Abstract
Background
The emergence of SARS-CoV-2 variants in places where the virus is uncontained poses a global threat from the perspective of public health and vaccine efficacy. Travel has been important factor for the easy spread of SARS-CoV-2 variants worldwide. India has also observed the importation of SARS-CoV-2 variants through international travelers.
Methods
In this study, we have collected the oropharyngeal and nasopharyngeal swab specimens from 58 individuals with travel history from United Arab Emirates (UAE), East, West and South Africa, Qatar, Ukraine and Saudi Arabia arrived in India during February–March 2021. The clinical specimens were initially screened for SARS-CoV-2 using Real time RT-PCR. All the specimens were inoculated on to Vero CCL-81 cells for virus isolation. The viral isolates were further sequenced using Next-Generation Sequencing.
Results
All 58 cases were tested positive for SARS-CoV-2 using Real time RT-PCR. Four specimens showed progressive infectivity with fusion of the infected cells with neighboring cells leading to large mass of cells. Replication competent virus was confirmed from culture supernatant of the passage 2 using Real time RT-PCR. Two plaque purified SARS-CoV-2 isolates demonstrated high viral RNA load of 3.8–7.5 × 1011 and 1.1–1.6 × 1011 at passage 4 and 5 respectively. Nucleotide variations along with amino acid changes were also observed among these two isolates at passage 2–5. All four cases were male with no symptoms and co-morbidity. The sequence analysis has shown two different clusters, first cluster with nucleotide deletions in the ORF1ab and the spike, while second cluster with deletions in spike region. The viral isolates demonstrated 99.88–99.96% nucleotide identity with the representative sequences of Beta variant (B.1.351).
Conclusion
These findings suggest easier transmission of SARS-CoV-2 variants with human mobility through international travel. The isolated Beta variant would be useful to determine the protective efficacy of the currently available and upcoming COVID-19 vaccines in India.
Keywords: Variant of concern, SARS-CoV-2, Beta variant, Traveler, United Arab Emirates, India
Abbreviations: SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2
Introduction
Since its emergence in 2019, SARS-CoV-2 has effectively evolved accumulating the deleterious mutations in its genome i.e., Alpha (B.1.1.7), Beta (B.1.351), Gamma (B.1.1.28 P1), Delta (B.1.617.2). Within the five months of January–May 2021, multiple SARS-CoV-2 variants have been detected across the globe which are more transmissible, possibly escapes the natural and vaccine-induced immunity, and could lead to increased SARS-CoV-2 infection [1,2]. Of these, SARS-CoV-2 Beta variant is the most dreadful variant reported to emerge from South Africa in December 2020. However, its earliest detection was traced back to October 2020. It has been now the most prevalent lineage in South Africa and has also been reported from 115 countries [1]. Beta variant has 21 mutations with 9 spike protein mutations in the genome. The key mutations beyond N501Y are E484K, K417N, orf1b deletion in the Receptor binding domain (RBD) and L18F, D80A, D215G, Δ242–244, R264I, A701V in the N terminal domain [3]. Pearson et al. had estimated that the Beta variant could be highly transmissible than the earlier circulating strains of SARS-CoV-2. It has been observed that Beta variant accounted for about 40% of new SARS-CoV-2 infections compared to only 20% for Alpha variant. An in vitro study on the monoclonal antibodies and convalescent plasma samples of COVID-19 cases demonstrated reduced activity against Beta compared to Alpha [4]. The findings of the NVX-CoV2373 clinical trial have shown post hoc vaccine efficacy of 51% in South Africa where Beta variant was prevalent [5]. The ChAdOx1 nCoV-19 clinical trials results didn’t show protection against mild-moderate infection with Beta variant [6]. Besides this, Beta variant is also reported to be less susceptible to the currently available vaccines i.e., ChAdOx1 nCoV-19, mRNA-1273, BNT162b2, NVX-CoV2373 [[6], [7], [8], [9], [10]]. It is presumed that natural and vaccine-induced immunity would not protect against Beta variant.
With its worldwide occurrence, Beta variant has been an addition to the gruesome situation of the SARS-CoV-2 pandemic. The presence of Beta variant in India was initially detected from the clinical specimens of the South African travelers during late December 2020 [11]. With ongoing genomic surveillance, the country has also reported the presence of other SARS-CoV-2 variants of interest (VOI) such B.1.1.28.2, Eta (B.1.525) and Kappa (B.1.617.1) and the variants of concern (VOC) Alpha and Delta and neutralization potential with currently available vaccines in India [[11], [12], [13], [14], [15], [16], [17], [18], [19], [20]]. Recent study by Gupta et al. demonstrated major dominance of Delta variant, along with presence of Alpha, Kappa, Delta AY.1, and Delta AY.2 during the second wave in India [19].
With the emergence of all these new SARS-CoV-2 variants, a continuous effort for detection and isolation of SARS-CoV-2 variants was made from COVID-19 positive individuals who traveled from foreign countries to India. Here, we report the isolation and characterization of Beta variant from the United Arab Emirate (UAE) travelers who arrived in India.
Methods
Ethical approval
The study was approved by the Institutional Biosafety Committee and Institutional Human Ethics Committee of ICMR-NIV, Pune, India under project ‘Propagation of new SARS-CoV-2 variant isolate and characterization in cell culture and animal model’.
Clinical specimens
With ongoing process of SARS-CoV-2 screening of travelers from foreign countries to India, the oropharyngeal and nasopharyngeal swab specimens were collected from 58 individuals with travel history from UAE (n = 39), East, West and South Africa (n = 10), Qatar (n = 5), Ukraine (n = 3) and Saudi Arabia (n = 1) arrived at New Delhi International airport in India during February–March 2021. All the subjects were asymptomatic and found to be SARS-CoV-2 positive by real time RT-PCR (Table 1S) [21,22].
Virus isolation and titration
Vero CCL-81 cells were grown to confluent monolayer in 24-well plate maintained in Eagle’s Minimum essential medium (MEM) supplemented with 10 % fetal bovine serum (FBS) (HiMedia, Mumbai), penicillin (100 U/ml) and streptomycin (100 mg/ml). After decanting the growth medium, one hundred microliter volume of clinical specimens of 58 subjects were inoculated onto 24-well cell culture monolayer of Vero CCL-81. The cells were incubated for one hour at 37 °C to allow virus adsorption, with rocking every 10 min for uniform inoculum distribution. After the incubation, the inoculum was removed and the cells were washed with 1× phosphate-buffered saline (PBS). The MEM supplemented with two percent FBS was added to each well. The culture was incubated further in incubator at 37 °C with 5% CO2 and observed daily for cytopathic effects (CPEs) under an inverted microscope (Nikon, Eclipse Ti, Japan) [12]. The virus titration was carried out using the cell culture supernatants of the clinical specimens displaying CPE in cell culture. Median tissue culture infective dose (TCID50) value was calculated using the Reed and Muench method [23].
Genomic characterization of SARS-CoV-2 isolates
Genomic characterization of the virus isolates was carried out using Next-Generation Sequencing (NGS) with the quantified RNA. Briefly, the ribosomal RNA depletion was performed using Nebnext rRNA depletion kit (Human/mouse/rat) followed by cDNA synthesis using the first strand and second synthesis kit. The RNA libraries were prepared using TruSeq Stranded Total RNA library preparation kit. The amplified RNA libraries were quantified and loaded on the Illumina sequencing platform after normalization [24]. The reads generated from the machine were analyzed in CLC Genomics Workbench v20. The reference-based assembly method in the workbench was used to retrieve the SARS-CoV-2 sequences. Wuhan Hu-1 was taken as the reference sequence (NC_045512). The phylogenetic tree was generated using the MEGAX software (https://www.megasoftware.net/). The mismatches in the different isolates were obtained using the online highlighter tool (https://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html). Further the basic variant tool of the CLC genomics workbench was used to retrieve the variants and illustrated using the Graph Pad prism v 8 (https://www.graphpad.com/scientific-software/prism/).
Results
Virus isolation and titration
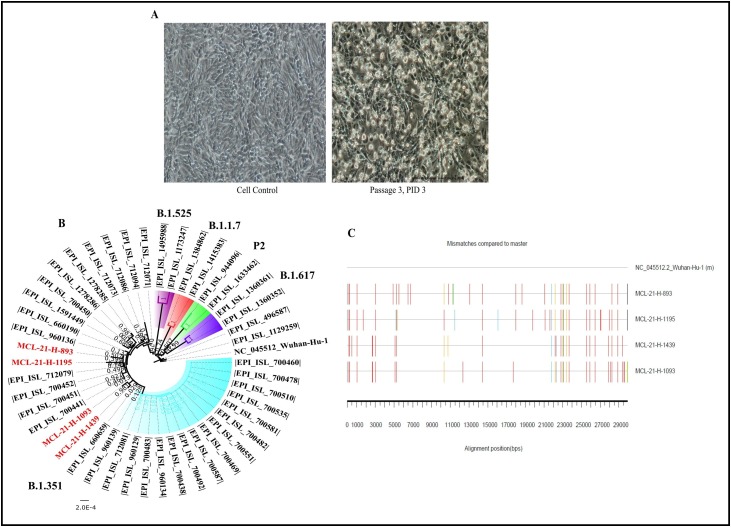
Cytopathic effect (CPE) was observed in four out of fifty-eight specimens with the syncytial formation on 2nd post-infection day (PID). The progressive infectivity was observed with the fusion of the infected cells with neighboring cells leading to the generation of the large mass of cells on 3rd PID (Fig. 1 A). The presence of the replication-competent virus was confirmed by Real-time RT-PCR that demonstrated a higher viral load of 4.6 × 109–1.8 × 1010 in the cell culture medium at passage 2 than inoculated specimens. Two plaque purified SARS-CoV-2 isolates demonstrated high viral RNA load of 3.8–7.5 × 1011 and 1.1–1.6 × 1011 at passage 4 and 5 respectively. Culture that shown CPE were centrifuged at 4815×g for 10 min at 4 °C; the supernatants were processed immediately or stored at −80 °C. The four virus isolates titrated at passage 3 demonstrated the virus titer of 105.5‒105.66 TCID50/ml respectively.
Fig. 1.
Isolation and bioinformatics analysis of SARS-CoV-2 Beta variant sequences: (A) cytopathic effect observed on the Vero CCL-81 cell line for the 3rd passage at 3rd PID along with the control (left). (B) Neighbor joining tree of the isolate sequences with reference sequences downloaded from GISAID with Tamura’s 3 Parameter model and a bootstrap replication of 1000 cycles. The sequences retrieved in study are colored red. (D) SARS-CoV-2 Beta variant sequences retrieved in this study are aligned with reference isolate of Wuhan-HU-1 (Accession No.: NC_045512.2). The nucleotide changes are generated using the highlighter plot available at https://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.htm.
The mismatches are marked in different colors adenine (A) green; cytosine (C) blue; guanine (G) orange, thymine (T) red and gap black color.
Clinical history of the cases
All four cases were male, asymptomatic, and belonged to the age group of 27, 32, 42, and 68 years (Table 1S). None of them had any co-morbid conditions. All these cases recovered completely with no new symptoms or complications. Currently, there is no evidence to suggest that this variant has caused any disease severity in these cases.
Genomic characterization of SARS-CoV-2 isolates
The neighbor-joining tree demonstrated a separate cluster each consisting of two SARS-CoV-2 sequences; cluster 1: MCL-21-H-1195 (EPI_ISL_2014135) and MCL-21-H-893 (EPI_ISL_2014131) and cluster 2: MCL-21-H-1093 (EPI_ISL_2014132) and MCL-21-H-1439 (EPI_ISL_2014133) (Fig. 1B). EPI_ISL_2014132 and EPI_ISL_2014133 shared common mutations at genomic positions A2692T, C5100T, G27870T, and C29358T. Fig. 1C depicts the nucleotide mismatches observed in the isolates. All the four SARS-CoV-2 isolate sequences retrieved had common spike mutation (D80A, D215G, L242_L244del, K417N, E484K, N501Y, D614G and A701V) (Table 2S). These isolates also have common mutations in the 5′ UTR (G174T, C241T), ORF1ab (C3037T) and ORF8 (28,253). The L18F and R246I amino acid mutations described by Tegally et al. [3], were absent in the spike protein in each isolate (Table 2S).
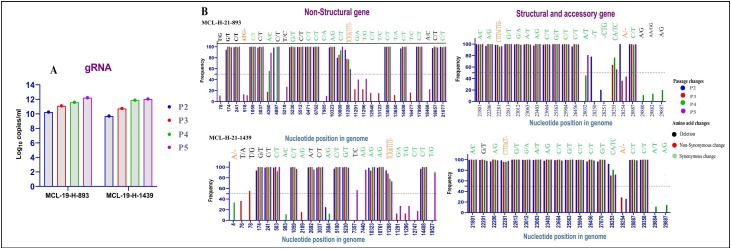
The variations in nucleotides for MCL-21-H-893 and MCL-21-H-1439 isolates were analyzed at different passages levels in Vero CCL-81 cell lines. The viral copy number at different passages is depicted in Fig. 2 A. Fig. 2B depicts the nucleotide changes over four passages observed in two isolates. Few quasi-species are observed in the two isolates majority of them being observed in 3′ UTR region. However, conservation in the spike nucleotide region in observed at different passage level of each isolate. This isolates also have unique nucleotide changes that are also conserved across genome.
Fig. 2.
Nucleotide changes of SARS-CoV-2 for two isolate in different genes: (A) the viral copy number for the isolates at passage 2–5 levels using real-time RT-PCR. (B) The nucleotide changes for the isolate MCL-19-H-893 is marked in the upper panel for passage 2–5 and for isolate MCL-19-H-1493 in the lower panel. The x axis represents the different passages and y axis depicts the frequency of their observation. A cut-off of 50% defines the consensus sequences nucleotide changes while below it are quasi-species for the isolates. Non-synonymous changes are marked in red color; synonymous changes are marked in green color; deletions of nucleotide are marked in black color.
The analysis of South African Beta lineage sequences demonstrated the presence of two different clusters having a deletion of nine nucleotides each in the ORF1ab (GP:11288–11296) and the spike (GP:22287–22295); while the second clusters with deletion of nine nucleotides in spike (GP:22287−22295) region. The percent nucleotide similarity demonstrated a 99.88–99.96% similarity of the isolates with the representative Beta lineage sequences (Table 3S).
Discussion
With its worldwide occurrence and probable immune escape, Beta variant has been an addition to the gruesome situation of the SARS-CoV-2 pandemic. The presence of Beta variant was first reported from India during December 2020 [11] Various SARS-CoV-2 variants i.e., Alpha, B.1.1.28.2, Eta, Kappa, Delta, Delta AY.1 have been detected from India [[12], [13], [14], [15], [16], [17], [18], [19], [20]]. The SARS-CoV-2 genomic surveillance was continued to detect the new SARS-CoV-2 variants specifically from the foreign travelers arrived in India. This has led to the isolation and characterization of Beta variant from COVID-19 positive individuals who traveled from UAE to India. The emergence of these new variants has been a serious threat to the COVID-19 vaccination program. Hence, it is necessary to determine the neutralizing capacity of the newly emerging SARS-CoV-2 VUI and VOC variants against the available vaccines. There has been many reports suggesting the reduction in the susceptibility of Beta variant to the currently available vaccines [8,10].
Beta variant is known to be highly transmissible SARS-CoV-2 variant. However, little information is available on the replication kinetics of the Beta variant. It is the subject of further exploration, whether the acquired mutations and in vitro/in vivo replication dynamics of Beta variant correlate with higher rate of transmissibility compared to other variants of concern. Pyke et al., have demonstrated quick replication of the Beta variant peaking at 48 h post infection with considerably higher levels of virus replication compared to Alpha variant [25]. Apparently, the affinity of Beta RBD binding to ACE2 receptors is also found to be 2.7 fold higher than that of Alpha variant [26,27]. The presence of triple mutation N501Y, E484K and K417N are likely to increase the interaction of RBD-ACE2 receptors and support the escape of the Beta variant from immune response. The studies on replication kinetics and genomic characterization of emerging SARS-CoV-2 variants would help to correlate their transmissibility potential. In summary, the available Beta variant isolate could be used to assess the efficacy of the currently available and upcoming COVID-19 vaccines in India. It will be also significant for developing new vaccine, diagnostic or antiviral testing in near future.
Conclusion
The recent emergence of SARS-CoV-2 variant has created major public health problem in the emerging country. This has ultimately hampered the globe with easy transmission of these variants with international travel. The second wave of COVID-19 pandemic has also raised main concern about the immune escape of these new variants against the currently approved vaccines. In depth studies are needed to ascertain the humoral response, cellular immune response and breakthrough infections post vaccination. The data generated from such studies would help in the development of new or tweaked vaccine with improved vaccine efficacy. To conclude, active surveillance of new SARS-CoV-2 variants and breakthrough infections would be required to monitor the genome evolution and vaccine efficacy.
Authors’ contributions
PDY and NG contributed to study design, data analysis, writing and critical review. PS, SP, SB and AK performed the laboratory experiments, interpretation, and data analysis. AS, RRS, DAN, DYP, HK, NA, VP, SM and AR contributed to data collection, interpretation, writing and critical review.
Funding
Financial support was provided by the Indian Council of Medical Research(ICMR), New Delhi at ICMR-National Institute of Virology, Pune under intramural funding ‘COVID-19’.
Competing interests
None declared.
Ethical approval
Not required.
Acknowledgments
The author gratefully acknowledges the contribution of Mr. Rajen Lakra, Mrs. Savita Patil, Mrs. Triparna Majumdar, Mr. Hitesh Dighe, Ms. Manisha Dudhmal, Mr. Yash Joshi, Mr. Vishwajit Dhanore from Maximum Containment Facility, ICMR-NIV, Pune. Authors would also like to acknowledge Prof. Priya Abraham, Director, ICMR-NIV, Pune for her constant support.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jiph.2021.12.011.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.PANGO lineages. https://cov-lineages.org/descriptions.html. [Accessed 25 May 2020].
- 2.Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 variants—clinical, public health, and vaccine implications. N Engl J Med. 2021;384(19):1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 4.Pearson C.A.B., Russel T.W., Davies N., Kucharski A.J. 2020. Estimates of severity and transmissibility of novel South Africa SARS-CoV-2 variant 501Y.V2.https://cmmid.github.io/topics/covid19/sa-novel-variant.html . [Accessed 25 May 2020] [Google Scholar]
- 5.Shinde V., Bhikha S., Hossain Z., Archary M., Bhorat Q., Fairlie L., et al. Preliminary efficacy of the NVX-CoV2373 Covid-19 vaccine against the B.1.351 variant. medRxiv. 2021 doi: 10.1101/2021.02.25.21252477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384(20):1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen X., Tang H., Pajon R., Smith G., Glenn G.M., Shi W., et al. Neutralization of SARS-CoV-2 variants B.1.429 and B.1.351. N Engl J Med. 2021;384(24):2352–2354. doi: 10.1056/NEJMc2103740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med. 2021;27(4):620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., et al. SARS-CoV-2 variants B.1.351 and P. 1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G.L., Wang Z.Y., Duan L.J., Meng Q.C., Jiang M.D., Cao J., et al. Susceptibility of circulating SARS-CoV-2 variants to neutralization. N Engl J Med. 2021;384(24):2354–2356. doi: 10.1056/NEJMc2103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yadav P.D., Nyayanit D.A., Sahay R.R., Shete A.M., Majumdar T., Patil S., et al. Imported SARS-CoV-2 V501Y. V2 variant (B.1.351) detected in travelers from South Africa and Tanzania to India. Travel Med Infect Dis. 2021;41 doi: 10.1016/j.tmaid.2021.102023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkale P., Patil S., Yadav P.D., Nyayanit D.A., Sapkal G., Baradkar S., et al. First isolation of SARS-CoV-2 from clinical samples in India. Indian J Med Res. 2020;151:244. doi: 10.4103/ijmr.IJMR_1029_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav P.D., Nyayanit D.A., Sahay R.R., Sarkale P., Pethani J., Patil S., et al. Isolation and characterization of the new SARS-CoV-2 variant in travellers from the United Kingdom to India: VUI-202012/01 of the B.1.1.7 lineage. J Travel Med. 2021;28(2):taab009. doi: 10.1093/jtm/taab009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yadav P.D., Nyayanit D.A., Majumdar T., Patil S., Kaur H., Gupta N., et al. An epidemiological analysis of SARS-CoV-2 genomic sequences from different regions of India. Viruses. 2021;13(5):925. doi: 10.3390/v13050925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yadav P., Sapkal G.N., Abraham P., Ella R., Deshpande G., Patil D.Y., et al. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. bioRxiv. 2021 doi: 10.1101/2021.04.23.441101. [DOI] [PubMed] [Google Scholar]
- 16.Sapkal G., Yadav P.D., Ella R., Abraham P., Patil D.Y., Gupta N., et al. Neutralization of B.1.1.28 P2 variant with sera of natural SARS-CoV-2 infection and recipients of inactivated COVID-19 vaccine Covaxin. J Travel Med. 2021 doi: 10.1093/jtm/taab077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yadav P., Sapkal G.N., Abraham P., Deshpande G., Nyayanit D., Patil D.Y., et al. Neutralization potential of Covishield vaccinated individuals sera against B.1.617.1. bioRxiv. 2021 doi: 10.1101/2021.05.12.443645. [DOI] [PubMed] [Google Scholar]
- 18.Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9(7):1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta N., Kaur H., Yadav P.D., Mukhopadhyay L., Sahay R.R., Kumar A., et al. Clinical characterization and genomic analysis of samples from COVID-19 breakthrough infections during the second wave among the various states of India. Viruses. 2021;13(9):1782. doi: 10.3390/v13091782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sapkal G.N., Yadav P.D., Ella R., Deshpande G.R., Sahay R.R., Gupta N., et al. Inactivated COVID-19 vaccine BBV152/COVAXIN effectively neutralizes recently emerged B 1.1. 7 variant of SARS-CoV-2. J Travel Med. 2021;28(4):taab051. doi: 10.1093/jtm/taab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choudhary M.L., Vipat V., Jadhav S., Basu A., Cherian S., Abraham P., et al. Development of in vitro transcribed RNA as positive control for laboratory diagnosis of SARS-CoV-2 in India. Indian J Med Res. 2020;151(2–3):251. doi: 10.4103/ijmr.IJMR_671_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 23.Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 24.Yadav P.D., Nyayanit D.A., Shete A.M., Jain S., Majumdar T.P., Chaubal G.Y., et al. Complete genome sequencing of Kaisodi virus isolated from ticks in India belonging to Phlebovirus genus, family Phenuiviridae. Ticks Tick Borne Dis. 2019;10(1):23–33. doi: 10.1016/j.ttbdis.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Pyke A.T., Nair N., van den Hurk A.F., Burtonclay P., Nguyen S., Barcelon J., et al. Replication kinetics of B.1.351 and B.1.1.7 SARS-CoV-2 variants of concern including assessment of a B.1.1.7 mutant carrying a defective ORF7a gene. Viruses. 2021;13(6):1087. doi: 10.3390/v13061087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dejnirattisai W., Zhou D., Ginn H.M., Duyvesteyn H.M., Supasa P., Case J.B., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184(8):2183–2200. doi: 10.1016/j.cell.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant from naturally acquired and vaccine induced antibody immunity. Cell. 2021;184:2201–2211. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.