Abstract
The natural product citral has previously been demonstrated to possess antifungal activity against Magnaporthe oryzae. The purpose of this study was to screen and annotate genes that were differentially expressed (DEGs) in M. oryzae after treatment with citral using RNA sequencing (RNA-seq). Thereafter, samples were reprepared for quantitative real-time PCR (RT-qPCR) analysis verification of RNA-seq data. The results showed that 649 DEGs in M. oryzae were significantly affected after treatment with citral (100 μg/mL) for 24 h. Kyoto Encyclopedia of Genes and Genomes (KEGG) and a gene ontology (GO) analysis showed that DEGs were mainly enriched in amino sugar and nucleotide sugar metabolic pathways, including the chitin synthesis pathway and UDP sugar synthesis pathway. The results of the RT-qPCR analysis also showed that the chitin present in M. oryzae might be degraded to chitosan, chitobiose, N-acetyl-D-glucosamine, and β-D-fructose-6-phosphate following treatment with citral. Chitin degradation was indicated by damaged cell-wall integrity. Moreover, the UDP glucose synthesis pathway was involved in glycolysis and gluconeogenesis, providing precursors for the synthesis of polysaccharides. Galactose-1-phosphate uridylyltransferase, which is involved in the regulation of UDP-α-D-galactose and α-D-galactose-1-phosphate, was downregulated. This would result in the inhibition of UDP glucose (UDP-Glc) synthesis, a reduction in cell-wall glucan content, and the destruction of cell-wall integrity.
Keywords: Magnaporthe oryzae, citral, RNA-seq, antifungal mechanism, gene expression
1. Introduction
The fungus M. oryzae causes rice blast, which is the most widespread and serious disease affecting rice [1,2] and one of the most destructive and economically harmful diseases affecting rice production worldwide [3,4]. At present, the methods of disease-resistant variety breeding and chemical control are mainly used for the prevention and control of M. oryzae in China [5]. As a result of the shortcomings of traditional breeding of disease-resistant varieties, such as long breeding cycles, easy-to-lose resistance, and the high cost of research and development in the field of chemical pesticides, pesticide residues, and drug resistance [6], a move away from the traditional excessive dependence on chemical pesticides during crop management became necessary. Under the premise of stabilizing production, intensive efforts were needed to develop biological pesticide technology with low toxicity and low residue characteristics.
The discovery of antifungal and germicidal compounds from plants has led to the efficient creation of new pollution-free pesticides [7,8]. Many plants have antifungal activities, which are related to their antifungal constituents, including alkaloids, terpenes, polysaccharides, esters, ketones, and quinones [9,10,11], including lemongrass, cilantro, cassia, and cinnamon [12]. In particular, citral is an open-chain monoterpene extracted from Litsea cubeba oil, which has good antifungal, insecticidal, and antioxidant activities [13].
Our previous studies demonstrated that citral has significant biological activity against M. oryzae in vitro and in vivo, suggesting its potential application in controlling rice blast [14,15]. Citral may act by disrupting the cell-wall integrity and membrane permeability of M. oryzae, thus resulting in physiological changes and cytotoxicity [16]. Importantly, the inhibitory effect of citral on M. oryzae appears to be associated with its effects on reducing sugar, soluble protein, pyruvate, and malondialdehyde contents, and chitinase activity, in mycelia [15,17]. However, the gene targeted by citral and thus responsible for its activity in M. oryzae has not been identified. Recent advances in high-throughput genomic technology, including transcriptomics, have provided a new method for studying fungal growth, pathogenesis, and the genes involved in resistance [18,19,20]. Thus, we decided to study the gene expression profile of citral-treated M. oryzae to gain some insight into its mode of action. In this research, the objectives were (i) to screen and annotate the differentially expressed genes (DEGs) of M. oryzae after treatment with citral using RNA-seq, (ii) to ascertain the expression profiles of these M. oryzae DEGs, and (iii) to use the information about DEGs and their expression profiles to construct a model diagram illustrating the mechanism of citral acting on M. oryzae. Furthermore, the samples were reprepared for real-time fluorescence quantitative verification of the RNA-seq data. The inhibitory targets of citral in M. oryzae were obtained through the changes in the DEGs levels and RT-qPCR analysis verification results. As far as we know, this is the first report that uses the RNA-seq method to study the mechanism of citral in M. oryzae. Studies of the closely related response genes and signaling pathways can help elucidate the molecular mechanism of citral acting on M. oryzae, and provide a scientific basis for its mode of action and a valuable reference for the control of M. oryzae.
2. Materials and Methods
2.1. Strain and Growth Conditions
M. oryzae strain ZB15 was isolated from diseased rice panicles in Guizhou Province, China, and stored on potato dextrose agar (PDA) at 4 °C for 7 days. A 5 mm diameter agar disc was taken from the colony margin using a punch, inoculated on a cellophane sheet-covered PDA plate, and cultured at 28 °C for 15 days. The hyphae were then made into a suspension containing citral at concentrations of 0, 50, and 100 µg/mL. The inoculum was cultured at 28 °C with stirring at 180 rpm. The mycelia were collected by filtering through two layers of cotton gauze and washed with distilled deionized water at 0, 9, 12, and 24 h post incubation [21].
2.2. RNA Extraction and Preparation of cDNA Library
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The yield and purity of each RNA sample were quantified using a NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). The RNA integrity was assessed as a function of the RNA integrity number (RIN) >7.0, using the Agilent 2100 bioanalyzer.
Poly (A) RNA was purified from total RNA (5 µg) using poly-T oligo-attached magnetic beads using two rounds of purification. Then, the poly (A) RNA was fragmented into small pieces using divalent cations under high temperature. Thereafter, the cleaved RNA fragments were reverse-transcribed to create the cDNA, which were then used to synthesize U-labeled second-stranded DNAs with E. coli DNA polymerase I. RNase H and dUTP A-bases were added to the blunt ends of each strand, preparing them for T-base overhang adapter ligation. Single or dual index adapters were ligated to the fragments, and size selection was performed with AMPureXP beads. After heat-labile UDG enzyme treatment of the U-labeled second-stranded DNAs, the ligated products were amplified using PCR under the following conditions: initial denaturation at 95 °C for 3 min, followed by 8 cycles of denaturation at 98 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 30 s; with a final extension at 72 °C for 5 min. The average insert size for the final cDNA library was 300 bp (±50 bp). Finally, the 150 bp paired-end sequencing was performed on an Illumina Hiseq 4000 (LC Bio, Hangzhou, China). The bioinformatics analysis pipeline is shown in Supplementary Figure S1.
2.3. Sequence Assembly and Annotation
The raw data generated by sequencing needed to be preprocessed. Cutadapt was used to filter out unqualified sequences and obtain valid data before further analysis [22]. Reads with an adaptor containing more than 5% of N (N indicates that base information cannot be determined) were removed. Low quality reads (more than 20% of the total read exhibiting a mass value of Q ≤ 10) were removed. The original sequencing amount, the effective sequencing amount, Q20, Q30, and GC contents were counted and comprehensively evaluated. Thereafter, sequence quality was verified using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed on 21 November 2021). The known annotation information in the species database (https://www.ncbi.nlm.nih.gov/genome/?term=Magnaporthe%20oryzae, accessed on 21 November 2021) was analyzed, including chromosome number, gene number, transcript number, GO annotation (http://geneontology.org, accessed on 21 November 2021), and KEGG annotation (http://www.kegg.jp/kegg, accessed on 21 November 2021).
2.4. Gene Expression Analysis
The algorithm feature Counts version 1.5.0-p3 was used for the FPKM calculation of each gene, according to the length of the gene and the read count mapped to this gene [23]. Expression levels of different samples of M. oryzae treated with citral were obtained by calculating a whole-genome model [24,25]. We used edgeR to finish the StringTie assembly and quantitative genetic variance analysis (significant difference threshold for |log2 foldchange| ≥ 1, p < 0.05) [26]. Finally, the volcano figure, GO, and KEGG enrichment analyses were performed on the DEGs.
2.5. Quantitative Real-Time PCR Analysis
The total RNA for RT-qPCR analysis was extracted using a 2× SG Fast qPCR Master Mix (Sangon, Shanghai, China), and 1.0 μg of RNA was used for reverse transcription with an M-MuLV First-Strand cDNA Synthesis Kit (Sangon, Shanghai, China) in a 20 μL reaction volume according to the manufacturer’s instructions. Gene-specific primer pairs were designed using the Sangon primer design and synthesis online platform (https://www.sangon.com/newPrimerDesign, accessed on 21 November 2021) on the basis of the transcript sequence and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) (Table 1). The actin gene was used as an internal control [27]. The PCR was performed using SYBR Green’s method and a Bio-Rad CFX96 real-time PCR system (Bio-Rad, CA, USA) with a total of 96 well plates, each sample was set up with three biological replicates. The final volume for each reaction was 20 μL with the following components: 2 μL cDNA template (1 ng/μL), 10 μL 2× SG Fast qPCR Master Mix (Sangon, Shanghai, China), 0.4 μL forward primer (200 nM), 0.4 μL reverse primer (200 nM), and 7.2 μL PCR-grade water. The reaction was conducted under the following conditions: 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 10 s and annealing/extension at 56 °C for 30 s. The melting curve was obtained by heating the amplicon from 65 °C to 95 °C at increments of 0.5 °C per 5 s. The relative quantification of gene expression was computed using the 2−ΔΔCt method.
Table 1.
Primer design for RT-qPCR.
| Gene | Primer Name | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|---|
| actin | MGG_03982 | CTGGCACCGTCGTCGATGAAG | AAGGTCCGCTCTCGTCGTACTC |
| cell-wall glucanosyltransferase | MGG_00592 | TCACCGCCATCTACGAGTCGATC | TGTCACCATCCTTGCTGCTGTTG |
| thioredoxin | MGG_04236 | AATCCGGTCGCTTGCATCATCG | TCCTCTGGTGGTTGTTGCTGATTG |
| aldehyde reductase | MGG_16375 | GCGGCGAGGTTGACGTGATC | CTTGGTGGCAGGCAGGTTCATG |
| uncharacterized protein | MGG_00167 | CCTGACCTGACGCACTTCTACAAG | TCCTCCGAGTTCCACCAGTAAGAC |
| cAMP-dependent protein kinase regulatory subunit | MGG_07335 | GCGGCTTCACCAGTCCATTCG | GATTCCGTCGGCCTCCTCCTC |
| CAMK/CAMK1 protein kinase | MGG_08547 | CCAGCCATCCTCAACAACCTCAAC | TACTCGCCACTCTCATCGCTATCG |
| GPI-anchored cell wall β-1,3-endoglucanase EglC | MGG_10400 | CGCTTCTACGACGGTCTGAACAAG | GCCACAGCCTGGTTGATCTGC |
| Bacteriodes thetaiotaomicron symbiotic chitinase | MGG_01336 | GGCTGGCTATTGCGGTACATCTG | GCGGCAACGGCTTGGTAGTAG |
2.6. Gene Expression of the Genes Related to Amino Sugar and Nucleotide Sugar Metabolism
In order to verify the effect of amino sugar- and monosaccharide metabolism-associated gene expression on the cell wall, eight DEGs from the amino sugar and monosaccharide metabolism pathways were selected (Supplementary Table S1), and their effects on the cell wall were verified according to the method described in Section 2.5. The differential expression of M. oryzae genes was determined by RT-qPCR after treatment with citral (100 µg/mL) for 24 h.
3. Results
3.1. Read Generation and De Novo Assembly
Thirty-six cDNA libraries prepared from M. oryzae were sequenced using the Illumina Novaseq™ 6000 platform. A total of 16.08 million raw reads were generated. After removing adapters and filtering the low-quality sequences, a total of 1,330,379,916 clean reads were generated from the libraries of M. oryzae treated with citral of 0 µg/mL (a0, A2, A3, A4), 50 µg/mL (b0, B2, B3, B4), and 100 µg/mL (c0, C2, C3, C4). The sequencing depth of each sample was 4.68–6.69 Gb. The proportion of clean reads ranged from 75.23% to 88.76% (Supplementary Table S2). More than 91.54% clean reads could be mapped to the reference genome. Among the mapped reads, more than 85.33% were uniquely mapped, and more than 97.81% were mapped to gene exon regions (Supplementary Table S3). The results showed that the sequencing quality of mapped reads was higher, the assembly integrity was better, and the credibility was higher than for clean reads. Therefore, they could be used for subsequent quantitative expression and annotation analysis.
3.2. Differentially Expressed Gene Analysis
The DEGs from the 36 transcriptome libraries were analyzed. We used log10 (FPKM + 1) for gene expression display. The maximum gene expression FPKM value was 36,563.15 (Supplementary Table S4 and Figure S2). The global analysis of M. oryzae genes that were differentially expressed in response to treatment with citral was conducted using criteria of |log2 ratio| ≥ 1 and p < 0.05, and the results were visualized using a volcano plot and Venn diagram. There were 25 and 73 DEGs that overlapped across the four time points in M. oryzae after treatment with 50 and 100 μg/mL citral, respectively (Figure 1A,B). As shown in Figure 1C, the number of DEGs increased after treatment with 100 μg/mL citral compared with the 50 μg/mL citral treatment for 24 h. We identified 1224 upregulated genes and 3431 downregulated genes after treatment with 50 μg/mL citral for 24 h and 1009 upregulated genes and 2084 downregulated genes after treatment with 100 μg/mL citral for 24 h (Figure 2 and Figure 3). The results show that the numbers of common DEGs (2301) for all concentrations and time points of citral treatment and specific DEGs (649) in M. oryzae after treatment with 100 μg/mL citral for 24 h were the largest among all the treatment groups.
Figure 1.
Venn diagram of DEGs in the evaluated libraries. (A) Venn map of all DEGs in M. oryzae after treatment with citral (50 μg/mL) for the indicated times (h). (B) Venn map of all DEGs in M. oryzae after treatment with citral (100 μg/mL) for the indicated times (h). (C) Venn map of specific DEGs in M. oryzae after treatment with citral for 24 h.
Figure 2.
Volcanic map of DEGs in M. oryzae after treatment with citral at different concentrations. (A) Volcanic map analysis of DEGs in M. oryzae after treatment with citral (50 μg/mL) for 9 h. (B) Volcanic map analysis of DEGs in M. oryzae after treatment with citral (50 μg/mL) for 12 h. (C) Volcanic map analysis of DEGs in M. oryzae after treatment with citral (50 μg/mL) for 24 h. (D) Volcanic map analysis of DEGs in M. oryzae after treatment with citral (100 μg/mL) for 9 h. (E) Volcanic map analysis of DEGs in M. oryzae after treatment with citral (100 μg/mL) for 12 h. (F) Volcanic map analysis of DEGs in M. oryzae after treatment with citral (100 μg/mL) for 24 h. Red represents upregulated significantly differentially expressed genes, blue represents downregulated significantly differentially expressed genes, and gray dots represent non significantly differentially expressed genes.
Figure 3.
Volcanic map analysis of DEGs in M. oryzae after treatment with citral for 24 h. (A) Volcanic map analysis of DEGs in M. oryzae after treatment with citral (0 μg/mL and 50 μg/mL) for 24 h. (B) Volcanic map analysis of DEGs in M. oryzae after treatment with citral (0 μg/mL and 100 μg/mL) for 24 h.Red represents upregulated significantly differentially expressed genes, blue represents downregulated significantly differentially expressed genes, and gray dots represent non significantly differentially expressed genes.
3.3. Functional Annotation and Classification
The possible functions of the identified DEGs were predicted using the gene function classification system GO, which has three ontologies, namely, molecular functions, cellular components, and biological processes. We used GO analysis to classify the functions of genes in M. oryzae. A total of 10,537 annotated genes were categorized into 50 GO functional groups (Figure 4C). Most of the genes were enriched in the molecular function category, followed by the biological process and cellular component categories, as shown in Figure 4C. Within the biological process category, “mycelium development”, and “conidium formation” featured the highest number of genes. On the other hand, “cellular component”, “nucleus”, and “integral component of membrane” were the most enriched terms in the cellular component category. The “zinc ion binding” term was the most prevalent in the molecular function category (Figure 4A). For KEGG analysis, a total of 4073 annotated genes were used for mapping in M. oryzae; the main active pathways involved were: ribosome biogenesis in eukaryotes, glyoxylate and dicarboxylate metabolism, lysine degradation, and glycolysis. The largest category was purine metabolism (181, 4.4%), followed by biosynthesis of amino acids (175, 4.2%), meiosis in yeast (169, 4.1%), cell cycle in yeast (163, 4.0%), and carbon metabolism (159, 3.9%). These annotations provide a further understanding of the transcriptome data, as well as their functions and pathways, in M. oryzae.
Figure 4.
Summary of the annotations of DEGs in M. oryzae. (A) GO classifications. (B) Scatter plot of KEGG enrichment of DEGs. (C) Enrichment of GO genes in M. oryzae after treatment with citral (100 μg/mL) for 24 h.
3.4. RNA-Seq Validation by RT-qPCR
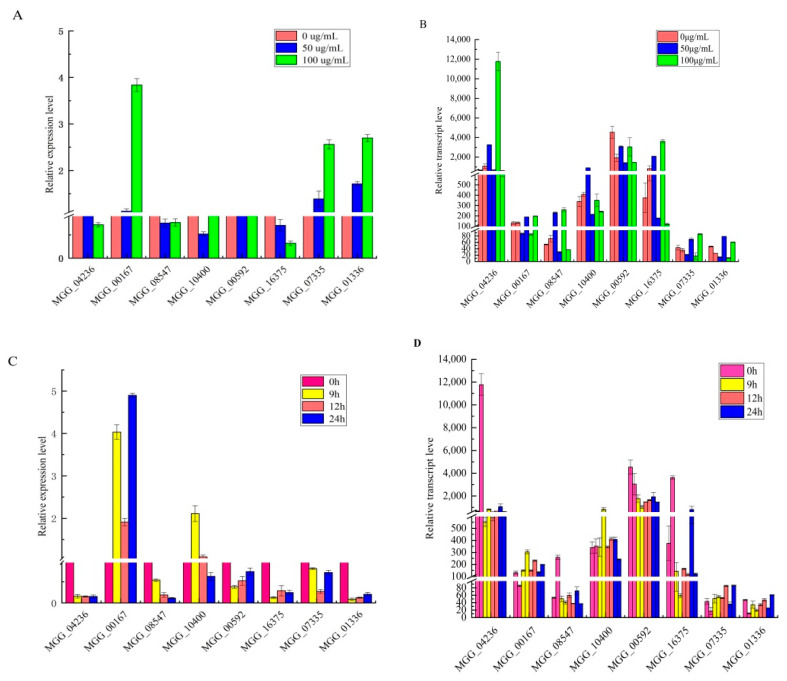
To confirm the accuracy of the sequencing results, we selected eight genes and used RT-qPCR to determine their relative expression levels in M. oryzae under the corresponding treatments. Eight pairs of primers were designed, and actin was used as an internal control for normalization (Table 1). The RT-qPCR results indicated that all eight genes were regulated in a manner similar to that revealed from the RNA-Seq data (Figure 5). The RT-qPCR results indicated that the RNA-Seq data were reliable and could be used for subsequent analysis.
Figure 5.
RT-qPCR verification of key genes of resistance to citral in M. oryzae. (A,B) The expression trend of genes as characterized by qPCR was consistent with that of RNA-seq analysis after 9, 12, and 24 h of treatment with citral (100 μg/mL). (C,D) The expression trend of genes as characterized by qPCR was consistent with that of the RNA-seq analysis after treatment with 0, 50, and 100 μg/mL of citral for 24 h. All values are based on three technical repeats and presented as means ± SD. Different characters indicate a statistically significant difference at p < 0.05 by t-test.
3.5. Identification of Genes in Amino Sugar and Nucleotide Sugar Metabolism
Functional annotation results for DEGs related to amino sugar and nucleotide sugar metabolism were identified. The pathways of amino sugar and nucleotide sugar metabolism were assigned using KEGG analysis, and they were found to influence the differential expression of enzyme nodes in the chitin formation and UDP glucose synthesis pathways. Nine related enzyme nodes were significantly differentially expressed in aminosaccharide and nucleotide sugar metabolism pathways according to the annotation results. In particular, five enzyme nodes were discovered for the chitin formation pathway: EC:3.2.1.14 (enzyme or protein chitosan (chitobiose)), EC:3.2.1.52 (N-acetyl glucosamine (GlcNAc)), EC:3.5.99.6 (glucosamine 6-phosphate isomerase (GlcN-6P)), EC:2.4.1.16 (chitin synthase), and EC:1.6.2.2 (ferrous cytochrome B5). On the other hand, EC:5.4.2.2 (phosphoglucomutase) and EC:2.7.7.12 (galactose-1-phosphate uridylyltransferase) were discovered for the UDP glucose synthesis pathway (Supplementary Figure S3). The other two related enzyme nodes were EC:3.2.1.55 (α-N-arabinofuranosidase) and EC:3.2.1.37 (β-xylosidase). However, the eight DEGs were related to the chitin formation and UDP glucose synthesis pathways according to the enzyme node enrichment analysis. The results show that enzyme nodes in the pathways of amino sugar and nucleotide sugar metabolism were differentially expressed in M. oryzae after treatment with citral, which was related to chitin formation, along with a certain effect on the cell wall. Thus, the results suggest that the mechanism of citral action against M. oryzae may involve damage to the cell wall.
3.6. Effect of Citral on Amino Sugar and Nucleotide Sugar Biosynthesis in M. oryzae
In order to further verify the regulatory effect of citral on the chitin synthesis pathway in M. oryzae, we selected eight DEGs (six upregulated genes and two downregulated genes) identified in the amino sugar and nucleotide sugar metabolism pathways of M. oryzae after treatment with citral. The changes in trends were observed to be similar between RNA-seq and qRT-PCR assays, although several genes presented a difference in expression at the same time point (Figure 6). There were three DEGs upregulated upstream of the chitin synthesis pathway compared with the control group, namely, MGG_10333 (class III chitinase), MGG_01336 (Bacteriodes thetaiotaomicron symbiotic chitinase), and MGG_00625 (glucosamine-6-phosphate isomerase), with gene expression upregulated by 17.52-, 3.75-, and 0.07-fold, respectively. The expression levels of MGG_01605 and MGG_03920, downstream of the chitin synthesis pathway, were also upregulated 0.41- and 2.69-fold, respectively, whereas the expression of MGG_09551 (ferrocytochrome B5) downstream of the chitin synthesis pathway was downregulated by 0.65-fold. The expression of MGG_04495 was upregulated in the UDP glucose synthesis pathway, involved in the regulation of α-d-glucose 6-phosphate synthesis. Moreover, the expression of MGG_05098 was downregulated, involved in the regulation of UDP-α-d-galactose and α-d-galactose-1-phosphate synthesis. The results show that the antifungal mechanism of citral against M. oryzae occurs via regulating the expression of key genes involved in the chitin and UDP glucose synthesis pathways.
Figure 6.
RT-qPCR analysis of key genes in the amino and nucleotide sugar pathways in response to citral treatment of M. oryzae after 24 h of treatment with citral (100 μg/mL). Significance is denoted by *.
4. Discussion
M. oryzae causes huge economic losses around the world. Although the wide-spread use of traditional fungicides can reduce crop loss, it may also potentially harm the environment and human health [28,29,30,31]. Therefore, it is very important to develop safe and environmentally friendly natural pesticide products. At present, microorganisms, microbial products, and biological fertilizers are being developed as biological control agents [32].
Transcriptomics technology allows the changes in RNA levels in micro-organisms to be analyzed under different environments at a molecular level from a macro perspective [33,34,35]. For example, on the basis of the results of RNA-seq, the effects of sodium pheophorbide, a (SPA) treatment, on the expression of three cell-wall-degrading enzyme-related genes in Pestalotiopsis neglecta hypha were detected using qRT-PCR, thereby revealing the molecular mechanism underlying the action of SPA on P. neglecta. SPA inhibited TCA-related enzyme activity and the expression of three cell-wall-degrading enzyme-related genes [36]. The genome and transcriptome of the pine saprophyte (Ophiostoma piceae) were compared with the genome and transcriptomes of the mountain pine beetle-associated pathogen Grosmannia clavigera. The differences revealed a major class of pine tree defense compound [37]. Camptothecin (CPT) is a botanical compound primarily isolated from Camptotheca acuminata Decne, which is an important medicinal plant in China. CPT is a promising lead compound for developing fungicides against rice blast, as it may bind to the DNA topoisomerase I complex of M. oryzae, thus affecting translation and carbohydrate metabolism/energy metabolism, leading to cell death [38]. Our group previously studied citral extracted from Litsea cubeba fruit, which had properties consistent with those of leaf oil extracted from Eucalyptus and showed a good inhibitory effect on M. oryzae growth [39]. In addition, further research reported the significant antibacterial activity of citral in Aspergillus ochraceus, Penicillium, Penicillium roqueforti, and so on [40,41,42]. In this study, the transcriptome analysis method based on RNA-seq was used to analyze the RNA levels of genes in M. oryzae treated with citral. The results show that citral might act to disrupt cell-wall integrity and the cytoplasmic membrane by causing changes in membrane permeability, leading to a loss of intracellular substances and the introduction of wrinkles and depressions on the cell surface.
Chitin is a β-1,4-linked polymer of N-acetylglucosamine and a major component of the fungal cell wall [43,44]. Fungi can successfully invade the host by destroying the conserved chitin in the fungal cell wall. Chitin hydrolase CcCti1 inhibited conidial germination and appressoria formation in M. oryzae, thereby affecting its normal growth [45]. In this study, most DEGs were enriched in biological processes related to mycelium development and conidia formation. The polar germination of conidia and the growth of infected hyphae of M. oryzae were related to dynamic changes in the M. oryzae cell wall. It was found that the expressions of MGG_10333 (class III chitinase), MGG_01336 (bacteriodes thetaiotaomicron symbiotic chitinase), and MGG_00625 (glucosamine-6-phosphate isomerase) were upregulated in M. oryzae after treatment with citral. Thus, the chitin present in M. oryzae might be degraded to chitosan, chitobiose, N-acetyl-D-glucosamine, and β-D-fructose-6-phosphate following treatment with citral. The expression of FAD-binding domain-containing protein MGG_01605 was upregulated in M. oryzae, and this gene was found downstream of the chitin synthesis pathway and was involved in the regulation of chitin. The expression of inositolphosphorylceramide-B C-26 hydroxylase MGG_03920 was upregulated, involved in ferrocytochrome B5 synthesis. Furthermore, the expression of chitin synthase MGG_09551 was downregulated, which inhibited chitin synthesis. Chitin degradation is indicated by damaged cell-wall integrity, as it is an important component of the cell wall in M. oryzae [46]. The KEGG pathway analysis showed that citral could affect chitin synthesis and UDP glucose synthesis in the amino sugar and nucleotide metabolic pathways of M. oryzae (Figure 7). UDP glucose is a donor in most polysaccharide synthesis pathways and the precursor of glucan synthesis [47]. The UDP glucose synthesis pathway is involved in glycolysis and gluconeogenesis, providing precursors for the synthesis of polysaccharides. The expression of phosphoglucomutase MGG_04495 was upregulated in M. oryzae after treatment with citral, which was involved in catalyzing the mutual transformation of glucose-6-phosphate and glucose-1- phosphate in the UDP glucose synthesis pathway. Galactose-1-phosphate uridylyl transferase MGG_05098, involved in the regulation of UDP-α-D-galactose and α-D-galactose-1-phosphate, was downregulated, which would result in the inhibition of UDP glucose (UDP-Glc) synthesis, a reduction in cell-wall glucan content, and the destruction of cell-wall integrity.
Figure 7.
Model diagram illustrating the citral’s mechanism against the cell wall of M. oryzae.
In conclusion, the lipophilic characteristic of citral facilitates its passage and access such that it can disrupt cell-wall integrity and the cytoplasmic membrane by causing changes in membrane permeability, leading to the loss of intracellular substances and the introduction of wrinkles and depressions on the cell surface [48]. The model diagram shows that the gene expression of chitin synthase was downregulated in the chitin formation pathway of M. oryzae after treatment with citral, leading to an inhibition of chitin synthesis. On the other hand, the gene expression of chitinase was upregulated, resulting in a reduction in chitin content in the cell wall of M. oryzae, leading to damage. However, the expressions of the glucosamine-6-phosphate deaminase and UDP glucose pyrophosphorylase genes were downregulated, resulting in an inhibition of UDP glucose synthesis and a reduction in the cell-wall glucan content, thereby disrupting the M. oryzae cell-wall integrity and inhibiting mycelial growth. This study identified the key genes related to cell-wall integrity in M. oryzae, which exhibited a response to citral. This further revealed that citral’s excellent antifungal activity against M. oryzae occurs by affecting the chitin content, thus providing a scientific basis for its mode of action. The regulatory molecular mechanisms of the transcription factors that mediate chitinase in M. oryzae following treatment with citral still require further study.
5. Conclusions
An RNA-seq analysis identified the effects of citral concentration on M. oryzae. The number of DEGs in M. oryzae reached the maximum value after treatment with citral (100 μg/mL) for 24 h. Most DEGs were enriched in biological processes related to mycelium development and conidia formation. KEGG pathway analysis showed that genes in the amino sugar and nucleotide sugar metabolism pathways of M. oryzae were differentially expressed following treatment with citral, and these pathways are related to chitin formation. In the chitin synthesis pathway of M. oryzae, chitin synthase gene expression was downregulated, resulting in an inhibition of chitin synthesis. Moreover, upregulation of chitinase gene expression can lead to an increase in chitin degradation, thus resulting in a decrease in chitin content and destruction of the cell-wall structure in M. oryzae. In the UDP glucose synthesis pathway of M. oryzae, UDP glucose synthesis was inhibited, which could reduce the cell-wall glucan content and disrupt the cell-wall integrity. This study further revealed that citral had excellent antifungal activity against M. oryzae by affecting the content of chitin, thus providing a scientific basis for elucidating its mode of action. At present, our research group is constructing silencing vectors for the target genes that were screened in this study. Moreover, we are observing the genotype expression and other related mechanisms in the later stages of this process.
Acknowledgments
The authors thank R.L. for her help with the statistical analyses and M.L. for her help with the methodology.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof7121023/s1, Figure S1: Experimental flow, Figure S2: Systematic clustering of differentially expressed genes (DEGs) in 36 transcriptome libraries, Figure S3: Pathways of amino and nucleotide sugar metabolism in M. oryzae, Table S1: Primer design of eight genes for RT-qPCR, Table S2: Summary of reference genetic data from processed samples, Table S3: Comparison of transcriptome sequencing and reference genome of M. oryzae under citral stress, Table S4: Distribution statistics of reference gene table for transcriptome sequencing of M. oryzae under citral stress.
Author Contributions
Conceptualization, R.L.; methodology and formal analysis, X.S., Q.Z., A.Z. and X.W.; funding acquisition, R.L. and M.L.; writing—review and editing, X.S. and R.L.; visualization, X.L. and T.X. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32160657, 32160658), Agricultural Research Projects of the Science and Technology Department of Guizhou Province (No. [2019]3001), and the Cultivation Project of Guizhou University (No. [2019]43).
Data Availability Statement
Sequencing data are available from https://www.ncbi.nlm.nih.gov/genome/?term=Magnaporthe%20oryzae, accessed on 21 November 2021.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yang Y.H., Yang D.S., Lei H.M., Li C.Y., Li G.H., Zhao P.J. Griseaketides A-D, New Aromatic Polyketides from the Pathogenic Fungus Magnaporthe grisea. Molecules. 2019;25:72. doi: 10.3390/molecules25010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryder L.S., Dagdas Y.F., Kershaw M.J., Chandrasekhar V., Madzvamuse A., Yan X., Cruz-Mireles N., Soanes D.M., Oses-Ruiz M., Styles V., et al. A sensor kinase controls turgor-driven plant infection by the rice blast fungus. Nature. 2019;574:423–427. doi: 10.1038/s41586-019-1637-x. [DOI] [PubMed] [Google Scholar]
- 3.Kongcharoen N., Kaewsalong N., Dethoup T. Efficacy of fungicides in controlling rice blast and dirty panicle diseases in Thailand. Sci. Rep. 2020;10:16233. doi: 10.1038/s41598-020-73222-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X.Y., Chen Y., Li C.G., Zhang X., Tan X.Q., Lv L., Liu Y., Zhang D. GroEL protein from the potential biocontrol agent Rhodopseudomonas palustris enhances resistance to rice blast disease. Pest Manag. Sci. 2021;77:5445–5453. doi: 10.1002/ps.6584. [DOI] [PubMed] [Google Scholar]
- 5.Xin W.J., Mao Y.S., Li T., Wang J.X., Duan Y.B., Zhou M.G. In vitro fungicidal activity and in planta control efficacy of coumoxystrobin against Magnaporthe oryzae. Pestic. Biochem. Physiol. 2020;162:78–85. doi: 10.1016/j.pestbp.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Breseghello F., Coelho A.S. Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.) J. Agric. Food Chem. 2013;61:8277–8286. doi: 10.1021/jf305531j. [DOI] [PubMed] [Google Scholar]
- 7.Vicentini C.B., Forlani G., Manfrini M., Carlo R., Mares D. Development of New Fungicides against Magnaporthe grisea: Synthesis and Biological Activity of Pyrazolo[3,4-d][1,3]thiazine, Pyrazolo[1,5-c][1,3,5]thiadiazine, and Pyrazolo[3,4-d]pyrimidine Derivatives. J. Agric. Food Chem. 2002;50:4839–4945. doi: 10.1021/jf0202436. [DOI] [PubMed] [Google Scholar]
- 8.Ouyang Q., Liu Y., Oketch O.R., Zhang M., Shao X., Tao N. Citronellal Exerts Its Antifungal Activity by Targeting Ergosterol Biosynthesis in Penicillium digitatum. J. Fungi. 2021;7:432. doi: 10.3390/jof7060432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabit F.T., Komolafe N.T., Tshikalange T.E., Nyila M.A. Phytochemical Constituents and Antioxidant and Antimicrobial Activity of Selected Plants Used Traditionally as a Source of Food. J. Med. Food. 2016;19:324–329. doi: 10.1089/jmf.2015.0099. [DOI] [PubMed] [Google Scholar]
- 10.Elshafie H.S., Camele I. An Overview of the Biological Effects of Some Mediterranean Essential Oils on Human Health. Biomed. Res. Int. 2017;2017:9268468. doi: 10.1155/2017/9268468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elshafie H.S., Aliberti L., Amato M., Feo V.D., Camele I. Chemical composition and antimicrobial activity of chia (Salvia hispanica L.) essential oil. Eur. Food Res. Technol. 2018;244:1675–1682. doi: 10.1007/s00217-018-3080-x. [DOI] [Google Scholar]
- 12.Powers C.N., Osier J.L., McFeeters R.L., Brazell C.B., Olsen E.L., Moriarity D.M., Satyal P., Setzer W.N. Antifungal and Cytotoxic Activities of Sixty Commercially-Available Essential Oils. Molecules. 2018;23:1549. doi: 10.3390/molecules23071549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thielmann J., Theobald M., Wutz A., Krolo T., Buergy A., Niederhofer J., Welle F., Muranyi P. Litsea cubeba fruit essential oil and its major constituent citral as volatile agents in an antimicrobial packaging material. Food Microbiol. 2021;96:103725. doi: 10.1016/j.fm.2020.103725. [DOI] [PubMed] [Google Scholar]
- 14.Li R.Y., Wu X.M., Yin X.H., Liang J.N., Li M. The Natural Product Citral Can Cause Significant Damage to the Hyphal Cell Walls of Magnaporthe grisea. Molecules. 2014;19:10279–10290. doi: 10.3390/molecules190710279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R.Y., Wu X.M., Yin X.H., Long Y.H., Li M. Naturally produced citral can significantly inhibit normal physiology and induce cytotoxicity on Magnaporthe grisea. Pestic. Biochem. Physiol. 2015;118:19–25. doi: 10.1016/j.pestbp.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Elshafie H.S., Armentano M.F., Carmosino M., Bufo S.A., Feo V.D., Camele I. Cytotoxic Activity of Origanum vulgare, L. on Hepatocellular Carcinoma Cell Line HepG2 and Evaluation of Its Biological Activity. Molecules. 2017;22:1435. doi: 10.3390/molecules22091435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Q.J., Ding Y., Song X.C., Liu S.J., Li M., Li R.Y., Ruan H. Proteomic analysis reveals that naturally produced citral can significantly disturb physiological and metabolic processes in the rice blast fungus Magnaporthe oryzae. Pestic. Biochem. Physiol. 2021;175:104835. doi: 10.1016/j.pestbp.2021.104835. [DOI] [PubMed] [Google Scholar]
- 18.Gowda M., Venu R., Raghupathy M.B., Nobuta K., Li H., Wing R., Stahlberg E., Couglan S., Haudenschild C.D., Dean R., et al. Deep and comparative analysis of the mycelium and appressorium transcriptomes of Magnaporthe grisea using MPSS, RL-SAGE, and oligoarray methods. BMC Genom. 2006;7:310. doi: 10.1186/1471-2164-7-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu S., Wang B., Li L., Zhou Q., Tian M., Zhao X.Z., Peng J., Liu F., Chen Y., Xu Y., et al. Effects of camptothecin on the rice blast fungus Magnaporthe oryzae. Pestic. Biochem. Physiol. 2019;163:108–116. doi: 10.1016/j.pestbp.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Wang P.Y., Wu H.B., Zhao G.W., He Y.H., Kong W.H., Zhang J., Liu S., Liu M., Hu K., Liu L., et al. Transcriptome analysis clarified genes involved in resistance to Phytophthora capsici in melon. PLoS ONE. 2020;15:e0227284. doi: 10.1371/journal.pone.0227284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martins M.P., Silva L.G., Rossi A., Sanches P.R., Souza L.D.R., Martinez-Rossi N.M. Global Analysis of Cell Wall Genes Revealed Putative Virulence Factors in the Dermatophyte Trichophyton rubrum. Front. Microbiol. 2019;10:2168. doi: 10.3389/fmicb.2019.02168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:2226–6089. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 23.Liao Y., Smyth G.K., Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 24.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 26.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S., An Y.Q., McDowell J.M., McKinney E.C., Meagher R.B. The Arabidopsis ACT11 actin gene is strongly expressed in tissues of the emerging inflorescence, pollen, and developing ovules. Plant Mol. Biol. 1997;33:125–139. doi: 10.1023/A:1005741514764. [DOI] [PubMed] [Google Scholar]
- 28.Camele I., Elshafie H.S., Caputo L., Sakr S.H., Feo V.D. Bacillus mojavensis: Biofilm formation and biochemical investigationo of its bioactive metabolites. J. Biol. Res. 2019;92:39–45. doi: 10.4081/jbr.2019.8296. [DOI] [Google Scholar]
- 29.Pepa T.D., Elshafie H.S., Capasso R., Feo V.D., Camele I., Nazzaro F., Scognamiglio M.R., Caputo L. Antimicrobial and phytotoxic activity of Origanum heracleoticum and O. majorana essential oils growing in Cilento (Southern Italy) Molecules. 2019;24:2576. doi: 10.3390/molecules24142576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gruľová D., Caputo L., Elshafie H.S., Baranová B., Martino L.D., Sedlák V., Gogaľová Z., Poráčová J., Camele I., de Feo V. Thymol Chemotype Origanum vulgare L. Essential Oil as a Potential Selective Bio-Based Herbicide on Monocot Plant Species. Molecules. 2020;25:595. doi: 10.3390/molecules25030595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elshafie H.S., Camele I., Sofo A., Mazzone G., Caivano M., Masi S., Canianic D. Mycoremediation effect of Trichoderma harzianum strain T22 combined with ozonation in diesel-contaminated sand. Chemosphere. 2020;252:126597. doi: 10.1016/j.chemosphere.2020.126597. [DOI] [PubMed] [Google Scholar]
- 32.An J., Shen X.F., Ma Q.B., Yang C.Y., Liu S.M., Chen Y. Transcriptome profiling to discover putative genes associated with paraquat resistance in goosegrass (Eleusine indica L.) PLoS ONE. 2014;9:e99940. doi: 10.1371/journal.pone.0099940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao Z.M., Li Y.J., Zhang X.R., Chu J., Ma J.H., Liu Z.X., Wang J., Sheng S., Wu F.-A. Identification and Functional Study of Chitin Metabolism and Detoxification-Related Genes in Glyphodes pyloalis Walker (Lepidoptera: Pyralidae) Based on Transcriptome Analysis. Int. J. Mol. Sci. 2020;21:1904. doi: 10.3390/ijms21051904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng J.W., Liu H., Shen M.C., Chen R.H., Li J.G., Dong Y.H. The inhibitory effects of different types of Brassica seed meals on the virulence of Ralstonia solanacearum. Pest. Manag. Sci. 2021;77:5129–5138. doi: 10.1002/ps.6552. [DOI] [PubMed] [Google Scholar]
- 35.Yang J., Ji J.Y., Zhang B.W., Chen Y.Z., Wang S.R., Zhang G.C., Zhang J. Transcriptome and cell wall degrading enzyme-related gene analysis of Pestalotiopsis neglecta in response to sodium pheophorbide a. Pestic. Biochem. Physiol. 2020;169:104639. doi: 10.1016/j.pestbp.2020.104639. [DOI] [PubMed] [Google Scholar]
- 36.Zhou L.J., Li F.R., Huang L.J., Yang Z.R., Yuan S., Bai L.H. Antifungal Activity of Eucalyptus Oil against Rice Blast Fungi and the Possible Mechanism of Gene Expression Pattern. Molecules. 2016;21:621. doi: 10.3390/molecules21050621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haridas S., Wang Y., Lim L., Massoumi A.S., Jackman S., Docking R., Robertson G., Birol I., Bohlmann J., Breuil C. The genome and transcriptome of the pine saprophyte Ophiostoma piceae, and a comparison with the bark beetle-associated pine pathogen Grosmannia clavigera. BMC Genom. 2013;14:373. doi: 10.1186/1471-2164-14-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong L.A., Yang J., Li G.T., Qi L.L., Zhang Y.J., Wang C.F., Zhao W.-S., Xu J.-R., Peng Y.-L. Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2012;8:e1002526. doi: 10.1371/journal.ppat.1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouzai Y., Mochizuki S., Nakajima K., Desaki Y., Hayafune M., Miyazaki H., Yokotani N., Ozawa K., Minami E., Kaku H., et al. Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol. Plant Microbe Interact. 2014;27:975–982. doi: 10.1094/MPMI-03-14-0068-R. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., Lin W., Yan H., Neng J., Zheng Y., Yang K., Xing F., Sun P. iTRAQ proteome analysis of the antifungal mechanism of citral on mycelial growth and OTA production in Aspergillus ochraceus. J. Sci. Food Agric. 2021;101:4969–4979. doi: 10.1002/jsfa.11140. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., Feng K., Yang H., Zhang Z., Yuan Y., Yue T. Effect of Cinnamaldehyde and Citral Combination on Transcriptional Profile, Growth, Oxidative Damage and Patulin Biosynthesis of Penicillium expansum. Front. Microbiol. 2018;9:597. doi: 10.3389/fmicb.2018.00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ju J., Xie Y.X., Yu H., Guo Y., Cheng Y.L., Zhang R.R., Yao W. Major components in Lilac and Litsea cubeba essential oils kill Penicillium roqueforti through mitochondrial apoptosis pathway. Ind. Crops Prod. 2020;149:112349. doi: 10.1016/j.indcrop.2020.112349. [DOI] [Google Scholar]
- 43.Reese S., Chelius C., Riekhof W., Marten M.R., Harris S.D. Micafungin-Induced Cell Wall Damage Stimulates Morphological Changes Consistent with Microcycle Conidiation in Aspergillus nidulans. J. Fungi. 2021;7:525. doi: 10.3390/jof7070525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai M.D., Wu M., Li Y., Su Z.Z., Lin F.C., Liu X.H. The chitin deacetylase PoCda7 is involved in the pathogenicity of Pyricularia oryzae. Microbiol. Res. 2021;248:126749. doi: 10.1016/j.micres.2021.126749. [DOI] [PubMed] [Google Scholar]
- 45.Li Z.K., Xia C.Y., Wang Y.X., Li X., Qian Y., Li C.Y., Zhou J., Zhang L., Ye X., Huang Y., et al. Identification of an endo-chitinase from Corallococcus sp. EGB and evaluation of its antifungal properties. Int. J. Biol. Macromol. 2019;132:1235–1243. doi: 10.1016/j.ijbiomac.2019.04.056. [DOI] [PubMed] [Google Scholar]
- 46.Kumari P., Arora N., Chatrath A., Gangwar R., Pruthi V., Poluri K.M., Prasad R. Delineating the Biofilm Inhibition Mechanisms of Phenolic and Aldehydic Terpenes against Cryptococcus neoformans. ACS Omega. 2019;4:17634–17648. doi: 10.1021/acsomega.9b01482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rocha M.C., Fabri J.H.T.M., Simões I.T., Silva R.R., Hagiwara D., da Cunha A.F., Goldman G.H., Cánovas D., Malavazi I. The cell wall integrity pathway contributes to the early stages of Aspergillus fumigatus asexual development. Appl. Environ. Microbiol. 2020;86:e02347-19. doi: 10.1128/AEM.02347-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J.B., Chen Y.C., Urban P.L. On-target labeling of intracellular metabolites combined with chemical mapping of individual hyphae revealing cytoplasmic relocation of isotopologues. Anal. Chem. 2012;84:5110–5116. doi: 10.1021/ac300903x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available from https://www.ncbi.nlm.nih.gov/genome/?term=Magnaporthe%20oryzae, accessed on 21 November 2021.