/P134S polypeptide. (A) Stable populations of IMR90 cells synthesizing either FLAG-tagged HCF-1N1011 (lanes 4 to 6), HCF-1N1011/P134S (lanes 7 to 9), or no recombinant HCF-1 protein (lanes 1 to 3) were subjected to biochemical fractionation as described for Fig. 1A. S2 (lanes 1, 4, and 7), S3 (lanes 2, 5, and 8), and P3 (lanes 3, 6, and 9) fractions were analyzed by immunoblotting with αFLAG antiserum. (B) The same IMR90 cell populations synthesizing FLAG-tagged HCF-1N1011 (b, e, and h), HCF-1N1011/P134S (c, f, and i), or no recombinant HCF-1 protein (a, d, and g) were subjected to immunofluorescence analysis with purified αHCF-1N antibodies in the presence of the peptide (N18) used to raise the antiserum (g to i) or of a nonspecific peptide (FLAG) (d to f). The nuclei in the cells shown in images d to f were identified by staining with DAPI (a to c). WT, wild type.
For the immunofluorescence analysis, the αFLAG antiserum was not sufficiently sensitive to detect the recombinant HCF-1N proteins. We therefore used the αHCF-1N antiserum to detect both endogenous and recombinant HCF-1N proteins in control IMR90 cells (Fig. 3B, panels a, d, and g) or IMR90 cells synthesizing wild-type (b, e, and h) or P134S mutant (c, f, and i) HCF-1N1011 protein. The cells were stained with DAPI to identify nuclei (Fig. 3B, panels a to c) and with the αHCF-1N antipeptide serum in the absence (d to f) or presence (g to i) of specific peptide to establish the specificity of the HCF-1N staining; indeed, the αHCF-1N antiserum produces a low level of nonspecific staining (g to i). The control cells (Fig. 3B, panel d) exhibit the typical predominantly nuclear immunofluorescence pattern of endogenous HCF-1 (11). When the wild-type recombinant HCF-1N1011 subunit is present in the IMR90 cells, there is an increase in nuclear staining (compare Fig. 3B, panels d and e), whereas when the mutant HCF-1N1011/P134S subunit is present, there is an increase in cytoplasmic staining (compare Fig. 3B, panels d and f). These results are consistent with nuclear localization of the wild-type but not the P134S mutant HCF-1N1011 subunit and suggest that the predominantly cytoplasmic localization of the wild-type HCF-1N subunit observed previously during transient overexpression results from the very high levels of transient HCF-1N subunit synthesis. Perhaps an HCF-1N receptor protein(s) can bring the HCF-1N subunit to the nucleus and chromatin and this receptor is saturated by HCF-1N overexpression.
HCF-1 is an abundant protein.
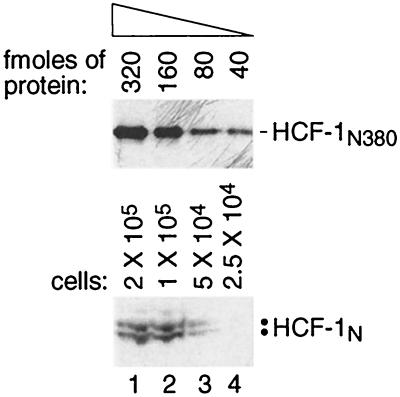
The experiments described above suggest that HCF-1 is targeted to chromatin through protein-protein association. Purification of HCF-1 in a previous study has suggested that it is an abundant protein (29), but the number of molecules in a cell is unknown. Such a number would indicate how many chromatin-bound receptor proteins are required to recruit the HCF-1N subunit to chromatin if they associate stoichiometrically as in the VP16-induced complex. To determine the number of HCF-1 molecules in a cell, we compared the immunoblot signal of endogenous HCF-1 from a known number of cells to that of a known concentration of HCF-1N380 protein synthesized in E. coli (see Materials and Methods). From the comparison, whose results are shown in Fig. 4, we obtained a minimal estimate of the number of HCF-1 molecules per cell by estimating that 105 cells (lane 2, lower panel) contain 80 fmol of protein (lane 3, upper panel) or about 5 × 105 HCF-1 molecules per HeLa cell. We estimated about half that amount per IMR90 cell (data not shown). Thus, even if our number is not absolutely precise, HCF-1 is an abundant protein probably present at more than 100,000 molecules per HeLa or IMR90 cell. This result suggests that, if only one or a few proteins target HCF-1 to chromatin, one or more of them should be very abundant. Alternatively, a large number of less abundant proteins may target HCF-1 to chromatin.
FIG. 4.
HeLa cells contain over 100,000 molecules of HCF-1 per cell. The immunoblot signal of a twofold titration of bacterially synthesized HCF-1N380 protein of known molar concentration (upper panel; the number of femtomoles of HCF-1N380 protein in each lane is indicated) is directly compared to the immunoblot signal of a twofold titration of HCF-1 from a known number of HeLa cells (lower panel; the number of cells used for each lane is indicated).
HCF-1 chromatin association is temperature sensitive in tsBN67 cells.
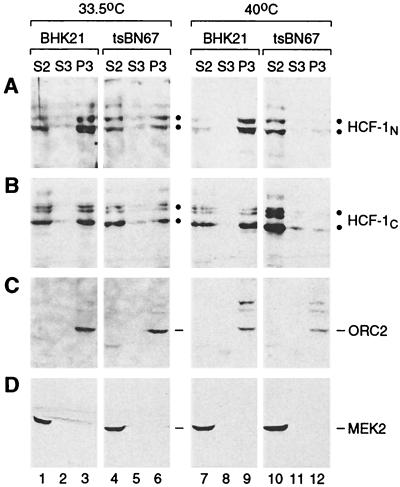
The tsBN67 P134S mutation prevents chromatin association of HCF-1N subunits in HeLa cells grown at 37°C (Fig. 2C). The same mutation causes temperature-sensitive proliferation arrest in tsBN67 cells at 40°C. To determine whether there is a relationship between HCF-1 chromatin association and the temperature-sensitive defect in tsBN67 cell proliferation, we performed biochemical fractionation on four sets of cells: BHK21 and tsBN67 cells grown at permissive (33.5°C) and nonpermissive (40°C) temperatures for 64 h.
To monitor the fractionation of the tsBN67 cell extracts, the distribution of chromatin-bound ORC2 and cytosolic MEK2 in the different fractions was assayed as shown in Fig. 5C and D. In both cell lines and at both temperatures, ORC2 was present exclusively in the P3 chromatin pellet (Fig. 5C, lanes 1 to 12), indicating that the soluble fractions were not contaminated with chromatin-associated proteins, and MEK2 was solely recovered in the cytosolic fraction S2 (Fig. 5D, lanes 1 to 12), indicating that the extraction of cytosolic proteins was complete.
FIG. 5.
The HCF-1–chromatin association is temperature sensitive in tsBN67 cells. BHK21 (lanes 1 to 3 and 7 to 9) and tsBN67HR1 (lanes 4 to 6 and 10 to 12) cells grown at 33.5°C (lanes 1 to 6) or 40°C (lanes 7 to 12) for 64 h were biochemically fractionated as described for Fig. 1A, and the resulting S2 (lanes 1, 4, 7, and 10), S3 (lanes 2, 5, 8, and 11), and P3 (lanes 3, 6, 9, and 12) fractions were analyzed by immunoblotting with αHCF-1N (A), αHCF-1C (B), αORC2 (C), and αMEK2 (D) antisera. The protein contents of separate fractionations were normalized by Bradford assay of each S2 fraction. The positions of the relevant polypeptides are indicated.
In the same extracts, the HCF-1N and HCF-1C subunits behaved similarly in this assay. At 33.5°C, most, albeit (unlike with HeLa, 293, and IMR90 cells grown at 37°C) not all, of the wild-type HCF-1 in BHK21 cells was chromatin associated (Fig. 5A and B, lanes 1 to 3). In tsBN67 cells grown at 33.5°C, about half of the HCF-1 was chromatin bound (Fig. 5A and B, lanes 4 to 6). Thus, in contrast to the recombinant HCF-1N1011/P134S protein in HeLa cells grown at 37°C, the P134S mutant HCF 1 protein can bind chromatin in tsBN67 cells grown at 33.5°C. At 40°C, however, whereas the HCF-1 from BHK21 cells was still predominantly chromatin bound (lanes 7 to 9), the HCF-1N and HCF-1C subunits from tsBN67 cells were largely absent from the chromatin-containing P3 fraction (lanes 10 to 12). Although still probably being nuclear owing to the HCF-1C NLS, the tsBN67 HCF-1 subunits were recovered instead in the soluble S2 fraction, probably because soluble nuclear proteins leak out of the nucleus during the fractionation procedure. Whatever the reason, however, these results indicate that there is a correlation between HCF-1–chromatin association and tsBN67 cell proliferation. Furthermore, the lack of HCF-1C subunit association with chromatin in temperature-arrested tsBN67 cells indicates that the association of HCF-1C subunits with chromatin is dependent on the VP16 interaction domain of the HCF-1N subunit.
Dissociation of tsBN67 HCF-1 from chromatin at 40°C is not immediate but precedes cell proliferation arrest.
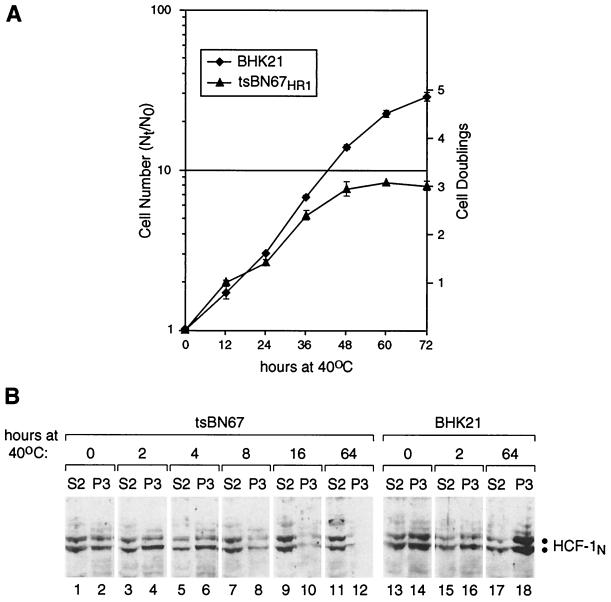
A characteristic property of tsBN67 cells is that they cease to proliferate at the nonpermissive temperature of 40°C after a lag of a few cell divisions (7). To characterize the dissociation of HCF-1 from chromatin in tsBN67 cells at 40°C, we asked how the timing of HCF-1–chromatin dissociation correlates with the timing of the tsBN67 cell proliferation arrest. As shown in Fig. 6A, as with the original tsBN67 cells (7), the subclone of tsBN67 cells used in this study (called tsBN67HR1, see Materials and Methods) stopped proliferating only after a lag of 36 to 48 h at 40°C. In contrast, BHK21 cells continued to proliferate at 40°C. Remarkably, biochemical fractionation of tsBN67 cells harvested at different time points after the shift to 40°C revealed that the P134S mutant HCF-1 subunits were still bound to chromatin after 4 h (Fig. 6B, compare lanes 2, 4, and 6) but began to dissociate markedly from chromatin by 8 h (lane 8) and were largely dissociated from chromatin by 16 h (lane 10). More refined time points have indicated that complete HCF-1N–chromatin dissociation occurs after about 12 to 18 h at 40°C (data not shown). In contrast, in BHK21 cells, HCF-1 was present in the chromatin-containing P3 fraction throughout the course of the experiment (lanes 14, 16, and 18). Thus, in tsBN67 cells grown at 40°C, temperature-sensitive HCF-1 dissociates from chromatin in the middle of the 36- to 48-h lag in cell proliferation arrest, preceding the arrest by about 20 to 30 h.
FIG. 6.
Dissociation of HCF-1 from chromatin precedes tsBN67 cell proliferation arrest. (A) tsBN67HR1 cells possess temperature-induced growth arrest characteristics similar to those of parental tsBN67 cells. BHK21 and tsBN67HR1 cells were shifted to 40°C at 0 h, harvested, and counted at the indicated time points as described in Materials and Methods. N0, number of cells at time of transfer to 40°C; Nt, number of cells at time of harvest. Each point is the average of results from two samples, with the deviation shown. (B) Timing of tsBN67 HCF-1P134S dissociation from chromatin at 40°C. BHK21 (lanes 13 to 18) and tsBN67HR1 (lanes 1 to 12) cells were grown at 40°C for the indicated times and subsequently harvested and fractionated as described for Fig. 1A. The resulting S2 (odd-numbered lanes) and P3 (even-numbered lanes) fractions were analyzed by immunoblotting with the αHCF-1N antiserum. The positions of the HCF-1N polypeptides are indicated.
Arrest of tsBN67 cell proliferation before transfer to 40°C prevents temperature-induced dissociation of HCF-1 from chromatin.
The aforementioned results indicate that HCF-1 dissociates from chromatin in tsBN67 cells at 40°C but only after a delay of up to 18 h. We hypothesize two mechanisms for this delay: (i) a proliferation-independent mechanism in which it may simply take time for the elevated temperature to dissociate the temperature-sensitive HCF-1 from chromatin or (ii) a proliferation-dependent mechanism in which proliferation at 40°C is required to dissociate the mutant HCF-1 from chromatin. To discriminate between these two mechanisms, we starved tsBN67 cells of serum at 33.5°C for 24 h to arrest cell proliferation and then transferred the arrested cells to 40°C, maintaining the low concentration of serum, and harvested them for biochemical fractionation at different time points for up to 30 h. At the time of transfer to 40°C, DNA content analysis showed that the serum-starved tsBN67 cells were arrested with a G1/G0 DNA content (data not shown). As shown in Fig. 7 (upper panels), the temperature-sensitive HCF-1 did not dissociate from chromatin in serum-starved cells throughout the 30-h time course. In contrast, as expected, with an asynchronously proliferating population, HCF-1 was not detectable in the P3 chromatin-containing fraction by 18 h postshifting to 40°C (Fig. 7, lower panel). This analysis provides two conclusions. First, because HCF-1 is bound to chromatin in the cells arrested by serum starvation (Fig. 7, upper panel), HCF-1 can be chromatin bound in nonproliferating cells. Second, dissociation of the mutant HCF-1 from chromatin after transfer to the nonpermissive temperature is proliferation dependent, suggesting that HCF-1 dissociation from chromatin is an active process during the course of tsBN67 cell proliferation.
DISCUSSION
The study presented here clarifies our understanding of HCF-1 structure and function. They show that HCF-1—an abundant protein—is brought to DNA, probably indirectly, through a single structural element—its VP16 interaction domain—which is implicated solely in protein-protein and not protein-DNA interactions.
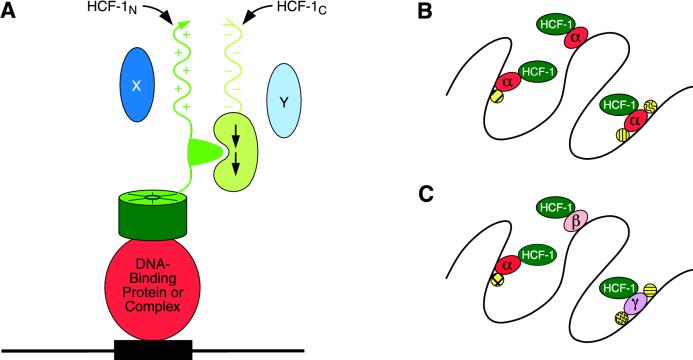
That the same region of HCF-1 is involved in both VP16-induced complex assembly and chromatin association suggests that the VP16-induced complex represents a model for how HCF-1 binds to chromatin naturally. Consistent with this hypothesis, VP16 and the two cellular proteins that have been shown to associate with the HCF-1 VP16 interaction domain share the tetrapeptide motif (D/E)HXY for HCF-1 binding, and in each case the P134S tsBN67 mutation prevents protein association (5, 15, 17). By analogy to the VP16-induced complex, Fig. 8A illustrates how we believe HCF-1 associates with chromatin. We hypothesize that the HCF-1 VP16 interaction (or Kelch) domain brings HCF-1 to the DNA by contacting a solitary DNA-bound protein, such as LZIP, or a member of a complex of DNA-bound proteins as occurs in the VP16-induced complex.
FIG. 8.
Models of HCF-1 binding to DNA in chromatin. (A) Hypothetical structure of HCF-1 bound to chromatin in cartoon form. In this model, HCF-1 is targeted to the DNA (black) in chromatin by its VP16 interaction or Kelch domain (dark green) through protein-protein interaction with a DNA-binding protein or protein complex (red). Other regions of HCF-1—the basic region (+), acidic region (−), HCF-1N–HCF-1C association region (light green), and HCF-1C–HCF-1N association region (kidney shape with arrows)—are shown in shades of green. Hypothetical effectors of HCF-1, proteins X and Y, are shown in shades of blue. (B and C) Alternative models for HCF-1 binding to chromatin. (B) HCF-1 associates with a single abundant species labeled α (red) through its VP16 interaction domain. A multiplicity of other factors (yellow) may associate with and perhaps stabilize the association with the α species in chromatin. (C) HCF-1 associates with different DNA-binding proteins (three, α, β, and γ, are shown in the figure), any one member of which need not be abundant, but may bind DNA in different transcription factor (yellow) contexts.
This view of HCF-1 function suggests that, by targeting HCF-1 to specific DNA-bound regulatory proteins, the HCF-1 VP16 interaction domain is responsible for providing regulatory specificity. Other regions of HCF-1 are also essential, however, for its activity. For example, the HCF-1 basic region is necessary to promote cell proliferation (28). Because there is no evidence from our studies that these other regions of HCF-1 naturally bind to DNA either directly or indirectly, we suggest that they function by associating with other non-DNA-binding proteins (e.g., X and Y in Fig. 8A). An attractive model is that hypothetical basic region- and HCF-1C-associated proteins are enzymes that regulate, for example, chromatin structure or transcription factor function. With the HCF-1C subunit, such a candidate effector protein is the cell cycle-regulatory protein phosphatase 1, which can bind to the carboxy-terminal HCF-1C subunit (1). The basic region has not been shown to associate with any such enzymatic activities but instead is thought to associate with the DNA-binding proteins GABP (26) and Sp1 (8). Because the basic region is not required for HCF-1 binding to chromatin, however, HCF-1 association with GABP or Sp1 is probably not essential for HCF-1 to bind to chromatin. Nevertheless, it may associate with these two proteins once brought to the DNA through its VP16 interaction domain or a minor population of HCF-1 not detected in our study may associate with chromatin through these proteins.
The findings that a single structural element of HCF-1—its VP16 interaction domain—probably brings HCF-1 to the DNA by stoichiometric association with a DNA-binding protein(s) and that HCF-1 is very abundant in the cell—over 100,000 copies in a typical tissue culture cell—have important implications for our understanding of the nature of the molecular target or targets of HCF-1. If the HCF-1 VP16 interaction domain associates with a single species in the cell, as suggested by the hypothetical α protein in Fig. 8B, this species must be equally abundant. Proteins such as LZIP and ZF, which are present at very low levels in tissue culture cells (14, 15), do not satisfy this criterion. We favor a second model shown in Fig. 8C, in which HCF-1 associates with a large family of DNA-binding proteins, of which any one member need not be abundant. Such a model would be consistent with functional association of HCF-1 with proteins such as LZIP and ZF, which share the short and degenerate (D/E)HXY HCF-1-binding motif. If true, this model would suggest that HCF-1 may function like coregulatory proteins such as the coactivators p300 and CREB-binding protein and the corepressor mSin3, each of which associates with many different DNA-binding transcriptional regulators (see references 2, 6, and 24).
Whichever model is correct, the finding that the wild-type HCF-1N subunit lacking a canonical NLS (12) is both nuclear and chromatin bound suggests that, as with VP16, HCF-1 can associate with its DNA-binding target protein(s) off of the DNA and—perhaps in contrast to VP16 (see reference 12)—bring HCF-1 to the nucleus. Consistent with this hypothesis, the HCF-1N/P134S subunit, which fails to associate with putative target proteins, is apparently neither chromatin bound nor nuclear (Fig. 3). The chromatin association of wild-type HCF-1N subunits explains why the HCF-1N1011Δ382–450 protein, which lacks the NLS and does not associate with the HCF-1C subunit, can still rescue the temperature-induced tsBN67 cell proliferation defect (28).
Surprisingly, it is the very ability of HCF-1 to bind to chromatin that is temperature sensitive in the tsBN67 cell line, and loss of chromatin association precedes temperature-induced arrest of tsBN67 cell proliferation. These results suggest that the temperature-sensitive cell proliferation defect of tsBN67 cells results from the loss of HCF-1P134S association with chromatin at the nonpermissive temperature. Both the loss of HCF-1P134S chromatin association (Fig. 7) and the delayed loss of cell proliferation (7), however, require cell proliferation itself, suggesting that they are each the result of an active growth process in the mutant tsBN67 cells.
The different time courses of HCF-1P134S dissociation from chromatin (up to 18 h) and cell proliferation arrest (36 to 48 h) upon transfer of tsBN67 cells to the nonpermissive temperature begin to reveal a cascade of events that leads to the arrest of tsBN67 cell proliferation. Perhaps HCF-1 dissociation from chromatin is delayed in asynchronous cells at the nonpermissive temperature because it occurs during a particular phase of the tsBN67 cell cycle. Such a property would explain why proliferation is required for dissociation of HCF-1 from chromatin and why there is a lag of about one cell cycle (i.e., about 12 to 18 h) for full loss of HCF-1 chromatin association (Fig. 6).
The results described here, together with the role of HCF-1 in HSV-1 VP16-induced transcriptional activation, suggest that HCF-1 is a new player in transcriptional control of cell proliferation. HCF-1 may well have activities similar to those of better-understood regulators of cell proliferation that, like HCF-1, are recruited to the DNA by sequence-specific DNA-binding proteins. Two prominent examples are the Rb protein, which associates with E2F and recruits histone deacetylases (4, 19, 22), and mSin3, which associates with numerous DNA-binding proteins and also recruits deacetylases (2, 24). Unlike these complexes, however, which repress cell proliferation, a putative HCF-1 complex promotes cell proliferation because loss of HCF-1 function leads to an arrest of cell proliferation. Thus, similar to its function as a viral transcriptional coactivator, HCF-1 may be a coactivator of transcription in uninfected cells. Consistent with this hypothesis, HCF-1 can enhance transcriptional activation by LZIP in transient-expression assays (17, 18).
Although the experiments described here have revealed similarities between HCF-1 and transcriptional regulatory proteins such as Rb, the association of VP16 with HCF-1 and the DNA tumor virus early proteins such as the adenovirus E1A protein and simian virus 40 T-antigen proteins with Rb suggest opposing strategies of virus-host cell interaction. The DNA tumor virus early proteins are synthesized de novo upon infection, and by stoichiometric association they inactivate Rb to promote infected-cell entry into S phase. In contrast, VP16 is not synthesized de novo early in infection but instead is brought in by the infecting virion(s), where there are only 500 to 1,000 molecules of VP16 per virion (9) compared to the estimated 100,000 molecules of HCF-1. Thus, unlike how E1A or T antigen inactivate the Rb protein early in infection, VP16 probably does not inactivate endogenous HCF-1 early in infection and indeed may have little effect on its normal functions. Thus, rather than commandeer the cell, as do E1A and T antigen, VP16 may associate with HCF-1 surreptitiously and use productive association as one indication for whether the cellular environment is appropriate to promote lytic infection.
ACKNOWLEDGMENTS
We are indebted to J. Méndez for many helpful discussions concerning protein binding to chromatin and the small-scale chromatin isolation procedure; J. Lees for advice on the quantitation of HCF-1; T. Nishimoto for BHK21 and tsBN67 cells; members of the Herr laboratory for helpful discussions; N. Hernandez, E. Julien, J. Méndez, and W. Tansey for critical readings of the manuscript; J. Duffy and P. Renna for photography; and J. Reader for help in preparation of the manuscript.
This study was funded by U.S. Public Health Service grants GM54598 and CA13106.
REFERENCES
- 1.Ajuh P M, Browne G J, Hawkes N A, Cohen P T, Roberts S G, Lamond A I. Association of a protein phosphatase 1 activity with the human factor C1 (HCF) complex. Nucleic Acids Res. 2000;28:678–686. doi: 10.1093/nar/28.3.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 3.Bork P, Doolittle R F. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 4.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 5.Freiman R N, Herr W. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman R H, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 7.Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 8.Gunther M, Laithier M, Brison O. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol Cell Biochem. 2000;210:131–142. doi: 10.1023/a:1007177623283. [DOI] [PubMed] [Google Scholar]
- 9.Heine J W, Honess R W, Cassai E, Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974;14:640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herr W. The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation. Cold Spring Harbor Symp Quant Biol. 1998;63:599–607. doi: 10.1101/sqb.1998.63.599. [DOI] [PubMed] [Google Scholar]
- 11.Kristie T M, Pomerantz J L, Twomey T C, Parent S A, Sharp P A. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 12.La Boissiere S, Hughes T, O'Hare P. HCF-dependent nuclear import of VP16. EMBO J. 1999;18:480–489. doi: 10.1093/emboj/18.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Boissiere S, Walker S, O'Hare P. Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol Cell Biol. 1997;17:7108–7118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R, Misra V. Potential role for luman, the cellular homologue of herpes simplex virus VP16 (alpha gene trans-inducing factor), in herpesvirus latency. J Virol. 2000;74:934–943. doi: 10.1128/jvi.74.2.934-943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R, Misra V. Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res. 2000;28:2446–2454. doi: 10.1093/nar/28.12.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu R, Yang P, O'Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor Mol. Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu R, Yang P, Padmakumar S, Misra V. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF. J Virol. 1998;72:6291–6297. doi: 10.1128/jvi.72.8.6291-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luciano R L, Wilson A C. N-terminal transcriptional activation domain of LZIP comprises two LxxLL motifs and the host cell factor-1 binding motif. Proc Natl Acad Sci USA. 2000;97:10757–10762. doi: 10.1073/pnas.190062797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 20.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgenstern J P, Land H. Advanced mammalian gene transfer high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 23.O'Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- 24.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 25.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 26.Vogel J L, Kristie T M. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J. 2000;19:683–690. doi: 10.1093/emboj/19.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson A C, Boutros M, Johnson K M, Herr W. HCF-1 amino- and carboxy-terminal subunit association through two separate sets of interaction modules: involvement of fibronectin type 3 repeats. Mol Cell Biol. 2000;20:6721–6730. doi: 10.1128/mcb.20.18.6721-6730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson A C, Freiman R N, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson A C, LaMarco K, Peterson M G, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 30.Wilson A C, Peterson M G, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- 31.Xue F, Cooley L. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]