TCR signal strength determines T cell differentiation and function in tumors, with high-signal-strength interactions resulting in dysfunction and low-signal-strength interactions resulting in functional inertness. Fine-tuning TCR signal strength to an intermediate range improves anti-tumor activity in vivo.
Abstract
T cell receptor (TCR) signal strength is a key determinant of T cell responses. We developed a cancer mouse model in which tumor-specific CD8 T cells (TST cells) encounter tumor antigens with varying TCR signal strength. High-signal-strength interactions caused TST cells to up-regulate inhibitory receptors (IRs), lose effector function, and establish a dysfunction-associated molecular program. TST cells undergoing low-signal-strength interactions also up-regulated IRs, including PD1, but retained a cell-intrinsic functional state. Surprisingly, neither high- nor low-signal-strength interactions led to tumor control in vivo, revealing two distinct mechanisms by which PD1hi TST cells permit tumor escape; high signal strength drives dysfunction, while low signal strength results in functional inertness, where the signal strength is too low to mediate effective cancer cell killing by functional TST cells. CRISPR-Cas9–mediated fine-tuning of signal strength to an intermediate range improved anti-tumor activity in vivo. Our study defines the role of TCR signal strength in TST cell function, with important implications for T cell–based cancer immunotherapies.
Introduction
The affinity of the interaction between TCR and peptide-bound MHC (pMHC) determines the kinetics and magnitude of the T cell response (Skokos et al., 2007; Denton et al., 2011; King et al., 2012; Zikherman and Au-Yeung 2015; Ozga et al., 2016; Conley et al., 2016). In vitro studies demonstrated that ligand affinity determines the frequency and rate at which naive T cells become activated, but not the course of subsequent differentiation; while high-affinity ligands activate more T cells, all activated T cells commit to the same effector differentiation program and achieve the same cytolytic capacity, regardless of ligand affinity (Altan-Bonnet and Germain, 2005; Balyan et al., 2017; Richard et al., 2018). In support of these findings, during acute infections, high-affinity interactions lead to increased T cell expansion, but both low- and high-affinity interactions result in memory T cell formation (Zehn et al., 2009).
The tumor-reactive CD8 T cell repertoire is highly diverse, with specificity for self-proteins and tumor-specific (mutant and viral) neoantigens. The TCR affinity of T cells specific to tumor self-antigens is generally low (McMahan et al., 2006; Buhrman and Slansky, 2013; Hollingsworth and Jansen, 2019; Hoffmann and Slansky, 2020), and efforts to isolate higher-affinity T cell clones or engineer affinity-enhanced TCR have improved anti-tumor efficacy, albeit with increased risk of on-target and off-target toxicity (Lyman et al., 2005; Schmid et al., 2010; Parkhurst et al., 2011; Bos et al., 2012; Morgan et al., 2013; Zhong et al., 2013; Cameron et al., 2013; Stone et al., 2015; Schmitt et al., 2015; Rapoport et al., 2015; Schmitt et al., 2017; Chapuis et al., 2019). In contrast, tumor-specific T cells (TST cells) recognize neoantigens generally with high-affinity TCR, yet TST cells become dysfunctional, allowing tumors to develop and progress (Willimsky and Blankenstein, 2005; Zhou et al., 2005; Aleksic et al., 2012; Lu et al., 2013; Tran et al., 2014; Gros et al., 2014; Blankenstein et al., 2015; Leisegang et al., 2016; Gros et al., 2016; Schietinger et al., 2016; Stevanović et al., 2017; Philip et al., 2017; Bobisse et al., 2018; Thommen and Schumacher 2018; Azizi et al., 2018; Sade-Feldman et al., 2018; Li et al., 2019; Gros et al., 2019). Here, we assessed how TCR signal strength impacts TST cell differentiation and dysfunction and how it contributes to the phenotypic and functional heterogeneity of the TST cells pool in tumors. Using in vivo mouse models, we reveal that (1) TST cell dysfunction is a composite of TCR affinity–dependent and TCR affinity–independent transcriptional and epigenetic modules, and (2) TCR signal strength drives two distinct mechanisms of TST cell hyporesponsiveness and tumor escape in vivo: high-affinity interactions lead to rapid dysfunction, while low-affinity interactions drive functional inertness, a state defined by the lack of in vivo anti-tumor effector function despite maintenance of a cell-intrinsic functional program. CRISPR-Cas9–mediated fine-tuning of TCR signal strength of TST cells to an intermediate range (between dysfunction and functional inertness) significantly enhances anti-tumor immunity in vivo and responsiveness to immunotherapeutic interventions.
Results
Generation of an APL model to modulate TCR signal strength in tumor-specific CD8 T cells
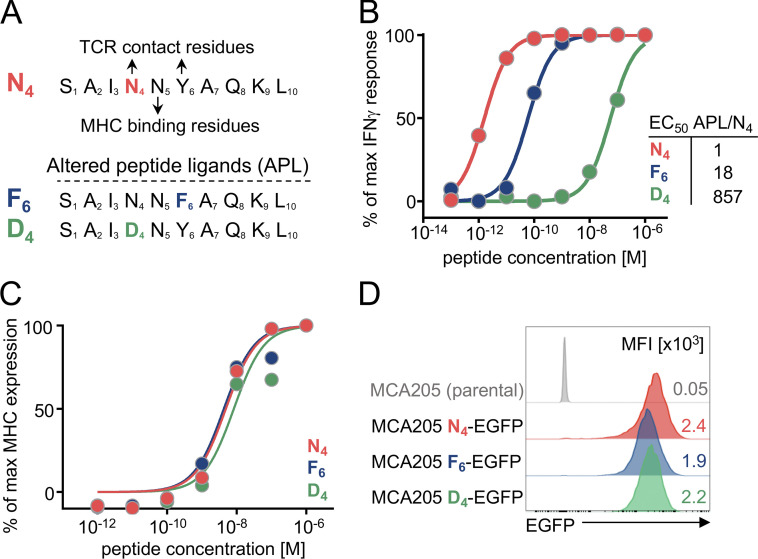
To investigate the impact of TCR signal strength on TST cell differentiation and dysfunction during tumor development, we generated a cancer mouse model with a defined tumor-specific antigen, SV40 large T antigen epitope I (TAG), that is recognized by TAG-specific CD8+ T cells (TCRTAG cells; Tanaka et al., 1989; Schietinger et al., 2016; Philip et al., 2017). We generated altered peptide ligands (APLs) of the native TAG peptide, SAINNYAQKL (N4) through single-amino-acid substitutions of the TCR contact residues at positions 4 and 6 (Fig. 1 A); APL model systems have been used extensively to understand the impact of TCR signal strength on T cell development (thymic selection) and differentiation in vitro and in vivo (e.g., infections; Evavold and Allen, 1991; Zehn et al., 2009; Daniels et al., 2006; Richard et al., 2018; Martínez-Usatorre et al., 2018). We chose two APLs that spanned the functional avidity range of human tumor antigens, generated through a tyrosine to phenylalanine substitution at position 6 (F6) or an asparagine to aspartate substitution at position 4 (D4; Fig. 1 A). Compared with the native N4 peptide, F6 and D4 peptides were 18-fold and 857-fold less potent in their ability to stimulate effector TCRTAG cells, as measured by IFN-γ production (functional avidity; Fig. 1 B; Zehn et al., 2009). To ensure that the APL bound equally well to MHC class I (H-2Db), we pulsed varying concentrations of N4, F6 and D4 peptides on TAP2-deficient RMA-S cells and assessed their ability to stabilize surface H-2Db (Townsend et al., 1989; Zehn et al., 2009; Daniels et al., 2006). N4, F6, and D4 peptides stabilized surface H-2Db equally well, suggesting these peptides do not differ in their binding to MHC class I (Fig. 1 C). We define affinity of TCR–pMHC interaction as a relative parameter determining the functional outcome when (1) pMHC affinity, (2) antigen concentration on target cells, and (3) TCR density on T cells are held constant.
Figure 1.
Generation of the SV40 TAG APL tumor model. (A) TAG APLs were generated through single-amino-acid substitutions of the TCR contact residues of the native TAG peptide (N4, red) at positions 4 and 6, generating F6 (blue) and D4 (green) APLs. (B) Functional avidity measured as IFN-γ production by effector TAG-specific CD8+ T cells (TCRTAG cells) after 4-h stimulation with antigen-presenting cells pulsed with the indicated peptide concentrations. The ratio of APL peptide concentration required to induce half-maximum IFN-γ response (EC50) relative to native N4 peptide is shown. Data are represented as mean of n = 2 technical replicates per condition and representative of two independent experiments. (C) Dose–response curves of surface expression of MHC class I (H-2Db) on RMA-S cells incubated with N4, F6, or D4 peptides at the indicated concentrations. Data show mean of n = 2 technical replicates per condition and are representative of two independent experiments. Value points for the highest peptide concentration (10−6 M) reach y = 100% and are masked by the N4 peptide. (D) EGFP expression levels of MCA205 N4-, F6-, and D4-EGFP tumor cells; parental MCA205 cell line is shown in gray. Inset numbers represent mean fluorescence intensity (MFI) of EGFP. Data are representative of two independent experiments.
Low- and high-affinity interactions drive robust T cell activation and effector differentiation in tumor-draining LNs (dLNs)
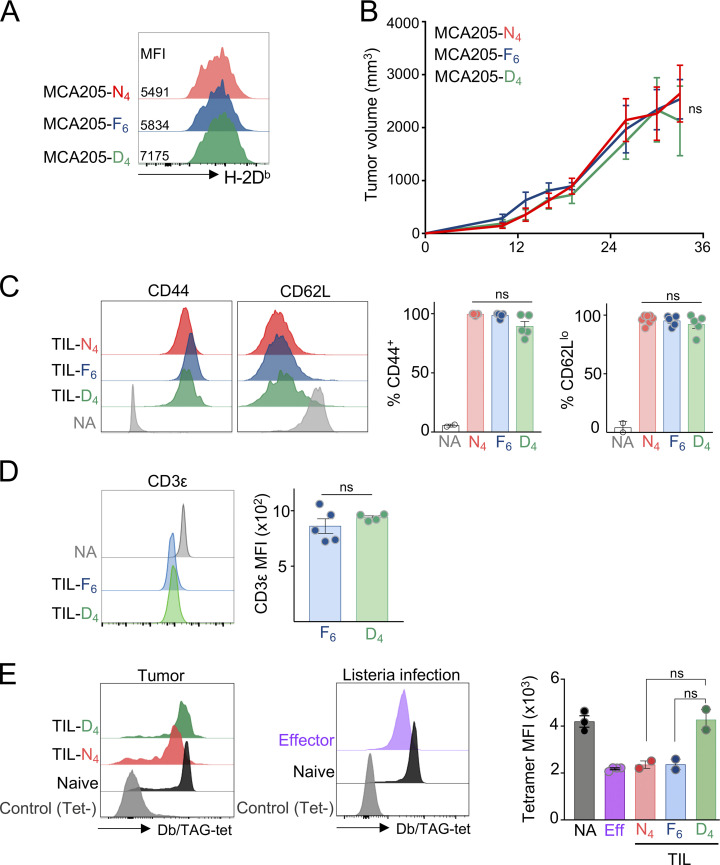
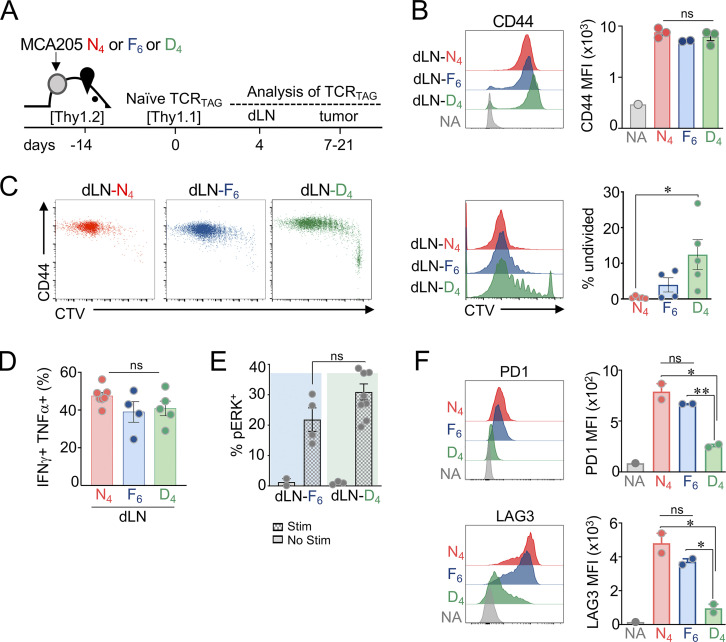
Retroviral vectors were constructed with genes encoding each APL epitope fused to EGFP. MCA205, a C57BL/6-derived fibrosarcoma cell line that expresses high levels of MHC class I (Fig. S1 A), was transduced with retroviral vectors encoding N4-, F6-, or D4-EGFP. MCA-APL cell lines were sorted for EGFP levels to ensure comparable APL expression (Fig. 1 D). APL-expressing MCA cells injected subcutaneously into mice had similar growth rates in vivo (Fig. S1 B). Once tumors were established (∼2 wk after tumor inoculation), we assessed the differentiation dynamics of adoptively transferred naive TCRTAG cells (Fig. 2 A). First, we asked whether TCR signal strength impacts T cell priming and activation in dLNs. CellTrace Violet (CTV)–labeled naive, congenically marked (Thy1.1+) TCRTAG cells were adoptively transferred into MCA-APL tumor-bearing hosts. 4 d later, transferred T cells were isolated from the dLN. All MCA-APL tumors elicited robust activation and proliferation of TCRTAG cells, as evidenced by the expression of activation marker CD44 and dilution of CTV (Fig. 2, B and C). Approximately 10% of TCRTAG cells in the dLN of MCA-D4 tumor-bearing mice were still undivided and CD44lo (Fig. 2 C), consistent with previous reports demonstrating that TCR signal strength impacts the frequency and rate of T cell activation and proliferation (Martínez-Usatorre et al., 2018; Zehn et al., 2009; Richard et al., 2018). All TCRTAG cells produced similarly high levels of the effector cytokines IFN-γ and TNF-α (Fig. 2 D) and phosphorylated ERK in response to ex vivo stimulation (Fig. 2 E). Interestingly, despite their similar activation and effector function profiles, TCRTAG cells encountering high-affinity N4 and F6 antigens expressed higher levels of the inhibitory receptors (IRs) PD1 and LAG3, in contrast to TCRTAG cells in the dLN of low-affinity MCA-D4 tumors, which were PD1lo and LAG3lo (Fig. 2 F). Thus, both high- and low-affinity TCR–pMHC interactions trigger the activation, proliferation, and effector differentiation of naive TST cells in dLN, despite differences in IR expression levels.
Figure S1.
MCA205 APL tumor model. (A) Flow cytometric analysis of MHC class I (H-2Db) expression of MCA-APL cell lines. Histograms are gated on APL-EGFP–expressing MCA205 cancer cells. Data are representative of two independent experiments. Inset numbers show MFI. (B) Tumor outgrowth of MCA-APL cell lines in TCROT1 hosts. Data show mean ± SEM of n = 5–8 mice per APL; ns, three-way ANOVA. (C–E) Phenotypic characterization of TIL-TCRTAG cells isolated from MCA-APL tumors. Flow cytometric analysis of CD44, CD62L (C), CD3ε (D), and TCR expression levels through tetramer staining (E) of TIL-TCRTAG cells isolated 14 d after AT into MCA-APL tumor-bearing hosts. Each symbol represents an individual mouse. NA, naive TCRTAG; control, tetramer-negative (Tet−) endogenous CD8+ T cells; Eff, TCRTAG effector CD8 T cells at the peak of response 5 d after Listeria (LmTAG) infection. Data are representative of two independent experiments. Data show mean ± SEM; ns, unpaired two-tailed Student’s t test.
Figure 2.
Low- and high-affinity interactions lead to robust activation and differentiation of naive TST cells in dLNs. (A) Experimental scheme. MCA-APL cell lines were injected subcutaneously into TCROT1 (Thy1.2) mice. 2 wk later, naive, CTV-labeled congenically marked (Thy1.1+) TCRTAG cells were adoptively transferred. Transferred T cells were reisolated from dLN and tumors at indicated time points. (B) CD44 expression levels of TCRTAG from dLN (MFI shown for dividing cells). n = 2–3 per APL; data are representative of three independent experiments. (C) CTV dilution and CD44 expression of TCRTAG cells isolated from dLN of APL tumor-bearing mice. Percentages of undivided TCRTAG cells (CTVhi) are shown (right). (D) Production of effector cytokines IFN-γ and TNF-α by TCRTAG cells isolated from dLN and stimulated with N4 peptide (0.5 µg/ml). Each symbol represents an individual mouse. For C and D, n = 4–5 per APL; data are representative of three independent experiments. (E) Flow cytometric analysis of phospho-ERK (pERK) by TCRTAG cells isolated 4 d after AT from dLN of MCA-F6 (dLN-F6) and MCA-D4 (dLN-D4) tumor-bearing mice. T cells were simulated ex vivo with MCA-N4 tumor cells (see Materials and methods). Each circle is an individual mouse. n = 4 for dLN-F6, and n = 9 for dLN-D4. Data are representative of two independent experiments. (F) PD1 and LAG3 expression levels on TCRTAG cells from dLN (n = 2 per APL). Data are representative of two independent experiments. (B–F) Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ns, P > 0.05; unpaired two-tailed Student’s t test.
High-affinity interactions cause TST cell dysfunction, while low-affinity interactions preserve a cell-intrinsic functional state in TST cells
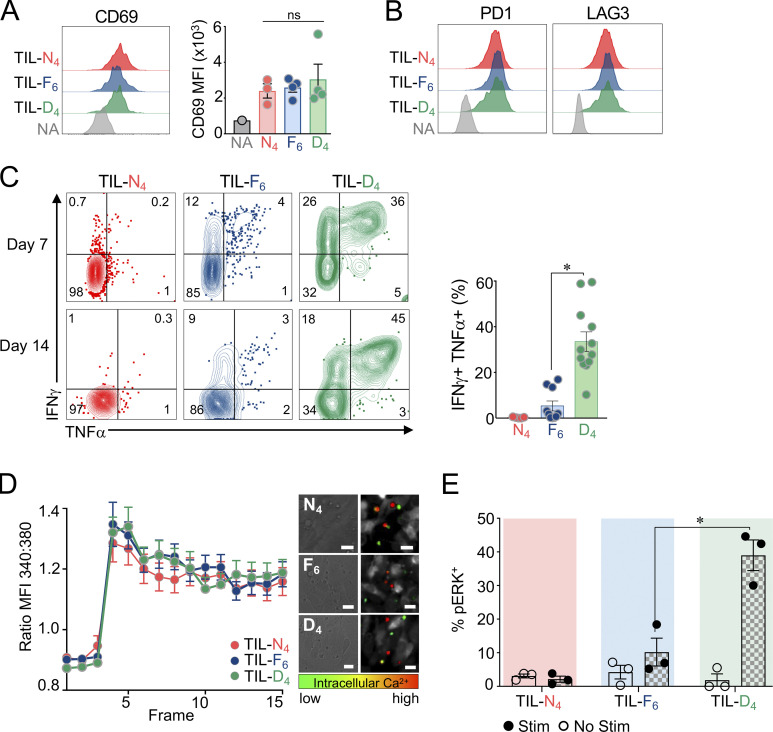
Next, we asked whether TCR signal strength determines TST cell functional states within tumors, the site of chronic tumor antigen encounter and TCR stimulation. Naive, congenically marked (Thy1.1+) TCRTAG cells were adoptively transferred into MCA-APL tumor-bearing hosts and isolated from tumors 7–14 d later. Tumor-infiltrating TCRTAG cells were uniformly CD44hiCD62Llo (Fig. S1 C) and expressed similar levels of the activation marker CD69 (Fig. 3 A). In contrast to what we observed in the dLN, tumor-infiltrating lymphocytes (TILs) from all MCA-APL tumors expressed similarly high levels of PD1 and LAG3 (Fig. 3 B), demonstrating that even very-low-affinity TCR–pMHC interactions within the tumor lead to high expression of these canonical IRs. Strikingly, despite their similar activation and immunophenotype, high-affinity N4 and F6 TCR–pMHC interactions led to the rapid loss of IFN-γ and TNF-α production, in contrast to low-affinity TIL-D4, which remained functional (Fig. 3 C). The loss of effector cytokine production in high-affinity TILs was not due to TCR down-regulation (Fig. S1, D and E). Together, our data reveal that (1) TIL functional states in tumors are dictated by TCR signal strength, and (2) canonical IRs, such as PD1 and LAG3, are expressed in an affinity-independent fashion within the tumor and do not reflect the cell-intrinsic functional state of TILs.
Figure 3.
Tumor-infiltrating TST cells encountering low-affinity antigens preserve a cell-intrinsic functional state. (A) CD69 expression levels on TCRTAG isolated from MCA-APL tumors 14 d after AT. Data are representative of two independent experiments with n = 3–5 per APL. (B) PD1 and LAG3 expression levels by tumor-infiltrating TCRTAG cells (TILs) 14 d after AT. Data are representative of four independent experiments (n = 4–6 per APL). (C) Intracellular IFN-γ and TNF-α production of TCRTAG TILs isolated from APL tumors 7 (top) and 14 (bottom) d after AT, assessed after 4-h peptide stimulation with N4 peptide (0.5 µg/ml). Each symbol represents an individual mouse (n = 4–5 per APL). Data are representative of four independent experiments. (D) Calcium flux of TCRTAG TILs isolated from MCA-APL tumors 14 d after AT and loaded with the calcium-sensing dye Fura-2AM. Time-lapse microscopy was performed with T cells encountering MCA-N4 tumor cells. Shown is the ratio of the emission at 340 nm to 380 nm. Data are shown as mean ± SEM and are representative of two independent experiments (n = 20–50 cells per condition). (E) Flow cytometric analysis of phospho-ERK (pERK) by TCRTAG TIL (isolated 14 d after AT) following TCR stimulation with MCA-N4 tumor cells. n = 3 per APL. (A–E) Data are shown as mean ± SEM. *, P < 0.001, unpaired two-tailed Student’s t test.
We then sought to investigate affinity-dependent alterations in TCR signaling in TST cells. Two pathways downstream of the TCR have been implicated in affinity sensing: calcium and MAPK signaling pathways (Daniels et al., 2006; Conley et al., 2016). TCRTAG cells were sorted from MCA-APL tumors 14 d after adoptive transfer (AT) and assayed for their ability to flux calcium and phosphorylate ERK (a readout for MAPK activity) in response to TCR stimulation ex vivo. While TCRTAG cells from all MCA-APL tumors were able to flux calcium (Fig. 3 D), only low-affinity TIL-D4 cells were able to phosphorylate ERK (Fig. 3 E), suggesting potential negative feedback on the MAPK signaling pathway in response to chronic, high-affinity TCR stimulation.
TCR signal strength drives distinct transcriptional and epigenetic programs in TILs
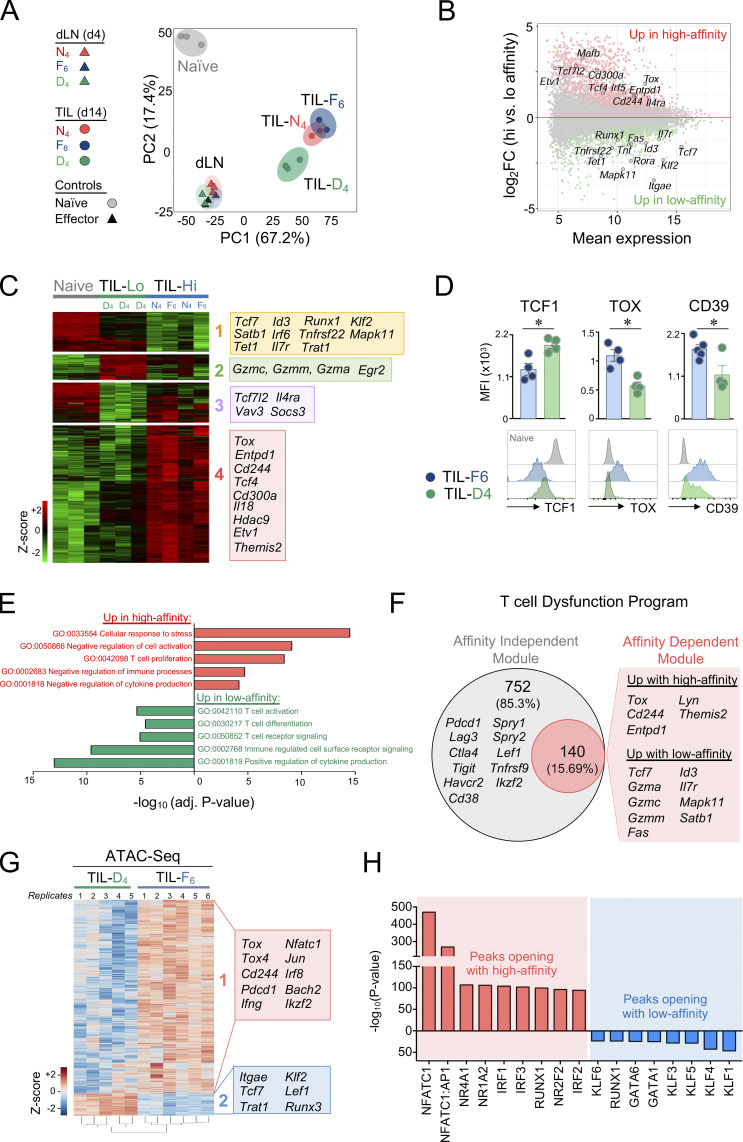
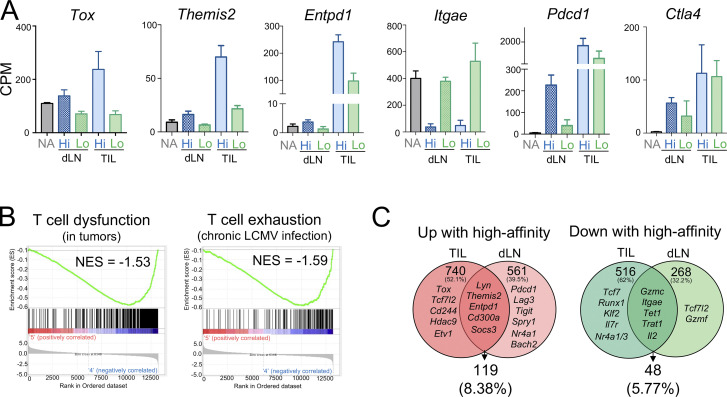
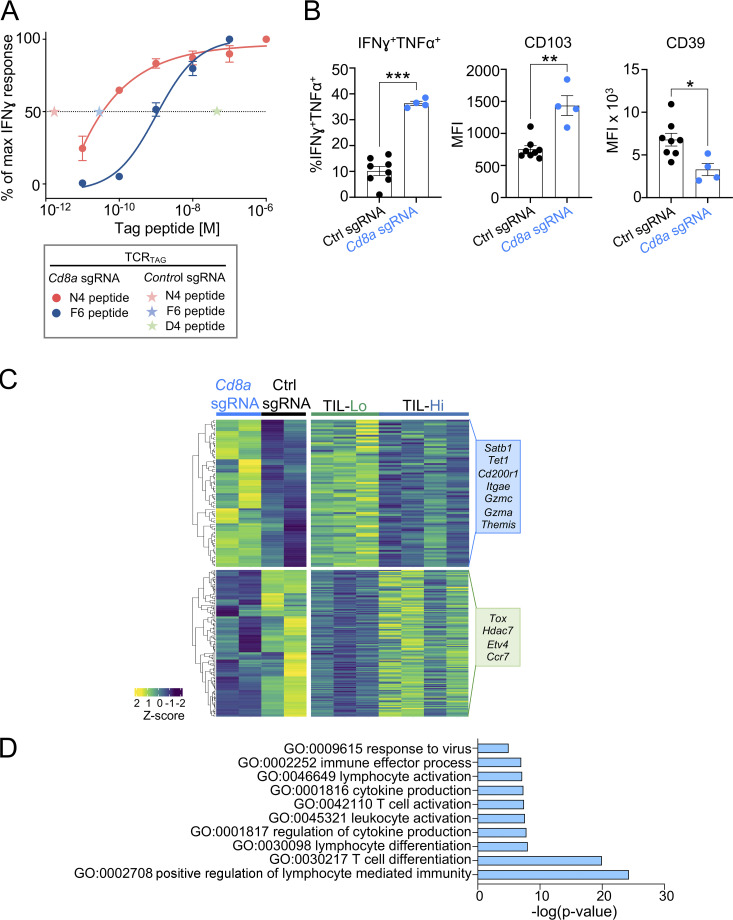
We previously demonstrated that TST cell dysfunctional states are defined by transcriptional and epigenetic programs (Schietinger et al., 2016; Philip et al., 2017; Scott et al., 2019; Philip and Schietinger, 2021). To understand if TCR signal strength regulates transcriptional programs, we performed RNA sequencing (RNA-seq) on TCRTAG cells isolated from dLN (dLN-N4, -F6, and -D4) and tumors (TIL-N4, -F6, -D4) of MCA-APL tumor-bearing hosts; as controls, we included naive TCRTAG cells, as well as effector TCRTAG cells isolated from LN of mice immunized with a recombinant Listeria monocytogenes strain expressing the high-affinity N4 epitope. Principal-component analysis revealed that TCRTAG cells from dLN-N4, -F6, -D4, and effector TCRTAG cells clustered together and were markedly distinct from their TIL counterparts (Fig. 4 A). High-affinity TIL-N4 and -F6 clustered separately from low-affinity TIL-D4, with 2,300 genes differentially expressed between high- and low-affinity TILs (Fig. 4, B and C). High-affinity TCR–pMHC interactions led to the up-regulation of critical transcription factors and IRs associated with T cell dysfunction and exhaustion, including Tox, Mafb, Tcf4, Etv1, Cd244 (2B4), and Entpd1 (CD39), and down-regulation of genes associated with stem-like progenitor and memory differentiation states, such as Tcf7 (TCF1), Runx1, Id3, and Il7r (Fig. 4, B–D; and Fig. S2 A). Gene ontology (GO) classification revealed that pathways associated with negative regulation of T cell activation and effector function were enriched in TILs encountering the high-affinity tumor antigens N4 and F6, but not the low-affinity antigen D4 (Fig. 4 E). Moreover, gene programs associated with tumor-specific T cell dysfunction (Philip et al., 2017) or T cell exhaustion during chronic infections (West et al., 2011) were negatively enriched in low-affinity TIL-D4 (Fig. S2 B). Thus, PD1hi TILs that encounter tumor-specific antigens with lower TCR signal strength retain transcriptional and functional features of effector/memory T cell states. Interestingly, 86% of the genes differentially expressed between high- and low-affinity TILs were not differentially expressed in the dLN (Fig. S2 C), suggesting that the observed transcriptional differences were established de novo at the tumor site and not preestablished during the priming phase in the dLN.
Figure 4.
TCR signal strength drives distinct transcriptional and epigenetic programs in TST cells. (A) TCRTAG cells isolated from dLN (4 d after AT) and tumors (14 d after AT) were subjected to RNA-seq; naive TCRTAG and effector TCRTAG cells isolated from LN 4 d after infection with a L. monocytogenes strain expressing TAG (N4) epitope were used as controls. Principal-component (PC) analysis of RNA-seq data. Each symbol represents a biological replicate, and each component is indicated with the amount of variation that it explains. (B) MA plot of RNA-seq data showing the relationship between average expression and expression changes between high-affinity TIL-N4/F6 and low-affinity TIL-D4. Statistically significantly DEGs are shown in red and green, with select genes highlighted for reference. (C) Hierarchical clustering of genes differentially expressed (log2 fold change >1, false discovery rate <0.1) in high-affinity TILs (from MCA205-N4 and MCA205-F6 tumors; TIL-Hi) versus low-affinity TILs (from MCA205-D4; TIL-Lo). Expression in naive TCRTAG cells is shown as a control. Selected genes within each cluster are shown. (D) TCF1, TOX, and CD39 expression levels from TIL-F6 and TIL-D4 14 d after AT (n = 4–5 biological replicates per APL). All values are mean ± SEM. *, P < 0.05, unpaired two-tailed Student’s t test. (E) Selected GO terms enriched in genes up-regulated in response to high-affinity (red) or low-affinity (green) TCR stimulation in TILs. (F) Affinity-dependent (red) and affinity-independent (gray) modules of the tumor-specific T cell dysfunction program. Select genes of each module are highlighted. (G) Heatmap of log2-transformed normalized read counts per regions with differential chromatin accessibility comparing TIL-F6 and TIL-D4. Genes associated with the two major clusters are highlighted. (H) Top 17 most-significantly enriched transcription factor motifs in peaks with increased accessibility in high-affinity TIL-F6 (red) or low-affinity TIL-D4 (blue).
Figure S2.
TCR affinity drives distinct molecular programs of tumor-specific T cells. (A) mRNA expression levels of select genes in TCRTAG isolated from the dLN (day 4 after AT) and tumors (TILs; day 10–14 after AT) from high-affinity (blue) or low-affinity (green) MCA-APL tumor-bearing mice. Expression in naive (NA) TCRTAG cells is shown as control. (B) Enrichment of gene sets described for tumor-specific T cell dysfunction (from Philip et al., 2017; left), and T cell exhaustion during chronic viral infection (from West et al., 2011 [GSE30962]; right) in TIL-Lo. NES, normalized enrichment score. (C) Venn diagrams showing the degree of overlap between genes up- or down-regulated in TCRTAG isolated from dLN or tumors (TILs). Examples of genes in each category are shown.
To examine genes and pathways controlling TIL functional states, we categorized the transcriptional program of T cell dysfunction into TCR affinity–dependent and affinity–independent modules. Of the 892 genes that we previously identified as differentially expressed in dysfunctional TILs compared with functional effectors (Philip et al., 2017), ∼15% (140 genes) were specifically regulated by TCR affinity and likely dictate the functional state of TST cells (Fig. 4 F). The remaining dysfunction-associated genes, including Pdcd1, Ctla4, Lag3, Havcr2 (TIM3), Cd38, Lef1, and Ikzf2, were expressed independently of TCR affinity and thus are likely not associated with TIL functionality.
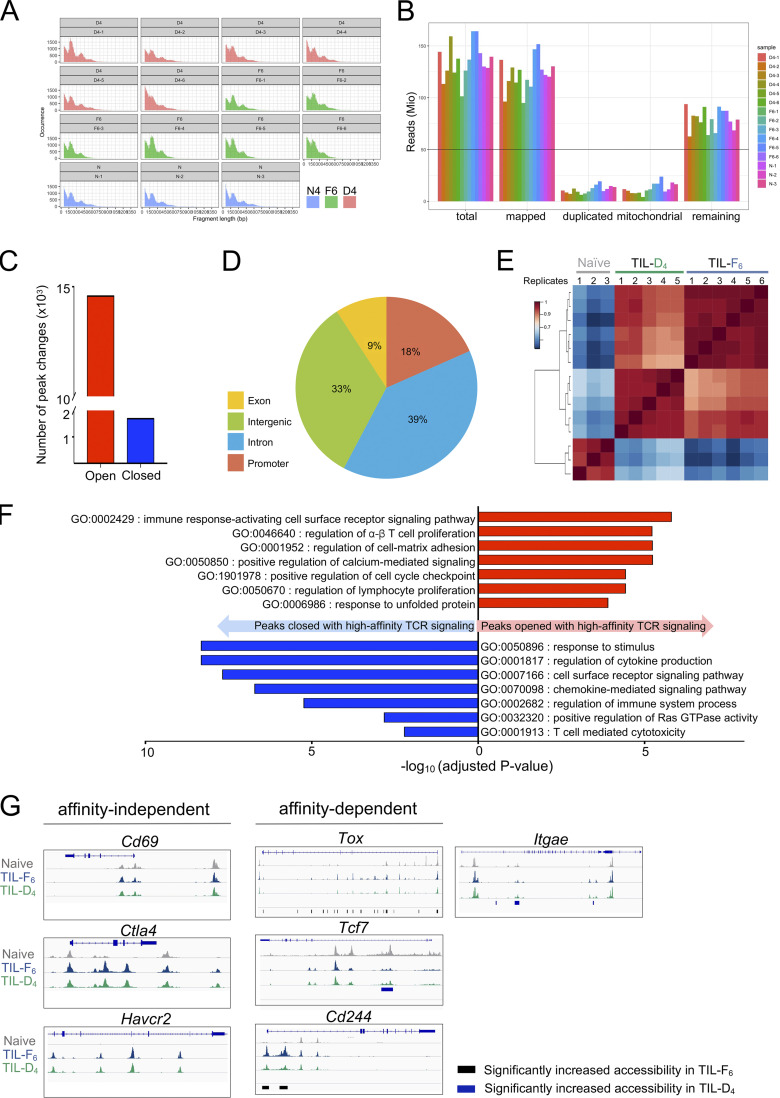
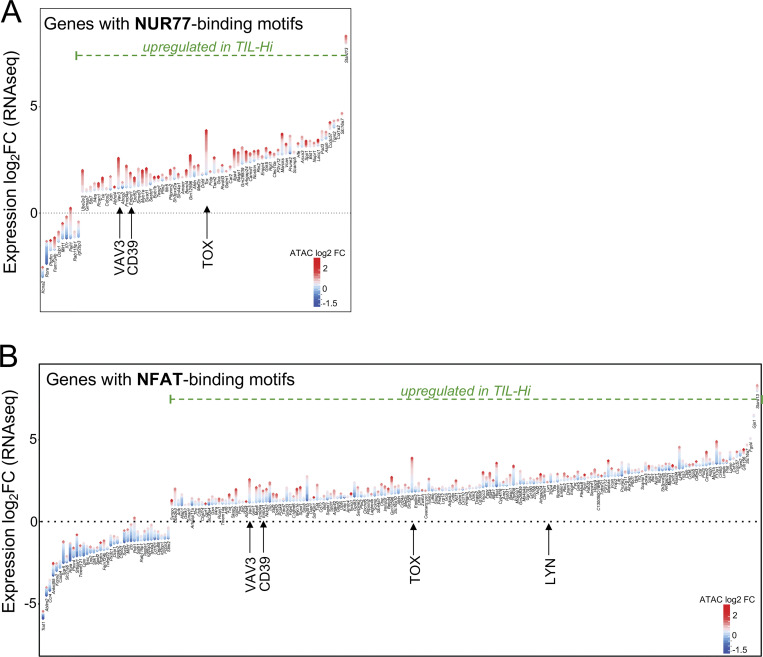
TST cell dysfunction is epigenetically encoded (Philip et al., 2017; Mognol et al., 2017; Sade-Feldman et al., 2018; Bengsch et al., 2018). To understand whether TCR affinity alters TST cell epigenetic programs, we performed an assay for transposase-accessible chromatin with sequencing (ATAC-seq) on TIL from high- (TIL-F6) and low-affinity (TIL-D4) MCA-APL tumors (Fig. S3, A and B). 16,264 regions were differentially accessible between TIL-F6 and TIL-D4, with the majority of these peaks (89%) opening in response to high-affinity TCR stimulation (Fig. 4 G; and Fig. S3, C–E). These loci were enriched for genes associated with cell cycle regulation and proliferation, while loci with decreased accessibility were enriched for genes critical for T cell effector function and cell surface receptor signaling (Fig. S3 F). Interestingly, many of the affinity-dependent differentially expressed genes (DEGs; e.g., Tox, Tcf7, Cd244, and Itgae) were also found to have differential chromatin accessibility patterns, while genes with affinity-independent expression profiles (e.g., Cd69, Ctla4, and Havcr2) maintained similar accessibility profiles in high- versus low-affinity TILs (Fig. S3 G). Transcription factor motif analysis revealed that peaks with increased accessibility in response to high-affinity TCR stimulation were enriched for Nfatc1 (NFAT2) and Nr4a1 (NUR77) binding motifs (Fig. 4 H), and this was accompanied by up-regulation of their respective target genes (Fig. S4). The maintained ability of TILs to flux calcium, combined with their loss of MAPK activity (Fig. 3, D and E), supports the enrichment of Nfatc1-binding motifs, as calcium signaling is required for nuclear localization of NFAT, but colocalization of binding partner AP1 is contingent on MAPK activation. While the role of TCR affinity in regulating NUR77 and NFAT activity in thymocytes and mature T cells has been demonstrated (Marangoni et al., 2013; Dolmetsch et al., 1997; Baldwin and Hogquist, 2007; Moran et al., 2011), our data suggest that TCR affinity also regulates the activity of these key transcription factors in the context of tumors. Thus, TCR signal strength drives distinct transcriptional and epigenetic programs that underlie T cell functional heterogeneity in tumors.
Figure S3.
Chromatin accessibility changes in response to varying TCR signal strength in TST cells. (A and B) Quality control plots for ATAC-seq samples. (A) Fragment length distribution plots (base pairs on x axis and read count on y axis). (B) Number of aligned reads per sample showing the number remaining after mapping and removing duplicated and nonmitochondrial reads. The ENCODE guideline is indicated by the horizontal line. (C) Number of chromatin accessibility changes in TIL encountering high- versus low-affinity tumor antigen. (D) Pie chart showing the proportion of reproducible ATAC-seq peaks in exonic, intronic, intergenic, and promoter regions. (E) Correlation heatmap of peaks that are differentially accessible (false discovery rate <0.05) between TIL-F6 versus TIL-D4. (F) Selected GO terms enriched in peaks opened (red) or closed (blue) in response to high-affinity TCR stimulation in TILs. (G) ATAC-seq signal profiles of loci of affinity-dependent genes, including Tox, Tcf7, Cd244, and Itgae, and affinity-independent genes, including Cd69, Ctla4, and Havcr2. Vertical bars at the bottom of each plot represent regions with statistically significant changes in accessibility in TIL-F6 versus TIL-D4.
Figure S4.
Transcription factor binding motif analysis for peaks with differential accessibility based on TCR signal strength. (A and B) Gains and losses of regulatory elements for the most DEGs containing NUR77 (A) or NFAT (B) binding motifs. Plots are divided into top and bottom genes with the highest and lowest respective log2 fold change (FC) of gene expression (shown on the y axis). Each gene is illustrated by a stack of diamonds representing peaks gained (red) or lost (blue) in high-affinity TILs.
Tumor escape results from two distinct mechanisms in the tumor-reactive PD1hi TIL repertoire, depending on TCR signal strength: T cell dysfunction and functional inertness
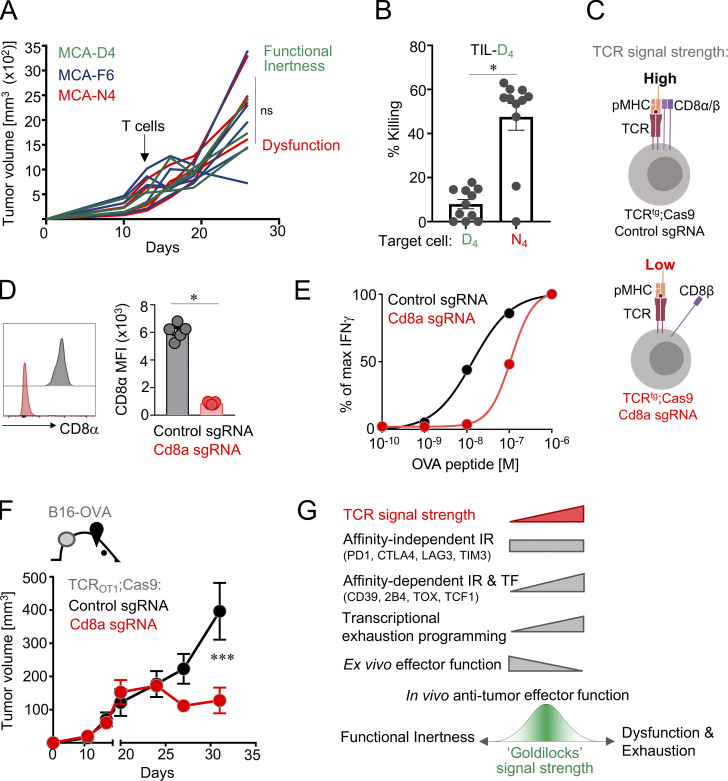
Given that TST cells encountering neoantigens with low TCR signal strength preserved a cell-intrinsic functional molecular program and the ability to produce effector cytokines ex vivo (Figs. 3 and 4), we hypothesized that these characteristics of T cell function would also correlate with enhanced anti-tumor immunity in vivo. Surprisingly, in vivo anti-tumor effector function of low-affinity TILs was no better than that of dysfunctional, exhausted TILs encountering high-affinity tumor antigens (Fig. 5 A). These results suggest that tumor escape can result from two fundamentally distinct mechanisms operating within the tumor-reactive PD1hi TIL repertoire; high TCR signal strength leads to T cell dysfunction (Figs. 3 and 4; Schietinger et al., 2016; Scott et al., 2019; Philip et al., 2017), while low TCR signal strength results in functional inertness, a state defined by lack of in vivo anti-tumor function, despite retention of an effector-like functional molecular program. To further define the characteristics of functionally inert low-affinity PD1hi TST cells, we isolated TIL-D4 10 d after AT and assessed their ability to kill target cells presenting the cognate D4 or the high-affinity N4 antigen ex vivo. TIL-D4 were unable to kill D4-target cells but could efficiently kill N4 targets (Fig. 5 B). Thus, low-affinity functionally inert PD1hi TST cells are functional by most phenotypic and transcriptional metrics, but their TCR–pMHC affinity is too low to effectively mediate cancer cell killing.
Figure 5.
Optimal anti-tumor efficacy requires an intermediate range of TCR signal strength. (A) Tumor outgrowth of MCA-APL tumor-bearing mice receiving AT of naive TCRTAG cells at day 14. Data show mean ± SEM of n = 5–7 mice per APL. ns, three-way ANOVA. Data are representative of two independent experiments. (B) Functional inertness of TIL-D4. TIL-D4 fail to kill D4 targets but can eliminate N4 targets. TIL-D4 were sorted from MCA-D4 tumors 10 d after AT and incubated with MCA-D4 or MCA-N4 tumor cells in vitro at a 1:10 effector to target ratio. Killing of tumor cells was assessed 18 h later by flow cytometry (see Materials and methods for technical details). Each circle represents an individual mouse (n = 11). Values are mean ± SEM. *, P < 0.0001, unpaired two-tailed Student's t test. Data are representative of two independent experiments. (C) CRISPR-Cas9–mediated deletion of Cd8a in high-affinity Cas9;TCRtg T cells to partially lower TCR signal strength. (D) CD8α expression on TCROTI;Cas9 T cells transduced with Cd8a sgRNA (red) or control sgRNA (black) and reisolated at 30 d after AT into B16-OVA tumor-bearing hosts. Values are mean ± SEM. Each symbol represents an individual mouse. *, P < 0.01, unpaired two-tailed Student's t test. Data are representative of two independent experiments. (E) Functional avidity measured as production of IFN-γ by TCROT1;Cas9 T cells transduced with Cd8a sgRNA (red) or control sgRNA (black) after 4-h stimulation with SIINFEKL peptide at the indicated concentrations. Data represent mean of technical replicates n = 2 and are representative of two independent experiments. (F) Lowering TCR signal strength through CRISPR-Cas9–mediated deletion of Cd8a in TCROTI enhances anti-tumor efficacy in vivo. B16-OVA tumor outgrowth in B6 mice that received congenically marked CD8 T cells (Thy1.1/Thy1.2) from TCROTI;Cas9 mice transduced with Cd8a or control sgRNA, sorted based on CD8α and sgRNA reporter (red fluorescence protein) expression and treated with anti-PD1 and anti-PDL1 antibodies, starting 4 d after T cell transfer, every other day. Data are representative of two independent experiments (n = 6 mice). Values are mean ± SEM. Significance was calculated by two-way ANOVA. (G) Summary and conclusions of the study. Phenotypic, functional, and transcriptional characteristics of TST cells encountering antigens with distinct TCR signal strength. TF, transcription factor. We propose a Goldilocks signal strength range that allows effective anti-tumor immunity in vivo. TST cells with affinity beyond this range are dysfunctional due to exhaustion (for high-affinity TST cells) or functionally inert (for T cells specific to low-affinity neoantigens and tumor/self-antigens). Affinity tuning for immunotherapeutic interventions through lowering signal strength of high-affinity TST cells or high-affinity chimeric antigen receptor T cells or signal strength enhancement of low-affinity T cells could result in increased anti-tumor effector function.
Therapeutic fine-tuning of TCR signal strength empowers TST cells to eliminate tumors in vivo
Next, we asked whether there is an intermediate “Goldilocks” level of TCR affinity (between dysfunction/exhaustion and functional inertness) that maintains TST cell function and mediates anti-tumor activity. Given that tumor antigen affinity in patients cannot be modified and that lowering TCR affinity of the various clones through manipulation of complementarity-determining regions is unfeasible, we explored alternative and potentially therapeutically applicable strategies to alter TCR signal strength. The CD8αβ coreceptors increase peptide sensitivity by stabilizing the interaction between the TCR and pMHC and by recruiting LCK (Artyomov et al., 2010; Holler and Kranz, 2003); we hypothesized that genetic deletion of Cd8a could decrease TCR signal strength without the need to discern or alter the sequence of the TCR itself. We generated CD8α-deficient TCRTAG cells by transducing CD8+ splenocytes of TCRTAG;Cas9 mice with either a single guide RNA (sgRNA) targeting Cd8a (Cd8a sgRNA) or a control (control sgRNA; Fig. 5 C). Indeed, CD8α-deficient TCRTAG cells had significantly lower functional avidity for N4 and F6, as measured by IFN-γ production in vitro, compared with control T cells (Fig. S5 A). CD8α-deficient TCRTAG cells were more functional, producing IFN-γ+ and TNF-α+ cells ex vivo and expressing higher levels of CD103 and lower levels of CD39 (Fig. S5 B). RNA-seq revealed that a large number of genes that were differentially expressed between T cells encountering low- versus high-affinity APL tumors were also differentially expressed between Cd8a KO and control TCRTAG cells (Fig. S5 C), and genes expressed in Cd8a KO TILs were enriched for pathways involved in T cell activation, differentiation, and effector function, similar to the TIL-D4 counterparts (Fig. S5 D).
Figure S5.
Lowering TCR signal strength of high-affinity TCRTAG enhances anti-tumor effector function. (A) Functional avidity measured as production of IFN-γ by TCRTAG;Cas9 CD8 T cells deficient of Cd8a (transduced with Cd8a sgRNA) after 4-h stimulation with N4 (red) or F6 (blue) peptides at the indicated concentrations. EC50 of control CD8α-sufficient TCRTAG (transduced with control sgRNA) encountering each APL is shown with asterisks. Data represent mean of technical replicates (n = 2) and two independent experiments. (B) TCRTAG cells were transduced with either Cd8α-targeting or control sgRNA to generate CD8α-deficient (blue) or CD8α-sufficient control (black) TCRTAG T cells. CD8α-deficient or control TCRTAG cells were injected into MCA-F6 tumor-bearing mice. Ex vivo cytokine production and expression levels of CD103 and CD39 of TCRTAG cells isolated from tumors were assessed 10–11 d after transfer. MCA-F6 tumor cells were used in this experiment, because the EC50 of CD8α-deficient T cells to F6 is between that of the CD8α-sufficient control to D4 and F6. Each dot represents an individual mouse. Values are mean ± SEM. Significance is calculated by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Hierarchical clustering of genes differentially expressed (log2 fold change >1) both in CD8α-deficient TCRTAG cells versus control cells and in low-affinity TILs (TCRTAG cells isolated from MCA205-D4; TIL-Lo) versus high-affinity TILs (TCRTAG cells isolated from MCA205-N4 and MCA205-F6 tumors; TIL-Hi). Selected genes within each cluster are shown. (D) Selected GO terms enriched in genes up-regulated in CD8α-deficient TCRTAG TILs.
To validate our findings in another tumor/neoantigen model, we used murine B16F10 (B16) melanoma cells expressing OVA (B16-OVA), a model neoantigen recognized by OVA-specific OT1 CD8+ T cells (TCROT1); TCROT1 cells recognize the H-2Kb-restricted OVA peptide SIINFEKL with high affinity. Similar to the TAG system, CD8α-deficient TCROT1 had significantly lower functional avidity for H-2Kb/SIINFEKL compared with control TCROT1 (Fig. 5, D and E). To investigate the anti-tumor efficacy of CD8α-deficient TCROT1 in vivo, B16-OVA cells were injected subcutaneously into wild-type C57BL/6J mice. Once tumors were established, CD8α-deficient TCROT1 or control T cells were adoptively transferred and mice were treated with PD1/PDL1 checkpoint blockade. Strikingly, we observed that CD8α-deficient TCROT1 significantly slowed tumor growth compared with control TCROT1 (Fig. 5 F). Thus, there is a critical range of TCR signal strength for TST cells, which preserves cell-intrinsic functional molecular programs and mediates anti-tumor activity in vivo (Fig. 5 G). Below this Goldilocks affinity range, tumor-reactive PD1hi TILs are functionally inert, while above this range, T cells enter a differentiation state associated with dysfunction and exhaustion.
Discussion
Our study identifies several novel insights into TST cells differentiation and dysfunction. First, we reveal that TST cell dysfunction is the composite of affinity-dependent and affinity-independent molecular programs. Surprisingly, several canonical IRs, including PD1 and LAG3, were similarly expressed on high- and low-affinity TST cells and TILs. While these IRs have been associated with T cell exhaustion, we show that they can also be associated with functional inertness and do not correlate with the intrinsic functional state of TILs. In contrast, the expression of exhaustion markers such as 2B4 and CD39 is dependent on TCR signal strength. This finding is relevant to biomarker-based research using CD39 to demarcate tumor-reactive (CD39+) from nontumor reactive, bystander (CD39lo/−) T cells within tumors (Simoni et al., 2018); our data suggest that CD39lo/− TILs can be part of the tumor-reactive TIL repertoire that encounters tumor antigens with lower TCR signal strength.
Second, both high- and low-signal-strength interactions lead to failed tumor control in vivo, uncovering two opposing mechanisms within the tumor-reactive PD1hi TIL repertoire that drive tumor evasion: (1) T cell dysfunction and exhaustion in response to high TCR signal strength, and (2) functional inertness in response to low TCR signal strength, characterized as lack of in vivo effector function despite effector/memory-like transcriptional and epigenetic programs. While low signal strength is sufficient to lead to activation, proliferation, and the up-regulation of certain IRs (PD1 and LAG3), it is too low to mediate in vivo cancer cell killing. Thus, molecular signatures of tumor-reactive PD1hi TILs do not necessarily correlate with or predict T cell anti-tumor efficacy in vivo. While T cell dysfunction and exhaustion has been generally considered to be the major underlying mechanism of PD1hi tumor-specific T cells permitting tumor evasion, we show that functional inertness represents another mechanism of the PD1hi TIL repertoire contributing to tumor escape.
Third, T cells within an intermediate range of TCR signal strength exhibit effective anti-tumor responses, and lowering TCR signal strength in high-affinity TST cells and/or targeting neoantigens within an intermediate range, instead of those with highest affinity, could improve T cell–based immunotherapeutic interventions.
In line with this, a recent study assessing the immunogenicity of over 500 predicted neoantigen-derived peptides in lung adenocarcinoma and melanoma patients showed that the epitopes with highest affinity were nonimmunogenic, and the vast majority of the immunogenic peptides were found within an intermediate affinity range (Wells et al., 2020). In addition, another study characterizing successful T cell clones in an adoptive T cell transfer setting targeting KRAS-G12D mutations showed that TCR–pMHC affinity inversely correlates with in vivo persistence in cancer patients. Specifically, the T cell clones with the highest TCR–pMHC affinity, which made up half of the infusion product, were undetectable after 40 d post-infusion. On the other hand, the clone with the lowest TCR–pMHC affinity that only made up 20% of the infusion product was maintained at 10% in circulation at 9 mo after infusion (Sim et al., 2020). Interestingly, in chronic viral infections, another setting of persistent antigen and TCR stimulation, high-affinity virus-specific T cells more readily enter a state of T cell exhaustion, undergo deletion, or become senescent (Probst et al., 2003; Ueno et al., 2004; Lin and Welsh, 1998; Schober et al., 2020). Together, these findings demonstrate that in settings of chronic antigen stimulation, high-affinity neoantigen-specific T cells and high-affinity virus-specific T cells more rapidly differentiate to dysfunction/exhaustion or do not persist, and that paradoxically decreasing signal strength might improve effector function in vivo. Indeed, lowering TCR signal strength through CRISPR/Cas9-mediated deletion of Cd8a improved anti-tumor immunity in vivo. Interestingly, a recent study showed that lowering the signal strength of chimeric antigen receptor T cells, which generally bind to their target tumor antigen with high affinity, led to enhanced anti-tumor effector function in vivo (Feucht et al., 2019). Thus, we propose that there is a critical Goldilocks range of TCR signal strength (Slansky and Jordan, 2010), which allows TST cells to maintain a cell-intrinsic functional molecular program while licensing T cells to execute anti-tumor effector functions in vivo (Fig. 5 G). Outside of this window, T cells are either functionally inert or dysfunctional and exhausted. Future studies into the phenotype and function of human TILs within and outside of this range are needed in order to accurately delineate the range that maximizes anti-tumor function and better guide therapeutic fine-tuning of TCR signal strength.
Materials and methods
Mice
TCRTAG transgenic mice (B6.Cg-Tg(TcraY1,TcrbY1)416Tev/J), TCROT1 mice (C57BL/6-Tg(TcraTcrb)1100Mjb/J), Rosa26-Cas9 mice (Gt(ROSA)26Sortm1.1(CAG-cas9*,-EGFP)Fezh/J), B6 Thy1.1 mice (B6.PL-Thy1a/CyJ), and B6 mice (C57BL/6J) were purchased from the Jackson Laboratory. TCRTAG mice and Rosa26-Cas9 mice were each crossed to Thy.1.1 mice to generate TCRTAG Thy1.1 and Cas9 Thy1.1 mice, respectively. TCROT1 (Thy1.2) mice were bred to Cas9 Thy1.1 mice to generate TCROT1;Cas9 Thy1.1/Thy1.2 progeny. TCR transgenic mice were not on a RAG-deficient background. Both female and male mice were used for experimental studies. Donor and host mice were age and sex matched; mice were 6–12 wk old. All mice were bred and maintained in the animal facility at Memorial Sloan Kettering Cancer Center (MSKCC). Experiments were performed in compliance with the MSKCC Institutional Animal Care and Use Committee regulations.
Antibodies and reagents
The native SV40 large T antigen epitope I (TAG) peptide [SAINNYAQKL (N4)] and TAG APL [SAINNFAQKL (F6) and SAIDNYAQKL (D4)] were purchased from GenScript. Fluorochrome-conjugated antibodies were purchased from BD Biosciences, eBioscience, Cell Signaling Technology, and BioLegend.
Intracellular cytokine staining and CTV labeling
Intracellular cytokine staining was performed using the Cytofix/Cytoperm Plus kit (BD Biosciences) following the manufacturer’s protocol. Briefly, T cells isolated from spleens, LNs, or tumors were mixed with 2 × 106 congenically marked B6 splenocytes and incubated with 0.5 µg/ml TAG peptide for 4 h at 37°C in the presence of GolgiPlug (BD Biosciences). After staining for cell surface molecules, cells were fixed, permeabilized, and stained with antibodies against IFN-γ (XMG1.2) and TNF-α (MP6-XT22). To assess cell proliferation in vivo, naive TCRTAG cells were incubated with 5 µM CTV (Thermo Scientific). Excess extracellular dye was quenched with FBS, and cells were washed twice with serum-free RPMI before transfer into host mice.
Flow cytometric analysis
Flow cytometric analyses were performed using Fortessa X20. Cells were sorted using BD FACS Aria (BD Biosciences) at the MSKCC Flow Core Facility. Flow data were analyzed with FlowJo v.10 software (Tree Star).
Generation of plasmids and tumor cell lines
pMFG-APL-EGFP
pMFG-N4-EGFP, pMFG-F6-EGFP, and pMFG-D4-EGFP were constructed by inserting annealed oligonucleotides encoding triple SAINNYAQKL-AAY, SAINNFAQKL-AAY, or SAIDNYAQKL-AAY repeats into the NcoI-linearized pMFG-EGFP vector. Restriction enzymes were purchased from New England Biolabs. All constructs were verified by sequence analysis. Packaging cells (ATCC) were transfected with APL constructs; supernatants were used to transduce MCA205 cells to generate MCA205-N4-EGFP, MCA205-F6-EGFP, and MCA205-D4-EGFP, respectively, as previously described (Schietinger et al., 2010). Transduced bulk cell lines were sorted for similar EGFP expression levels.
pMIUR
A 1.142-kb fragment encoding turbo RFP in the 5′ to 3′ orientation and an inverted U6-gRNA-scaffold cassette in the 3′ to 5′ orientation was obtained from IDT and cloned into pMIG (Addgene; catalog no. 9044) using standard restriction enzyme–based methods. Briefly, the fragment was amplified using primers that add EcoRI and SalI restriction sites on the 5′ and 3′ regions, respectively, and subsequently digested and cloned into linearized pMIG backbone harboring EcoRI and SalI cloning overhangs. All constructs were sequence verified using Sanger sequencing. The following primer sequences were used: FSR-158, 5′-GGCCAAGAATTCGCCACCATGGTGTCTAAGGGC-3′; FSR-153: 5′-GGCCAAGTCGACGAGGGCCTATTTCCCATGATT-3′; vector, EcoRI-tRFP-NoBbsI-INV-U6-gRNA-BbsI-SalI cassette.
Listeria infection
The L. monocytogenes ΔactA ΔinlB strain (Brockstedt et al., 2004) expressing the TAG N4 epitope (SAINNYAQKL; LmTAG) was generated by Aduro BioTech as previously described (Philip et al., 2017, Schietinger et al., 2016). Experimental vaccination stocks were prepared by growing bacteria to early stationary phase and washing in PBS, formulated at ∼1 × 1010 CFU/ml and stored at −80°C. Mice were infected with 5 × 106 cfu of LmTAG intraperitoneally.
Adoptive T cell transfer
For the generation of effector TCRTAG CD8+ T cells, 105 CD8+ splenocytes from TCRTAG Thy1.1 transgenic mice were adoptively transferred into B6 (Thy1.2) mice. 1 d later, mice were infected with 5 × 106 CFU LmTAG. For in vitro TCR functional avidity measurements, adoptively transferred effector TCRTAG cells were isolated from the spleens of B6 host mice at 7 d after LmTAG immunization. For AT studies of naive TCRTAG cells into MCA-APL tumor-bearing hosts, 2 × 106 CD8+ splenocytes from TCRTAG Thy1.1 transgenic mice were adoptively transferred (i.v.) into tumor-bearing TCROT1 (Thy1.2) mice ∼2 wk after tumor implantation, when tumors reached an average size of 400 mm3.
APL tumor model
3 × 106 MCA-N4, MCA-F6, or MCA-D4 cells were injected subcutaneously into TCROT1 mice, whose CD8+ T cell compartment is restricted to an irrelevant antigen (OVA), thereby preventing immune selection by the host with resultant changes in antigen load and avidity. Once tumors were established, naive TCRTAG CD8+ T cells were adoptively transferred and reisolated at the indicated time points from dLNs and tumors.
T cell isolation for subsequent analyses
Mice were euthanized by cervical dislocation. LNs were mechanically disrupted with the back of a 3-ml syringe and filtered through a 100-µm strainer, and RBCs were lysed with ammonium chloride potassium buffer. Cells were washed twice with cold RPMI 1640 media supplemented with 2 µM glutamine, 100 U/ml penicillin/streptomycin, and 3% FBS. Tumor tissue was mechanically disrupted with a glass pestle and a 150-µm metal mesh in 5 ml cold HBSS with 3% FBS. Cell suspension was filtered through a 100-µm strainer. Tumor homogenate was spun down at 400 g for 5 min at 4°C. Pellet was resuspended in 15 ml HBSS with 3% FBS, 500 µl (500 U) heparin, and 8 ml isotonic Percoll (GE); mixed by several inversions; and spun at 500 g for 10 min at 4°C. Pellet was lysed with ammonium chloride potassium buffer, and cells were further processed for downstream applications.
Sample preparation for RNA-seq and ATAC-seq
Samples were isolated as follows: (1) naive TCRTAG Thy1.1+ T cells were isolated from the spleens of TCRTAG Thy1.1 transgenic mice and stained for CD8α (eBioscience; clone 53–6.7), Thy1.1 (BD Biosciences; clone OX-7), and CD44 (BD Biosciences; clone IM7). CD8+CD90.1+CD44lo naive cells were sorted by FACS. (2) Day 4 effector TCRTAG Thy1.1+ T cells were isolated from lymph nodes of infected B6 (Thy1.2+) host mice (see above) at 4 d after LmTAG immunization; cells were sorted for a CD8+CD90.1+CD44hi phenotype. (3) Day 4 TCRTAG Thy1.1+ cells from tumor-bearing mice were isolated from the tumor-draining (inguinal) LNs; cells were stained and sorted for a CD8+CD90.1+CD44hi phenotype. (4) Day 10–14 TCRTAG Thy1.1+ cells from tumor-bearing mice were isolated from tumors (see above); cells were sorted for a CD8+CD90.1+CD44hi phenotype. Samples for RNA-seq were directly sorted into Trizol LS reagent (Invitrogen; catalog no. 10296010) and stored at −80°C. Samples for ATAC-seq were resuspended in FBS with 10% DMSO and stored at −80°C.
Calcium flux imaging
8-well chamber slides were coated with 2 µg fibronectin and incubated at 37°C for 1 h. Wells were subsequently washed twice with PBS. MCA-N4 tumor cells were seeded at 30,000 cells per well in complete DMEM (5% FBS and 100 U/ml penicillin/streptomycin) and incubated at 37°C overnight. The following day, TCRTAG cells were sorted from MCA-APL tumors by FACS and loaded with Fura2-AM (Thermo Fisher Scientific; catalog no. F1221) at 4 µM for 30 min. T cells were then washed and resuspended in phenol red–free complete RPMI (5% FBS and 100 U/ml penicillin/streptomycin) and incubated for an additional 30 min to allow full de-esterification of the intracellular esters. Microplates were washed with phenol red–free complete RPMI and T cells were added to the well immediately before the start of imaging. Imaging was performed with a 20× objective lens (Olympus) using 340-nm and 380-nm excitation every 30 s for 30 min. Emission in the GFP channel was also acquired to localize tumor cells.
Image analysis
Using SlideBook image analysis software (https://www.intelligent-imaging.com/slidebook), the ratio of 340-nm to 380-nm excitation for each cell in the field of view was calculated. Responses of all cells were aligned based on the initial time of influx and then averaged across the dataset.
ERK activation assay
96-well plates were seeded with 100,000 of MCA-N4 tumor cells or parental MCA205 cells as control 4–5 h before the start of the experiment. T cells isolated from tumors (see above) were allowed to rest in prewarmed complete RPMI at 37°C for 1 h. 100,000 T cells were added to each well and incubated at 37°C for 20 min. Ice-cold 4% PFA was then added for a final concentration of 1.6% PFA, and cells were fixed on ice for 10 min. Cells were then spun down and permeabilized with ice-cold 90% MeOH for 20 min on ice. Plates were spun down and washed with FACS buffer (PBS with 3% FBS) and stained for 30 min at room temperature with anti-phospho-p44/42 MAPK (Cell Signaling Technology; clone E10), BV650-conjugated anti-CD8α (BioLegend; clone 53–6.7), and FITC-conjugated anti-Thy1.1 (eBioscience; clone HIS51). Primary antibodies were then washed off before the addition of Alexa Fluor 647–conjugated goat anti-mouse IgG1 (Invitrogen). Cells were incubated for 30 min at room temperature before being washed and analyzed by flow cytometry.
RMA-S MHC class I binding assay
TAP-deficient RMA-S cells were incubated with a 1-nM to 1-µM concentration of N4, F6, or D4 peptides for 2 h at 37°C. Cells were then washed and stained for surface expression of H-2Db.
RNA-seq
RNA from sorted cells was extracted using the RNeasy Mini Kit (Qiagen; catalog no. 74104) according to instructions provided by the manufacturer. After RiboGreen quantification and quality control by an Agilent BioAnalyzer, total RNA underwent amplification using the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech), with 12 cycles of amplification for 2 ng input. Subsequently, 10 ng amplified cDNA was used to prepare libraries with the KAPA Hyper Prep Kit (Kapa Biosystems) using eight cycles of PCR. Samples were barcoded and run on a HiSeq 2500 in a 50-bp/50-bp paired-end run with the HiSeq SBS Kit v4 (Illumina). An average of 53 million paired reads were generated per sample.
ATAC-seq
Profiling of chromatin was performed by ATAC-seq as previously described (Buenrostro et al., 2013). Briefly, 38,000–64,000 viably frozen T cells were washed in cold PBS and lysed. The transposition reaction was incubated at 42°C for 45 min. The DNA was cleaned with the MinElute PCR Purification Kit (Qiagen; catalog no. 28004), and material was amplified for five cycles. After evaluation by real-time PCR, 7–13 additional PCR cycles were done. The final product was cleaned by AMPure XP beads (Beckman Coulter, catalog no. A63882) at a 1× ratio, and size selection was performed at a 0.5× ratio. Libraries were sequenced on a HiSeq 2500 or HiSeq 4000 in a 50-bp/50-bp paired-end run using the TruSeq SBS Kit v4, HiSeq Rapid SBS Kit v2, or HiSeq 3000/4000 SBS Kit (Illumina). An average of 60 million paired reads were generated per sample.
Bioinformatics methods
The quality of the sequenced reads was assessed with FastQC and QoRTs (for RNA-seq samples; Hartley and Mullikin, 2015; Andrews, 2010). Unless stated otherwise, plots involving high-throughput sequencing data were created using R version 3.4.1 (see https://github.com/abcwcm/Shakiba2020 for the code; R Core Team, 2017) and ggplot2 (Wickham, 2016).
RNA-seq data
DNA sequencing reads were aligned with default parameters to the mouse reference genome (GRCm38) using STAR (Dobin et al., 2013). Gene expression estimates were obtained with featureCounts using composite gene models (union of the exons of all transcript isoforms per gene) from Gencode (version M16; Liao et al., 2014).
DEGs
Based on similarity in phenotype, function (Figs. 2 and 3), and transcriptional profile (Fig. 4 A) of TIL-N4 and TIL-F6, they were treated as replicates for downstream DEG analysis. DEGs were determined with DESeq2 with a q-value cutoff of 0.1.
Pathway and GO term enrichment analyses
Gene set enrichment analyses were done using GSEA (Subramanian et al., 2005) on reads per kilobase million values (the seed was set to 149).
GO analysis was performed on up- and down-regulated DEGs using the goseq R package (Young et al., 2010). Only GO categories overenriched using a 0.05 false discovery rate cutoff were considered.
Heatmaps
Heatmaps in Fig. 4 C were created using log2-transformed read counts per million of genes identified as differentially expressed by DESeq2. Rows were centered and scaled.
ATAC-seq data
Alignment and identification of open chromatin regions
The data were processed following the recommendations of the ENCODE consortium (https://www.encodeproject.org/atac-seq/). Reads were aligned to the mouse reference genome (version GRCm38) with BWA-backtrack (Li and Durbin, 2009). Postalignment filtering was done with samtools and Picard tools (Li et al., 2009) to remove unmapped reads, improperly paired reads, nonunique reads, and duplicates. To identify regions of open chromatin represented by enrichments of reads, peak calling was performed with MACS2 (Liu, 2014). For every replicate, the narrowpeak results of MACS2 were used after filtering for adjusted P values smaller than 0.01. Filtered peaks were annotated using the ChIPseeker package in R (Yu et al., 2015).
Differentially accessible regions
Regions where the chromatin accessibility changed between different conditions were identified with DiffBind, with the following options: minOverlap = 5, bUseSummarizeOverlaps = T, minMembers = 3, and bFullLibrarySize = TRUE. 16,264 differentially accessible peaks were identified between high- and low-affinity TST cells. A q-value cutoff of <0.05 was used.
GO term enrichment analyses
Enrichment of GO terms was calculated on hyper- or hypoaccessible peaks separately using GREAT v3 (Genomic Regions Enrichment of Annotations Tool) with default parameters (McLean et al., 2010). The consensus peakset identified by DiffBind was used as the background set.
Coverage files
Individual coverage files per replicate normalized for differences in sequencing depths between the different samples were generated with bamCoverage of the deepTools suite using the following parameters: -bs 10–normalizeUsing RPGC–effectiveGenomeSize 2652783500–blackListFileName mm10.blacklist–ignoreForNormalization chrX chrY–ignoreDuplicates–minFragmentLength 40 -p 1. To create merged coverage files of replicates of the same condition, we used multiBigwigSummary to obtain the sequencing-depth-normalized coverage values for 10-bp bins along the entire genome; i.e., for every condition, we obtained a table with the coverage values in every replicate within the same bin. Subsequently, we chose the mean value for every bin to represent the coverage in the resulting “merged” file (see https://github.com/abcwcm/Shakiba2020 for the actual code that was used). Merged coverage files were used for display in Integrated Genomics Viewer shown in Fig. S3 G.
Heatmaps
Heatmaps based on the differentially accessible peaks identified between high- and low-affinity TST cells as shown in Fig. 4 G were created using the dba.plotHeatmap function from the DiffBind package.
Motif analyses
To identify motifs enriched in regions with changes of chromatin accessibility, we ran v-4.9.1 (Heinz et al., 2010) with the following flags: -size given –mask. HOMER was run separately on hyper- or hypoaccessible peaks. The consensus peakset identified by DiffBind was used as the background set.
Combining RNA-seq and ATAC-seq data
The relationship between RNA-seq and ATAC-seq was explored via “diamond” plots for genes enriched for select transcription factor motifs. Each gene was represented by a stack of diamond-shaped points colored by that gene’s associated chromatin state (blue indicating closing and red indicating opening). The bottom-most point in each stack corresponds to the log2 fold change in expression of that gene.
Cloning Cd8a-targeting sgRNA into pMIUR
To target Cd8a locus, three sgRNA were designed using the Broad Institute GPP sgRNA Designer tool (Doench et al., 2016): sgRNA-1 (5′-TGGGTGAGTCGATTATCCTG-3′), sgRNA-2 (5′-ATCCCACAACAAGATAACGT-3′), and sgRNA-3 (5′-CACCCTGAACAAGTTCAGCA-3′). A sgRNA targeting a safe harbor on chromosome 8 was used as control (5′-GACATTTCTTTCCCCACTGG-3′). sgRNAs were cloned using standard restriction enzyme-based cloning strategies. Briefly, pMIUR was digested with BbsI-HF (New England Biolabs). Annealed and complementary sgRNA pairs were ligated to the linearized backbone. All constructs were sequence verified by Sanger sequencing.
Anti-tumor efficacy study with Cd8a KO T cells
For the TAG model, TCROTI mice were injected subcutaneously with 2 × 106 MCA-F6 tumor cells. For the OVA model, C57BL/6J mice were injected subcutaneously with 1 × 106 B16-OVA tumor cells. The sgRNAs targeting the Cd8a locus (Cd8a sgRNA) or a safe harbor on chromosome 8 (control sgRNA) were delivered to TCRTAG;Cas9 Thy1.1/1.2 or TCROT1;Cas9 Thy1.1/1.2 splenocytes using retroviral transduction. Briefly, Platinum-E cells (ATCC) were transfected with each construct using the Mirus TransIT-LT1 reagent (catalog no. 2305). Viral supernatant was supplemented with polybrene and added to splenocytes, and the cells were transduced via spinfection on two consecutive days. For splenocytes transduced with Cd8a sgRNA, live CD4−CD90.1+RFP+CD8− cells were FACS sorted 48 h after transduction for AT. For those transduced with control sgRNA, live CD4−CD90.1+RFP+CD8+ cells were FACS sorted. C57BL/6J tumor-bearing mice were treated with cyclophosphamide (250 mg/kg) 1 d before AT, and 230,000 T cells were transferred i.v. into each mouse (∼14 d after tumor implantation). Mice were treated with anti-PD1 and anti-PDL1 (200 µg each, per mouse) starting day 4 after AT and every other day thereafter. Tumors were measured manually with a caliper. Tumor volume was estimated with the formula (length × width × height)/2.
Data reporting
No statistical methods were used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment. Mice were excluded if donor or tumor-infiltrating T cells could not be found.
Statistical analyses
Statistical analyses on flow cytometric data were performed using unpaired two-tailed Student’s t tests (Prism 7.0, GraphPad Software). A P value of < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows MHC class I expression levels and growth kinetics of MCA-APL tumors, as well as phenotypic characteristics of tumor-specific T cells in MCA-APL tumors. Fig. S2 shows transcriptional features of tumor-specific T cells. Fig. S3 shows chromatin accessibility changes of MCA-APL tumor-infiltrating T cells. Fig. S4 shows gains and losses of regulatory elements for DEG containing NUR77- or NFAT2-binding motifs. Fig. S5 shows functional, phenotypic, and transcriptional differences between Cd8a-deficient and control T cells.
Acknowledgments
We thank the members of the Schietinger laboratory, Ming Li, and Steve Reiner for helpful discussions. We thank the MSKCC Flow Cytometry Core and the Integrated Genomics Operation Core for their outstanding support.
This work was supported by National Institutes of Health/National Cancer Institute grants DP2 CA225212 and R00 CA172371 (A. Schietinger), the V Foundation for Cancer Research (A. Schietinger), the Josie Robertson Young Investigator Award (A. Schietinger), the Cancer Research Institute Lloyd Old STAR Award (A. Schietinger), Pershing Square Foundation Sohn Cancer Research Alliance (A. Schietinger), the Weill Cornell Medicine Core Laboratories Center (P. Zumbo, F. Dündar, and D. Betel), the U.S. Department of Defense Cancer Horizon Award 17-1-0277 (M. Shakiba), the V Foundation for Cancer Research Scholar Award (M. Philip), and the Serodino Family Adventure Allee Fund (M. Philip). M.D. Hellmann is supported, in part, by the Damon Runyon Cancer Research Foundation (grant CI-98-18); M.D. Hellmann, D.K. Wells, N.A. Defranoux, and A. Schietinger are members of the Parker Institute for Cancer Immunotherapy. We acknowledge the use of the Integrated Genomics Operation Core, funded by a National Cancer Institute Cancer Center Support Grant (P30 CA08748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Author contributions: M. Shakiba, M. Philip, and A. Schietinger conceived and designed the study, carried out experiments, and analyzed and interpreted data. F. Dündar, P. Zumbo, and D. Betel performed all computational analyses presented in this paper. R.P. Koche, V.P. Reuter, and N.D. Socci contributed to initial computational analyses. G. Espinosa-Carrasco, L. Menocal, S. Carson, E.M. Bruno, and S. Camara assisted with experiments. M.D. Hellman, D.K. Wells, and N.A. Defranoux provided critical feedback on human TESLA dataset for discussion. F.J. Sanchez-Rivera and S.W. Lowe designed the sgRNA vectors. B. Whitlock, F. Tamzalit, and M. Huse assisted with microscopy experiments. M. Shakiba, M. Philip, and A. Schietinger wrote the manuscript, with all authors contributing to the writing and providing feedback.
Data availability
The RNA-seq and ATAC-seq data have been deposited in the Gene Expression Omnibus, SuperSeries GSE141818, including GSE141816 (ATAC-seq data) and GSE141817 (RNA-seq data). All data generated and supporting the findings of the study are available within the paper. Additional information and materials will be made available upon request.
References
- Aleksic, M., Liddy N., Molloy P.E., Pumphrey N., Vuidepot A., Chang K.M., and Jakobsen B.K.. 2012. Different affinity windows for virus and cancer-specific T-cell receptors: implications for therapeutic strategies. Eur. J. Immunol. 42:3174–3179. 10.1002/eji.201242606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan-Bonnet, G., and Germain R.N.. 2005. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 3:e356. 10.1371/journal.pbio.0030356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Artyomov, M.N., Lis M., Devadas S., Davis M.M., and Chakraborty A.K.. 2010. CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc. Natl. Acad. Sci. USA. 107:16916–16921. 10.1073/pnas.1010568107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi, E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M., et al. 2018. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. 174:1293–1308 e36. 10.1016/j.cell.2018.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, T.A., and Hogquist K.A.. 2007. Transcriptional analysis of clonal deletion in vivo. J. Immunol. 179:837–844. 10.4049/jimmunol.179.2.837 [DOI] [PubMed] [Google Scholar]
- Balyan, R., Gund R., Ebenezer C., Khalsa J.K., Verghese D.A., Krishnamurthy T., George A., Bal V., Rath S., and Chaudhry A.. 2017. Modulation of Naive CD8 T Cell Response Features by Ligand Density, Affinity, and Continued Signaling via Internalized TCRs. J. Immunol. 198:1823–1837. 10.4049/jimmunol.1600083 [DOI] [PubMed] [Google Scholar]
- Bengsch, B., Ohtani T., Khan O., Setty M., Manne S., O’Brien S., Gherardini P.F., Herati R.S., Huang A.C., Chang K.M., et al. 2018. Epigenomic-Guided Mass Cytometry Profiling Reveals Disease-Specific Features of Exhausted CD8 T Cells. Immunity. 48:1029–1045 e5. 10.1016/j.immuni.2018.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenstein, T., Leisegang M., Uckert W., and Schreiber H.. 2015. Targeting cancer-specific mutations by T cell receptor gene therapy. Curr. Opin. Immunol. 33:112–119. 10.1016/j.coi.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobisse, S., Genolet R., Roberti A., Tanyi J.L., Racle J., Stevenson B.J., Iseli C., Michel A., Le Bitoux M.A., Guillaume P., et al. 2018. Sensitive and frequent identification of high avidity neo-epitope specific CD8 (+) T cells in immunotherapy-naive ovarian cancer. Nat. Commun. 9:1092. 10.1038/s41467-018-03301-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, R., Marquardt K.L., Cheung J., and Sherman L.A.. 2012. Functional differences between low- and high-affinity CD8(+) T cells in the tumor environment. OncoImmunology. 1:1239–1247. 10.4161/onci.21285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockstedt, D.G., Giedlin M.A., Leong M.L., Bahjat K.S., Gao Y., Luckett W., Liu W., Cook D.N., Portnoy D.A., and Dubensky T.W. Jr. 2004. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc. Natl. Acad. Sci. USA. 101:13832–13837. 10.1073/pnas.0406035101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J.D., Giresi P.G., Zaba L.C., Chang H.Y., and Greenleaf W.J.. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10:1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhrman, J.D., and Slansky J.E.. 2013. Improving T cell responses to modified peptides in tumor vaccines. Immunol. Res. 55:34–47. 10.1007/s12026-012-8348-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, B.J., Gerry A.B., Dukes J., Harper J.V., Kannan V., Bianchi F.C., Grand F., Brewer J.E., Gupta M., Plesa G., et al. 2013. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci. Transl. Med. 5:197ra103. 10.1126/scitranslmed.3006034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis, A.G., Egan D.N., Bar M., Schmitt T.M., McAfee M.S., Paulson K.G., Voillet V., Gottardo R., Ragnarsson G.B., Bleakley M., et al. 2019. T cell receptor gene therapy targeting WT1 prevents acute myeloid leukemia relapse post-transplant. Nat. Med. 25:1064–1072. 10.1038/s41591-019-0472-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley, J.M., Gallagher M.P., and Berg L.J.. 2016. T Cells and Gene Regulation: The Switching On and Turning Up of Genes after T Cell Receptor Stimulation in CD8 T Cells. Front. Immunol. 7:76. 10.3389/fimmu.2016.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, M.A., Teixeiro E., Gill J., Hausmann B., Roubaty D., Holmberg K., Werlen G., Hollander G.A., Gascoigne N.R., and Palmer E.. 2006. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 444:724–729. 10.1038/nature05269 [DOI] [PubMed] [Google Scholar]
- Denton, A.E., Wesselingh R., Gras S., Guillonneau C., Olson M.R., Mintern J.D., Zeng W., Jackson D.C., Rossjohn J., Hodgkin P.D., et al. 2011. Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses. J. Immunol. 187:5733–5744. 10.4049/jimmunol.1003937 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34:184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch, R.E., Lewis R.S., Goodnow C.C., and Healy J.I.. 1997. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 386:855–858. 10.1038/386855a0 [DOI] [PubMed] [Google Scholar]
- Evavold, B.D., and Allen P.M.. 1991. Separation of IL-4 production from Th cell proliferation by an altered T cell receptor ligand. Science. 252:1308–1310. 10.1126/science.1833816 [DOI] [PubMed] [Google Scholar]
- Feucht, J., Sun J., Eyquem J., Ho Y.J., Zhao Z., Leibold J., Dobrin A., Cabriolu A., Hamieh M., and Sadelain M.. 2019. Calibration of CAR activation potential directs alternative T cell fates and therapeutic potency. Nat. Med. 25:82–88. 10.1038/s41591-018-0290-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros, A., Robbins P.F., Yao X., Li Y.F., Turcotte S., Tran E., Wunderlich J.R., Mixon A., Farid S., Dudley M.E., et al. 2014. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J. Clin. Invest. 124:2246–2259. 10.1172/JCI73639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros, A., Parkhurst M.R., Tran E., Pasetto A., Robbins P.F., Ilyas S., Prickett T.D., Gartner J.J., Crystal J.S., Roberts I.M., et al. 2016. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 22:433–438. 10.1038/nm.4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros, A., Tran E., Parkhurst M.R., Ilyas S., Pasetto A., Groh E.M., Robbins P.F., Yossef R., Garcia-Garijo A., Fajardo C.A., et al. 2019. Recognition of human gastrointestinal cancer neoantigens by circulating PD-1+ lymphocytes. J. Clin. Invest. 129:4992–5004. 10.1172/JCI127967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley, S.W., and Mullikin J.C.. 2015. QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics. 16:224. 10.1186/s12859-015-0670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M.M., and Slansky J.E.. 2020. T-cell receptor affinity in the age of cancer immunotherapy. Mol. Carcinog. 59:862–870. 10.1002/mc.23212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler, P.D., and Kranz D.M.. 2003. Quantitative analysis of the contribution of TCR/pepMHC affinity and CD8 to T cell activation. Immunity. 18:255–264. 10.1016/S1074-7613(03)00019-0 [DOI] [PubMed] [Google Scholar]
- Hollingsworth, R.E., and Jansen K.. 2019. Turning the corner on therapeutic cancer vaccines. NPJ Vaccines. 4:7. 10.1038/s41541-019-0103-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, C.G., Koehli S., Hausmann B., Schmaler M., Zehn D., and Palmer E.. 2012. T cell affinity regulates asymmetric division, effector cell differentiation, and tissue pathology. Immunity. 37:709–720. 10.1016/j.immuni.2012.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisegang, M., Kammertoens T., Uckert W., and Blankenstein T.. 2016. Targeting human melanoma neoantigens by T cell receptor gene therapy. J. Clin. Invest. 126:854–858. 10.1172/JCI83465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., van der Leun A.M., Yofe I., Lubling Y., Gelbard-Solodkin D., van Akkooi A.C.J., van den Braber M., Rozeman E.A., Haanen J., Blank C.U., et al. 2019. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell. 176:775–789 e18. 10.1016/j.cell.2018.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lin, M.Y., and Welsh R.M.. 1998. Stability and diversity of T cell receptor repertoire usage during lymphocytic choriomeningitis virus infection of mice. J. Exp. Med. 188:1993–2005. 10.1084/jem.188.11.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. 2014. Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol. Biol. 1150:81–95. 10.1007/978-1-4939-0512-6_4 [DOI] [PubMed] [Google Scholar]
- Lu, Y.C., Yao X., Li Y.F., El-Gamil M., Dudley M.E., Yang J.C., Almeida J.R., Douek D.C., Samuels Y., Rosenberg S.A., and Robbins P.F.. 2013. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J. Immunol. 190:6034–6042. 10.4049/jimmunol.1202830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman, M.A., Nugent C.T., Marquardt K.L., Biggs J.A., Pamer E.G., and Sherman L.A.. 2005. The fate of low affinity tumor-specific CD8+ T cells in tumor-bearing mice. J. Immunol. 174:2563–2572. 10.4049/jimmunol.174.5.2563 [DOI] [PubMed] [Google Scholar]
- Marangoni, F., Murooka T.T., Manzo T., Kim E.Y., Carrizosa E., Elpek N.M., and Mempel T.R.. 2013. The transcription factor NFAT exhibits signal memory during serial T cell interactions with antigen-presenting cells. Immunity. 38:237–249. 10.1016/j.immuni.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Usatorre, A., Donda A., Zehn D., and Romero P.. 2018. PD-1 Blockade Unleashes Effector Potential of Both High- and Low-Affinity Tumor-Infiltrating T Cells. J. Immunol. 201:792–803. 10.4049/jimmunol.1701644 [DOI] [PubMed] [Google Scholar]
- McLean, C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., and Bejerano G.. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28:495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan, R.H., McWilliams J.A., Jordan K.R., Dow S.W., Wilson D.B., and Slansky J.E.. 2006. Relating TCR-peptide-MHC affinity to immunogenicity for the design of tumor vaccines. J. Clin. Invest. 116:2543–2551. 10.1172/JCI26936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mognol, G.P., Spreafico R., Wong V., Scott-Browne J.P., Togher S., Hoffmann A., Hogan P.G., Rao A., and Trifari S.. 2017. Exhaustion-associated regulatory regions in CD8(+) tumor-infiltrating T cells. Proc. Natl. Acad. Sci. USA. 114:E2776–E2785. 10.1073/pnas.1620498114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289. 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, R.A., Chinnasamy N., Abate-Daga D., Gros A., Robbins P.F., Zheng Z., Dudley M.E., Feldman S.A., Yang J.C., Sherry R.M., et al. 2013. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J. Immunother. 36:133–151. 10.1097/CJI.0b013e3182829903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga, A.J., Moalli F., Abe J., Swoger J., Sharpe J., Zehn D., Kreutzfeldt M., Merkler D., Ripoll J., and Stein J.V.. 2016. pMHC affinity controls duration of CD8+ T cell-DC interactions and imprints timing of effector differentiation versus expansion. J. Exp. Med. 213:2811–2829. 10.1084/jem.20160206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst, M.R., Yang J.C., Langan R.C., Dudley M.E., Nathan D.A., Feldman S.A., Davis J.L., Morgan R.A., Merino M.J., Sherry R.M., et al. 2011. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol. Ther. 19:620–626. 10.1038/mt.2010.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip, M., and Schietinger A.. 2021. CD8+ T cell differentiation and dysfunction in cancer. Nat. Rev. Immunol. 10.1038/s41577-021-00574-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip, M., Fairchild L., Sun L., Horste E.L., Camara S., Shakiba M., Scott A.C., Viale A., Lauer P., Merghoub T., et al. 2017. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 545:452–456. 10.1038/nature22367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst, H.C., Tschannen K., Gallimore A., Martinic M., Basler M., Dumrese T., Jones E., and van den Broek M.F.. 2003. Immunodominance of an antiviral cytotoxic T cell response is shaped by the kinetics of viral protein expression. J. Immunol. 171:5415–5422. 10.4049/jimmunol.171.10.5415 [DOI] [PubMed] [Google Scholar]
- R Core Team . 2017. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. [Google Scholar]
- Rapoport, A.P., Stadtmauer E.A., Binder-Scholl G.K., Goloubeva O., Vogl D.T., Lacey S.F., Badros A.Z., Garfall A., Weiss B., Finklestein J., et al. 2015. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat. Med. 21:914–921. 10.1038/nm.3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, A.C., Lun A.T.L., Lau W.W.Y., Gottgens B., Marioni J.C., and Griffiths G.M.. 2018. T cell cytolytic capacity is independent of initial stimulation strength. Nat. Immunol. 19:849–858. 10.1038/s41590-018-0160-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman, M., Yizhak K., Bjorgaard S.L., Ray J.P., de Boer C.G., Jenkins R.W., Lieb D.J., Chen J.H., Frederick D.T., Barzily-Rokni M., et al. 2018. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell. 175:998–1013 e20. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schietinger, A., Philip M., Liu R.B., Schreiber K., and Schreiber H.. 2010. Bystander killing of cancer requires the cooperation of CD4(+) and CD8(+) T cells during the effector phase. J. Exp. Med. 207:2469–2477. 10.1084/jem.20092450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schietinger, A., Philip M., Krisnawan V.E., Chiu E.Y., Delrow J.J., Basom R.S., Lauer P., Brockstedt D.G., Knoblaugh S.E., Hammerling G.J., et al. 2016. Tumor-Specific T Cell Dysfunction Is a Dynamic Antigen-Driven Differentiation Program Initiated Early during Tumorigenesis. Immunity. 45:389–401. 10.1016/j.immuni.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, D.A., Irving M.B., Posevitz V., Hebeisen M., Posevitz-Fejfar A., Sarria J.C., Gomez-Eerland R., Thome M., Schumacher T.N., Romero P., et al. 2010. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. J. Immunol. 184:4936–4946. 10.4049/jimmunol.1000173 [DOI] [PubMed] [Google Scholar]
- Schmitt, T.M., Stromnes I.M., Chapuis A.G., and Greenberg P.D.. 2015. New Strategies in Engineering T-cell Receptor Gene-Modified T cells to More Effectively Target Malignancies. Clin. Cancer Res. 21:5191–5197. 10.1158/1078-0432.CCR-15-0860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, T.M., Aggen D.H., Ishida-Tsubota K., Ochsenreither S., Kranz D.M., and Greenberg P.D.. 2017. Generation of higher affinity T cell receptors by antigen-driven differentiation of progenitor T cells in vitro. Nat. Biotechnol. 35:1188–1195. 10.1038/nbt.4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober, K., Voit F., Grassmann S., Muller T.R., Eggert J., Jarosch S., Weissbrich B., Hoffmann P., Borkner L., Nio E., et al. 2020. Reverse TCR repertoire evolution toward dominant low-affinity clones during chronic CMV infection. Nat. Immunol. 21:434–441. 10.1038/s41590-020-0628-2 [DOI] [PubMed] [Google Scholar]
- Scott, A.C., Dundar F., Zumbo P., Chandran S.S., Klebanoff C.A., Shakiba M., Trivedi P., Menocal L., Appleby H., Camara S., et al. 2019. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 571:270–274. 10.1038/s41586-019-1324-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim, M.J.W., Lu J., Spencer M., Hopkins F., Tran E., Rosenberg S.A., Long E.O., and Sun P.D.. 2020. High-affinity oligoclonal TCRs define effective adoptive T cell therapy targeting mutant KRAS-G12D. Proc. Natl. Acad. Sci. USA. 117:12826–12835. 10.1073/pnas.1921964117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni, Y., Becht E., Fehlings M., Loh C.Y., Koo S.L., Teng K.W.W., Yeong J.P.S., Nahar R., Zhang T., Kared H., et al. 2018. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 557:575–579. 10.1038/s41586-018-0130-2 [DOI] [PubMed] [Google Scholar]
- Skokos, D., Shakhar G., Varma R., Waite J.C., Cameron T.O., Lindquist R.L., Schwickert T., Nussenzweig M.C., and Dustin M.L.. 2007. Peptide-MHC potency governs dynamic interactions between T cells and dendritic cells in lymph nodes. Nat. Immunol. 8:835–844. 10.1038/ni1490 [DOI] [PubMed] [Google Scholar]
- Slansky, J.E., and Jordan K.R.. 2010. The Goldilocks model for TCR-too much attraction might not be best for vaccine design. PLoS Biol. 8:e1000482. 10.1371/journal.pbio.1000482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanović, S., Pasetto A., Helman S.R., Gartner J.J., Prickett T.D., Howie B., Robins H.S., Robbins P.F., Klebanoff C.A., Rosenberg S.A., and Hinrichs C.S.. 2017. Landscape of immunogenic tumor antigens in successful immunotherapy of virally induced epithelial cancer. Science. 356:200–205. 10.1126/science.aak9510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, J.D., Harris D.T., and Kranz D.M.. 2015. TCR affinity for p/MHC formed by tumor antigens that are self-proteins: impact on efficacy and toxicity. Curr. Opin. Immunol. 33:16–22. 10.1016/j.coi.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., Anderson R.W., Maloy W.L., and Tevethia S.S.. 1989. Localization of an immunorecessive epitope on SV40 T antigen by H-2Db-restricted cytotoxic T-lymphocyte clones and a synthetic peptide. Virology. 171:205–213. 10.1016/0042-6822(89)90527-8 [DOI] [PubMed] [Google Scholar]
- Thommen, D.S., and Schumacher T.N.. 2018. T Cell Dysfunction in Cancer. Cancer Cell. 33:547–562. 10.1016/j.ccell.2018.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, A., Ohlen C., Bastin J., Ljunggren H.G., Foster L., and Karre K.. 1989. Association of class I major histocompatibility heavy and light chains induced by viral peptides. Nature. 340:443–448. 10.1038/340443a0 [DOI] [PubMed] [Google Scholar]
- Tran, E., Turcotte S., Gros A., Robbins P.F., Lu Y.C., Dudley M.E., Wunderlich J.R., Somerville R.P., Hogan K., Hinrichs C.S., et al. 2014. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 344:641–645. 10.1126/science.1251102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno, T., Tomiyama H., Fujiwara M., Oka S., and Takiguchi M.. 2004. Functionally impaired HIV-specific CD8 T cells show high affinity TCR-ligand interactions. J. Immunol. 173:5451–5457. 10.4049/jimmunol.173.9.5451 [DOI] [PubMed] [Google Scholar]
- Wells, D.K., van Buuren M.M., Dang K.K., Hubbard-Lucey V.M., Sheehan K.C.F., Campbell K.M., Lamb A., Ward J.P., Sidney J., Blazquez A.B., et al. 2020. Key Parameters of Tumor Epitope Immunogenicity Revealed Through a Consortium Approach Improve Neoantigen Prediction. Cell. 183:818–834 e13. 10.1016/j.cell.2020.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, E.E., Youngblood B., Tan W.G., Jin H.T., Araki K., Alexe G., Konieczny B.T., Calpe S., Freeman G.J., Terhorst C., et al. 2011. Tight regulation of memory CD8(+) T cells limits their effectiveness during sustained high viral load. Immunity. 35:285–298. 10.1016/j.immuni.2011.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. 2016. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag. https://ggplot2.tidyverse.org
- Willimsky, G., and Blankenstein T.. 2005. Sporadic immunogenic tumours avoid destruction by inducing T-cell tolerance. Nature. 437:141–146. 10.1038/nature03954 [DOI] [PubMed] [Google Scholar]
- Young, M.D., Wakefield M.J., Smyth G.K., and Oshlack A.. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang L.G., and He Q.Y.. 2015. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 31:2382–2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Zehn, D., Lee S.Y., and Bevan M.J.. 2009. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 458:211–214. 10.1038/nature07657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, S., Malecek K., Johnson L.A., Yu Z., Vega-Saenz de Miera E., Darvishian F., McGary K., Huang K., Boyer J., Corse E., et al. 2013. T-cell receptor affinity and avidity defines antitumor response and autoimmunity in T-cell immunotherapy. Proc. Natl. Acad. Sci. USA. 110:6973–6978. 10.1073/pnas.1221609110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Dudley M.E., Rosenberg S.A., and Robbins P.F.. 2005. Persistence of multiple tumor-specific T-cell clones is associated with complete tumor regression in a melanoma patient receiving adoptive cell transfer therapy. J. Immunother. 28:53–62. 10.1097/00002371-200501000-00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikherman, J., and Au-Yeung B.. 2015. The role of T cell receptor signaling thresholds in guiding T cell fate decisions. Curr. Opin. Immunol. 33:43–48. 10.1016/j.coi.2015.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA-seq and ATAC-seq data have been deposited in the Gene Expression Omnibus, SuperSeries GSE141818, including GSE141816 (ATAC-seq data) and GSE141817 (RNA-seq data). All data generated and supporting the findings of the study are available within the paper. Additional information and materials will be made available upon request.