Abstract
CXCL12 expression was significantly lower in tumor samples than in corresponding normal samples. CXCL12 expression was significantly positively related to the infiltration levels of T cells, dendritic cells (DCs), immature DCs, cytotoxic cells, Tfh cells, mast cells, B cells, Th1 cells, natural killer (NK) cells, pDCs, neutrophils, and T helper cells (Spearman correlation coefficient > 0.5, P < 0.001) and negatively correlated with the infiltration level of NK CD56bright cells. In addition, pancreatic hTERT-HPNE cells treated with three diverse CXCL12 isoforms exhibited changes mainly in the regulation of the epithelial-mesenchymal transition activation pathway.
Keywords: CXCL12, Pancreatic cancer, Splicing isoforms, Bioinformatics analysis, Tumor microenvironment, Pathway
Core Tip: CXCL12 expression was significantly lower in tumor samples than in normal samples. CXCL12 expression was significantly positively associated with the infiltration levels of 12 immune cells, especially T cells, which may encourage further exploration of the effect of CXCL12 in pancreatic ductal adenocarcinoma immunotherapy. In addition, treating pancreatic hTERT-HPNE cells with three diverse CXCL12 isoforms mainly affected the regulation of the epithelial-mesenchymal transition activation pathway.
TO THE EDITOR
We read with interest the article by Cecati et al[1]. They investigated the specific roles of α, β, and γ CXCL12 isoforms in pancreatic ductal adenocarcinoma (PDCA) onset by microarray analysis of hTERT-HPNE cells cured by three diverse isoforms of CXCL12, which indicated that CXCL12 isoforms have different roles in PDAC pathogenesis.
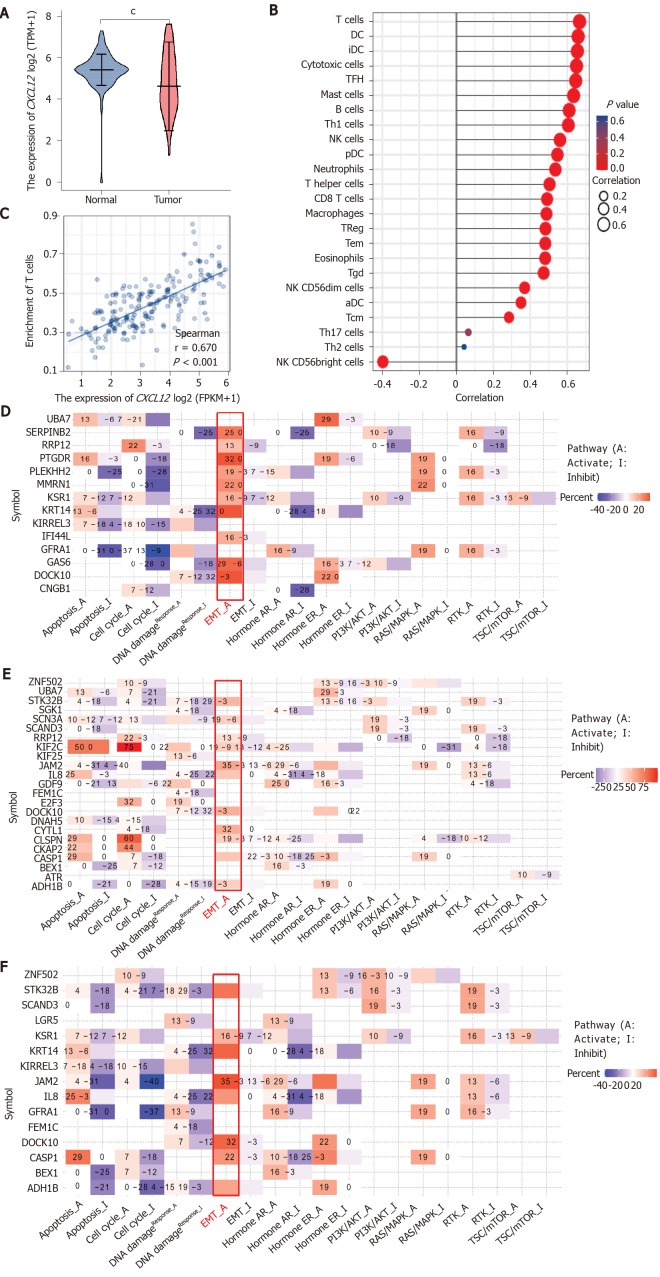
We appreciate the unique perspective provided by the authors’ exploration of the roles of the different isomers of CXCL12 in PDAC. However, the results might be made more meaningful if the authors built on this by presenting the differential expression of CXCL12 in normal and tumor tissues of PDCA as a whole, such as through a bioinformatics analysis of PDCA cases in The Cancer Genome Atlas (TCGA) database or their own data. We discovered that the CXCL12 expression was significantly lower in tumor samples than in normal samples (Figure 1A). Detailed statistical results are described in Table 1.
Figure 1.
The effect of CXCL12 in the development of pancreatic ductal adenocarcinoma. A: The differential CXCL12 expression in pancreatic ductal adenocarcinoma (PDCA) and normal samples. The expression level of CXCL12 in tumor tissues is indicated in orange, and that in normal tissues is indicated in purple. Data source: UCSC XENA (https://xenabrowser.net/datapages/) RNAseq data in TPM format for The Cancer Genome Atlas (TCGA) and GTEx processed uniformly through the Toil process[4]. PAAD (pancreatic cancer) data were extracted from TCGA, and corresponding normal sample data were from GTEx. Significance markers: NS, P ≥ 0.05, aP < 0.05, bP < 0.01, cP < 0.001; B: The expression level of CXCL12 and its relationship to 24 immune cell infiltration levels in PDCA. Data source: RNAseq data and clinical data in level 3 HTSeq-FPKM format from the TCGA (https://portal.gdc.cancer.gov/) PAAD (pancreatic cancer) project. Data filtering: Removal of paraneoplastic tissue; C: The expression level of CXCL12 and its relationship to the T cell infiltration level in PDCA; D and E: Pathway analysis of differentially expressed genes under all treatment conditions (α, β, and γ CXCL12 isoforms); D: CXCL12 α isoform vs control; E: CXCL12 β isoform vs control; F: CXCL12 γ isoform vs control.
Table 1.
Detailed statistical results of CXCL12 differential expression analysis in pancreatic ductal adenocarcinoma (mean ± SD)
|
Group
|
Number
|
Minimum
|
Maximum
|
Median
|
IQR
|
Lower quartile
|
Upper quartile
|
Mean
|
|
SE
|
| Normal | 171 | 0 | 7.296 | 5.433 | 0.756 | 5.028 | 5.784 | 5.403 | 0.88 | 0.067 |
| Tumor | 179 | 1.333 | 7.629 | 4.632 | 2.134 | 3.727 | 5.861 | 4.803 | 1.445 | 0.108 |
IQR: Interquartile distance; SE: Standard error.
The tumor microenvironment (TME), mediated by interactions between stromal cells and pancreatic epithelial/carcinoma cells, is essential for PDCA progression and has been associated with failure of chemotherapy, radiotherapy, and immunotherapy[2]. The formation of the microenvironment requires interactions between pancreatic cancer cells and stromal cells. A pancreatic cancer microenvironment composition that favors demyelination and immunosuppression is related to poor prognosis[3-5]. Although immunotherapy has transformed cancer therapy, patients with PDCA rarely respond to these regimens, and this failure is attributed to poor infiltration and activation of T cells in the TME. We found that CXCL12 expression was positively correlated with the level of infiltration of 22 immune cells, especially T cells (Figure 1B and C), which may encourage further exploration of the effect of CXCL12 in PDCA immunotherapy. Detailed information on the correlation between CXCL12 expression and immune cell infiltration is shown in Table 2.
Table 2.
Detailed information on the correlation between CXCL12 expression and immune cell infiltration
|
Gene
|
Cell
|
Correlation coefficient (Pearson)
|
P
value (Pearson)
|
Correlation coefficient (Spearman)
|
P
value (Spearman)
|
| CXCL12 | aDC | 0.355 | < 0.001 | 0.350 | < 0.001 |
| CXCL12 | B cells | 0.614 | < 0.001 | 0.610 | < 0.001 |
| CXCL12 | CD8 T cells | 0.508 | < 0.001 | 0.491 | < 0.001 |
| CXCL12 | Cytotoxic cells | 0.674 | < 0.001 | 0.650 | < 0.001 |
| CXCL12 | DC | 0.668 | < 0.001 | 0.658 | < 0.001 |
| CXCL12 | Eosinophils | 0.488 | < 0.001 | 0.480 | < 0.001 |
| CXCL12 | iDC | 0.639 | < 0.001 | 0.654 | < 0.001 |
| CXCL12 | Macrophages | 0.488 | < 0.001 | 0.487 | < 0.001 |
| CXCL12 | Mast cells | 0.635 | < 0.001 | 0.634 | < 0.001 |
| CXCL12 | Neutrophils | 0.554 | < 0.001 | 0.535 | < 0.001 |
| CXCL12 | NK CD56bright cells | -0.411 | < 0.001 | -0.397 | < 0.001 |
| CXCL12 | NK CD56dim cells | 0.376 | < 0.001 | 0.369 | < 0.001 |
| CXCL12 | NK cells | 0.566 | < 0.001 | 0.560 | < 0.001 |
| CXCL12 | pDC | 0.558 | < 0.001 | 0.546 | < 0.001 |
| CXCL12 | T cells | 0.682 | < 0.001 | 0.666 | < 0.001 |
| CXCL12 | T helper cells | 0.511 | < 0.001 | 0.504 | < 0.001 |
| CXCL12 | Tcm | 0.337 | < 0.001 | 0.285 | < 0.001 |
| CXCL12 | Tem | 0.483 | < 0.001 | 0.481 | < 0.001 |
| CXCL12 | TFH | 0.668 | < 0.001 | 0.645 | < 0.001 |
| CXCL12 | Tgd | 0.364 | < 0.001 | 0.472 | < 0.001 |
| CXCL12 | Th1 cells | 0.594 | < 0.001 | 0.605 | < 0.001 |
| CXCL12 | Th17 cells | 0.057 | 0.453 | 0.065 | 0.387 |
| CXCL12 | Th2 cells | 0.069 | 0.357 | 0.032 | 0.675 |
| CXCL12 | TReg | 0.493 | < 0.001 | 0.482 | < 0.001 |
aDC: Activated DC; DC: Dendritic cells; iDC: immature DC; pDC: Plasmacytoid DC; Tfh: T follicular helper; Tgd: T gamma delta; NK: Natural killer.
We agree with Cecati et al[1], who reported that all CXCL12 isoforms influenced cell migration, adhesion, and cytoskeleton-associated gene expression. In our study, we found that treating pancreatic hTERT-HPNE cells with three diverse CXCL12 isoforms mainly affects the regulation of the EMT activation pathway (Figure 1D-F), which confirms that the work done by Cecati et al[1] is worthy of recognition and that our findings can be a supplement to their study. In the future, we should investigate the role played by CXCL12 in the PDCA immune microenvironment in depth.
Statistical analysis
Software: R (version 3.6.3) was used to perform statistical analysis and visualization results. Differential expression of CCXL12 between pancreatic cancer tissues and normal tissues was adopted by the Wilcoxon rank-sum test and visualized results using R-package "ggplot2". Immune cell algorithm: ssGSEA (built-in algorithm of GSVA package[6]). Correlation test using Spearman's correlation coefficient. Pathway analysis was performed by the online tool GSCALite (http://bioinfo.life.hust.edu.cn/web/GSCALite/)[7].
ACKNOWLEDGEMENTS
Yan-Dong Miao would like to give particularly grateful to Wu-Xia Quan for her care, patience, and support over the past years and for her contributions to this study.
Footnotes
Conflict-of-interest statement: No conflict of interest associated with any of the senior authors or other coauthors contributed their efforts in this manuscript.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: June 22, 2021
First decision: July 4, 2021
Article in press: December 2, 2021
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Nagaraju GP S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
Contributor Information
Yan-Dong Miao, The First Clinical Medical College, Lanzhou University, Lanzhou 730000, Gansu Province, China. miaoyd19@lzu.edu.cn.
Jiang-Tao Wang, The First Clinical Medical College, Lanzhou University, Lanzhou 730000, Gansu Province, China.
Xiao-Long Tang, The First Clinical Medical College, Lanzhou University, Lanzhou 730000, Gansu Province, China.
Deng-Hai Mi, The First Clinical Medical College, Lanzhou University, Lanzhou 730000, Gansu Province, China; Dean's office, Gansu Academy of Traditional Chinese Medicine, Lanzhou 730000, Gansu Province, China.
References
- 1.Cecati M, Giulietti M, Righetti A, Sabanovic B, Piva F. Effects of CXCL12 isoforms in a pancreatic pre-tumour cellular model: Microarray analysis. World J Gastroenterol. 2021;27:1616–1629. doi: 10.3748/wjg.v27.i15.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren B, Cui M, Yang G, Wang H, Feng M, You L, Zhao Y. Tumor microenvironment participates in metastasis of pancreatic cancer. Mol Cancer. 2018;17:108. doi: 10.1186/s12943-018-0858-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas D, Radhakrishnan P. Tumor-stromal crosstalk in pancreatic cancer and tissue fibrosis. Mol Cancer. 2019;18:14. doi: 10.1186/s12943-018-0927-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivian J, Rao AA, Nothaft FA, Ketchum C, Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD, Musselman-Brown A, Schmidt H, Amstutz P, Craft B, Goldman M, Rosenbloom K, Cline M, O'Connor B, Hanna M, Birger C, Kent WJ, Patterson DA, Joseph AD, Zhu J, Zaranek S, Getz G, Haussler D, Paten B. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35:314–316. doi: 10.1038/nbt.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34:3771–3772. doi: 10.1093/bioinformatics/bty411. [DOI] [PubMed] [Google Scholar]