Abstract
Emerging new sequencing technologies have provided researchers with a unique opportunity to study factors related to microbial pathogenicity, such as antimicrobial resistance (AMR) genes and virulence factors. However, the use of whole-genome sequence (WGS) data requires good knowledge of the bioinformatics involved, as well as the necessary techniques. In this study, a total of nine Escherichia coli and Klebsiella pneumoniae isolates from Norwegian clinical samples were sequenced using both MinION and Illumina platforms. Three out of nine samples were sequenced directly from blood culture, and one sample was sequenced from a mixed-blood culture. For genome assembly, several long-read, (Canu, Flye, Unicycler, and Miniasm), short-read (ABySS, Unicycler and SPAdes) and hybrid assemblers (Unicycler, hybridSPAdes, and MaSurCa) were tested. Assembled genomes from the best-performing assemblers (according to quality checks using QUAST and BUSCO) were subjected to downstream analyses. Flye and Unicycler assemblers performed best for the assembly of long and short reads, respectively. For hybrid assembly, Unicycler was the top-performing assembler and produced more circularized and complete genome assemblies. Hybrid assembled genomes performed substantially better in downstream analyses to predict putative plasmids, AMR genes and β-lactamase gene variants, compared to MinION and Illumina assemblies. Thus, hybrid assembly has the potential to reveal factors related to microbial pathogenicity in clinical and mixed samples.
Keywords: Oxford Nanopore, Illumina, short-read, long-read, hybrid assembly, antimicrobial resistance, virulence factors, clinical isolates, blood culture, plasmids
1. Introduction
The pathogenicity of bacteria is often associated with antimicrobial resistance genes and/or virulence factors. Antimicrobial resistance (AMR) is the ability of microorganisms to defy antimicrobials, such as antibiotics. Globally, infections due to AMR bacteria are increasing and considered a threat to modern health care [1,2]. During the last two decades, scientific communities have seen a growing trend towards using next-generation sequencing (NGS) technology such as Illumina sequencing to identify AMR genes and virulence factors. Although NGS provides high depth coverage data, the output reads from NGS platforms such as Illumina are only about a few hundred base pairs long. Therefore, constructing a genome assembly based on the short-reads often results in an incomplete and fragmented assembly, which makes downstream analyses challenging [3].
New sequencing technologies known as third-generation sequencing technologies have been developed to overcome the short-read sequencing limitations. Pacific Biosciences (PacBio) is one of the most successful platforms for generating long reads [4]. One of the latest examples of devices that benefit from the new sequencing technology is the MinION sequencer from Oxford Nanopore Technologies (ONT). It can produce reads up to 2.3 million bases in length [5], which is sufficient to satisfy the repetitive elements flanked to the AMR genes [6]. Despite the advantages of long reads, they suffer from a high sequencing error rate [7], mainly due to older flowcells, kits and base calling algorithms. It has been shown that such reads remained error prone, even after error correction and polishing [8]. These properties restrict the usage of long reads to the study of small plasmids, which might carry AMR genes [9]. However, recent developments in flowcells and MinION sequencing chemistry, as well as more accurate neural network models used for MinION base calling, have greatly reduced the error rate [10].
Considering the benefits and drawbacks of both short and long reads, several attempts have been made to apply a hybrid assembly approach, which uses both type of reads [3,9,11,12,13]. For this purpose, different assemblers, such as Unicycler [3], hybridSPAdes [14], and MaSurCa [15], have been developed. All these hybrid assemblers benefit from the greater depth of short reads and increased length of long reads. Hybrid assembly offers several advantages over de novo assembly solely using short or long reads. For instance, hybrid assembly makes the downstream analyses, mapping, and annotation of genomic features more accurate [16]. Furthermore, it has been shown that hybrid assembly provides a better resolution for studying tandem repeats as well as gene variants [17], and it is the ideal approach for predicting the plasmids and AMR genes [13].
In recent years, different long, short and hybrid assemblers have been developed and tested, mainly using environmental samples [18]. The performance of different assemblers and the success of different assembly approaches for clinical isolates, especially where multiple bacteria are present, is unclear. Therefore, this study specifically focused on clinical isolates as well as blood samples spiked with bacteria species to mimic realistic clinical scenarios. Here, we aimed to compare the different tools available for constructing the short, long and hybrid assemblies and identify the top-performing assemblers for each approach. Secondly, we intended to identify plasmids, potential AMR genes, and virulence factors in assemblies produced by the top-performing assemblers for each approach.
2. Materials and Methods
2.1. Sample Collection and Characterization
In the present research, nine isolates consisting of four E. coli (1–4) and five K. pneumoniae (1–5) isolates, isolated from blood specimens of Norwegian patients, were used. The bacteria were grown overnight on agar plates, as described previously [19]. An overview of the samples and culture system is presented in Supplementary Table S1.
2.2. Spiking the Blood Samples and Incubation of Blood Cultures
Spiking and culturing of the blood samples was performed using two K. pneumoniae and one E. coli isolates at Oslo University Hospital, as described previously [20]. In brief, human blood was obtained from healthy anonymous donors via the blood bank at Oslo University Hospital and were transformed to four BD BACTEC 40 mL flasks (Becton, Franklin Lakes, NJ, USA). Then, blood samples in the flasks were spiked with isolate E. coli 4 (A2-39) and isolates K. pneumoniae 4 (A2-23), K. pneumoniae 5 (A2-37) and both E. coli 4 and K. pneumoniae 5 (mixed culture sample). The flasks were incubated in a BD BACTEC FX blood culture instrument until the culture was flagged positive.
2.3. Library Preparation and Whole-Genome Sequencing
The bacterial DNA from four blood cultures and six fresh grown isolates (three K. pneumoniae and three E. coli) was isolated and the libraries for Nanopore sequencing were constructed according to previously published protocols [19,20]. In brief, purified DNA was barcoded using the Rapid Barcoding Sequencing kit SQK-RBK004 (Oxford Nanopore, Oxford, UK) and further purified using an Agencourt AMPure XP system (Beckman Coulter, Brea, CA, USA). Sequencing, data collection and base calling (high accuracy mode) were performed using MinION flow cells (R9.4.1 FLO-MIN106, Oxford Nanopore), MinKNOW software v3.6.5 and Guppy basecaller v3 (ONT), respectively. Human data were discarded, and reads were categorized based on the read quality score as pass (≥5) or fail (<5) by the basecaller. DNA libraries for Illumina sequencing were prepared using Illumina Nextera XT DNA sample preparation kit (Illumina, San Diego, CA, USA). Illumina libraries were sequenced in pair-end mode (2 × 300 bp) using the Illumina MiSeq platform.
2.4. Bioinformatic Analyses of Bacterial Genomics
2.4.1. Quality Control and Trimming of Illumina and Nanopore Reads
Illumina reads were quality checked using FastQC (v0.11.8 for Linux) [21], adapters were removed, and low-quality reads (Phred < 25) were filtered out using Trimmomatic with default parameters [22], integrated into OmicsBox (v1.4.11 for Linux) [23]. For MinION reads, adapter and barcode trimming were performed using Porechop (v0.2.4 for Linux) with default settings [11]. Long and high-quality reads were collected using Filtlong (v0.2.0 for Linux) with default parameters [24]. Before downstream analyses, basic quality and statistics of long reads were checked using NanoPlot [25].
2.4.2. Bacterial Whole-Genome Assembly and Visualization
In this study, genome assemblies from Illumina short-reads (hereafter referred to as IllumASM) were created using SPAdes (v3.11.1) [26], Unicycler (v0.4.9) [3] and ABySS (v2.3.0) [27] assemblers. For the assembly of MinION long-reads (hereafter referred to as MinIONASM), different assemblers, including Unicycler, Flye (v2.8.2) [28], Canu (v1.7.1) [29] and Miniasm (v0.3.0) [30], were tested. Later, Illumina short-reads and MinION long-reads were combined to construct hybrid assembly (hereafter referred to as HybASM) using Unicycler, hybridSPAdes [14] and MaSurCa [15] assemblers.
General assembly statistics and quality of the assembled genomes were calculated using QUAST (v4.6.0 for Linux) [31] and BUSCO, which evaluate assemblies for highly conserved genes and generate a completeness score for the genome [32]. Furthermore, assembly visualization was performed using Bandage (v0.8.1 for Windows) [33]. For each of the short, long and hybrid reads, only one assembly (based on QUAST and BUSCO results, as well as the circularity of assemblies) was considered for downstream analyses (in total, three assembles per isolate).
In addition to the isolates, we have considered the E. coli NCTC strain 13441 as a reference genome. This strain was cultured in blood and sequenced directly from blood using MinION, as described in Section 2.2 and Section 2.3. In order to create Illumina reads for E. coli strain NCTC 13441, assembly file for this strain, was downloaded from NCBI assembly database (https://www.ncbi.nlm.nih.gov/assembly/GCF_900119685.1, access date: 25 September 2021) and short Illumina MiSeq reads were re-generated in silico from assembly file using InSilicoSeq sequencing simulator [34]. Reference genome assemblies were created using the top-performing assemblers, which described and identified for other isolates. Reference genome assemblies were considered for all downstream analyses and the results were considered as ground truths for E. coli isolates. The basic information for sequence, assembly and downstream analyses for reference samples is presented in Supplementary Table S2.
2.4.3. Bacterial Whole-Genome Annotation
Genome assemblies for each isolate were annotated using Prokka (v1.14.5 for Linux) [35], and information regarding different genomic features, such as coding sequence (CDS), tRNA, rRNA, tmRNA, and repeat regions, was extracted
2.4.4. Bacterial Plasmid Identification
Generated assembly files for each isolate were used to identify plasmids. For this purpose, the PlasmidFinder online tool (software version: 2.0.1, database version: 2020-07-13) [36], with minimum identity 95% and coverage 60%, was utilized. Plasmid hits were further visually confirmed for circularity using assembly graphs constructed in Bandage.
2.4.5. Detection of Antimicrobial Resistance Genes
In this study, AMR genes associated with mobile elements on chromosome/plasmids were identified using ResFinder online tool (v4.1, software version: 2020-10-21, database version: 2020-12-01) [37]. Only hits showing ≥95% identity and length coverage were considered as true AMR genes. To identify AMR genes associated with a chromosomal point mutation, the PointFinder online tool (software version: 2020-10-21, database version: 2019-07-02) [38], with the same search criteria as ResFinder, was used.
2.4.6. Bacterial Virulence Factor Identification
To identify virulence factors (VFs) hosted either by plasmids or chromosomes, the nucleotide virulence factor database (VFDB) was downloaded (database version: 2020-11-18) [39]. Then, the assembled genomes were BLAST-searched against the downloaded VFDB. Only hits with identity and alignment coverage ≥95% and e-values of 0 were considered as virulence factors.
3. Results
3.1. Basic Statistics of Short and Long Reads
Basic read information for both MinION and Illumina reads is presented in Table 1. Isolates E. coli 3 and K. pneumoniae 1 showed low read coverage, and isolate K. pneumoniae 2 showed remarkably high read coverage for their respective MinION long reads. These isolates had comparable read coverage for their Illumina short reads. Overall, Illumina reads clearly had higher coverage compared to MinION reads for E. coli isolates, whereas for K. pneumoniae, an opposite trend was observed.
Table 1.
An overview of basic sequence information statistics and quality of reads after trimming and filtering. The mixed culture was obtained from the co-culturing of E. coli 4 and K. pneumoniae 5 isolates. Coverage of E. coli isolates was calculated by dividing the number of bp in each read over the number of bp in reference genome (E. coli NCTC 13441). Coverage of K. pneumoniae isolates was calculated by dividing the number of bp reads over the number of bp reads in the K. pneumoniae reference genome (median genome size of all K. pneumoniae isolates in NCBI database). Coverage of mixed culture sample was calculated by dividing the number of bp in mix culture sample over the sum of pb of E. coli NCTC 13441 and median genome size of all K. pneumoniae isolates in NCBI database.
| MinION Long Reads | Illumina Short Reads | |||||||
|---|---|---|---|---|---|---|---|---|
| Read Length N50 (bp) | Mean Read Quality (Q) | Number of Reads | Total bp |
Coverage (X) | Number of Reads | Total bp |
Coverage (X) | |
| E. coli 1 | 2520 | 11.4 | 63,036 | 92,626,571 | 17.4 | 670,985 | 91,989,902 | 17.2 |
| E. coli 2 | 1466 | 11.3 | 67,331 | 88,553,163 | 16.6 | 597,154 | 141,802,031 | 26.6 |
| E. coli 3 | 1384 | 11.5 | 41,103 | 39,979,342 | 7.5 | 1,786,471 | 396,985,933 | 74.4 |
| E. coli 4 | 5956 | 9.8 | 81,317 | 256,369,935 | 48.0 | 1,419,582 | 353,790,894 | 66.3 |
|
E. coli (mean ± SD) |
2832 ± 2147 | 11 ± 0.8 | 63,197 ± 16,669 | 119,382,253 ± 94,404,708 | 22.4 ± 18 | 1,118,548 ± 579,915 | 246,142,190 ± 151,648,580 | 46.1 ± 28 |
| K. pneumoniae 1 | 1428 | 11.3 | 13,694 | 25,125,702 | 4.5 | 889,410 | 222,836,627 | 39.8 |
| K. pneumoniae 2 | 7302 | 11.5 | 199,822 | 859,067,656 | 153.5 | 559,060 | 131,573,009 | 23.5 |
| K. pneumoniae 3 | 4250 | 11.5 | 51,624 | 136,843,964 | 24.5 | 744,422 | 111,911,073 | 20.0 |
| K. pneumoniae 4 | 2044 | 9.9 | 329,042 | 375,495,020 | 67.1 | 1,302,920 | 313,973,441 | 56.1 |
| K. pneumoniae 5 | 3941 | 9.3 | 48,463 | 64,316,017 | 11.5 | 712,218 | 178,050,866 | 31.8 |
|
K. pneumoniae (mean ± SD) |
3793 ± 2302 | 11 ± 1 | 128,529 ± 133,041 | 292,169,672 ± 344,844,995 | 52.2 ± 62 | 841,606 ± 283,335 | 191,669,003 ± 80,759,064 | 34.2 ± 14 |
| Mixed culture sample | 4200 | 9.8 | 143,076 | 387,311,832 | 35.4 | 2,131,800 | 531,841,759 | 48.7 |
3.2. Unicycler Performed Better Than SPAdes and ABySS for the Assembly of Short-Reads
In this study, short reads were assembled using Unicycler, SPAdes and ABySS assemblers. According to QUAST and BUSCO results, Unicycler and SPAdes performed similarly and better than ABySS (Supplementary Table S3 and Supplementary Figure S5). The coverage fraction of reference genome and N50 average value for E. coli isolates, indicated a better performance of SPAdes over Unicycler (85.5% genome fraction vs. 82.5% and 237,038 bp vs. 225,244 bp N50). However, for K. pneumoniae, an opposite trend was observed, and the average N50 value was higher for Unicycler as compared to SPAdes (292,361 vs. 259,498). Although the core algorithm in Unicycler for the assembly of short reads is still SPAdes, the Unicycler assembler produced better assemblies compared to SPAdes alone. For instance, the assemblies from Unicycler had fewer contigs (on average, 138 for E. coli and 78 for K. pneumoniae in Unicycler vs. 243 for E. coli and 585 for K. pneumoniae in SPAdes). Furthermore, more circularized chromosomes and/or plasmids were observed in assemblies from Unicycler and the number of dead ends (number of occurrences where an end of a node does not connect to any other nodes) was also fewer (on average, 4 for E. coli and 9 for K. pneumoniae in Unicycler vs. 440 for E. coli and 1654 for K. pneumoniae in SPAdes) (Supplementary Figure S1). A similar better performance of Unicycler over SPAdes was documented for mixed culture sample (Supplementary Table S3). Therefore, all downstream analyses for short reads were performed using assemblies from Unicycler.
3.3. Flye as a Top-Performing Assembler for MinION Long-Reads
We compared different assemblers to assemble the MinION long-reads. Based on the QUAST assembly statistics, Flye and Canu clearly outperformed the other assemblers. Although the E. coli-assembled genomes using Flye covered a smaller portion of the reference genome compared to assemblies made by Canu (68.2% for Canu and 55.5% for Flye), Flye statistics were higher compared to Canu for other parameters. For instance, the average N50 value for K. pneumoniae isolates was 1,996,100 bp (Flye) and 1,789,477 bp (Canu). For E. coli isolates, the average N50 value was 343,234 bp (Flye) and 435,539 bp (Canu) (Supplementary Table S3). Furthermore, after the visualization of assembly files, more circularized chromosomes and/or plasmids and fewer dead-ends (Supplementary Figure S2) were observed for Flye. The average dead-end number was 77 for E. coli and 44 for K. pneumoniae isolates for Flye vs. 287 for E. coli and 135 for K. pneumoniae isolates using Canu (Supplementary Figure S2). The BUSCO analyses (Supplementary Figure S5), showed that assemblies constructed using Flye had better average BUSCO results compared to Canu (27.7% complete, 22.6% fragmented and 49.7% missing for Flye vs. 22.7% complete, 25% fragmented and 53.3% missing for Canu). Therefore, all downstream analyses for long-read sequences were performed using assemblies from Flye.
3.4. Unicycler Produced Superior Hybrid Assemblies over hybridSPAdes and MaSurCa
To make hybrid assemblies, we have tested three different tools. According to QUAST and BUSCO results, Unicycler and hybridSPAdes showed comparable and better performance than MaSurCa (Supplementary Table S3 and Supplementary Figure S5). For the mixed sample, MaSurCa performed excellently and displayed a higher genome fraction and N50 as well as a lower number of contigs. For E. coli isolates, the fraction of the reference genome which was covered by E. coli isolates assembly, was marginally higher for hybridSPAdes compared to Unicycler (83.2% vs. 82.9%). The average N50 value for E. coli isolates was 1,474,667 bp for hybridSPAdes, followed by Unicycler (1,005,273 bp). For K. pneumoniae isolates, the average N50 value was 3,880,247 bp for Unicycler, followed by hybridSPAdes (3,737,967 bp). Moreover, Unicycler produced fewer fragmented assemblies as compared to hybridSPAdes. For instance, assembly graphs indicated more circularized chromosomes and plasmids for Unicycler assemblies compared with hybridSPAdes (Supplementary Figure S3). Furthermore, assemblies produced using Unicycler had fewer dead ends (four and one dead ends for E. coli and K. pneumoniae isolates using Unicycler vs. 6072 and 5143 dead ends using hybridSPAdes, respectively). Therefore, all downstream analyses for hybrid assemblies were performed using hybrid genomes assembled using Unicycler.
3.5. Assembly Comparison between the Top-Performing Long, Short and Hybrid Read Assemblers
An overview of assembly statistics for the best IllumASM (using Unicycler), MinIONASM (using Flye), and HybASM (using Unicycler) are presented in Table 2. Results for individual isolates can be found in Supplementary Table S4. Overall, HybASM provided more complete and circular genomes. For both mono- and mixed culture isolates, IllumASM was more fragmented (higher number of contigs), followed by MinIONASM and HybASM. Furthermore, the N50 value was higher in HybASM followed by MinIONASM and IllumASM.
Table 2.
An overview of statistics for different E. coli and K. pneumoniae assemblies produced by the top-performing assemblers. IllumASM was produced using Unicycler, MinIONASM using Flye and HybASM created using Unicycler. The top values are highlighted in bold. The mixed culture was obtained from the co-culturing of E. coli 4 and K. pneumoniae 5 isolates. Numbers show the average ± SD.
| Number of Dead Ends | Number of Contigs | Total Length (bp) | N50 (bp) | ||
|---|---|---|---|---|---|
| E. coli | IllumASM | 4 ± 4 | 138 ± 90 | 5,232,982 ± 335,084 | 225,244 ± 82,435 |
| MinIONASM | 77 ± 94 | 49 ± 47 | 3,870,499 ± 2,664,510 | 343,234 ± 504,598 | |
| HybASM | 4 ± 2 | 50 ± 28 | 5,317,286 ± 426,129 | 1,005,273 ± 476,961 | |
| K. pneumoniae | IllumASM | 10 ± 7 | 78 ± 13 | 5,577,253 ± 181,931 | 247,095 ± 138,114 |
| MinIONASM | 44 ± 48 | 35 ± 32 | 4,694,978 ± 2,235,357 | 1,996,101 ± 2,279,327 | |
| HybASM | 1 ± 3 | 20 ± 17 | 5,648,111 ± 211,443 | 3,880,248 ± 2,149,256 | |
| Mixed culture sample | IllumASM | 2 | 371 | 11,193,506 | 147,235 |
| MinIONASM | 65 | 120 | 11,827,293 | 344,695 | |
| HybASM | 0 | 117 | 11,495,693 | 1,245,846 |
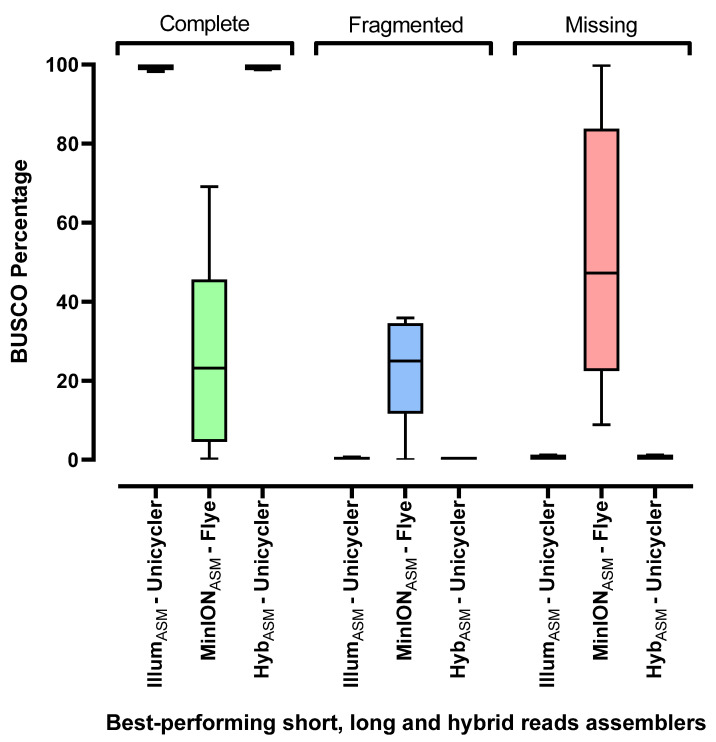
The BUSCO results (Figure 1) showed a similar performance of HybASM and IllumASM (on average, 0.6% BUSCO missing rate for both HybASM and IllumASM and 99.3% and 99.2% BUSCO complete for HybASM and IllumASM, respectively). Interestingly, MinIONASM performed worst in comparison to both HybASM and IllumASM. For instance, 22.6% of candidate genes in BUSCO were fragmented and only 27.7% were complete, whereas 49.7% of BUSCO genes were reported as missing in MinIONASM.
Figure 1.
Box plots for BUSCO results of the best-performing assemblers. IllumASM was produced using Unicycler, MinIONASM using Flye and HybASM was created using Unicycler. Each box extends from Min to Max values in each group and the middle black line in each box indicates the mean value. The BUSCO percentage for mixed samples is not included in the graph.
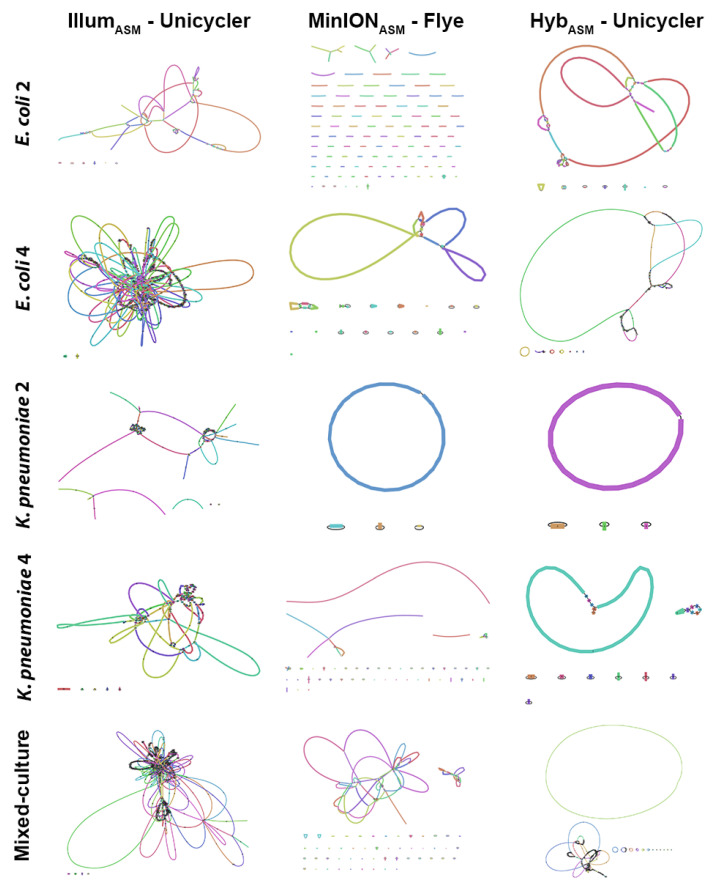
Using MinIONASM alone, we were able to close the chromosome for K. pneumoniae 2, similar to HybASM for the same isolate. In contrast, the IllumASM was fragmented for the same isolate (Figure 2). Overall, using HybASM, we were able to close the chromosome structure for three K. pneumoniae isolates (2, 3, and 4), whereas no circularized chromosome was obtained for E. coli isolates (Supplementary Figure S4). For the isolate from the mixed sample, two clear chromosomes were reconstructed using HybASM, including one circular chromosome. The circular contig sequence was BLAST-searched using PATRIC [40], and the results showed 94% identity to the complete K. pneumoniae subsp. pneumoniae genome.
Figure 2.
Representative assembly graphs for some of the isolates including E. coli 2 and 4, K. pneumoniae 2 and 4 as well as a mixed sample from the co-culturing of E. coli 4 and K. pneumoniae 5 isolates. The GFA files produced by the top-performing assemblers (Unicycler for Illumina short reads, Flye for MinION long reads and Unicycler for hybrid reads) were used to construct the assembly graphs using Bandage. Illumina assemblies were fragmented, and putative plasmids were limited. MinION produced much larger contigs and more putative plasmids. However, proper circular chromosomes were not observed for the majority of isolates using either IllumASM or MinIONASM. However, hybrid assemblies provided us with clear and close chromosome/putative plasmids.
3.6. Whole-Genome Annotation of the Short, Long and Hybrid Assemblies
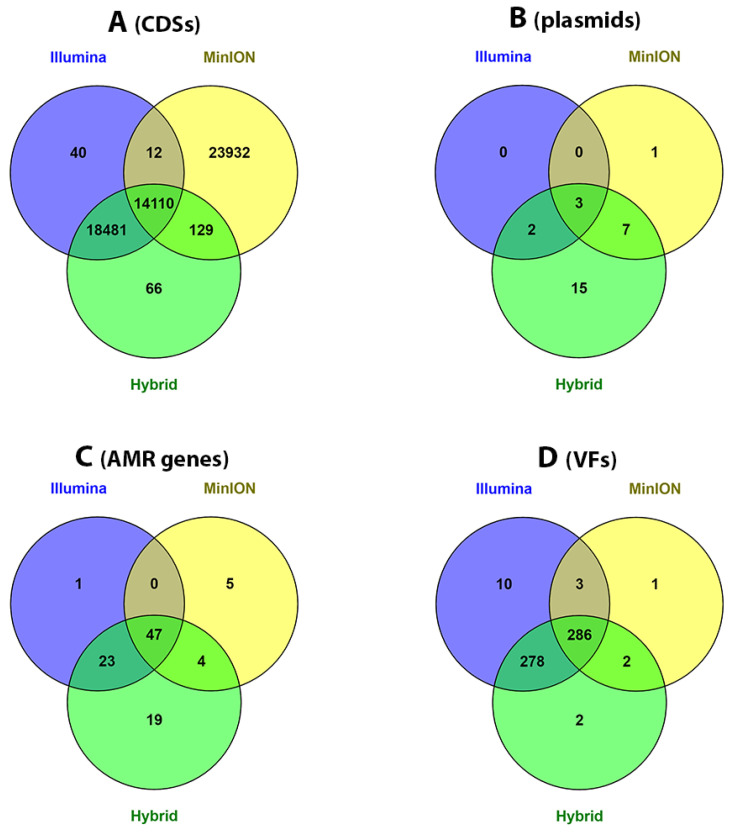
Results of the genome annotation are presented in Table 3 and Supplementary Table S5. The MinION data were not sufficient to capture all tRNA in the isolates as compared to IllumASM and HybASM. The overlaps between annotated CDSs from various assemblies for all the isolates (except the isolate from mixed culture) is presented in Figure 3A. Both IllumASM and HybASM exhibited comparable results, whereas MinIONASM showed divergent results compared to IllumASM and HybASM. For instance, using MinIONASM, a total number of 23,932 annotated CDSs were exclusively identified in isolates and, on average, MinION data had up to two times more annotated CDS. However, when we searched for which CDSs contributed to such high difference, the majority of these ‘extra’ CDSs belonged to duplicates of genes detected in HybASM (Supplementary Table S6). Annotations of IllumASM, MinIONASM and HybASM for E. coli isolates, on average, resulted in identifying of 3632, 4212 and 3684 CDSs, respectively (hypothetical and putative proteins were not considered). This corresponded to 0.55% fewer CDSs in IllumASM compared to the Illumina assembly of the reference genome (E. coli NCTC 13441) (Supplementary Table S2). Furthermore, on average, MinIONASM and HybASM predicted 3.8% and 0.21% more CDSs, respectively, as compared with corresponding assemblies of the reference genome. Results for annotated rRNA and tRNA indicated that HybASM showed closer association with corresponding data from the reference genome than IllumASM and MinIONASM.
Table 3.
Average values for annotating the genomic features of different assemblies from monocultures and mixed cultures of E. coli and K. pneumoniae isolates. IllumASM was produced using Unicycler, MinIONASM using Flye and HybASM was created using Unicycler. The mixed culture was obtained from the co-culturing of E. coli 4 and K. pneumoniae 5 isolates. Numbers show the average ± SD.
| CDS | rRNA | tRNA | tmRNA | ||
|---|---|---|---|---|---|
| E. coli | IllumASM | 4952 ± 392 | 5 ± 1 | 83 ± 5 | 1 ± 0 |
| MinIONASM | 6715 ± 4615 | 12 ± 10 | 63 ± 44 | 1 ± 1 | |
| HybASM | 5042 ± 532 | 15 ± 9 | 88 ± 11 | 1 ± 0 | |
| K. pneumoniae | IllumASM | 5201 ± 185 | 4 ± 1 | 79 ± 1 | 1 ± 0 |
| MinIONASM | 8120 ± 3933 | 20 ± 10 | 67 ± 36 | 1 ± 1 | |
| HybASM | 5261 ± 217 | 21 ± 8 | 84 ± 4 | 1 ± 0 | |
| Mixed culture sample | IllumASM | 10,660 | 8 | 164 | 2 |
| MinIONASM | 20,158 | 47 | 181 | 2 | |
| HybASM | 10,995 | 44 | 184 | 2 |
Figure 3.
An overview of downstream analysis results for different assemblies created using the top-performing assemblers. Venn diagrams prepared using the Venny online platform to plot differences in the number of annotations obtained, in which data for four E. coli and five K. pneumoniae isolates were merged. Numbers in the overlap area indicate the mutual hit names (hits identified in the exact same isolates). (A) The number of annotated CDSs (putative and hypothetical proteins not plotted). (B) The number of identified and confirmed plasmid contigs using PlasmidFinder and Bandage visualization tools, respectively. (C) The number of AMR genes, including both acquired and point mutations. (D) The number of identified VFs.
3.7. Plasmid Identification in Short, Long and Hybrid Assemblies
As can be seen from Figure 3B, more plasmids (confirmed using Bandage) were identified in HybASM (11 plasmids for E. coli isolates, 16 plasmids for K. pneumoniae isolates), followed by MinIONASM (3 plasmids for E. coli isolates, 8 plasmids for K. pneumoniae isolates) and IllumASM (3 plasmids for E. coli isolates, 2 plasmids for K. pneumoniae isolates). The majority of detected plasmids hosted IncF replicons in both E. coli and K. pneumoniae isolates. Only three plasmids (Col156, Col8282 and ColpVC), ranging from 1981 to 5146 bp in length, were detected in all three assembles. All three types of assembled genomes for the reference isolate (E. coli NCTC 13441), indicated that the reference genome (Supplementary Table S2) could have up to two plasmids (IncFIA and IncFII). The IllumASM, MinIONASM and HybASM results showed that E. coli isolates could have up to three, two and four putative plasmids, respectively. The complete list of plasmids and replicons is presented in Supplementary Table S7.
3.8. Identification of Acquired Antimicrobial Resistance Genes and Mutations
As shown in Figure 3C, using HybASM, we were able to identify more antimicrobial resistance genes (16 genes for E. coli isolates, 77 genes for K. pneumoniae isolates) than IllumASM (16 genes for E. coli isolates, 55 genes for K. pneumoniae isolates). MinIONASM demonstrated the worst performance in predicting the AMR genes (15 genes for E. coli isolates and 43 genes for K. pneumoniae isolates). Overall, 47% of identified AMR genes were found to be common between all types of assemblies.
Furthermore, we have identified chromosomal mutations conferring resistance to antibiotics for all the different assemblies. For all isolates from both mono- and mixed cultures, HybASM and IllumASM results were entirely identical (genes such as gyrA, parC, parE, acrR, ompK37 and ramR were identified at the identical isolates using both IllumASM and HybASM). Results from MinIONASM showed partial overlap (only ompK37 and ramR genes) with HybASM and/or IllumASM (Supplementary Table S8).
We were particularly interested in identifying different variants of β-lactamase genes in different assemblies. Using HybASM and not IllumASM or MinIONASM, we were able to identify a variety of β-lactamase genes mostly belonging to different variants of blaTEM (1C, 29, 55, 57, 122, 135, 141 and 209) and blaSHV (28, 31, 40, 56, 76, 79, 85, 89, 106, 164 and 172) genes. AMR genes such as blaTEM-1B, blaSHV-187, blaCTX-M (14, 15) and blaOXA-9 were the only β-lactamase genes identified in the same isolates using all types of assemblies (Supplementary Table S8).
Data from E. coli reference genome (Supplementary Table S2) showed that the reference genome could have up to 14 AMR genes (in Illumina and hybrid assemblies) and 9 AMR genes in the MinION assembly. Here, and on average, we identified four AMR genes per isolate (in each of the assemblies for E. coli isolates).
3.9. Identification of Virulence Factors in Short, Long and Hybrid Assemblies
Using the VFDB core database, we identified bacterial virulence factors. In all three different assemblies, the number of identified virulence factors was higher in E. coli than in K. pneumoniae. As shown in (Figure 3D), the majority of identified VFs were mutual between HybASM and IllumASM; therefore, IllumASM and HybASM showed almost similar performance. Similar results were observed for the reference sample (Supplementary Table S2). MinIONASM covered fewer VFs (136 VFs for E. coli, 156 VFs for K. pneumoniae). However, all the hits (291 VFs), except just one VF in MinIONASM, were detected using either hybrid or Illumina assemblies. Results for each individual isolate are presented in Supplementary Table S9.
The reference genome for E. coli showed 46 (based on long read assembly) and 85 (based on short read and hybrid assemblies) VFs (Supplementary Table S2). In comparison with the reference genome, and on average, we identified 76, 34 and 74 VFs for E. coli isolates using IllumASM, MinIONASM and HybASM, respectively.
3.10. HybASM Enables the Complete Recovery of Plasmid Replicons, AMR Genes, and Virulence Factors from the Mixed Culture Sample
According to MinION data, Isolate E. coli 4 and K. pneumoniae 5 possessed p0111 and IncFII replicons, respectively. In the mixed culture sample, the MinIONASM failed to recover the IncFII plasmid replicon from K. pneumoniae 5. In contrast, hybrid data revealed replicons such as IncHI2, IncHI2A, and p0111 in E. coli 4, as well as replicons such as IncFIA(HI1), IncFIB(K), IncFII, IncFII(pKP91) in K. pneumoniae 5. Interestingly, HybASM recovered all mentioned plasmid replicons in mixed samples too. Regarding recovering the AMR genes, although HybASM was able to recover all the AMRs identified in both E. coli 4 and K. pneumoniae 5 for mixed samples, IllumASM and MinIONASM each missed one gene (sul1 and 16S_rrsC in E. coli 4 for IllumASM and MinIONASM, respectively). Similar to recovering the AMR genes, HybASM recovered all the VFs (plus two more VFs) in the mixed culture sample. IllumASM also was able to recover complete VFs in the mixed sample, which were identified individually in E. coli 4 and K. pneumoniae 5. Although MinIONASM identified 19 unique VFs in the mixed culture sample, it missed 10 VFs in E. coli 4 (data corresponding to annotation, plasmid replicons, AMR and VF for E. coli 4, K. pneumoniae 5 and the mixed sample are presented in Supplementary Table S10).
4. Discussion
In the current study, we tested different short, long and hybrid read assemblers. The assembled genomes from the top-performing assemblers in each approach were subjected to downstream analyses.
For short read assembly, ABySS, Unicycler and SPAdes were tested; based on both QUAST and BUSCO results, ABySS performed worse than Unicycler and SPAdes. In line with our results, a better performance of SPAdes over ABySS previously has been documented for the de novo assembly of small RNA-Seq samples taken from plant species [41]. This observation might be explained by the fact that SPAdes takes advantages of various Kmer sizes simultaneously, whereas in ABySS, one must specify the Kmer cut-off size. In this study, assembly statistics and graphs indicated a slightly better performance of Unicycler over SPAdes to assemble the short reads. Although SPAdes is the main algorithm implemented in Unicycler, the better performance of Unicycler might be explained by the implementation of additional steps such as strict filtering steps, repeat resolution algorithm and polishing [3].
In this study, we observed a comparable result between Flye and Canu to assemble the long reads. However, a higher degree of genome circularization was observed in assemblies produced by the Flye assembler. Similar conclusions between Flye and Canu assemblers were made in previous research, where the authors tested different assemblers for prokaryote whole-genome sequencing [42]. Following the present results, previous studies have demonstrated that both Flye and Canu assemblers could be considered as the first choice to assemble not only prokaryote genomes, but also plant and crop genomes based on long reads [43,44]. We observed a low BUSCO score using long reads. A similar low BUSCO score for assemblies based on MinION reads has previously been observed [45]. This might be explained by the low coverage of ONT reads. Overall, according to the BUSCO results, the Flye assembler performed best. This might be explained by the five polishing steps performed using integrated Pilon software with Flye; prior studies have reported that polishing the MinION assembly increases the BUSCO completeness score [45,46]. Although the Canu assembler takes advantage of polishing using both Racon and Pilon (two rounds each), the BUSCO completeness score for Canu was considerably lower than Flye. At the same time, QUAST statistics were similar for both Canu and Flye. Therefore, to draw the conclusion regarding choosing the appropriate assembler for long reads, it may be necessary to evaluate the assemblies using both QUAST and BUSCO. Here, using both tools, we observed superior long-reads assembly for Flye as compared to Canu. Although the BUSCO score for Miniasm indicated an acceptable performance, the QUAST statistics demonstrated weak performance for this assembler. However, it must be kept in mind that Miniasm still is in the development phase, and it does not perform any polishing or read correction processes for MinION data [30].
Furthermore, we have tested three different tools for making a hybrid assembly. Both QUAST and BUSCO documented a similar performance in Unicycler and hybridSPAdes and a less efficient performance in MaSurCa. In accordance with the current study, previous research has revealed that both hybridSPAdes and Unicycler produce more accurate hybrid assemblies compared with MaSurCa [47]. In this study, Unicycler produced less fragmented HybASM as compared with hybridSPAdes. Similar observations have previously been reported for clinical samples [47]. Differences between Unicycler and hybridSPAdes might be partially explained by different integrated polishers (i.e., Unicycler uses Pilon and SPAdes uses Racon for polishing) and the step where polishing is implemented. The average N50 values for E. coli HybASM using both Unicycler and hybridSPAdes were remarkably lower compared to the average K. pneumoniae N50 value. This might be explained by the low coverage of ONT data for E. coli (22.4× compared to K. pneumoniae (52.2×) isolates. Surprisingly, in our study, the MaSurCa assembler provided remarkably lower quality hybrid assembly (10 times lower N50 values) for isolates E. coli 3 and K. pneumoniae 1 compared to both Unicycler and hybridSPAdes. Our results documented a low MinION coverage for mentioned isolates. Therefore, the current finding suggest that low-quality long reads could greatly affect the hybrid assembly produced by MaSurCa, and both Unicycler and hybridSPAdes can tolerate more low-quality long reads. It is worth mentioning that the application of MaSurCa for bacterial hybrid genome assembly is limited thus far; therefore, applications of the MaSurCa assembler for clinical sampling deserve further investigation.
In this study, we observed a considerable size variation in assembled genomes following the use of long reads from MinION. For instance, isolates E. coli 3 and K. pneumoniae 1 had a remarkably smaller genome size than HybASM or IllumASM (Supplementary Table S4). Inaccuracy in genome size using Nanopore technology has previously been reported for a conjugated test plasmid [13], and might be explained by the technology’s greater sequencing error [7,48]. In addition, inaccuracy in genome size can be explained by lower MinION coverage for the mentioned isolates. Lower MinION coverage might be related to a lower quantity and quality of isolated DNA. Due to the complexity of the samples, the DNA extraction could have compromised the recovery of long DNA molecules, thus affecting the N50 read length. Another reason which might explain the lower coverage for some of the samples is that the data for corresponding samples were generated during a rapid barcoding run, with six samples per run. According to rapid barcoding protocols, isolated DNA will not undergo PCR amplification during MinION library preparation. Notably, the E. coli 3 and K. pneumoniae 1 isolates showed remarkable examples where even a minimal quantity of long reads effectively contributed to improved HybASM results. The minimal quantity of long reads further reflected in QUAST results: E. coli 3 and K. pneumoniae 1 long reads only contributed as much as 46% and 3% in HybASM of corresponding isolates. However, and despite the minimal quantity of long reads, MinION data provided 74 and 248 Kb improvements in N50 (Supplementary Table S4) in the HybASM of mentioned isolates, respectively. These results provide proof for previous hypotheses suggesting that combining even a few long reads with short reads could be the most cost-effective way to map a complete bacterial genome [3].
Regarding the prediction of plasmids, more putative plasmids were detected using MinIONASM as compared with IllumASM. The poor performance of IllumASM to predict the plasmids is likely to be related to a higher level of fragmentation, which makes the reconstruction of plasmids difficult. The better performance of HybASM to resolve the putative plasmids in the current study agrees with a previous study, where small plasmids were absent from long-read assemblies but not from HybASM [49]. Furthermore, the superiority of hybrid assemblies (assembled using Unicycler) in the plasmid detection of clinical pathogens has previously been reported [50,51]. The numbers of both AMR genes and mutations, predicted here using HybASM for E. coli isolates (four AMRs/isolate), were lower than previously reported results for HybASM of clinical E. coli isolates in Canada (eight AMRs/isolate) [52] and less than the E. coli reference genome. This might be due to Norway’s lower antibiotic resistance occurrence; it is a country with one of the lowest drug resistance indexes [53].
Furthermore, our results showed that nanopore sequencing is not suitable for studying gene variants and/or predicting chromosomal mutations. For instance, using MinIONASM, we were not able to predict the AMR gene variants for β-lactamase genes (blaTEM and blaSHV variants), which only differ by one or a few base pairs. This is in line with our previous findings [20]. Although IllumASM performed marginally better in predicting these gene variants, HybASM performed best. MinIONASM results for predicting chromosomal mutations also indicated poor performance, whereas IllumASM and HybASM yielded similar results. It seems possible that these results are due to the low sensitivity and high error rate of nanopore sequencing technology [7,48].
VF predictions showed that both IllumASM and HybASM were performed similar and comparable, whereas MinIONASM performed worse in predicting the VFs. Therefore, one must interpret the data with care when studying the VFs solely using MinIONASM. Results for the current study are in contrast with previously published results for VFs detected in Shiga toxin-producing E. coli, where authors reported better performance for MinIONASM over IllumASM [54]. These differences could be explained by using different assemblers or technology used for library preparation and sequencing.
One must consider that AMR and VF profiles are not stable over the isolates and plasmid-mediated AMR genes can be horizontally transferred between isolates. Hence, identifying the exact number of AMR or VFs and comparing the results with reference samples might be challenging. Although here we included E. coli strain NCTC 13441 as a reference isolate and annotation results correlated well with the reference genome, this study was limited by the absence of reference genome for K. pneumoniae.
Translating the current finding for long reads to the output from other platforms such as PacBio is challenging. Previous research showed that PacBio generated both longer and more accurate reads compared to ONT [55,56]. However, the applicability of the long-read sequencer is largely depending on the type of the research. For instance, it has been shown that ONT performance for quantitative analyses such as transcriptome studies was better than PacBio [55]. Furthermore, the superiority of ONT over PacBio for the rapid identification of pathogens has been shown previously [57]. Our data show that MinION data assembly is faster compared to both IllumASM and HybASM. Our analyses indicated that all type of assemblies can be performed using a Linux machine with standard computational resources. We tested the elapsed time for the assembly of the reference genome (E. coli strain NCTC 13441). The data showed that the assembly took 1 h and 13 min for HybASM (using Unicycler), 40 min for IllumASM (using Unicycler) and 10 min for MinIONASM (using Flye). A shorter turnaround time for assembly, in parallel with a shorter turnaround time for MinION sequencing as compared to Illumina, which we have previously shown [20], make MinION the favorable sequencing platform, especially for field and diagnostic research.
In our previous research, we identified AMR genes and plasmids in clinical isolates, using solo plasmid assembly [58]. We concluded that results are heavily dependent on the database of choice therefore, HybASM might be a better approach for in depth analyses of WGS data. Although HybASM has been used to study hospital Mycobacterium chelonae infections [59], extraintestinal pathogenic E. coli isolates [52] and one pan-drug-resistant K. pneumoniae isolate [60], the application of HybASM for studying pathogenic factors in clinical samples is somewhat limited. In the current study, HybASM showed reliable results for studying the clinical samples, specifically for the mixed culture as compared to IllumASM and MinIONASM. Compared to MinIONASM and IllumASM, downstream analyses using HybASM was accurate and more informative. These findings are in agreement with previous research, where the authors suggested that combining the ONT and PacBio data with Illumina data and generating a hybrid assembly greatly improved the accuracy and mappability of long reads [56]. Promising results of HybASM have facilitated the annotation of clinically relevant genomic elements. Interestingly, similar conclusions using HybASM have previously been drawn for assemblies from environmental samples [18,61] and clinical samples [62,63].
Taken all together, prior to sequencing, one might consider the experiment need; for HybASM, the same sample needs to be sequenced in two different platforms, which is resource demanding. The MinION sequencer demonstrated an acceptable performance to study the genome of clinical isolates. However, solely long reads might not be ideal for predicting gene variants, point mutation, and virulence factors. Moreover, it seems possible that the source of the samples and the method for library preparation for long-read sequencing might play an important role in the quality and the amount of collected data. Nanopore technology is evolving, and one may be optimistic that the current weaknesses could be overcome with technology improving in the near future. Despite all uncertainty regarding the long reads, and although we did not use exactly the same DNA for both platforms, the data showed that even a low quantity of long reads in combination with short reads could greatly improve the assembly. Therefore, HybASM should be considered a successful approach to overcome uncertainty caused by Oxford Nanopore technology. Considering the cost of experiments, extended data analyses, and the possibility of mixed infections in clinical samples, application of HybASM for the study the genome, AMR and VF genes, and potential plasmid, could be justified. Otherwise, Illumina assembly could be considered as sufficient.
5. Conclusions
In conclusion, HybASM is a good approach for the in-depth analysis of clinically relevant samples, including blood cultures, as demonstrated here. It is recommended to benefit from the advantages of HybASM for genome study of complicated mixed isolates, the in-depth determination of pathogenicity and epidemiological studies. The present findings emphasize the fact that selecting the appropriate approach for sequencing and assembly could have great impact on the results and could indirectly shorten the times required to detect pathogenicity factors in clinical settings.
Acknowledgments
The authors would like to thank Arne Michael Taxt for selecting the clinical isolates and Stephan A. Frye for performing the WGS.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9122560/s1. Figure S1_Assembly graphs for IllumASM; Figure S2_Assembly graphs for MinIONASM.; Figure S3_Assembly graphs for HybASM.; Figure S4_Assembly graphs for assemblies from top performed assemblers; Figure S5_BUSCO graphs for all assemblies produced by different assemblers.; Table S1_Isolates ID, ENA accession number, source, and culture method; Table S2_Reference genome information; Table S3_Assembley statistics for all assemblies produced by all assemblers; Table S4_Assembly statistics for assemblies produced by top performing assemblers; Table S5_Summary of Prokka annotation results for top performing assemblers; Table S6_Full Prokka table for top performing assemblers; Table S7_Identified plasmids in assemblies produced by top performing assemblers; Table S8_Identified AMR genes in assemblies produced by top performing assemblers; Table S9_Identified VFs in assemblies produced by top performing assemblers; Table S10_Mixed cultured results.
Author Contributions
Conceptualization, R.A., A.K. and E.A.; methodology, A.K. and E.A.; software, A.K.; formal analysis, A.K. and E.A.; investigation, A.K. and E.A.; data curation, A.K.; writing—original draft preparation, A.K.; writing—review and editing, R.A., A.K. and E.A.; visualization–A.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Norwegian Research Council, grant number 273609, to AMR-Diag. The APC was funded by the AMR-Diag grant and from the Inland Norway University of Applied Sciences support.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from The European Nucleotide Archive (ENA) under primary accession number PRJEB45084 and secondary accession number ERP129212. An overview of submitted reads is provided in Supplementary Table S1.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cassini A., Hogberg L.D., Plachouras D., Quattrocchi A., Hoxha A., Simonsen G.S., Colomb-Cotinat M., Kretzschmar M.E., Devleesschauwer B., Cecchini M., et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn S.J., Connor C., McNally A. The evolution and transmission of multi-drug resistant Escherichia coli and Klebsiella pneumoniae: The complexity of clones and plasmids. Curr. Opin. Microbiol. 2019;51:51–56. doi: 10.1016/j.mib.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhoads A., Au K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Payne A., Holmes N., Rakyan V., Loose M. BulkVis: A graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics. 2018;35:2193–2198. doi: 10.1093/bioinformatics/bty841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:30. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laver T., Harrison J., O’Neill P.A., Moore K., Farbos A., Paszkiewicz K., Studholme D.J. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol. Detect. Quantif. 2015;3:1–8. doi: 10.1016/j.bdq.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loman N.J., Quick J., Simpson J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 9.Juraschek K., Borowiak M., Tausch S.H., Malorny B., Käsbohrer A., Otani S., Schwarz S., Meemken D., Deneke C., Hammerl J.A. Outcome of Different Sequencing and Assembly Approaches on the Detection of Plasmids and Localization of Antimicrobial Resistance Genes in Commensal Escherichia coli. Microorganisms. 2021;9:598. doi: 10.3390/microorganisms9030598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rang F.J., Kloosterman W.P., de Ridder J. From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018;19:90. doi: 10.1186/s13059-018-1462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 2017;3:e000132. doi: 10.1099/mgen.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayliss S.C., Hunt V.L., Yokoyama M., Thorpe H.A., Feil E.J. The use of Oxford Nanopore native barcoding for complete genome assembly. Gigascience. 2017;6:gix001. doi: 10.1093/gigascience/gix001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berbers B., Ceyssens P.J., Bogaerts P., Vanneste K., Roosens N.H.C., Marchal K., De Keersmaecker S.C.J. Development of an NGS-Based Workflow for Improved Monitoring of Circulating Plasmids in Support of Risk Assessment of Antimicrobial Resistance Gene Dissemination. Antibiotics. 2020;9:503. doi: 10.3390/antibiotics9080503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antipov D., Korobeynikov A., McLean J.S., Pevzner P.A. hybridSPAdes: An algorithm for hybrid assembly of short and long reads. Bioinformatics. 2015;32:1009–1015. doi: 10.1093/bioinformatics/btv688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimin A.V., Marçais G., Puiu D., Roberts M., Salzberg S.L., Yorke J.A. The MaSuRCA genome assembler. Bioinformatics. 2013;29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kancharla N., Jalali S., Narasimham J.V., Nair V., Yepuri V., Thakkar B., Reddy V.B., Kuriakose B., Madan N., Arockiasami S. De Novo Sequencing and Hybrid Assembly of the Biofuel Crop Jatropha curcas L.: Identification of Quantitative Trait Loci for Geminivirus Resistance. Genes. 2019;10:69. doi: 10.3390/genes10010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J.R., Zhou P., Mudge J., Gurtowski J., Lee H., Ramaraj T., Walenz B.P., Liu J., Stupar R.M., Denny R., et al. Hybrid assembly with long and short reads improves discovery of gene family expansions. BMC Genom. 2017;18:541. doi: 10.1186/s12864-017-3927-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown C.L., Keenum I.M., Dai D., Zhang L., Vikesland P.J., Pruden A. Critical evaluation of short, long, and hybrid assembly for contextual analysis of antibiotic resistance genes in complex environmental metagenomes. Sci. Rep. 2021;11:3753. doi: 10.1038/s41598-021-83081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avershina E., Sharma P., Taxt A.M., Singh H., Frye S.A., Paul K., Kapil A., Naseer U., Kaur P., Ahmad R. AMR-Diag: Neural network based genotype-to-phenotype prediction of resistance towards β-lactams in Escherichia coli and Klebsiella pneumoniae. Comput. Struct. Biotechnol. J. 2021;19:1896–1906. doi: 10.1016/j.csbj.2021.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taxt A.M., Avershina E., Frye S.A., Naseer U., Ahmad R. Rapid identification of pathogens, antibiotic resistance genes and plasmids in blood cultures by nanopore sequencing. Sci. Rep. 2020;10:7622. doi: 10.1038/s41598-020-64616-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online] [(accessed on 19 May 2019)]. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 22.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biobam OmicsBox—Bioinformatics Made Easy. BioBam Bioinformatics. [(accessed on 3 March 2019)]. Available online: https://www.biobam.com/omicsbox/
- 24.Wick R.R. Filtlong. [(accessed on 17 November 2020)]. Available online: https://github.com/rrwick/Filtlong.
- 25.De Coster W., D’Hert S., Schultz D.T., Cruts M., Van Broeckhoven C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics. 2018;34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J.M., Birol I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolmogorov M., Yuan J., Lin Y., Pevzner P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 29.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H. Minimap and miniasm: Fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2016;32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 33.Wick R.R., Schultz M.B., Zobel J., Holt K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gourlé H., Karlsson-Lindsjö O., Hayer J., Bongcam-Rudloff E. Simulating Illumina metagenomic data with InSilicoSeq. Bioinformatics. 2018;35:521–522. doi: 10.1093/bioinformatics/bty630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 36.Carattoli A., Zankari E., Garcia-Fernandez A., Voldby Larsen M., Lund O., Villa L., Moller Aarestrup F., Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bortolaia V., Kaas R.S., Ruppe E., Roberts M.C., Schwarz S., Cattoir V., Philippon A., Allesoe R.L., Rebelo A.R., Florensa A.F., et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zankari E., Allesoe R., Joensen K.G., Cavaco L.M., Lund O., Aarestrup F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017;72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., Jin Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis J.J., Wattam A.R., Aziz R.K., Brettin T., Butler R., Butler R.M., Chlenski P., Conrad N., Dickerman A., Dietrich E.M., et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020;48:D606–D612. doi: 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrero R.A., Napier K.R., Cunnington J., Liefting L., Keenan S., Frampton R.A., Szabo T., Bulman S., Hunter A., Ward L., et al. An internet-based bioinformatics toolkit for plant biosecurity diagnosis and surveillance of viruses and viroids. BMC Bioinform. 2017;18:26. doi: 10.1186/s12859-016-1428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wick R.R., Holt K.E. Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Research. 2019;8:2138. doi: 10.12688/f1000research.21782.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murigneux V., Rai S.K., Furtado A., Bruxner T.J.C., Tian W., Harliwong I., Wei H., Yang B., Ye Q., Anderson E., et al. Comparison of long-read methods for sequencing and assembly of a plant genome. GigaScience. 2020;9:giaa146. doi: 10.1093/gigascience/giaa146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung H., Jeon M.S., Hodgett M., Waterhouse P., Eyun S.I. Comparative Evaluation of Genome Assemblers from Long-Read Sequencing for Plants and Crops. J. Agric. Food Chem. 2020;68:7670–7677. doi: 10.1021/acs.jafc.0c01647. [DOI] [PubMed] [Google Scholar]
- 45.Vasudevan K., Devanga Ragupathi N.K., Jacob J.J., Veeraraghavan B. Highly accurate-single chromosomal complete genomes using IonTorrent and MinION sequencing of clinical pathogens. Genomics. 2020;112:545–551. doi: 10.1016/j.ygeno.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Miller D.E., Staber C., Zeitlinger J., Hawley R.S. Highly Contiguous Genome Assemblies of 15 Drosophila Species Generated Using Nanopore Sequencing. G3 Genes Genomes Genet. 2018;8:3131–3141. doi: 10.1534/g3.118.200160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z., Erickson D.L., Meng J. Benchmarking hybrid assembly approaches for genomic analyses of bacterial pathogens using Illumina and Oxford Nanopore sequencing. BMC Genom. 2020;21:631. doi: 10.1186/s12864-020-07041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahlin K., Medvedev P. Error correction enables use of Oxford Nanopore technology for reference-free transcriptome analysis. Nat. Commun. 2021;12:2. doi: 10.1038/s41467-020-20340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.George S., Pankhurst L., Hubbard A., Votintseva A., Stoesser N., Sheppard A.E., Mathers A., Norris R., Navickaite I., Eaton C., et al. Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: Assessment of MinION and MinION/Illumina hybrid data assembly approaches. Microb. Genom. 2017;3:e000118. doi: 10.1099/mgen.0.000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sydenham T.V., Overballe-Petersen S., Hasman H., Wexler H., Kemp M., Justesen U.S. Complete hybrid genome assembly of clinical multidrug-resistant Bacteroides fragilis isolates enables comprehensive identification of antimicrobial-resistance genes and plasmids. Microb. Genom. 2019;5:e000312. doi: 10.1099/mgen.0.000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Maio N., Shaw L.P., Hubbard A., George S., Sanderson N.D., Swann J., Wick R., AbuOun M., Stubberfield E., Hoosdally S.J., et al. Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb. Genom. 2019;5:e000294. doi: 10.1099/mgen.0.000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mattrasingh D., Hinz A., Phillips L., Carroll A.C., Wong A. Hybrid Nanopore-Illumina Assemblies for Five Extraintestinal Pathogenic Escherichia coli Isolates. Microbiol. Resour. Announc. 2021;10:e01027-20. doi: 10.1128/MRA.01027-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.NORM/NORM-VET . Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. Norwegian Institute of Public Health; Tromsø/Oslo, Norway: 2019. [Google Scholar]
- 54.Gonzalez-Escalona N., Allard M.A., Brown E.W., Sharma S., Hoffmann M. Nanopore sequencing for fast determination of plasmids, phages, virulence markers, and antimicrobial resistance genes in Shiga toxin-producing Escherichia coli. PLoS ONE. 2019;14:e0220494. doi: 10.1371/journal.pone.0220494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Udaondo Z., Sittikankaew K., Uengwetwanit T., Wongsurawat T., Sonthirod C., Jenjaroenpun P., Pootakham W., Karoonuthaisiri N., Nookaew I. Comparative Analysis of PacBio and Oxford Nanopore Sequencing Technologies for Transcriptomic Landscape Identification of Penaeus monodon. Life. 2021;11:862. doi: 10.3390/life11080862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weirather J., de Cesare M., Wang Y., Piazza P., Sebastiano V., Wang X., Buck D., Au K. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Research. 2017;6:100. doi: 10.12688/f1000research.10571.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loit K., Adamson K., Bahram M., Puusepp R., Anslan S., Kiiker R., Drenkhan R., Tedersoo L., Druzhinina I.S. Relative Performance of MinION (Oxford Nanopore Technologies) versus Sequel (Pacific Biosciences) Third-Generation Sequencing Instruments in Identification of Agricultural and Forest Fungal Pathogens. Appl. Environ. Microbiol. 2019;85:e01368-19. doi: 10.1128/AEM.01368-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khezri A., Avershina E., Ahmad R. Plasmid Identification and Plasmid-Mediated Antimicrobial Gene Detection in Norwegian Isolates. Microorganisms. 2020;9:52. doi: 10.3390/microorganisms9010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu C.H., Zhao C., Hofstaedter C., Tebas P., Glaser L., Baldassano R., Bittinger K., Mattei L.M., Bushman F.D. Investigating hospital Mycobacterium chelonae infection using whole genome sequencing and hybrid assembly. PLoS ONE. 2020;15:e0236533. doi: 10.1371/journal.pone.0236533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruan Z., Wu J., Chen H., Draz M.S., Xu J., He F. Hybrid Genome Assembly and Annotation of a Pandrug-Resistant Klebsiella pneumoniae Strain Using Nanopore and Illumina Sequencing. Infect. Drug Resist. 2020;13:199–206. doi: 10.2147/IDR.S240404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neal-McKinney J.M., Liu K.C., Lock C.M., Wu W.-H., Hu J. Comparison of MiSeq, MinION, and hybrid genome sequencing for analysis of Campylobacter jejuni. Sci. Rep. 2021;11:5676. doi: 10.1038/s41598-021-84956-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldstein S., Beka L., Graf J., Klassen J.L. Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing. BMC Genom. 2019;20:23. doi: 10.1186/s12864-018-5381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Z., Kuang D., Xu X., González-Escalona N., Erickson D.L., Brown E., Meng J. Genomic analyses of multidrug-resistant Salmonella Indiana, Typhimurium, and Enteritidis isolates using MinION and MiSeq sequencing technologies. PLoS ONE. 2020;15:e0235641. doi: 10.1371/journal.pone.0235641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from The European Nucleotide Archive (ENA) under primary accession number PRJEB45084 and secondary accession number ERP129212. An overview of submitted reads is provided in Supplementary Table S1.