Abstract
Proteases are involved in a broad range of physiological processes, including host invasion by fungal pathogens, and enzymatic inhibition is a key molecular mechanism controlling proteolytic activity. Importantly, inhibitors from natural or synthetic sources have demonstrated applications in biochemistry, biotechnology, and biomedicine. However, the need to discover new reservoirs of these inhibitory molecules with improved efficacy and target range has been underscored by recent protease characterization related to infection and antimicrobial resistance. In this regard, naturally-sourced inhibitors show promise for application in diverse biological systems due to high stability at physiological conditions and low cytotoxicity. Moreover, natural sources (e.g., plants, invertebrates, and microbes) provide a large reservoir of undiscovered and/or uncharacterized bioactive molecules involved in host defense against predators and pathogens. In this Review, we highlight discoveries of protease inhibitors from environmental sources, propose new opportunities for assessment of antifungal activity, and discuss novel applications to combat biomedically-relevant fungal diseases with in vivo and clinical purpose.
Keywords: proteases, protease inhibitors, fungal pathogens, natural compounds, biomedical applications, antimicrobial resistance
1. Introduction
Proteases hydrolyze the peptide bonds of polypeptides and proteins, with proteases accounting for 6% of total proteins in the human genome and 1–5% of microbial (e.g., bacteria, fungi, and virus) genomes [1]. Proteases are used by microorganisms in many processes, including stress response, nutrient acquisition, and protein maturation for cell division. Likewise, pathogens use these enzymes as important virulence factors in both direct and indirect damage of the host to: (i) gain access to nutrients [2]; (ii) destroy host cells and tissues to facilitate invasion and dissemination [3,4]; (iii) degrade host immune molecules for defense evasion [5,6,7]; (iv) promote pathogen propagation and maturation [8]; and (v) process self-molecules for pathogenicity [9,10]. Such roles promote the development of protease-based therapies [11] for pathogen-related diseases, including fungal meningitis [12], HIV/AIDS [13], candidiasis [14], aspergillosis [15], and COVID-19 [16].
Conversely, inhibition is one of the main molecular control mechanisms regulating proteolytic activity by which organisms use protease inhibitors to prevent self-damage [17], and provide protection against pathogens [18,19,20,21] or predators [22,23]. Currently, there are several protease inhibitors on the market for the management of human diseases, such as dabigatran and angiotensin converting enzyme inhibitors (ACEI) for the management of pulmonary embolism and hypertension, respectively [24,25]. Similarly, there are pharmaceuticals, such as bortezomib (clinically approved for the treatment of multiple myeloma by inhibition of proteasome complex) [26] with potential for applications against fungal pathogens. For example, in the widespread human fungal pathogen, Cryptococcus neoformans through regulation of virulence factor elaboration (i.e., polysaccharide capsule) [27,28]. However, such synthetic protease inhibitors can be plagued by low stability, high toxicity effects, or encounter resistance mechanisms, supporting the discovery of novel protease inhibitors from the natural environment [26,29]. Investigation of naturally-sourced protease inhibitors therefore, presents an alternative opportunity to expand our repertoire of antimicrobial agents and avoid such undesired features.
In this Review, we highlight the role of proteases related to fungal virulence and the impact of protease inhibition as an anti-virulence strategy. Next, we argue the benefits of naturally-derived protease inhibitors through presentation of representative examples derived from plants, invertebrates, and microbial sources with a focus on antifungal activity. Finally, we propose opportunities to expand our current repertoire of antifungals through discovery and characterization of naturally-sourced protease inhibitors with potential applications in emergent diseases. The goal is to aid researchers in finding effective strategies with greater target specificity that are less prone to the evolution of resistance.
2. Protease Inhibition Exerts Anti-Virulence Effects on Fungal Pathogens
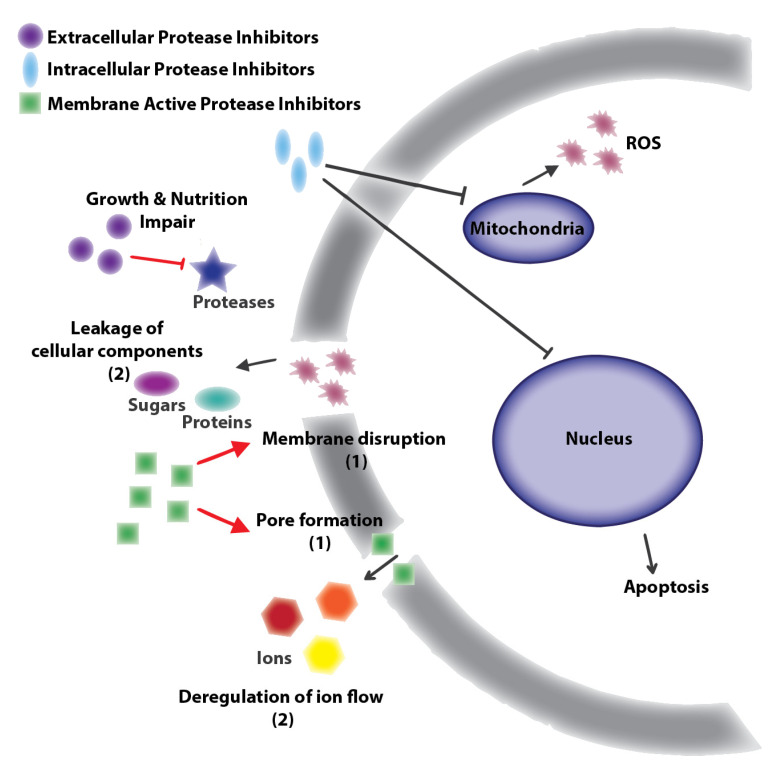
Several natural protease inhibitors exert anti-virulence effects by targeting extracellular proteases, impairing nutritional and/or growth functions [30,31,32], or hindering virulence mechanisms, such as tissue invasion (Figure 1) [33]. For instance, secreted aspartic proteases (SAPs), are involved in several virulence processes, including tissue invasion, growth, and immune system evasion among the important human fungal pathogens, Candida albicans and C. neoformans [34,35]. Additionally, SAPs have been assessed as antifungal targets using protease inhibitors with promising results for further exploration [36,37,38]. Other important anti-virulence mechanisms include cell wall disruption or membrane pore formation initiated by protease inhibitors to deregulate ion flow and/or membrane disruption to cause leakage of internal cellular components, affecting cell viability [30,31]. Further, endogenous, or intracellular fungal proteases are involved in important mechanisms, such as protein maturation for development or growth, and apoptosis regulation [39,40] and natural protease inhibitors have reported intracellular targets (e.g., mitochondria or nucleus), producing damage by oxidative stress or apoptosis deregulation, affecting pathogen survival [30,31,32]. Notably, C. neoformans uses intracellular proteases for resistance against current antifungal treatments (e.g., site-2 protease), which is required for virulence and survival in the presence of azole drugs [41]. Therefore, compounds capable of crossing fungal membranes and inhibiting endogenous proteases constitute potential antifungal agents and perhaps opportunities to overcome resistance. However, it is important to note that evidence of targeting intracellular organelles of fungal pathogens also poses a risk of off-target effects with toxicity towards human cells. Therefore, investigation into precise mechanism(s) of action and targets is needed to assess the potential and requirement for inhibitor optimization. Recognizing the promise of targeting proteases with protease inhibitors for treatment of fungal pathogens, we continue with the description, identification, and examples of naturally-sourced protease inhibitors.
Figure 1.
General targets of natural antifungal protease inhibitors: Protease inhibitors with extracellular targets produce nutrition or growth impairment by inhibition of nutrition related proteases [32,42,43,44]. Protease inhibitors with membrane cell targets cause disruption or pore formation leading to ion (e.g., Na+, K+, Ca2+ deregulation or leakage of cellular components [30,31]. Finally, protease inhibitors with intracellular targets inhibit mitochondria or nuclear proteases producing reactive oxygen species (ROS) or apoptosis [30,31]. Black lines correspond to antifungal compounds and red lines to molecules with similar antifungal or antibacterial effects.
3. Classification of Naturally-Derived Protease Inhibitors
Naturally-derived protease inhibitors are generally small molecules (15 to 60 amino acids or organic compounds) and contain a relatively high content of disulfide bridges, conferring higher stability [45,46]. They are classified according to enzymatic specificity, such as serine, aspartic, or cysteine protease inhibitors [47], or according to structural features. For instance, natural protease inhibitors can be classified as Bowman–Birk serine protease inhibitors, which are typically 1.5 to 20 kDa with several sulfide bridges, commonly displaying specific activity towards elastase, trypsin, and chymotrypsin [48,49]. Kunitz-type inhibitors, which are low molecular weight proteins with two or three disulfide bridges and one reactive site, showing specificity towards serine proteases [50]. Another example includes Kazal-type inhibitors, which are double-headed and inhibit trypsin and chymotrypsin simultaneously [51,52]. Compared to chemically synthesized products, natural inhibitors are often designated as safer with a specific mechanism of action, which leads to fewer off-target effects. This is a desirable trait for the development of novel antifungals based on the close evolutionary relationship between fungi and the mammalian host [53,54]. Additionally, natural compounds have evolved to possess physiochemical properties, including the ability to penetrate bacterial cells, unlike synthetic molecules not subject to such evolution. Although bacterial and fungal cells are highly distinct (e.g., cell wall composition, presence of organelles), there are several reports of protease inhibitors with biological activity against both types of cells, suggesting that protease inhibitors with antibacterial activity have the potential for similar properties against fungal cells [44,53,55,56,57,58]. Furthermore, the evolution towards resistance against environmentally-sourced protease inhibitors is often reduced given the drive by natural selection to interact with cellular targets with high efficiency and selectivity to avoid resistance and off-target effects [59]. Based on the variety of potential targets of protease inhibitors, and advantages afforded by naturally-occurring protease inhibitors, we explore examples derived from plants, invertebrates, and microbes.
4. Plant-Derived Protease Inhibitors
Natural compounds produced by plants are an important source of bioactive molecules with a wide range of biologic targets, including protease inhibitors with regulatory roles for endogenous proteases, storage, and defense [60,61,62,63]. Over the last 20 years, the number of identified plant-derived protease inhibitors with anti-virulence activity has increased, corresponding with a heightened importance in biomedicine (Table 1). Here, we outline inhibitor activity and provide insight into mechanisms of action and potential roles against fungal pathogens.
Table 1.
Protease inhibitors derived from plants with antimicrobial activity.
| Source | Protease Inhibitor Designation (Source) | Enzymatic Family | MW (kDa) | Activity (Mechanism of Action) |
Reference |
|---|---|---|---|---|---|
|
Fabaceae
(Leguminosae) |
IETI (Inga edulis) | Kunitz | 19.7 | Antifungal (Protease inhibition, membrane disruption and oxidative stress) | [31] |
| ILTI (Inga laurica) | 20 | [30] | |||
| ApTI (A, B, C) (Acacia plumosa) |
Kunitz | 20 | Antifungal (Secreted protease inhibition and nutrition impairment) | [32] | |
| API (Albizia amara) |
Unknown | 49 | Antifungal and Antibacterial | [55] | |
| Lupinine (Lupinus spp.) | Quinolizidine alkaloid | 0.17 | Anticryptococcal (secreted metallopeptidase inhibition) | [33] | |
| Diosgenin (Trigonella foenum-graecum) | Steroidal sapogenin | 0.41 | |||
| Solanaceae | Potide-G (S. tuberosum L. Cv. Golden Valley) |
Kunitz | 5.57 | Antibacterial and Antifungal (Secreted protease inhibition and nutrition impairment) | [44] |
| PG-2 (S. tuberosum L. Cv. Gogu Valley) | Kunitz | 3.2 | Antibacterial and Antifungal | [56] | |
| AFP-J (S. tuberosum L. Cv. L. Jopung) | Kunitz | 13.5 | Antifungal | [64] | |
| Rhamnaceae | RflP-1 (Rhamnus frangula) | Kunitz | 22.5 | Antibacterial and Antifungal |
[57,58] |
| Rutaceae | CLTI (Clausena lamsium) | Unknown | 54 | Anti-HIV-1 reverse transcriptase activity and Antifungal | [65] |
| Pinaceae | Abietic acid (Pinus spp.) | Abietane diterpenoid | 0.3 | Anticryptococcal (secreted metallopeptidase inhibition) | [33,66] |
MW: Molecular weight.
4.1. Fabaceae (Leguminosae) Family
Kunitz-type trypsin inhibitors, ILTI and IETI, were isolated from seeds of the tropical trees, Inga edulis and Inga laurica, respectively [30,31]. These inhibitors have antifungal activity, showing growth inhibition towards Candida tropicalis and Candida buinensis. This activity is mediated by several mechanisms, including protease inhibition, alteration of the plasma membrane causing ion flow deregulation, triggering of oxidative stress by a mitochondrial target, or triggering of apoptosis in yeasts that block important serine peptidases (e.g., metacaspases), and a nuclear mediator of apoptosis (Nma111p) [40,67]. Similarly, the Kunitz-type trypsin and chymotrypsin inhibitors, ApTIA, ApTIB, and ApTIC, isolated from seeds of the Brazilian plant Acacia plumosa possess antifungal activity against Aspergillus niger, Thielaviopsis paradoxa, and Colletotrichum sp. P10 is associated with inhibition of serine proteases secreted by the fungi in growth medium, impairing nutritional mechanisms [32]. Another example includes the protease inhibitor, API, which is derived from the seeds Albizia amara Boiv., possessing antibacterial activity against Pseudomonas aeruginosa and Bacillus subtilis [55] and antifungal activity against several pathogens, such as C. albicans with a minimal inhibitory concentration (MIC) value of 32 µg/mL (comparable to current antimicrobials). Although, the target and mechanisms of API have not been reported, the observed inhibitory roles and relative potency support further exploration against additional fungal pathogens or investigation of synergistic activity with known antifungals.
Lastly, Lupinine and Diosgenin are two plant derived compounds that possess antifungal properties against C. neoformans [33]. These compounds inhibit a secreted metallopeptidase relevant in brain invasion by cryptococcal cells causing meningoencephalitis, CnMpr-1 (Inhibitory concentration [IC50] 5.025 µM and 9.659 µM, respectively) [68]. Lupinine is a quinolizidine alkaloid found primarily within flowering plants of the Lupinus genus [69], whereas diosgenin is a plant steroidal sapogenin isolated from dietary fenugreek (Trigonella foenum-graecum) seeds [70]. Interestingly, these compounds impair fungal crossing of the blood–brain barrier without detrimental effects to the host [33]. Diosgenin also inhibits matrix metalloproteinases (e.g., MMP-2 and MMP-9) involved in matrix integrity or cell migration [71,72,73,74,75]. Together, these compounds highlight the potential of plant-derived sources for inhibition of proteases produced by cryptococcal cells and underscores an opportunity for synergistic assessment with known antifungals and extrapolation to additional fungal pathogens.
4.2. Solanaceae Family
Potatoes (Solanum tuberosum) are a worldwide food staple; however, their global distribution also contributes to pathogen spread, affecting crop quality, and productivity. Defense proteins and peptides with antifungal and antibacterial activities derived from potatoes represent a reservoir for disease protection against both agricultural and medical pathogens [76]. For example, the peptide, Potide-G, isolated from the tubers of the potato S. tuberosum L. Cv. Golden Valley, is a Kunitz-type serine protease inhibitor that inhibits growth of diverse pathogens, including C. albicans, Rhizoctonia solani, Staphylococcus aureus, and Listeria monocytogenes through regulation of extracellular enzymes related to nutrition [44]. Potide-G possesses MIC values less than 30 µg/mL, a similar potency to other plant protease inhibitors and known antibiotics [44]. Similarly, PG-2, a peptide isolated from potato tubers of cv. Gogu Valley exhibits antifungal and antibacterial activity against C. albicans, Clavibacter michiganensis ssp. michiganense, and S. aureus [56]. In addition, PG-2 exerts minimal cytotoxic effects against human red blood cells, making the compound an interesting option for further investigation of direct and indirect targets. Other protease inhibitors derived from potato tubers include AFP-J, a serine protease inhibitor belonging to the Kunitz family isolated from cv. L. Jopung [64]. This protein inhibits chymotrypsin, pepsin, and trypsin, possessing antifungal activity against several microorganisms, including C. albicans, Trichosporon beigelii, and Saccharomyces cerevisiae with antimicrobial potency (MIC 6.25 µg/mL) like other antibiotics, and with no known hemolytic activity. To date, no direct target has been reported for this compound, and therefore, these results support further investigation to define the mechanisms of action and to uncover additional pathogenic targets.
4.3. Rhamnaceae Family
Rhamnus frangula is a tall deciduous shrub in the family Rhamnaceae. Crude extracts of R. frangula leaves exhibit antioxidant, antimicrobial, and free radical scavenging activities with a Kunitz-type serine protease inhibitor, RflP-1, isolated from leaves. This inhibitor acts on serine proteases of commercial fungal, such as Aspergillus oryzae, and bacterial proteases isolated from B. licheniformis [57,58]. RflP-1 also possesses an appreciable antibacterial action against both Gram-positive and Gram-negative bacteria with similar effectiveness of ampicillin [57]. However, no direct targets have been identified to date, supporting exploration to define the mechanism of action and potential extrapolation to other pathogens.
4.4. Rutaceae Family
Clausena is a genus comprising approximately 14 species of evergreen trees and the Clausena lamsium trypsin inhibitor, CLTI, is a homodimer isolated from the seeds that exerts anti-HIV activity (i.e., Anti-HIV-1 reverse transcriptase activity) and antifungal activity against Physalospora piricola [65]. Importantly, no molecular targets have been described for CLT1 to explain the antifungal activity and, considering the common co-infection of C. neoformans within HIV/AIDS patients, this protease inhibitor, and its derivatives show promise for synergistic antifungal properties.
4.5. Pinaceae Family
Pinus is a genus of vascular plants, commonly known as pines possessing abietic acid, an abietane diterpenoid found primarily in pine resin with inhibitory properties against C. neoformans by blocking crossing of the blood–brain barrier through CnMpr-1 inhibition (IC50 5.143 µM) [33,77]. Similarly, some of the derivatives possess antimycotic and antibacterial activities [66], highlighting the potential of this compound as an important antifungal with broad reaching activity.
5. Invertebrate-Derived Protease Inhibitors
Invertebrates are a heterogeneous group of animals (about 1.3 million species) found ubiquitously within the environment, requiring strong defenses (e.g., production of chemicals) to adapt and survive against predators and pathogens, including protease inhibitors as self-defense systems [78,79]. For instance, many compounds with therapeutic potential detected from invertebrates show inhibition profiles against proteases with biotechnological and biomedical interest; although, many more remain to be studied [80,81,82,83]. Here, we present protease inhibitors derived from invertebrates and explore their described antimicrobial properties (Table 2).
Table 2.
Protease inhibitors derived from invertebrates with antimicrobial activity.
| Source | Protease Inhibitor Designation (Source) | Family/Chemical Class | MW (kDa) | Activity (Mechanism of Action) | Reference |
|---|---|---|---|---|---|
| Arthropoda | MjSerp1 (Marsupenaeus japonicas) | Serpin | 46.3 | Antibacterial | [84] |
| SWDPm2 (Penaeus monodon) | Type III crustin | 7.38 | [85] | ||
| BmoSPI51 (Bombyx mori) | Kunitz-type | 14 | Antifungal | [86] | |
| Mollusk | Peptides (Crassostrea gigas) |
Unknown | Unknown | HIV protease inhibitor (Competitive inhibition) | [87] |
MW: Molecular weight.
5.1. Arthropoda Phylum
Within Marsupenaeus japonicas (a shrimp), a serpin type protease inhibitor, MjSerp1, exhibits inhibitory activity against microbial serine proteases, such as subtilisin A and proteinase K and, also inhibits the growth of Gram-positive (e.g., S. aureus, B. subtilis, and Bacillus megaterium) and Gram-negative bacteria (e.g., Escherichia coli, Klebsiella pneumoniae, and Vibrio anguillarum) [84]. Similarly found within this phylum, are the single WAP (whey acidic protein) domain (SWD)-containing protein, SWDPm2, which is a Type III crustin isolated from the black tiger shrimp, Penaeus monodon [85]. This molecule is a potent competitive-type inhibitor of subtilisin A, a typical member of the S8 family, which is widely distributed among all kingdoms and in several human pathogens [88]. The primary functions of SWDPm2 include antimicrobial action and inhibition of bacterial peptidase to limit microbial infection and pathogenesis, as well as antibacterial activity against several Gram-positive bacteria (e.g., S. aureus, Aerococcus viridans, and B. megaterium). Although a mechanism of action remains to be defined, a potential for antifungal activity represents a new avenue of study as some human fungal pathogens, such as C. neoformans also use extracellular subtilisin-like proteases in their pathogenic mechanisms (e.g., Cerevisin and Pqp1) [35,89,90]. Lastly, BmoSPI51 is Kunitz-type trypsin inhibitor isolated from silkworm (Bombyx mori) cocoon with inhibitory growth properties against fungi, including S. cerevisiae and C. albicans [86,91]. Following fungal infection, BmoSPI51 production increases in B. mori supporting a role in immunity, such as protecting silk fibroin proteins from degradation by fungal enzymes [92]. Additionally, approximately 80 potential protease inhibitors from several families (e.g., TIL-type, Kunitz-type, and Kazal inhibitors) have been reported in the silkworm using genomic approaches [93], highlighting this organism as a rich source of new protease inhibitors with potential antifungal properties.
5.2. Mollusk Phylum
Mollusks present a wealth of natural compounds displaying antimicrobial activity, including 19 within the global marine pharmaceutical clinical pipeline and four approved by the US Food and Drug Administration to date [94]. Notably, over half of the secondary metabolites produced by mollusks have yet to be evaluated for bioactivity, representing a plethora of new avenues to pursue for in vitro, in vivo, and clinical studies [83,95]. For instance, protease inhibitors have been reported from oysters, such as Crassostrea gigas peptides, which are competitive inhibitors of HIV-1 protease with an inhibitory constant (ki) between 10 and 13 nM [87]. Inhibitory potency of these compounds is like the first generation of synthetic HIV-1 protease inhibitors, such as Indinavir, but lower than second generation options, such as Atazanavir (ki = 10 pM) [96,97]. Although the potency of these peptides can be improved through development and optimization of synthetic versions, the initial discovery and activity of naturally-produced compounds from mollusks shows great promise for new avenues of exploration. Furthermore, several HIV-1 protease inhibitors possess antifungal activity, mainly through inhibition of SAPs [98,99,100]; highlighting the potential of these peptides as future antifungal compounds and warranting further investigation.
6. Bacterial Protease Inhibitors
Protease inhibitors produced by microorganisms have protective roles against endogenous proteases. Conversely, secreted microbial protease inhibitors may modulate external proteolytic degradation to benefit the producer. For example, a microbe may secrete a protease inhibitor to regulate their own bacterial proteases (i.e., self-defense), defend against other microbes and infections, protect from predation, or in response to host proteases produced during invasion. Due to the importance of protease inhibitors produced by bacterial species, they have been extensively studied with the intent for developing novel therapeutic drugs [101,102]. Here, we highlight bacterial sources of protease inhibitors and discuss their relevance as antimicrobial strategies against other pathogens (Table 3).
Table 3.
Protease inhibitors derived from bacteria with antimicrobial activity.
| Source | Protease Inhibitor Designation (Source) | Family/Chemical Class | MW (kDa) | Activity (Mechanism of Action) | Ref. |
|---|---|---|---|---|---|
| Actinomycetaceae | Pepstatin A (Actinomycetes spp.) | Hexapeptide | 0.68 | Antifungal (Secreted protease inhibition) and HIV protease inhibitor | [42] |
| Bacillaceae | ATBI (Bacillus spp.) |
Heptapeptide | 1.1 | HIV-1 protease inhibitor (Competitive inhibition) | [103] |
MW: Molecular weight.
6.1. Actinomycetaceae Family
Pepstatin A is a microbial hexapeptide produced by Actinomycetes spp. and a potent inhibitor of almost all types of aspartic proteases, including SAPs [104]. This inhibitor modulates virulence of the SAP family and inhibits cell proliferation and adhesion to abiotic and biotic structures of Candida spp. showing promise as an antifungal therapeutic [42]. However, when administered intravenously, pepstatin A is ineffective in systemic infections, due to its unfavorable pharmacokinetic properties, underscoring the relevance and need for optimization [104,105,106]. Structural modifications may, therefore, present an opportunity for the design of novel potent and SAP inhibitors with antifungal properties.
6.2. Bacillaceae Family
Isolated from the extremophile Bacillus spp., ATBI, is a peptide and potent inhibitor of several aspartic proteases, including recombinant HIV-1 protease, pepsin, and fungal Aspergillus saitoi (F-Prot) aspartic protease [103]. ATBI binds within the active site of the HIV-1 protease (competitive inhibition), leading to inactivation of the enzyme, and thereby suggesting pharmaceutical potential as a drug for the treatment of AIDS. As described above, compounds such as HIV-1-protease inhibitors (e.g., indinavir or ritonavir) possess antifungal properties [36,37,38,99], highlighting the need for more research using ATBI against human fungal pathogens, such as Candida spp. and C. neoformans.
7. Future Directions and Conclusions
Our presentation of representative protease inhibitors derived from natural sources, including plants, invertebrates, and microbes underscores the immense potential of not only identifying and characterizing new natural compounds from these sources and others, but also, outlines opportunities for synthetic compound design based on informed observations. As identified here, important areas for further exploration include the search for natural compounds that mimic current synthetic compounds with anti-HIV activity. For example, the beneficial effects of anti-HIV protease inhibitors on the incidence of disease and the subsequent outcome of opportunistic fungal infections, such as candidiasis [36,37,98] and cryptococcosis [38,99,100,107,108]. This includes the off-target effects of anti-HIV aspartic protease inhibitors (e.g., saquinavir, indinavir and ritonavir) against hydrolytic enzymes (e.g., SAPs in C. albicans), which correspond with reduced fungal infections in HIV-infected patients [36,37,98,109]. Additionally, the HIV aspartic protease inhibitor, indinavir, selectively inhibits the production of proteases and urease by C. neoformans, interfering with capsule formation and resulting in heightened susceptibility of fungal cells to intracellular killing by natural effector cells [99]. In addition, prolonged incubation of C. neoformans with indinavir inhibits fungal growth, reducing virulence, and enhancing susceptibility to the endogenous antimicrobial activity of natural effector cells [108]. These unintended benefits of treating a viral infection led to increased host response and protection against fungal infections. Another avenue includes extrapolating the success of anti-bacterial protease inhibitors towards fungal proteases. For instance, the Euphorbiaceae family non-competitive trypsin inhibitor, JcTI-I demonstrates inhibitory activity against proteases from S. aureus and Salmonella enteric [43]. This inhibition is with high potency and low cytotoxicity making JcTI-I a pharmacologically interesting and valuable drug for the design of a novel antibiotic, but observations against fungal pathogens have not been reported [43].
Over the last 20 years, natural protease inhibitors and their biological activities have been reported from diverse sources (see Table 1, Table 2 and Table 3) with plants and invertebrates being rich reservoirs of compounds with biomedical applications. However, the isolation of microbes from the environment fails to capture the relationships among microbes within that environment (e.g., soil microbiome). Such interactions may drastically alter the production and abundance of proteins, as well as differences in protein profiles conveyed by microbes in the laboratory vs. the natural setting. This makes a comprehensive identification and appreciation of the intricacies of microbe–microbe interactions nearly impossible to replicate, suggesting that our current observations and discoveries are incomplete. Moreover, the selection criteria within clinical trials for efficacy, bioavailability, resistance, safety, and cost are critical to monitor and assess potential harmful outcomes for the host [29,96]. Considering the need for balanced specificity, which promotes potency of the inhibitor but allows for potential off-target effects, beneficial properties not previously anticipated (e.g., anti-HIV protease inhibitors described above) could therefore, be uncovered.
As emphasized in this Review, the potential for exploration of diverse protease inhibitors against fungal pathogens exposes our limited knowledge of defined mechanisms of antifungal activity. This Review provides insight into selectivity and off-target effects to move the described in vitro studies from the lab bench and into the clinic [110]. Finally, while we focus on protease inhibitors with relevance against biomedical fungal pathogens, opportunities and applications presented in this Review extend, through crosstalk and cross-reactivity of protease inhibitors, to the plethora of fungal pathogens currently impacting the agricultural sector and threatening global food security.
Acknowledgments
We thank members of the Geddes-McAlister lab and Jason A. McAlister for their critical reading and feedback on this manuscript.
Author Contributions
J.G.-M. and D.G.-G. conceptualized the topic. D.G.-G. prepared the first manuscript draft. D.G.-G. provided the figure. D.G.-G. and J.G.-M. edited and prepared the final manuscript version. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported, in part, by the University of Guelph, Canadian Foundation of Innovation (JELF 38798), New Frontiers Research Fund: Exploration, Banting Research Foundation—Jarislowsky Discovery Award, and Canadian Institutes of Health Research—Project Grant for J.G.-M.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of this manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barrett A.J. Bioinformatics of proteases in the MEROPS database. Curr. Opin. Drug Discov. 2004;7:334–341. [PubMed] [Google Scholar]
- 2.Siezen R.J., Leunissen J.A.M. Subtilases: The superfamily of subtilisin-like serine proteases. Protein Sci. 1997;6:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams C.M., Eckenroth B.E., Putnam E.E., Doublie S., Shen A. Structural and Functional Analysis of the CspB Protease Required for Clostridium Spore Germination. PLoS Pathog. 2013;9:e1003165. doi: 10.1371/journal.ppat.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L.C., Pirofski L.A., Casadevall A. Extracellular proteins of Cryptococcus neoformans and host antibody response. Infect. Immun. 1997;65:2599–2605. doi: 10.1128/iai.65.7.2599-2605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L., Blank E.S., Casadevall A. Extracellular Proteinase Activity of Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 1996;3:570–574. doi: 10.1128/cdli.3.5.570-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jansen H., Grenier D., Van der Hoeven J.S. Characterization of immunoglobulin G-degrading proteases of Prevotella intermedia and Prevotella nigrescens. Oral Microbiol. Immunol. 1995;10:138–145. doi: 10.1111/j.1399-302X.1995.tb00134.x. [DOI] [PubMed] [Google Scholar]
- 7.Kaminishi H., Miyaguchi H., Tamaki T., Suenaga N., Hisamatsu M., Mihashi I., Matsumoto H., Maeda H., Hagihara Y. Degradation of humoral host defense by Candida albicans proteinase. Infect. Immun. 1995;63:984–988. doi: 10.1128/iai.63.3.984-988.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A.C., Park Y., Tortorici M.A., Wall A., Mcguire A.T., Veesler D., Walls A.C., Park Y., Tortorici M.A., Wall A., et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coutard B., Valle C., De Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin- like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drag M., Salvesen G.S. Emerging principles in protease-based drug discovery. Nat. Rev. Drug Discov. 2010;9:690–701. doi: 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajasingham R., Smith R.M., Park B.J., Jarvis J.N., Govender N.P., Chiller T.M., Denning D.W., Loyse A., Boulware D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017;17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brik A., Wong C.-H. HIV-1 protease: Mechanism and drug discovery. Org. Biomol. Chem. 2003;1:5–14. doi: 10.1039/b208248a. [DOI] [PubMed] [Google Scholar]
- 14.Santos A.L.S., Braga-silva L.A. Aspartic Protease Inhibitors: Effective Drugs against the Human Fungal Pathogen Candida albicans. Med. Chem. (Los. Angeles) 2013;13:155–162. [PubMed] [Google Scholar]
- 15.Markaryan A., Morozova I., Yu H., Kolattukudy P.E. Purification and characterization of an elastinolytic metalloprotease from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect. Immun. 1994;62:2149–2157. doi: 10.1128/iai.62.6.2149-2157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemunaitis J., Stanbery L., Senzer N. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection: Let the virus be its own demise. Future Virol. 2020;15:381–395. doi: 10.2217/fvl-2020-0068. [DOI] [Google Scholar]
- 17.Neurath H. Proteolytic enzymes, past and future. Proc. Natl. Acad. Sci. USA. 1999;96:10962–10963. doi: 10.1073/pnas.96.20.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakao Y., Fusetani N. Enzyme Inhibitors from Marine Invertebrates. J. Nat. Prod. 2007;70:689–710. doi: 10.1021/np060600x. [DOI] [PubMed] [Google Scholar]
- 19.Sabotic J., Kos J. Microbial and fungal protease inhibitors—Current and potential applications. Appl. Microbiol. Biotechnol. 2012;93:1351–1375. doi: 10.1007/s00253-011-3834-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armstrong P.B., Quigley J.P. α2-macroglobulin: An evolutionarily conserved arm of the innate immune system. Dev. Comp. Immunol. 1999;23:375–390. doi: 10.1016/S0145-305X(99)00018-X. [DOI] [PubMed] [Google Scholar]
- 21.Xue Q. Pathogen proteases and host protease inhibitors in molluscan infectious diseases. J. Invertebr. Pathol. 2019;166:107214. doi: 10.1016/j.jip.2019.107214. [DOI] [PubMed] [Google Scholar]
- 22.Singh S., Singh A., Kumar S., Mittal P., Singh I.K. Protease inhibitors: Recent advancement in its usage as a potential biocontrol agent for insect pest management. Insect Sci. 2020;27:186–201. doi: 10.1111/1744-7917.12641. [DOI] [PubMed] [Google Scholar]
- 23.Sabotič J., Bleuler-Martinez S., Renko M., Caglič P.A., Kallert S., Štrukelj B., Turk D., Aebi M., Kos J., Künzler M. Structural basis of trypsin inhibition and entomotoxicity of cospin, serine protease inhibitor involved in defense of Coprinopsis cinerea fruiting bodies. J. Biol. Chem. 2012;287:3898–3907. doi: 10.1074/jbc.M111.285304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Nisio M., Middeldorp S., Büller H.R. Direct thrombin inhibitors. N. Engl. J. Med. 2005;353:1028–1040. doi: 10.1056/NEJMra044440. [DOI] [PubMed] [Google Scholar]
- 25.Agbowuro A.A., Huston W.M., Gamble A.B., Tyndall J.D.A. Proteases and protease inhibitors in infectious diseases. Med. Res. Rev. 2017:1–37. doi: 10.1002/med.21475. [DOI] [PubMed] [Google Scholar]
- 26.Merin N.M., Kelly K.R. Clinical use of proteasome inhibitors in the treatment of multiple myeloma. Pharmaceuticals. 2015;8:1–20. doi: 10.3390/ph8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geddes J.M.H., Caza M., Croll D., Stoynov N., Foster L.J., Kronstad J.W. Analysis of the protein kinase a-regulated proteome of Cryptococcus neoformans identifies a role for the ubiquitin-proteasome pathway in capsule formation. MBio. 2016;7:1–15. doi: 10.1128/mBio.01862-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geddes J.M.H., Croll D., Caza M., Stoynov N., Foster L.J., Kronstad J.W. Secretome profiling of Cryptococcus neoformans reveals regulation of a subset of virulence-associated proteins and potential biomarkers by protein kinase A. BMC Microbiol. 2015;15:1–26. doi: 10.1186/s12866-015-0532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pawar S.D., Freas C., Weber I.T., Harrison R.W. Analysis of drug resistance in HIV protease. BMC Bioinform. 2018;19:362. doi: 10.1186/s12859-018-2331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macedo M.L., Ribeiro S.F.F., Taveira G.B., Gomes V., De Barros K.M.C.A., Maria-Neto S. Antimicrobial Activity of ILTI, a Kunitz-Type Trypsin Inhibitor from Inga laurina (SW.) Willd. Curr. Microbiol. 2016;72:538–544. doi: 10.1007/s00284-015-0970-z. [DOI] [PubMed] [Google Scholar]
- 31.Dib H.X., De Oliveira D.G.L., De Oliveira C.F.R., Taveira G.B., Mello E.D.O., Verbisck N.V., Chang M., Junior D.C., Gomes V.M., Macedo M.L.R. Biochemical characterization of a Kunitz inhibitor from Inga edulis seeds with antifungal activity against Candida spp. Arch. Microbiol. 2019;201:223–233. doi: 10.1007/s00203-018-1598-8. [DOI] [PubMed] [Google Scholar]
- 32.Lopes J.L.S., Valadares N.F., Moraes D.I., Rosa J.C., Araújo H.S.S., Beltramini L.M. Physico-chemical and antifungal properties of protease inhibitors from Acacia plumosa. Phytochemistry. 2009;70:871–879. doi: 10.1016/j.phytochem.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Aaron P.A., Vu K., Gelli A. An Antivirulence Approach for Preventing Cryptococcus neoformans from Crossing the Blood-Brain Barrier via Novel Natural Product Inhibitors of a Fungal Metalloprotease. MBio. 2020;11:e01249-20. doi: 10.1128/mBio.01249-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naglik J.R., Challacombe S.J., Hube B. Candida albicans Secreted Aspartyl Proteinases in Virulence and Pathogenesis. Microbiol. Mol. Biol. Rev. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clarke S.C., Dumesic P.A., Homer C.M., O’Donoghue A.J., La Greca F., Pallova L., Majer P., Madhani H.D., Craik C.S. Integrated Activity and Genetic Profiling of Secreted Peptidases in Cryptococcus neoformans Reveals an Aspartyl Peptidase Required for Low pH Survival and Virulence. PLoS Pathog. 2016;12:e1006051. doi: 10.1371/journal.ppat.1006051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eldesouky H.E., Salama E.A., Lanman N.A., Hazbun T.R. Potent Synergistic Interactions between Lopinavir and Azole Antifungal Drugs against Emerging Multidrug-Resistant Candida auris. Antimicrob. Agents Chemother. 2021;65:e00684-20. doi: 10.1128/AAC.00684-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos A., Braga-Silva L., Gonçalves D., Ramos L., Oliveira S., Souza L., Oliveira V., Lins R., Pinto M., Muñoz J., et al. Repositioning Lopinavir, an HIV Protease Inhibitor, as a Promising Antifungal Drug: Lessons Learned from Candida albicans—In Silico, In Vitro and In Vivo Approaches. J. Fungi. 2021;7:424. doi: 10.3390/jof7060424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kryštůfek R., Ŝácha P., Starková J., Brynda J., Hradilek M., Tloušt’ová E., Grzymska J., Rut W., Boucher M.J., Drag M., et al. Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery. J. Med. Chem. 2021;64:6706–6719. doi: 10.1021/acs.jmedchem.0c02177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fahrenkrog B., Sauder U., Aebi U. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J. Cell Sci. 2004;117:115–126. doi: 10.1242/jcs.00848. [DOI] [PubMed] [Google Scholar]
- 40.Carmona-Gutierrez D., Eisenberg T., Büttner S., Meisinger C., Kroemer G., Madeo F. Apoptosis in yeast: Triggers, pathways, subroutines. Cell Death Differ. 2010;17:763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- 41.Bien C.M., Chang Y.C., Nes W.D., Kwon-chung K.J., Espenshade P.J. Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Mol. Microbiol. 2009;74:672–690. doi: 10.1111/j.1365-2958.2009.06895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cutfield S.M., Dodson E.J., Anderson B.F., Moody P.C.E., Marshall C.J., Sullivan P.A., Cutfield J.F. The crystal structure of a major secreted aspartic proteinase from Candida albicans in complexes with two inhibitors. Curr. Biol. 1995;3:1261–1271. doi: 10.1016/S0969-2126(01)00261-1. [DOI] [PubMed] [Google Scholar]
- 43.Costa H.P.S., Oliveira J.T.A., Sousa D.O.B., Morais J.K.S., Moreno F.B., Monteiro-Moreira A.C.O., Viegas R.A., Vasconcelos I.M. JcTI-I: A novel trypsin inhibitor from Jatropha curcas seed cake with potential for bacterial infection treatment. Front. Microbiol. 2014;5:5. doi: 10.3389/fmicb.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim M.-H., Park S.-C., Kim J.-Y., Lee S.Y., Lim H.-T., Cheong H., Hahm K.-S., Park Y. Purification and characterization of a heat-stable serine protease inhibitor from the tubers of new potato variety “Golden Valley”. Biochem. Biophys. Res. Commun. 2006;346:681–686. doi: 10.1016/j.bbrc.2006.05.186. [DOI] [PubMed] [Google Scholar]
- 45.Torres-Castillo J.A., Jacobo C.M., Blanco-Labra A. Characterization of a highly stable trypsin-like proteinase inhibitor from the seeds of Opuntia streptacantha (O. streptacantha Lemaire) Phytochemistry. 2009;70:1374–1381. doi: 10.1016/j.phytochem.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 46.Monteiro Júnior J.E., Valadares N.F., Pereira H.D., Dyszy F.H., da Costa Filho A.J., Uchôa A.F., de Oliveira A.S., da Silveira Carvalho C.P., Grangeiro T.B. Expression in Escherichia coli of cysteine protease inhibitors from cowpea (Vigna unguiculata): The crystal structure of a single-domain cystatin gives insights on its thermal and pH stability. Int. J. Biol. Macromol. 2017;102:29–41. doi: 10.1016/j.ijbiomac.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Losso J.N. The Biochemical and Functional Food Properties of the Bowman-Birk. Crit. Rev. Food Sci. Nutr. 2008;48:94–118. doi: 10.1080/10408390601177589. [DOI] [PubMed] [Google Scholar]
- 49.Qi R., Song Z., Chi C. Structural features and molecular evolution of Bowman-Birk protease inhibitors and their potential application. Acta Biochim. Biophys. Sin. (Shanghai) 2005;37:283–292. doi: 10.1111/j.1745-7270.2005.00048.x. [DOI] [PubMed] [Google Scholar]
- 50.Habib H., Fazili K.M. Plant protease inhibitors: A defense strategy in plants. Biotechnol. Mol. Biol. Rev. 2007;2:68–85. [Google Scholar]
- 51.Mistry R., Snashall P.D., Totty N., Briskin S., Guz A., Tetley T.D. Purification and characterization of a novel Kazal-type serine proteinase inhibitor of neutrophil elastase from sheep lung. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 1997;1342:51–61. doi: 10.1016/S0167-4838(97)00086-1. [DOI] [PubMed] [Google Scholar]
- 52.Rustgi S., Boex-Fontvieille E., Reinbothe C., von Wettstein D., Reinbothe S. The complex world of plant protease inhibitors Insights into a Kunitz type cysteine protease inhibitor of Arabidopsis thaliana. Commun. Integr. Biol. 2018;11:e1368599. doi: 10.1080/19420889.2017.1368599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Shea R., Moser H.E. Physicochemical properties of antibacterial compounds: Implications for drug discovery. J. Med. Chem. 2008;51:2871–2878. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- 54.Hussain I., Hawkins J., Harrison D., Hille C., Wayne G., Cutler L., Buck T., Walter D., Demont E., Howes C., et al. Oral administration of a potent and selective non-peptidic BACE-1 inhibitor decreases β-cleavage of amyloid precursor protein and amyloid-β production in vivo. J. Neurochem. 2007;100:802–809. doi: 10.1111/j.1471-4159.2006.04260.x. [DOI] [PubMed] [Google Scholar]
- 55.Dabhade A.R., Mokashe N.U., Patil U.K. Purification, characterization, and antimicrobial activity of nontoxic trypsin inhibitor from Albizia amara Boiv. Process Biochem. 2016 doi: 10.1016/j.procbio.2016.02.015. [DOI] [Google Scholar]
- 56.Kim J.-Y., Gopal R., Kim S.Y., Seo C.H., Lee H.B., Cheong H., Park Y. PG-2, a potent AMP against pathogenic microbial strains, from potato (Solanum tuberosum L cv. Gogu Valley) tubers not cytotoxic against human cells. Int. J. Mol. Sci. 2013;14:4349–4360. doi: 10.3390/ijms14024349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bacha A.B., Jemel I., Moubayed N.M.S., Ben I. Purification and characterization of a newly serine protease inhibitor from Rhamnus frangula with potential for use as therapeutic drug. 3 Biotech. 2017;7:1–13. doi: 10.1007/s13205-017-0764-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manojlovic N.T., Solujic S., Sukdolak S., Milosev M. Antifungal activity of Rubia tinctorum, Rhamnus frangula and Caloplaca cerina. Fitoterapia. 2005;76:244–246. doi: 10.1016/j.fitote.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 59.Wright G.D. Opportunities for natural products in 21st century for antibiotic disease. Nat. Prod. Reports. 2017;34:694–701. doi: 10.1039/C7NP00019G. [DOI] [PubMed] [Google Scholar]
- 60.Kim J., Park S., Hwang I., Cheong H. Protease Inhibitors from Plants with Antimicrobial Activity. Int. J. Mol. Sci. 2009;10:2860–2872. doi: 10.3390/ijms10062860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valueva T.A., Mosolov V. V Role of inhibitors of proteolytic enzymes in plant defense against phytopathogenic microorganisms. Biochemical. 2004;69:1305–1309. doi: 10.1007/s10541-005-0015-5. [DOI] [PubMed] [Google Scholar]
- 62.Lawrence P.K., Koundal K.R. Plant protease inhibitors in control of phytophagous insects. Electron. J. Biotechnol. 2002;5:5–6. doi: 10.2225/vol5-issue1-fulltext-3. [DOI] [Google Scholar]
- 63.Ryan C.A. Protease inhibitors in plants: Genes for improving defenses against insects and pathogens. Annu. Rev. Phytopathol. 1990;28:425–449. doi: 10.1146/annurev.py.28.090190.002233. [DOI] [Google Scholar]
- 64.Park Y., Choi B.H., Kwak J.-S., Kang C.-W., Lim H.-T., Cheong H.-S., Hahm K.-S. Kunitz-type serine protease inhibitor from potato (Solanum tuberosum L. cv. Jopung) J. Agric. Food Chem. 2005;53:6491–6496. doi: 10.1021/jf0505123. [DOI] [PubMed] [Google Scholar]
- 65.Ng T.B., Lam S.K., Fong W.P. A Homodimeric Sporamin-Type Trypsin Inhibitor with Antiproliferative, HIV Reverse Transcriptase-Inhibitory and Antifungal Activities from Wampee (Clausena lansium) Seeds. Biol. Chem. 2003;384:289–293. doi: 10.1515/BC.2003.032. [DOI] [PubMed] [Google Scholar]
- 66.Gonzalez M., Perez-Guaita D., Correa-Royero J., Zapata B., Agudelo L., Mesa-Arango A.B.-G.L. Synthesis and biological evaluation of dehydroabietic acid derivatives. Eur. J. Med. Chem. 2010;45:811–816. doi: 10.1016/j.ejmech.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 67.Madeo F., Fröhlich E., Ligr M., Grey M., Sigrist S.J., Wolf D.H., Fröhlich K.U. Oxygen stress: A regulator of apoptosis in yeast. J. Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vu K., Tham R., Uhrig J., Thompson G.R., Na Pombejra S., Jamklang M., Bautos J.M., Gelli A. Invasion of the Central Nervous System by Cryptococcus neoformans Requires a Secreted Fungal Metalloprotease. mBio. 2014;5:e01101-14. doi: 10.1128/mBio.01101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drummond C.S., Eastwood R.J., Miotto S.T.S., Hughes C.E. Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): Testing for key innovation with incomplete taxon sampling. Syst. Biol. 2012;61:443–460. doi: 10.1093/sysbio/syr126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sethi G., Shanmugam M.K., Warrier S., Merarchi M., Arfuso F., Kumar A.P., Bishayee A. Pro-apoptotic and anti-cancer properties of diosgenin: A comprehensive and critical review. Nutrients. 2018;10:645. doi: 10.3390/nu10050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Endo K., Takino T., Miyamori H., Kinsen H., Yoshizaki T., Furukawa M., Sato H. Cleavage of syndecan-1 by membrane type matrix metalloproteinase-1 stimulates cell migration. J. Biol. Chem. 2003;278:40764–40770. doi: 10.1074/jbc.M306736200. [DOI] [PubMed] [Google Scholar]
- 72.Chen P., Shih Y., Huang H., Cheng H. Diosgenin, a steroidal saponin, inhibits migration and invasion of human prostate cancer PC-3 cells by reducing matrix metalloproteinases expression. PLoS ONE. 2011;6:e20164. doi: 10.1371/journal.pone.0020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y., Aoki T., Mori Y., Ahmad M., Miyamori H., Takino T., Sato H. Cleavage of lumican by membrane-type matrix metalloproteinase-1 abrogates this proteoglycan-mediated suppression of tumor cell colony formation in soft agar. Cancer Res. 2004;64:7058–7064. doi: 10.1158/0008-5472.CAN-04-1038. [DOI] [PubMed] [Google Scholar]
- 74.Koshikawa N., Giannelli G., Cirulli V., Miyazaki K., Quaranta V. Role of cell surface metalloprotease MT1-MMP in epithelial cell migration over laminin-5. J. Cell Biol. 2000;148:615–624. doi: 10.1083/jcb.148.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ratnikov B.I., Rozanov D.V., Postnova T.I., Baciu P.G., Zhang H., DiScipio R.G., Chestukhina G.G., Smith J.W., Deryugina E.I., Strongin A.Y. An alternative processing of integrin αv subunit in tumor cells by membrane type-1 matrix metalloproteinase. J. Biol. Chem. 2002;277:7377–7385. doi: 10.1074/jbc.M109580200. [DOI] [PubMed] [Google Scholar]
- 76.Bártová V. Antifungal and antimicrobial proteins and peptides of potato (Solanum tuberosum L.) tubers and their applications. Appl. Microbiol. Biotechnol. 2019;103:5533–5547. doi: 10.1007/s00253-019-09887-9. [DOI] [PubMed] [Google Scholar]
- 77.Wakamiya I., Newton R.J., Johnston J.S., Price H.J. Genome size and environmental factors in the genus Pinus. Am. J. Bot. 1993;80:1235–1241. doi: 10.1002/j.1537-2197.1993.tb15360.x. [DOI] [Google Scholar]
- 78.Guan R., Mariuzza R.A. Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol. 2007;15:127–134. doi: 10.1016/j.tim.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 79.Summer K., Browne J., Liu L. Molluscan Compounds Provide Drug Leads for the Treatment and Prevention of Respiratory Disease. Mar. Drugs. 2020;18:570. doi: 10.3390/md18110570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Molinski T.F., Dalisay D.S., Lievens S.L., Saludes J.P. Drug development from marine natural products. Nat. Rev. 2008;8:69–85. doi: 10.1038/nrd2487. [DOI] [PubMed] [Google Scholar]
- 81.Cragg G.M., Newman D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta (BBA)-General Subj. 2013;1830:3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dang V.T., Benkendorff K., Green T. Marine Snails and Slugs: A Great Place To Look for Antiviral Drugs. J. Virol. 2015;89:8114–8118. doi: 10.1128/JVI.00287-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.González L., Sánchez R.E., Rojas L., Pascual I., García-fernández R., Chávez M.A., Betzel C. Screening of Protease Inhibitory Activity in Aqueous Extracts of Marine Invertebrates from Cuban Coast. Am. J. Anal. Chem. 2016;7:319–331. doi: 10.4236/ajac.2016.74030. [DOI] [Google Scholar]
- 84.Zhao Y., Xu Y., Jiang H., Xu S., Zhao X., Wang J. Antibacterial activity of serine protease inhibitor 1 from kuruma shrimp Marsupenaeus japonicus. Dev. Comp. Immunol. 2014;44:261–269. doi: 10.1016/j.dci.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 85.Amparyup P., Donpudsa S., Tassanakajon A. Shrimp single WAP domain (SWD)-containing protein exhibits proteinase inhibitory and antimicrobial activities. Dev. Comp. Immunol. 2008;32:1497–1509. doi: 10.1016/j.dci.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 86.Zhang X., Guo K., Dong Z., Chen Z., Zhu H., Zhang Y., Xia Q., Zhao P. Kunitz-type protease inhibitor BmSPI51 plays an antifungal role in the silkworm cocoon. Insect Biochem. Mol. Biol. 2020;116:103258. doi: 10.1016/j.ibmb.2019.103258. [DOI] [PubMed] [Google Scholar]
- 87.Lee T., Maruyama S. Isolation of HIV-1 Protease-Inhibiting Peptides from Thermolysin Hydrolysate of Oyster Proteins. Biochem. Biophys. Res. Commun. 1998;253:604–608. doi: 10.1006/bbrc.1998.9824. [DOI] [PubMed] [Google Scholar]
- 88.Gongora D.G., Perez L.R., Muñoz A.C., del Rivero Antigua M.A. Biomedical and Biological relevance of subtilases from the S8 family, diverse and widely distributed serine peptidases. Rev. Cuba. Cienc. Biológicas. 2021;9:1–13. [Google Scholar]
- 89.Eigenheer R.A., Lee Y.J., Blumwald E., Phinney B.S., Gelli A. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Res. 2007;7:499–510. doi: 10.1111/j.1567-1364.2006.00198.x. [DOI] [PubMed] [Google Scholar]
- 90.Homer C.M.M., Summers D.K., Goranov A.I., Nielsen K., Craik C.S.S., Madhani H.D.D. Intracellular Action of a Secreted Peptide Required for Fungal Virulence. Cell Host Microbe. 2016;19:849–864. doi: 10.1016/j.chom.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang X., Ni Y., Guo K., Dong Z., Chen Y., Zhu H., Xia Q., Zhao P. The mutation of SPI51, a protease inhibitor of silkworm, resulted in the change of antifungal activity during domestication. Int. J. Biol. Macromol. 2021;178:63–70. doi: 10.1016/j.ijbiomac.2021.02.076. [DOI] [PubMed] [Google Scholar]
- 92.Kurioka A., Yamazaki M., Hirano H. Primary structure and possible functions of a trypsin inhibitor of Bombyx mori. Eur. J. Biochem. 1999;259:120–126. doi: 10.1046/j.1432-1327.1999.00030.x. [DOI] [PubMed] [Google Scholar]
- 93.Zhao P., Dong Z., Duan J., Wang G., Wang L., Li Y., Xiang Z., Xia Q. Genome-wide identification and immune response analysis of serine protease inhibitor genes in the silkworm, Bombyx mori. PLoS ONE. 2012;7:e31168. doi: 10.1371/journal.pone.0031168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abadines I.B., Le K., Newman D.J., Glaser K.B., Mayer A.M. The marine pharmacology and pharmaceuticals pipeline in 2018. FASEB J. 2019;33:501–504. doi: 10.1096/fasebj.2019.33.1_supplement.504.1. [DOI] [Google Scholar]
- 95.Benkendor K. Chemical diversity in molluscan communities: From natural products to chemical ecology. In: Cosmo A.D., Winlow W., editors. Neuroecology and Neuroethology in Molluscs: The Interface between Behaviour and Environment. Nova Science Publishers; New York, NY, USA: 2014. pp. 13–41. [Google Scholar]
- 96.Abbenante G., Fairlie D.P. Protease Inhibitors in the Clinic. Med. Chem. (Los. Angeles) 2005;1:71–104. doi: 10.2174/1573406053402569. [DOI] [PubMed] [Google Scholar]
- 97.Randolph J.T., DeGoey D.A. Peptidomimetic inhibitors of HIV protease. Curr. Top. Med. Chem. 2004;4:1079–1095. doi: 10.2174/1568026043388330. [DOI] [PubMed] [Google Scholar]
- 98.Cassone A., De Bernardis F., Torosantucci A., Tacconelli E., Tumbarello M., Cauda R. In Vitro and In Vivo Anticandidal Activity of Human Immunodeficiency Virus Protease Inhibitors. J. Infect. Dis. 1999;180:448–453. doi: 10.1086/314871. [DOI] [PubMed] [Google Scholar]
- 99.Blasi E., Colombari B., Francesca C., Pinti M., Troiano L., Cossarizza A., Esposito R., Peppoloni S., Mussini C., Neglia R. The human immunodeficiency virus (HIV) protease inhibitor indinavir directly affects the opportunistic fungal pathogen Cryptococcus neoformans. FEMS Immunol. Med. Microbiol. 2004;42:187–195. doi: 10.1016/j.femsim.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 100.Manfredi R., Calza L., Chiodo F. AIDS-associated Cryptococcus infection before and after the highly active antiretroviral therapy era: Emerging management problems. Int. J. Antimicrob. Agents. 2003;22:449–452. doi: 10.1016/S0924-8579(03)00113-4. [DOI] [PubMed] [Google Scholar]
- 101.Quintero D., Bermudes D. A culture-based method for determining the production of secreted protease inhibitors. J. Microbiol. Methods. 2014;100:105–110. doi: 10.1016/j.mimet.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Supuran C.T., Scozzafava A., Clare B.W. Bacterial Protease Inhibitors. Med. Res. Rev. 2002;22:329–372. doi: 10.1002/med.10007. [DOI] [PubMed] [Google Scholar]
- 103.Vathipadiekal V., Umasankar P.K., Patole M.S., Rao M. Molecular cloning, over expression, and activity studies of a peptidic HIV-1 protease inhibitor: Designed synthetic gene to functional recombinant peptide. Peptides. 2010;31:16–21. doi: 10.1016/j.peptides.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 104.Wolber G., Hell M., Heinrich I.E., Cadicamo C.D., Michel D., Semlin L., Berger U., Korting H.C., Ho H., Koksch B., et al. Design, synthesis, inhibition studies, and molecular modeling of pepstatin analogues addressing different secreted aspartic proteinases of Candida albicans. Biochem. Pharmacol. 2013;85:881–887. doi: 10.1016/j.bcp.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 105.Bondaryk M., Staniszewska M., Zielinska P., Urbanczyk-Lipkowska Z., Zieli P. Natural Antimicrobial Peptides as Inspiration for Design of a New Generation Antifungal Compounds. J. Fungi. 2017;3:46. doi: 10.3390/jof3030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ruchel R., Ritter B. Modulation of experimental systemic murine candidosis by intravenous pepstatin. Zentralbl. Bakterio. 1996;273:391–403. doi: 10.1016/S0934-8840(11)80443-3. [DOI] [PubMed] [Google Scholar]
- 107.Sidrim J.J.C., Perdigão-Neto L.V., Cordeiro R.A., Brilhante R.S.N., Leite J.J.G., Teixeira C.E.C., Monteiro A.J., Freitas R.M.F., Ribeiro J.F., Mesquita J.R.L., et al. Viral protease inhibitors affect the production of virulence factors in Cryptococcus neoformans. Can. J. Microbiol. 2012;58:932–936. doi: 10.1139/w2012-075. [DOI] [PubMed] [Google Scholar]
- 108.Monari C., Pericolini E., Bistoni G., Cenci E., Bistoni F., Vecchiarelli A. Influence of Indinavir on Virulence and Growth of Cryptococcus neoformans. J. Infect. Dis. 2005;191:307–311. doi: 10.1086/426828. [DOI] [PubMed] [Google Scholar]
- 109.Naglik J., Albrecht A., Bader O., Hube B. Candida albicans proteinases and host/pathogen interactions. Cell. Microbiol. 2004;6:915–926. doi: 10.1111/j.1462-5822.2004.00439.x. [DOI] [PubMed] [Google Scholar]
- 110.Muselius B., Durand S., Geddes-McAlister J. Proteomics of Cryptococcus neoformans: From the Lab to the Clinic. Int. J. Mol. Sci. 2021;22:12390. doi: 10.3390/ijms222212390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.