Genome-wide CRISPR screens carried out with and without NPC1 function identify shared pathways that coordinately control lysosomal cholesterol and bis(monoacylglycero)phosphate. ER-localized SNX13 protein plays an unexpected regulatory role in modifying NPC1 function and forming membrane contacts to regulate cellular cholesterol localization.
Abstract
We report here two genome-wide CRISPR screens performed to identify genes that, when knocked out, alter levels of lysosomal cholesterol or bis(monoacylglycero)phosphate. In addition, these screens were also performed under conditions of NPC1 inhibition to identify modifiers of NPC1 function in lysosomal cholesterol export. The screens confirm tight coregulation of cholesterol and bis(monoacylglycero)phosphate in cells and reveal an unexpected role for the ER-localized SNX13 protein as a negative regulator of lysosomal cholesterol export and contributor to ER–lysosome membrane contact sites. In the absence of NPC1 function, SNX13 knockdown redistributes lysosomal cholesterol and is accompanied by triacylglycerol-rich lipid droplet accumulation and increased lysosomal bis(monoacylglycero)phosphate. These experiments provide unexpected insight into the regulation of lysosomal lipids and modification of these processes by novel gene products.
Introduction
Cellular lipid homeostasis is maintained by complex and dynamic interorganelle communication processes that coordinate uptake, biosynthesis, and degradation of >1,000 lipid species. Among all the organelles involved in lipid regulation, the lysosome plays a central role. Lysosomes are the final station at which endocytosed lipoprotein particles and membranes derived from intralumenal budding and autophagy undergo a series of degradative reactions to yield unesterified cholesterol and other lipid precursors (Gruenberg, 2020; Ballabio and Bonifacino, 2020). Free cholesterol is then exported out of the lysosome and either recycled for de novo synthesis of biological membranes and other sterol products or esterified and stored in lipid droplets (LDs).
One of the most studied lipid storage disorders is Niemann-Pick type C (NPC) disease, caused by genetic defects in the lysosomal cholesterol transport system, Niemann-Pick C1 and C2 proteins (NPC1 and NPC2). NPC mutant cells accumulate cholesterol and glycosphingolipids in lysosomes, leading to neurodegeneration and premature death (Pentchev, 2004). Despite recent advances in understanding of how transmembrane NPC1 and lumenal NPC2 export cholesterol from lysosomes (Pfeffer, 2019), the precise molecular events that regulate this process and function downstream of NPC1 are still unclear (Das et al., 2014; Infante and Radhakrishnan, 2017). Once cholesterol exits lysosomes, membrane contact sites between the lysosome surface and other juxtaposed compartments deliver cholesterol to the ER, a process that involves transit via the plasma membrane (PM; Infante and Radhakrishnan, 2017).
An important player in cholesterol regulation is bis(monoacylglycero)phosphate (BMP; also known as lysobisphosphatidic acid), which is found almost exclusively in intralumenal vesicles of multivesicular endosomes (MVEs; Gruenberg, 2020). Elevation of BMP levels occurs in many lipid storage disorders, including NPC, and BMP plays important roles in lipid catabolism and lysosomal cholesterol egress (Chevallier et al., 2008; reviewed in McCauliff et al., 2019). In the absence of active NPC1, cells fed phosphatidylglycerol increase their BMP content, which decreases their lysosomal cholesterol levels by a process that requires NPC2 (McCauliff et al., 2019). These findings support the existence of NPC1-independent, relatively slow cholesterol export pathways.
Because much remains to be learned regarding the mechanisms of cholesterol transport and its regulation, we performed genome-wide CRISPR screens to identify regulators of lysosomal cholesterol and BMP homeostasis. We repeated these screens under conditions of NPC1 inhibition to identify cellular components that may function in parallel with the NPC1 pathway to accomplish cholesterol export; such gene products might offer pathways to benefit patients with NPC disease. This strategy allowed us to confirm known components that regulate cholesterol transport and metabolism and revealed other previously unrecognized players. As one example, we show here that SNX13 is an ER-resident inter-organelle tether that regulates lysosomal cholesterol export. Remarkably, SNX13-depleted cells are able to redistribute cholesterol to the PM and other compartments, despite the absence of NPC1 function.
Results
Genome-wide screens to identify regulators of endolysosomal cholesterol
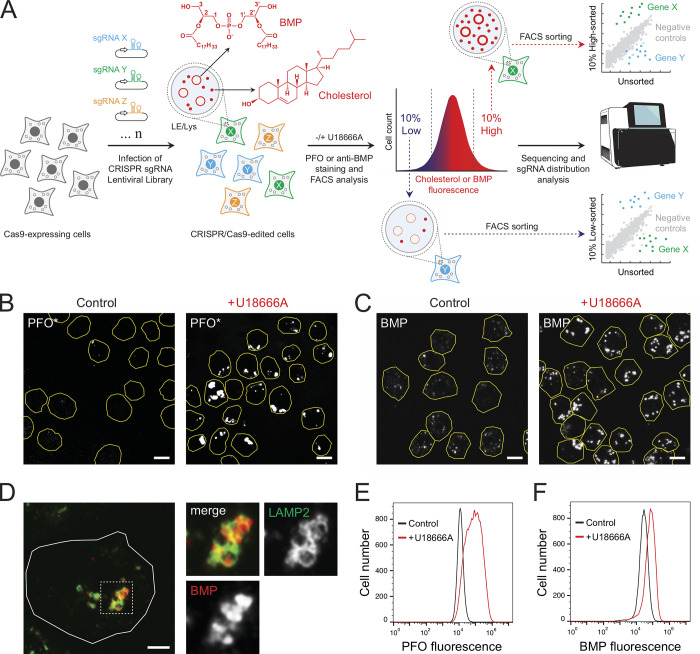
We established two screening protocols to monitor changes in either cholesterol or BMP using fluorescently tagged perfringolysin O* (PFO*) to label accessible cholesterol (Das et al., 2013) or an mAb to detect BMP in K562 cells in conjunction with flow cytometry (Fig. 1 A). Briefly, Cas9-expressing K562 cells were infected with libraries containing 10 sgRNA guides per gene and grown for 10 d before analysis for their content of cholesterol or BMP. In addition, both screens were also performed in parallel under conditions in which the lysosomal cholesterol exporter, NPC1, was inhibited by addition of U18666A to identify modifiers of NPC1 protein function (Lu et al., 2015). Cells were labeled and sorted by flow cytometry, and cells displaying the highest or lowest 10% signals for each marker were collected and sequenced (Fig. 1 A).
Figure 1.
Flow cytometry–based screens to identify cholesterol and BMP homeostatic regulators. (A) A genome-wide CRISPR sgRNA lentiviral library (10 sgRNAs/gene) was used to infect Cas9-expressing human K562 cells such that, after infection and selection, each cell expresses only a single sgRNA. CRISPR/Cas9-edited cells were then stained with either anti-BMP antibodies or fluorescently conjugated, cholesterol binding PFO*. LE, late endosome. (B) Immunofluorescence of K562 cells labeled with fluorescent PFO* with or without U18666A for 16 h. Cell boundaries (yellow) are shown and were determined by mCherry expression. (C) K562 cells stained with anti-BMP antibodies and circled as in B. (D) K562 cells with anti-LAMP2 (green) and anti-BMP antibodies (red). (E and F) Flow cytometry of cells labeled with either fluorescent PFO* or anti-BMP as in B and C with or without U18666A. Scale bars, 10 μm (B and C); 5μm (D).
As shown in Fig. 1 B, WT K562 cells showed little PFO* staining unless NPC1 was inhibited using U18666A. Moreover, endolysosomal cholesterol accumulation seen with U18666A was accompanied by increased BMP that was localized (as expected) in LAMP2-positive structures (Fig. 1, C and D). These phenotypes were easily scored by flow cytometry (Fig. 1, E and F) where U18666A treated cells were easily resolved by their cholesterol or BMP content. Note that this analysis monitors PFO* staining, which likely detects so-called active cholesterol pools (compare Das et al., 2014).
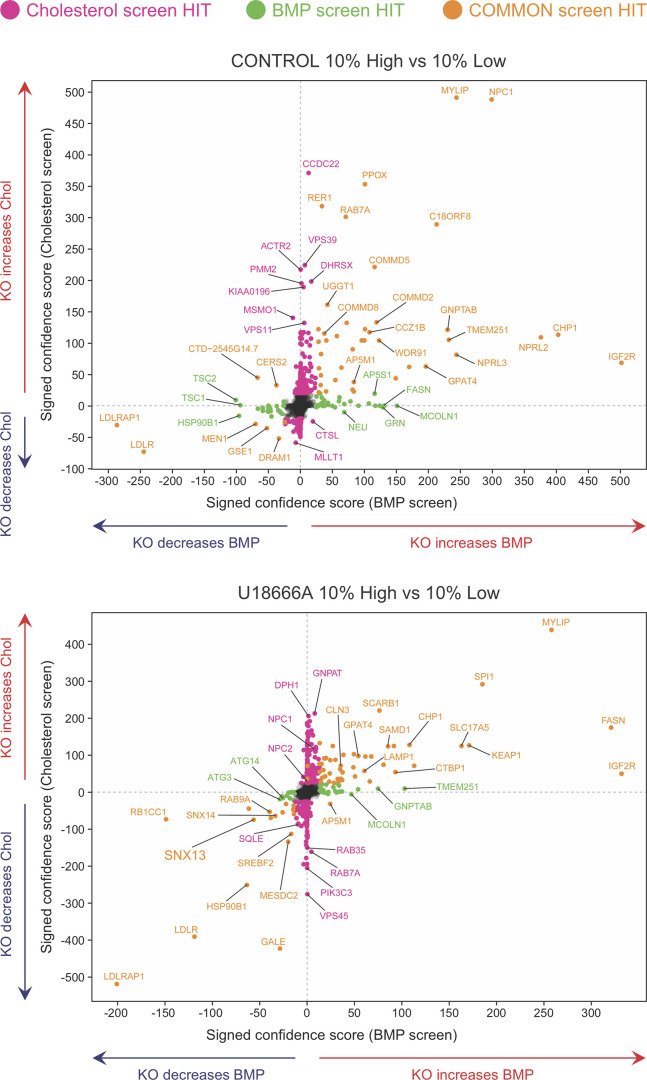
Fig. 2 presents the compiled effects of gene depletions on PFO*-stained cholesterol, BMP, and both of those lipids with and without U18666A, performed twice (see also Figs. S1, S2, and S3). Genes in the upper left and right quadrants of Fig. 2 increase PFO*-stained cholesterol upon knockout; genes in the right top and bottom quadrants increase BMP antibody staining upon knockout. Importantly, as expected, the strongest hits triggering cholesterol accumulation under control conditions were NPC1 and MYLIP, a ubiquitin ligase that regulates low-density lipoprotein receptor (LDLR) levels and would be expected to increase cholesterol uptake upon deletion. Similarly, knockdown of LDLR or the LDLRAP1 LDLR-specific endocytic adaptor decreased endolysosomal PFO*-stained cholesterol and BMP, both with and without U18666A treatment. This recapitulates prior studies showing that cholesterol accumulation in lysosomes is accompanied by increases in antibody-detected BMP (Gruenberg, 2020). Knockdown of the IGF2R mannose 6-phosphate receptor had a much greater effect on BMP levels than PFO*-stained cholesterol levels (Fig. 2, far right). This suggests that cells may up-regulate BMP synthesis to compensate for a defect in lysosomal enzyme delivery; alternatively, it may reflect the requirement for a mannose 6–phosphorylated enzyme(s) to carry out BMP degradation. It is also of interest that most of the hits were located in the upper right or lower left quadrants of the graphs, with no hits increasing cholesterol while simultaneously decreasing BMP and conversely no hits increasing BMP and decreasing cholesterol.
Figure 2.
Bivariate analyses comparing hits in relation to changes in cholesterol and BMP. Indicated genes presented in relation to their phenotypes determined by PFO* and BMP detection and flow cytometry, presented as signed confidence scores. Genes that appeared in both analyses are shown in goldenrod; those seen only in the cholesterol or BMP screens are shown in pink or green, respectively. The panels represent hits discovered in the top 10% versus bottom 10% comparisons with or without U18666A as indicated. Chol, cholesterol; KO, knockout.
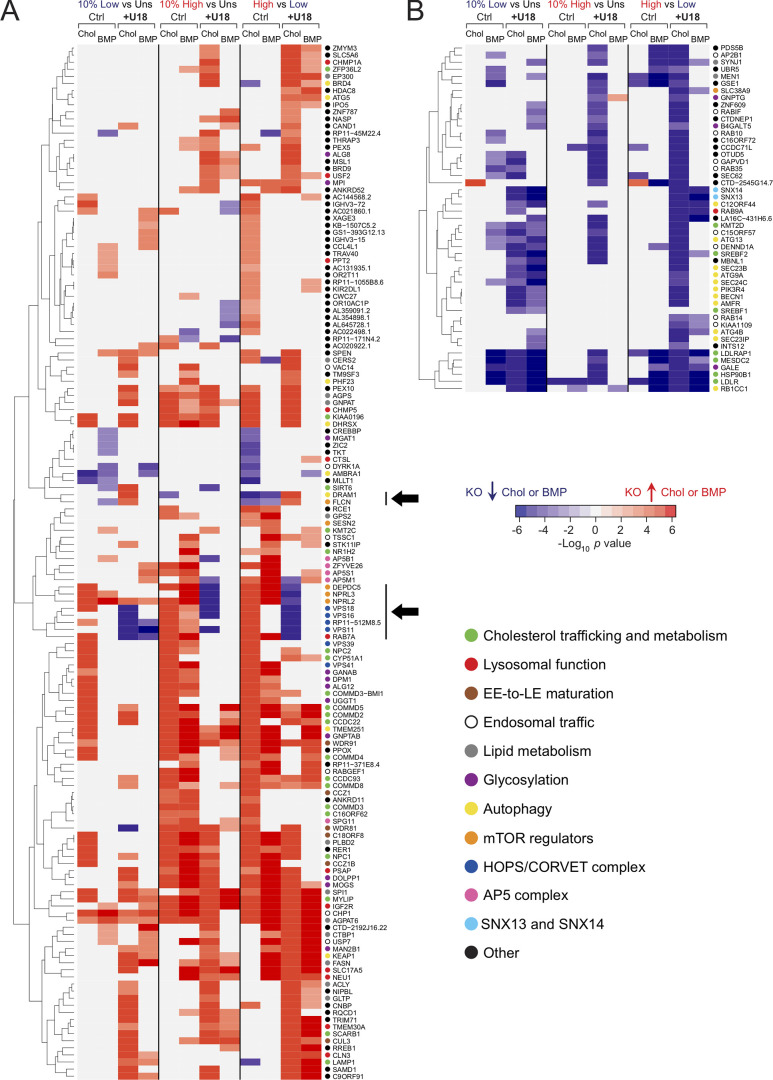
Figure S1.
Hierarchical clustering analysis comparing 10% low and 10% high populations. (A and B) Two main categories of genes; those increasing (A) or decreasing (B) cholesterol (Chol) and/or BMP are shown. For each category, the enrichment for cholesterol or BMP is shown as a heat map value. Responses under control (Ctrl) and U18666A (U18) conditions are also shown. The three types of screen data analysis performed (10% high versus unsorted [Uns], 10% low versus Uns, and 10% high versus 10% low) are indicated at the top. Only genes that were detected in both cholesterol and BMP screens are shown. Color intensity is presented using a logarithmic scale heat map proportional to the signed P value, as indicated. Individual hits in the 10% high pool are generally enriched in that population but may also be detected in the 10% low pool, explaining why red hits appear more populated on the 10% high versus Uns columns in A and blue hits on the 10% low versus Uns columns in B. In general, the behavior is consistent. Black arrows indicate hits that, upon deletion, yielded opposite phenotypes in control versus with U18666A conditions. Individual gene functions are shown using colored circles as indicated. EE, early endosome; KO, knockout.
Figure S2.
Subcellular localization of screen hits and their phenotypes. Selected top hits are annotated in colored circles that show increases (red) or decreases (blue) in cholesterol (Chol). Upper left, BMP; lower left, cholesterol with U18666A (upper right) or BMP with U18666A (lower right) as indicated in the legend at the bottom. Black arrows indicate trafficking routes. Green arrows indicate activation; inhibition signs are also displayed in green. COP, coat proteins; EE, early endosome; KO, knockout; LE, late endosome.
Figure S3.
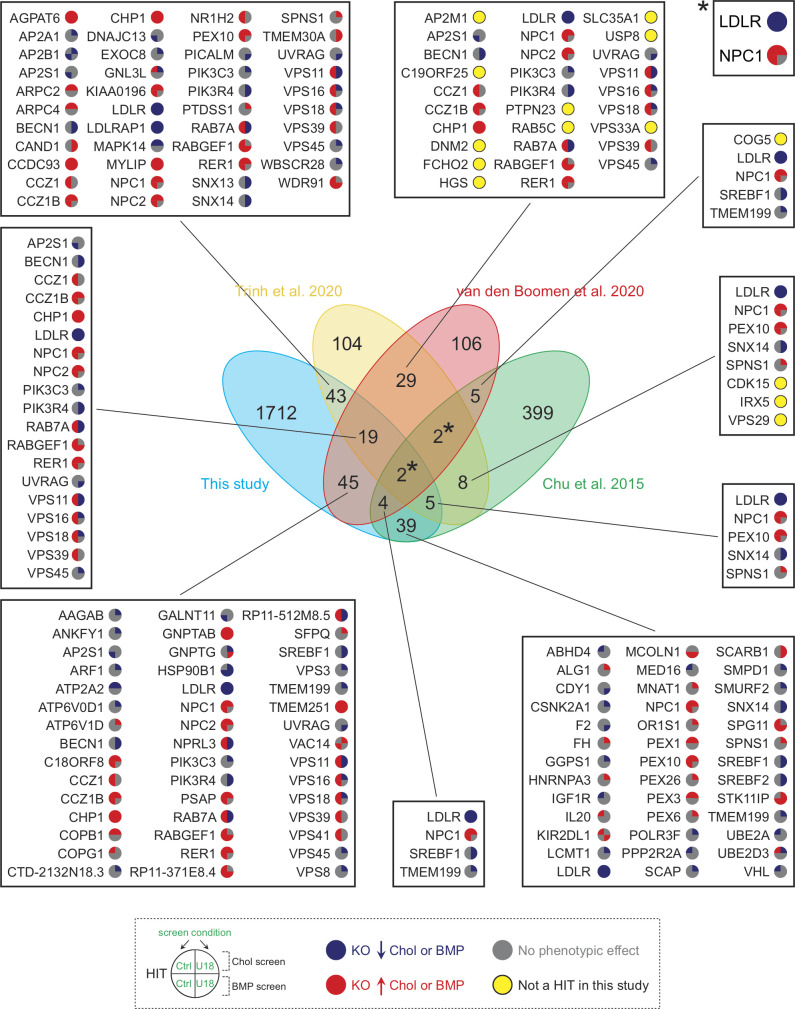
Comparison of hits from four independent studies analyzing cholesterol homeostasis. The central Venn diagram displays overlaps in hits from the indicated referenced screens. Numbers in the nonintersecting areas indicate the number of hits identified in each study, whereas numbers in the intersecting areas correspond to overlapping hits between the different referenced screens. Specific gene hits from this screen are shown in adjacent boxes with colored circles according to an increase (red) or decrease (blue) in cholesterol or BMP. Genes that were identified here but only under one or another condition are shown with a gray circle; those not detected at all are highlighted with a yellow circle. Asterisks refer to LDLR and NPC1 genes (box on the top right). Chol, cholesterol; KO, knockout.
The presence of U18666A to inhibit NPC1 should increase lysosomal cholesterol accumulation; yet, knockout of SNX13 and, to a lesser extent, SNX14 decreased both BMP- and PFO*-stained cholesterol in the K562 cells used for the screen (Fig. 2, lower panel, lower left quadrant). SNX14 is an ER-resident protein that accumulates within a subdomain of the ER surrounding LDs in oleic acid–fed cells (compare Henne et al., 2015; Datta et al., 2019, 2020; Ugrankar et al., 2019). Further studies on the role of SNX13 follow below.
These screens generated an enormous amount of data and were analyzed by multiple means. Shown in Fig. S1 is a hierarchical classification of 195 hits that were identified in both cholesterol and BMP screens, displayed in terms of their phenotypes. Two main categories of hits were revealed: hits that either increase (Fig. S1 A) or decrease (Fig. S1 B) PFO*-stained cholesterol or antibody-stained BMP upon CRISPR deletion. A minor, third category of hits that yielded opposite results in each category was also observed (black arrows in Fig. S1 A). This analysis revealed coordinate behavior of RAB7 and the homotypic fusion and vacuole protein sorting (HOPS) complex VPS11, VPS16, and VPS18 subunits, which, when knocked out in control conditions, led to increased cholesterol and BMP but in the presence of U18666A strongly decreased cholesterol (Fig. S1 A, navy dots). Perhaps loss of endosome fusion capacity leads to accumulation of cholesterol in much smaller structures. Alternatively, loss of these proteins may trigger MVE exocytosis. Similarly, the entire GATOR1 complex, composed of DEPDC5-, NPRL2-, and NPRL3-encoded subunits, negatively regulates mammalian target of rapamycin (mTOR) signaling and also decreases PFO*-stained cholesterol accumulation, but only when NPC1 function is blocked (Fig. S1 A, orange dots). On the other hand, the positive mTOR regulator, folliculin, which counteracts the activity of GATOR1, yields an opposite profile (Fig. S1 A, black arrows). Davis et al. (2021) have shown that NPC1 loss elevates mTORC1 signaling, and inhibition of mTORC1 can improve lysosome proteolytic capacity without correcting cholesterol accumulation. Here, loss of the negative regulatory activity of the GATOR1 complex may show a worse phenotype because of dysregulation of hyperactive mTORC1.
Autophagy-related genes dominated the group that decrease PFO*-stained cholesterol and/or antibody-stained BMP (yellow dots, Fig. S1 B, bottom right). Autophagocytosed membranes, derived from mitophagy, ER-phagy, or lipophagy, for example, represent a major source of lipid substrates in endolysosomes. Perhaps inhibition of autophagy decreases lysosomal catabolic burden and consequently the amount of BMP needed for lipolytic reactions (Kolter and Sandhoff, 2005), as well as intralysosomal cholesterol levels. Alternatively, upon decreased autophagy activity, endolysosomes may be smaller and more difficult to detect by flow cytometry. Finally, cholesterol homeostasis regulators (green dots), including LDLR, LDLRAP1, and the LDLR chaperones HSP90B1 and MESDC2, as well as SREBF1 and 2, are also prominent among genes that decrease cholesterol or BMP staining upon knockout, consistent with their known roles.
Four genome-wide screens for cholesterol regulators
Several groups have recently reported genome-wide screens to identify cholesterol regulators. Scott et al. (2015) presented an siRNA screen that detected a connection with Wnt signaling. Trinh et al. (2020) measured surface LDLR levels; van den Boomen et al. (2020) employed a synthetic reporter to monitor sterol regulatory element binding transcription factor 1 (SREBP) transcriptional activation; and Chu et al. (2015) used amphotericin to kill cells that succeeded in PM cholesterol delivery. Use of different approaches and/or cell types will surely yield different overall hit profiles. Shown in Fig. S3 is a comparison of three of these screens with the screens presented here. This comparison identified only two completely overlapping hits, LDLR and NPC1; thus, different approaches impact hit discovery. Nevertheless, a number of genes discovered here were also detected by others when compared individually. We uncovered roles for PTDSS1, SNX13, and SNX14 (Fig. S3; see also Figs. 2 and S2), all of which were among the top 104 hits of Trinh et al. (2020). Moreover, several factors that regulate LDLR trafficking, including different subunits of the AP2, Arp2/3, and CCC complexes or the ubiquitin ligase MYLIP, appear in this comparison. Finally, only a few genes are shared between previous screens but not ours (yellow dots, Fig. S3). This comparison confirms the comprehensive nature of the present screens and the importance of independent evaluations of lipid regulatory pathways.
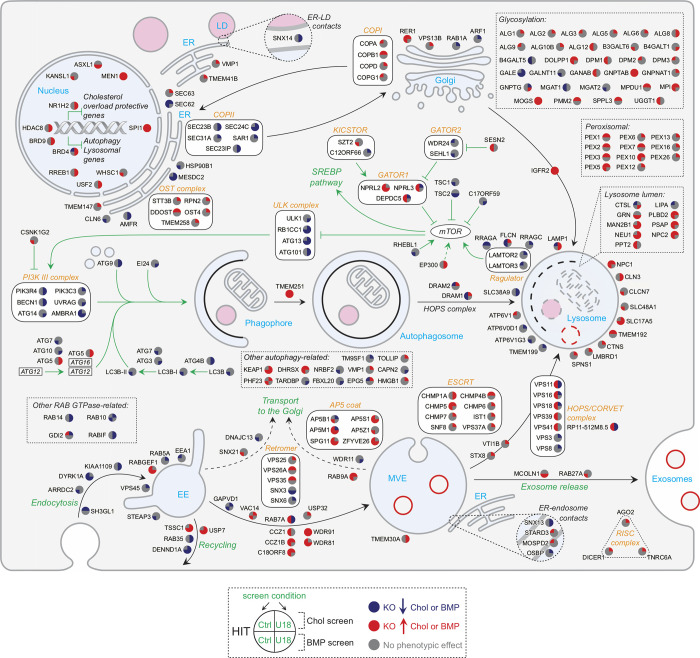
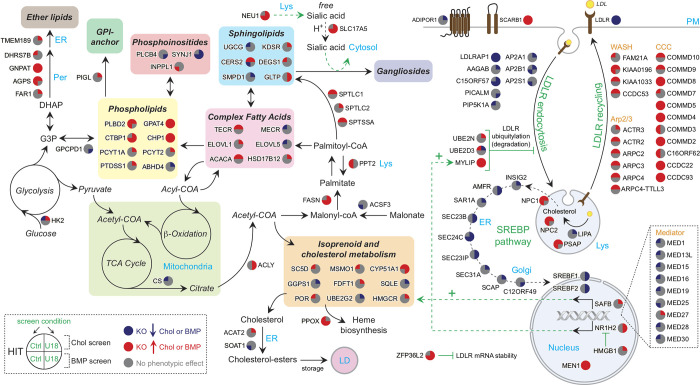
All hits identified in both cholesterol and BMP screens revealed the most represented intracellular pathways and processes controlling cholesterol and BMP homeostasis (Figs. S2 and 3). We used colors to indicate decreases (blue) or increases (red) of staining for these two lipids upon knockout, with quadrants of the individual gene nodes showing results for each gene with and without NPC1 function. As expected, many high-confidence interacting hits are involved in LDLR trafficking, SREBP pathway regulation, or early/late endosomal function. Other well-represented functional gene clusters involve autophagy and mTOR signaling, known to play critical roles in cellular lipid homeostasis (Figs. S1 and S2; Thelen and Zoncu, 2017). A large number of interacting hits are involved in transcriptional regulation (Figs. 3 and S2). In addition, we obtained valuable information related to the coordinate function of individual subunits of protein complexes. We detected identical behavior for almost all subunits of the Mediator chromatin remodeling and RISC complexes (Figs. 3 and S2). Genes involved in LDLR recycling are all red (Fig. 3, upper right), suggesting, paradoxically, that PFO*-stained cholesterol accumulates when LDLR cannot recycle. Perhaps this reflects trafficking of another protein(s) that facilitates cholesterol egress or increased receptor degradation followed by LDLR gene up-regulation. Unexpectedly, glycosylation genes are also mostly red (Fig. S2, upper right), possibly reflecting the importance of glycosylation for folding and assembly of membrane proteins that function in cholesterol export.
Figure 3.
Lipid metabolic pathways revealed in the screens. Genes are annotated in colored circles that show increases (red) or decreases (blue) in cholesterol (upper left), BMP (lower left), cholesterol with U18666A (upper right), or BMP with U18666A (lower right) as indicated. Black arrows indicate trafficking, metabolic routes, and enzymatic reactions. COA, coenzyme A; Chol, cholesterol; GPI, glycophosphatidylinositol; KO, knockout; Lys, lysosome; Per, peroxisome; TCA, tricarboxylic acid.
Complexes involved in late endosomal maturation, such as the RAB7 GEF MON/CCZ1/C18ORF8 (van den Boomen et al., 2020) or the WDR91/81 complex (Casanova and Winckler, 2017), show increased cholesterol or BMP accumulation upon knockout (Fig. S2, bottom), while endosome tethering complexes such as HOPS and Rab7 are mixed (Fig. S2, bottom right): increasing cholesterol and BMP on knockout while decreasing cholesterol and BMP on knockout with U18666A. This phenotypic difference may reflect the different sized endolysosomal compartments generated under the two conditions and/or a possible up-regulation in MVE exocytosis in the presence of U18666A (Strauss et al., 2010).
Focusing on lipid metabolic pathways (Fig. 3), the majority of hits that catalyze lipid biosynthesis appear to increase either cholesterol or BMP or both. The screens recovered key enzymes required for de novo cholesterol biosynthesis, including HMGCR, MSMO1, FDFT1, SQLE, and CYP51A1, and also almost the entire SREBP pathway (Fig. 3, lower right) and the recently described pathway regulator, C12ORF49 (Loregger et al., 2020; Bayraktar et al., 2020; Aregger et al., 2020). Identification of additional factors, such as UBE2G2, which controls sterol-stimulated ubiquitylation and turnover of the rate-limiting HMGCR enzyme (Tan et al., 2019), or ZFP36L2 (Fig. 3, bottom right), an RNA-binding protein that promotes LDLR mRNA degradation (Adachi et al., 2014), further highlights the level of biological detail revealed here. In most cases, the hit phenotypes observed were consistent with their respective biological functions in the context of cholesterol homeostatic regulation. Finally, a considerable number of transcription factors that control expression of lipid metabolism and lysosomal genes were also efficiently detected as hits in addition to SREBFs (see Figs. 3 and S2), including liver X receptor member NR1H2 (Wang and Tontonoz, 2018), BRD4 (Sakamaki et al., 2017), SPI1 (Solomon et al., 2017), USF2 (Yamanaka et al., 2016), and the multisubunit transcriptional coactivator Mediator (Youn et al., 2016).
ER-localized SNX13 links to endolysosomes and LD domains
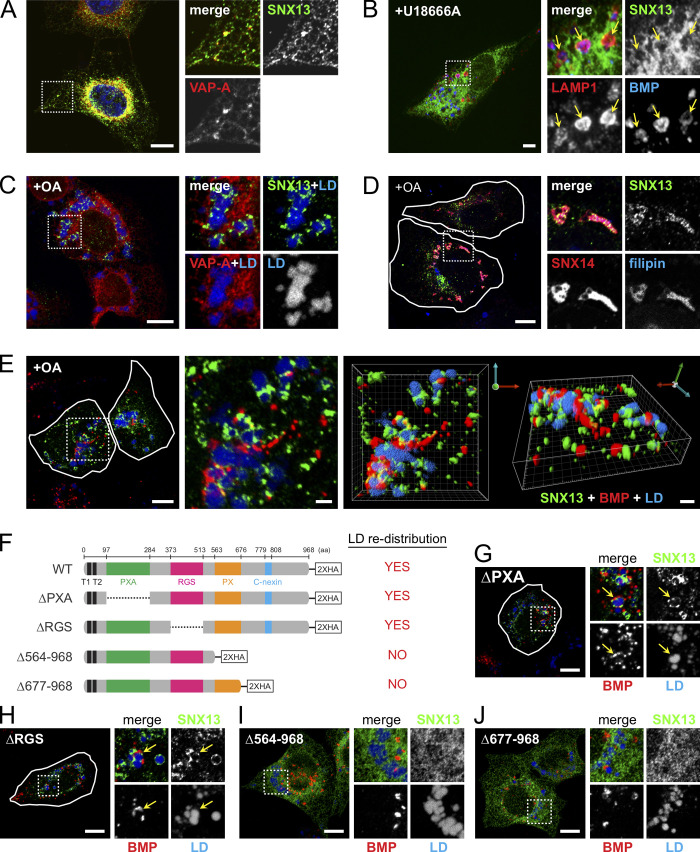
The goal of this screen was to identify modifiers of the NPC1 deletion–cholesterol accumulation phenotype, and SNX13 revealed itself as a gene that decreased PFO*-detected, accessible cholesterol and antibody-detected BMP in the presence of U18666A. Thus, we performed a cell biological characterization of the role of SNX13 in cholesterol regulation. Like SNX14, SNX13 is related to the single yeast MDM1 protein that mediates ER links to the yeast vacuole (Henne et al., 2015). SNX13 is a multidomain-containing protein composed of PXA, RGS, PX, and C-nexin domains (Fig. 4 F). In U2OS cells, we detected good colocalization of SNX13 with the ER-localized VAMP-associated protein A (VAPA; Fig. 4 A). When NPC1 was inhibited, SNX13 remained in the ER, and close apposition to lysosomes was detected (Fig. 4 B, retinal pigment epithelium [RPE] cell shown). Upon addition of oleic acid to induce LD formation, SNX13 redistributed to ER domains in contact with nascent LDs (Fig. 4 C), as shown recently for SNX14 (Fig. 4 D; Datta et al., 2019). Live-cell video microscopy revealed stable and prolonged association of SNX13 ER domains with LDs over time (Video 1), suggesting SNX13-mediated ER–LD tethering activity. Confocal microscopy (Fig. 4 E) showed intimate connections between SNX13 (green), BMP compartments (red), and LDs (blue) under these conditions. SNX13 truncation analysis showed that the C terminus is required for redistribution of SNX13 to LD-forming domains (Fig. 4, F–J), analogous to SNX14 (Datta et al., 2019).
Figure 4.
SNX13 is an ER-resident protein that associates with LDs via its C terminus. (A) Immunofluorescence microscopy of U2OS cells expressing SNX13-HA and VAPA-CFP. Proteins were detected with anti-HA antibodies or CFP fluorescence as indicated. Scale bar, 10 µm. (B) RPE cell expressing SNX13-GFP, treated with U18666A for 16 h. LAMP1 and BMP were detected using antibodies. Scale bar, 10 µm. (C and D) U2OS cells as in A, treated with oleic acid overnight. LDs were detected using LipidTOX, and cholesterol was detected using filipin. VAPA-CFP and SNX14-GFP were detected by intrinsic fluorescence. Shown in small boxes are enlargements of the boxed areas at left. Scale bars, 10 µm. (E) Cells as in A, visualized by confocal microscopy; green, SNX13; red, BMP; blue, LDs. Scale bars, 10 µm; enlarged inset and 3D rendering, 2 µm. (F) Schematic analysis of SNX13 constructs. (G–J) Localizations of constructs indicated in F. Cells were labeled as in E. Scale bars, 10 µm. Arrows indicate ring-like SNX13-ER domains in close contact with BMP-positive endolysosomes and LDs. PXA, PX-associated; RGS, regulator of G protein signaling; PX, phox homology.
Video 1.
Live-cell visualization SNX13-positive ER domains in contact with LDs. Time-lapse confocal video microscopy was performed using U2OS cells expressing SNX13-GFP and OFP-LD probe OFP-HPos. Images were acquired every 15 s; total video length is 59 min 45 s. Scale bar, 5 µm. Video plays at 15 seconds/frame.
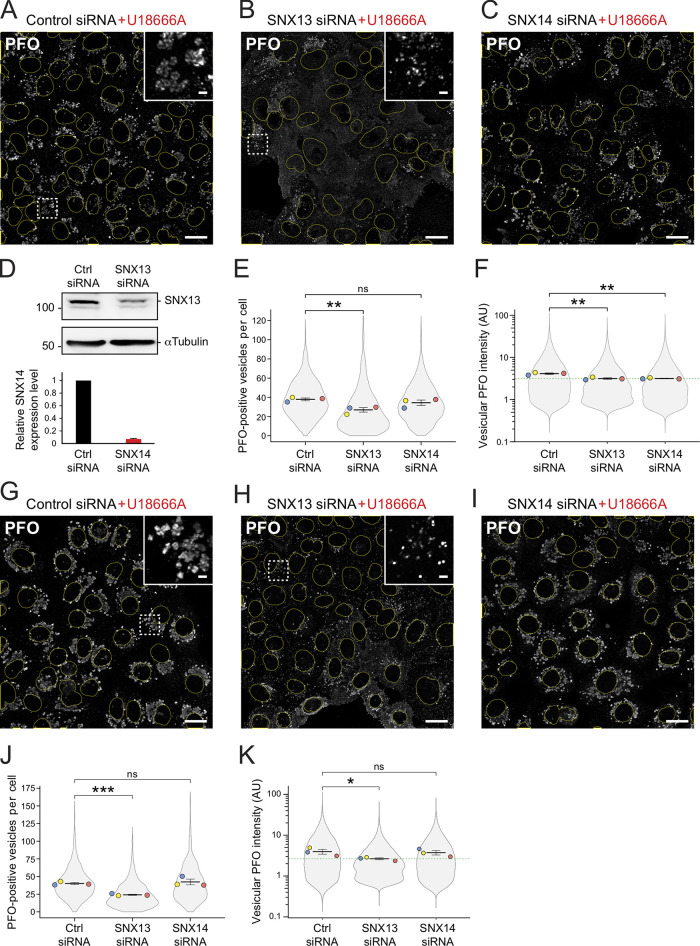
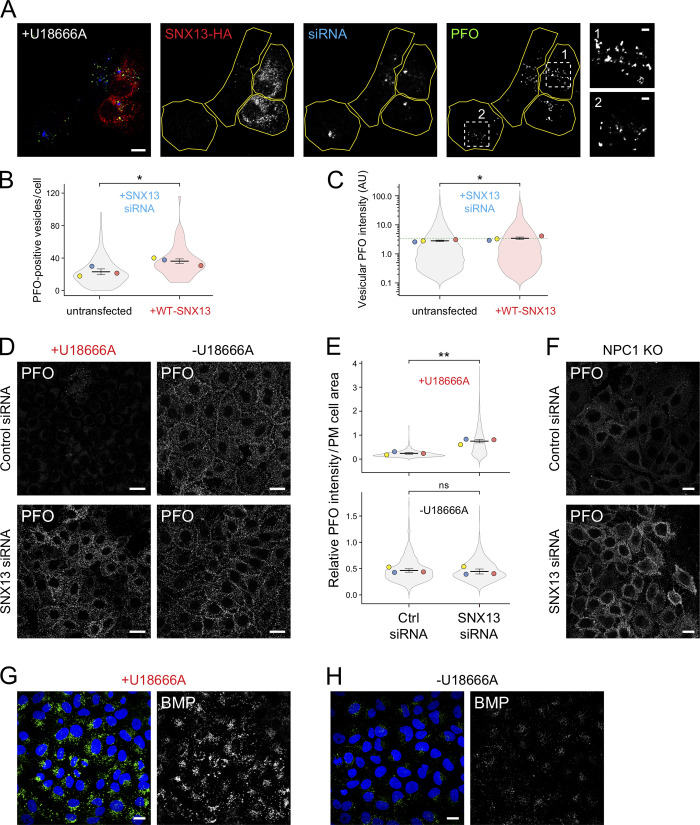
Immunofluorescence microscopy of U18666A-treated U2OS cells treated with SNX13 siRNA showed decreased cholesterol accumulation determined by PFO* staining compared with control siRNA (Fig. 5, A–D). Specifically, SNX13 siRNA-treated cells showed fewer PFO*-positive vesicles and decreased intensity of total vesicle-associated PFO* staining (Fig. 5, E and F). Similar results were obtained using U18666A-treated HeLa cells (Fig. 5, G–K). SNX14-depleted U18666A-treated U2OS cells (Fig. 5, C and F), but not HeLa cells (Fig. 5, I and K), behaved similarly to SNX13-depleted cells in terms of showing decreased vesicle-associated PFO* staining intensity, but SNX14-depleted cells did not show a decrease in PFO-positive vesicle number in either cell type (Fig. 5, E and J). Finally, cells ectopically expressing siRNA-resistant, HA-tagged WT SNX13 showed increased numbers of PFO-positive vesicles and higher PFO vesicular intensity compared with control transfected cells (Fig. S4, A–C), confirming the siRNA specificity.
Figure 5.
SNX13 depletion redistributes cellular cholesterol in the absence of NPC1 function. (A–C) U2OS cells treated with U18666A (+U18) for 16 h labeled with GST-PFO* detected using anti-GST antibodies 72 h after transfection with indicated siRNAs. Scale bar, 10 µm. Insets, enlargements of boxed areas; scale bars, 2 µm. (D) Immunoblot (at top) of SNX13 siRNA-treated cells as in A and B; 50 µg cell extract was analyzed. Molecular weight marker mobility is shown in kD here and in all subsequent figures. Below are relative SNX14 mRNA levels measured by quantitative PCR from U2OS cells from two independent experiments; error bar, SD. (E and F) Quantitation of PFO*-positive vesicle numbers and vesicular PFO* fluorescence intensity. Colored dots reflect means from independent experiments; >860 cells analyzed for each condition; significance determined by unpaired t test: **, P < 0.01. (G–K) Analysis of HeLa cells as described for A–F; >950 cells analyzed in each condition; significance determined by unpaired t test: *, P < 0.05; ***, P < 0.001. Actual P values: (E) Ctrl siRNA versus SNX13 siRNA, P = 0.0079; Ctrl siRNA versus SNX14 siRNA, P = 0.16; (F) Ctrl siRNA+U18 versus SNX13 siRNA+U18, P = 0.0057; Ctrl siRNA+U18 versus SNX14 siRNA+U18, P = 0.0033; (J) Ctrl siRNA+U18 versus SNX13 siRNA+U18, P = 0.00035; Ctrl siRNA+U18 versus SNX14 siRNA+U18, P = 0.31; (K) Ctrl siRNA+U18 versus SNX13 siRNA+U18, P = 0.038; Ctrl siRNA+U18 versus SNX14 siRNA+U18, P = 0.39. AU, arbitrary unit.
Figure S4.
SNX13 depletion redistributes cholesterol to the cell surface in the absence of NPC1 function. (A) U2OS cells were treated with SNX13 siRNA for 72 h. After 24 h, cells were transfected with plasmid encoding siRNA-resistant HA-tagged WT-SNX13. 16 h before fixation, cells were treated with U18666A for 16 h and then labeled with GST-PFO detected using anti-GST primary antibodies. Scale bar, 10 µm. Insets show enlargements of the boxed areas from cells rescued with siRNA-resistant SNX13-HA (box 1) or untransfected cells (box 2); scale bars, 2 µm. (B and C) Quantitation of PFO*-positive vesicles and vesicular PFO fluorescence intensity determined by CellProfiler. Colored dots reflect means from independent experiments; 64 rescued cells from a total of >300 cells were analyzed. Significance was determined by unpaired t test; *, P < 0.05. (D) Immunofluorescence microscopy of HeLa cells treated with U18666A for 16 h (left column images) or left untreated (right column images) and labeled in the absence of detergent permeabilization with GST-PFO detected using anti-GST primary antibodies 72 h after transfection with the indicated siRNAs. (E) Quantitation of PFO staining as a function of PM cell area in U18666A-treated (top plot) and untreated (bottom plot) cells determined using CellProfiler. Colored dots reflect means from independent experiments; >830 cells analyzed in each condition. Significance was determined by unpaired t test; **, P < 0.01. (F) Immunofluorescence microscopy of NPC1-knockout (KO) HeLa cells stained in the absence of detergent permeabilization with GST-PFO detected by anti-GST primary antibodies. (G and H) U2OS cells treated with U18666A for 16 h (G) or left untreated (H), labeled with anti-BMP antibodies, and imaged by confocal microscopy. Scale bars, 20 µm. Actual P values: (B) Untransfected + U18 versus + WT-SNX13 + U18, P = 0.023; (C) untransfected+U18 versus +WT-SNX13+U18, P = 0.047; (E) top: Ctrl siRNA+U18 versus SNX13 siRNA+U18, P = 0.0017; (E) bottom: Ctrl siRNA versus SNX13 siRNA, P = 0.377. AU, arbitrary unit.
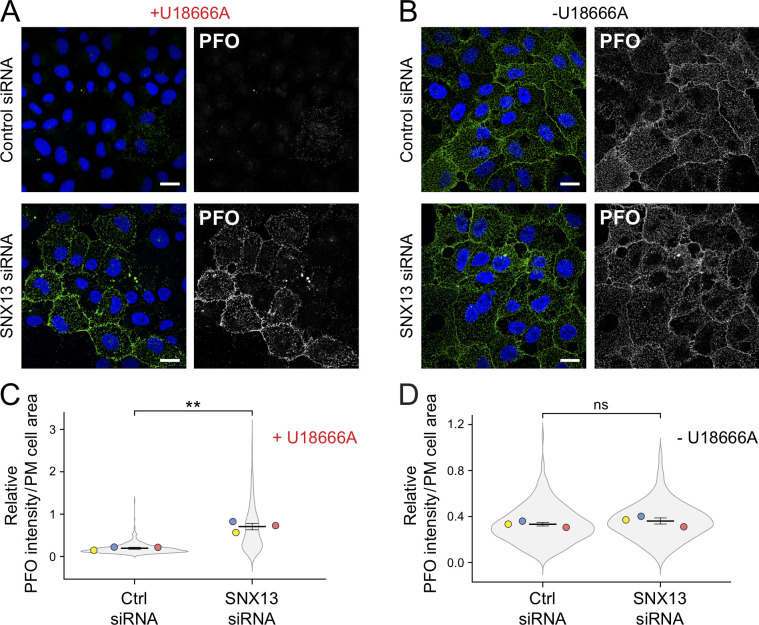
In addition to decreasing accessible lysosomal cholesterol, SNX13-depleted, U18666A-treated U2OS cells (Fig. 6, A–D) and HeLa cells (Fig. S4, D and E) showed clear, NPC1-independent redistribution of accessible cholesterol from endolysosomal compartments to the cell surface of nonpermeabilized cells that was not seen in control siRNA + U18666A-treated cells (Fig. 6 A and Fig. S4 D). Surface PFO* staining was comparable to that seen in the absence of U18666A treatment (Fig. 6, B and D). This was again unexpected, as NPC1 is thought to be required upstream of cholesterol delivery to the cell surface and would have been inhibited under U18666A treatment conditions. One possibility is that loss of SNX13 enhances formation of an endosome-PM and/or ER-PM contact sites that can bypass NPC1 (as seen by Höglinger et al., 2019). Importantly, depletion of SNX13 from NPC1-knockout HeLa cells yielded the same phenotype (Fig. S4 F), showing that this reflects a block in NPC1 function rather than an off-target effect of U18666A.
Figure 6.
SNX13 depletion redistributes cholesterol to the cell surface in the absence of NPC1 function. (A and B) U2OS cells treated with U18666A for 16 h (A) or untreated (B) and labeled without detergent permeabilization with GST-PFO* and anti-GST primary antibodies 72 h after transfection with the indicated siRNAs. (C and D) Quantitation of PFO* staining as a function of PM cell area in U18666A-treated (C) and untreated (D) cells. Colored dots reflect means from independent experiments; >630 cells analyzed per condition. Significance was determined by unpaired t test; **, P < 0.01. Scale bars, 20 µm. Actual P values: (C) Ctrl siRNA+U18 versus SNX13 siRNA+U18, P = 0.0015; (D) Ctrl siRNA versus SNX13 siRNA, P = 0.209.
Zoncu and colleagues have shown that blocking NPC1 function hyperactivates mTORC1 and leads to accumulation of accessible cholesterol on the outer leaflet of lysosomes, highlighting a cholesterol transfer pathway from the ER to the lysosome outer leaflet (Lim et al., 2019; Davis et al., 2021). Somehow, loss of SNX13 enables cholesterol transfer to the PM despite the absence of NPC1 function. This phenotype suggests a role for SNX13 as a negative regulator of cholesterol transport from lysosomes to the cell surface in coordination with NPC1.
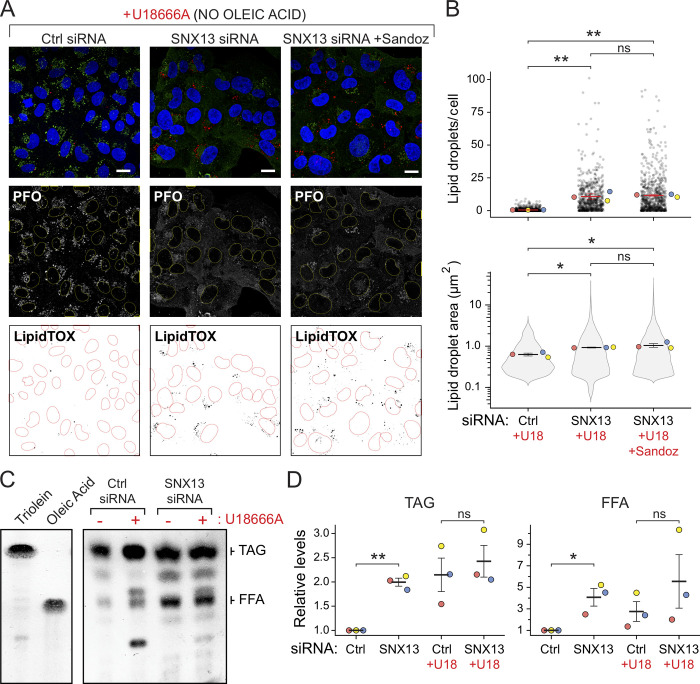
Even in the absence of oleic acid supplementation, SNX13 depletion increased the number of LDs per cell, as monitored by LipidTOX staining (Fig. 7, A and B). This increase was seen whether or not cells were treated with the Sandoz ACAT1 inhibitor, suggesting that these LDs contain primarily triglycerides rather than cholesterol esters. Total lipids were analyzed by TLC, which revealed elevated levels of both total triacylglycerol and free fatty acids, consistent with the increased NPC1-independent LD accumulation observed (Fig. 7, C and D).
Figure 7.
SNX13 depletion increases LD abundance in the absence of NPC1 function. (A) U2OS cells treated with U18666A for 16 h and labeled with GST-PFO* and anti-GST primary antibodies 72 h after transfection with the indicated siRNAs without oleic acid addition. Cells were also stained with LipidTOX. One set was also treated with ACAT1 inhibitor Sandoz 53-035. Scale bar, 10 µm. (B) Number of LDs and area determined as in A. Colored dots reflect means from independent experiments; >650 cells analyzed in each condition. Significance was determined by multiple t tests and Holm-Sidak method with α = 0.05. (C) TLC of lipids as in A. At left is mobility of indicated marker lipids. (D) Quantitation of triacylglycerol (TAG) and free fatty acid (FFA) levels determined by TLC. P values were determined by multiple t tests corrected by the Holm-Sidak method; *, P < 0.05; **, P < 0.01. Actual P values: (B) Top: Ctrl siRNA+U18 versus SNX13 siRNA+U18, P = 0.0052; Ctrl siRNA+U18 versus SNX13 siRNA+U18 + Sandoz, P = 0.0039; SNX13 siRNA versus SNX13 siRNA + Sandoz, P = 0.85; (B) bottom: Ctrl siRNA versus SNX13 siRNA, P = 0.0451; Ctrl siRNA versus SNX13 siRNA + Sandoz, P = 0.0144; SNX13 siRNA versus SNX13 siRNA + Sandoz, P = 0.382; (D) left: Ctrl siRNA versus SNX13 siRNA, P = 0.00683; Ctrl siRNA + U18 versus SNX13 siRNA + U18, P = 0.312; (D) right: Ctrl siRNA versus SNX13 siRNA, P = 0.0198; Ctrl siRNA + U18 versus SNX13 siRNA + U18, P = 0.351.
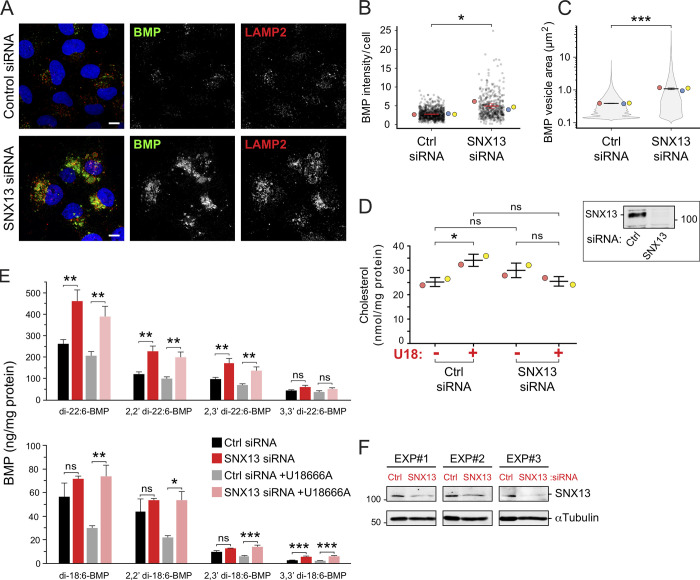
The initial flow cytometry screen using K562 cells indicated that SNX13 knockout had the strongest phenotype when NPC1 function was inhibited and that knockout concomitantly decreased BMP antibody staining (Figs. 2 and S1). Yet, immunofluorescence microscopy showed accumulation of antibody-detected BMP in NPC WT SNX13 siRNA-depleted U2OS cells (Fig. 8, A and B). This difference may be due to the fact that the screen relied on CRISPR deletion, whereas our validation relies on siRNA; alternatively, it could reflect differences between cell types. Many studies have now shown that cells subject to CRISPR modification adapt in diverse ways (Cerikan et al., 2016; Diofano et al., 2020; Salanga and Salanga, 2021). Nevertheless, SNX13 still bypassed NPC1 deficiency, and this could have been due to the unexpected increased BMP we detected that may be sufficient to facilitate cholesterol export from lysosomes; previous reports showed that cells fed phosphatidylglycerol display increases in BMP levels and rescue of the endolysosomal cholesterol accumulation typical of NPC1-deficient cells (McCauliff et al., 2019).
Figure 8.
SNX13 depletion in NPC1 WT U2OS cells increases BMP. (A) U2OS cells labeled with anti-BMP and anti-LAMP2 antibodies 72 h after transfection with the indicated siRNAs. Scale bar, 10 µm. (B and C) Quantitation of BMP-positive vesicle area (B) and total BMP intensity (C) per cell. Colored dots represent means from independent experiments; >480 cells analyzed in each condition. Significance was determined by unpaired t test; *, P < 0.05; ***, P < 0.001. (D) Total cell cholesterol in cells treated as in A. Samples were treated with cholesterol esterase before assay to cleave cholesterol esters. Colored dots reflect means from independent experiments. Significance was determined by uncorrected Dunn's test with Kruskal-Wallis one-way ANOVA; *, P < 0.05. Immunoblot at right shows endogenous SNX13 levels from the experiments in D. (E) MS determination of BMP isoforms normalized to protein content from cells treated as indicated. Data from three independent experiments (mean ± SEM). Significance was determined by two-way ANOVA with Tukey’s post hoc test; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (F) Immunoblot of the experiments in E; 50 µg of cell extract was analyzed. P values: (B) Ctrl siRNA versus SNX13 siRNA, P = 0.0171; (C) Ctrl siRNA versus SNX13 siRNA, P = 0.000305; (D) Ctrl siRNA without U18 versus Ctrl siRNA with U18, P = 0.025; Ctrl siRNA without U18 versus SNX13 siRNA without U18, P = 0.153; SNX13 siRNA without U18 versus SNX13 siRNA with U18, P = 0.31; Ctrl siRNA with U18 versus SNX13 siRNA with U18, P = 0.066; (E) top: di-22:6-BMP, P = 0.0024 and 0.0038; 2,2′ di-22:6-BMP, P = 0.0018 and 0.0026; 2,3′ di-22:6-BMP, P = 0.0025 and 0.004; 3,3′ di-22:6-BMP, P = 0.0812 and 0.157; (E) bottom: di-18:1-BMP, P = 0.247 and 0.0038; 2,2′ di-18:1-BMP, P = 0.517 and 0.011; 2,3′ di-18:1-BMP, P = 0.0948 and 0.0008; 3,3′ di-18:1-BMP, P = 0.0005 and 0.0001.
To complement our immunofluorescence data, we performed biochemical determination of total cholesterol content in cells with and without SNX13, with and without U18666A (Fig. 8 D). Samples were treated with cholesterol esterase before assay to cleave cholesterol esters. As expected, we detected a significant increase in total cholesterol upon U18666A treatment of control siRNA-transfected cells. In contrast, such an increase was not observed in SNX13-depleted cells under the same conditions (Fig. 8 D); no significant changes in total cholesterol content were detected between control and SNX13 siRNA-treated cells (Fig. 8 D). These data support the conclusion that loss of SNX13 in NPC1-deficient cells results in endolysosomal cholesterol redistribution to the PM without altering total cellular cholesterol levels.
Mass spectrometry (MS) was also performed to quantify BMP isoforms for cells with and without SNX13 with and without U18666A (Fig. 8, E and F). di-22:6 BMP was the predominant form in this cell type and increased significantly in SNX13 siRNA-treated cells, unlike the less abundant di-18:1 BMP forms (Fig. 8 E). Moreover, similar (but slightly decreased) levels of total BMP were seen in samples treated with U18666A, suggesting that any changes detected by microscopy reflect an antibody-accessible pool that accumulates when NPC1 function is blocked during the 16-h U18666A incubation time frame. Control experiments confirmed increased antibody-detected endolysosomal BMP levels in U18666A-treated U2OS cells (Fig. S4, G and H). Altogether, these data demonstrate that SNX13 function is tightly linked to regulation of BMP levels in U2OS cells.
SNX13 drives ER–endolysosomal membrane contacts
Despite the absence of NPC1 function, SNX13-depleted cells redistribute endolysosomal cholesterol to the PM (Figs. 7 A and S4 D). In addition, SNX13 ER domains colocalize with both LDs and endolysosomes (Fig. 4, B, E, G, and H), similar to parallel studies of SNX19 protein (Saric et al., 2021). Finally, the SNX13 yeast orthologue MDM1 mediates ER–vacuole contacts (Henne et al., 2015). These data prompted us to explore whether SNX13 drives ER–endolysosome (EL) membrane contacts to facilitate endosomal cholesterol egress.
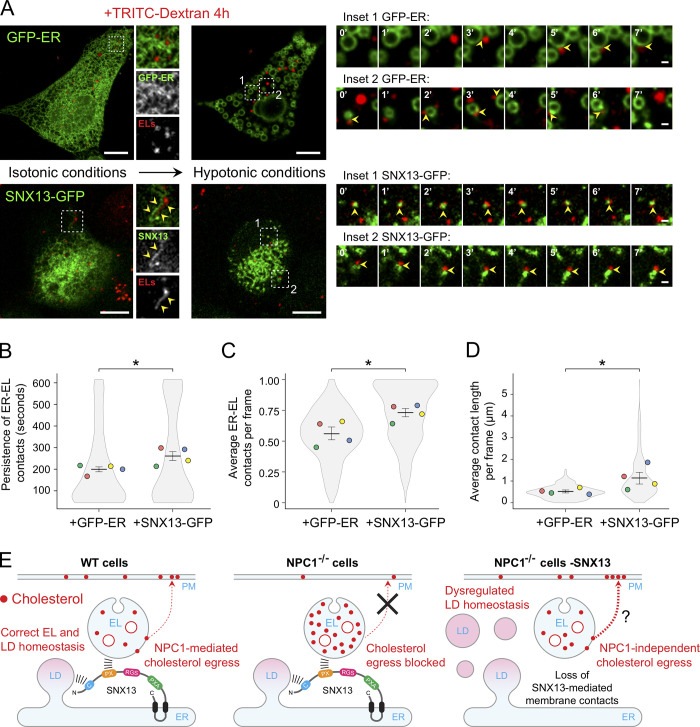
Lippincott-Schwartz and colleagues have recently used hypotonic swelling to facilitate detection of membrane contact sites (King et al., 2020). We used this method in conjunction with live-cell video microscopy to monitor possible changes in membrane contacts in cells overexpressing either SNX13-GFP or GFP-tagged ER (GFP-ER), an artificial ER-localized control fluorescent protein (Kassan et al., 2013). Endolysosomes were labeled by incubating cells for 4 h with TRITC-labeled dextran. Under isotonic conditions, ER–EL contacts were somewhat ambiguous (Fig. 9 A, left). Nevertheless, some SNX13 ER domains were seen in clear apposition to TRITC dextran–loaded ELs (Fig. 9 A, insets). In contrast, 10 min after addition of hypotonic media, cells swelled, ER morphology was drastically altered, and ER–EL contacts were readily observed (Fig. 9 A, center and insets). Interestingly, while most of the imaged control cells (overexpressing GFP-ER) showed consistent, large ER vacuoles under these conditions (Fig. 9 A; King et al., 2020), only ∼30% of SNX13-overexpressing cells showed this phenotype, and instead the ER appeared as discrete domains that resisted vacuolation. This suggests that SNX13 overexpression may stabilize overall ER structure and membrane contacts with other compartments. Despite many fewer large structures, hypotonic conditions revealed ER–EL contacts in both control and SNX13-overexpressing cells (Fig. 9, insets at right); swollen cells were imaged every 15 s for ∼10 min in hypotonic media to track contact sites (Videos 2 and 3).
Figure 9.
SNX13 overexpression induces ER–EL membrane contacts. (A) U2OS cells were transfected with the GFP-tagged ER control probe HNeu (GFP-ER) or SNX13-GFP. 24 h after transfection, ELs were loaded with TRITC-labeled dextran for 4 h at 37°C. Live images at left correspond to cells expressing GFP-ER (top) or SNX13-GFP (bottom) under isotonic conditions (left column) or hypotonic media (right column). Insets to the right of isotonic conditions show control (GFP-ER) or SNX13-labeled ER domains in proximity to ELs (yellow arrowheads). Consecutive image frames at right correspond to time-lapse recording of boxed areas (insets 1 and 2) from both control (top) and SNX13-expressing cells (bottom) under hypotonic conditions. Scale bars, 10 µm; enlarged insets, 1 µm. (B–D) Quantitation of persistence of ER–EL contacts (B), average number of ER–EL contacts per frame (C), and average ER–EL contact length per frame (D). Colored dots reflect means from independent experiments; 15 cells analyzed in each condition. Significance was determined by unpaired t test; *, P < 0.05. Actual P values: (B) +GFP-ER versus +SNX13-GFP, P = 0.02; (C) +GFP-ER versus +SNX13-GFP, P = 0.017; (D) +GFP-ER versus +SNX13-GFP, P = 0.035. (E) Model depicting a possible role for SNX13 in endosomal cholesterol egress regulation.
Video 2.
Live-cell imaging of ER–EL contacts in U2OS cells overexpressing the GFP-ER probe 10 min after incubation in hypotonic media. Yellow arrowheads indicate ER–EL contacts. Contact length is displayed in white. Images were acquired every 15 s; total video length is 10 min. Scale bars, 10 µm; enlarged insets, 1 µm. Video plays at 15 seconds/frame.
Video 3.
Live-cell imaging of ER–EL contacts in U2OS cells overexpressing SNX13-GFP 10 min after incubation in hypotonic media. Yellow arrowheads indicate ER–EL contacts. Contact length is displayed in white. Images were acquired every 15 s; total video length is 10 min. Scale bars, 10 µm; enlarged insets, 1 µm. Video plays at 15 seconds/frame.
We employed a recently described image analysis method (Bosch et al., 2020; Martín et al., 2021) to quantify ER–EL contacts over time. This analysis revealed increased persistence of ER–EL contacts in SNX13-overexpressing cells compared with control cells overexpressing the GFP-ER probe (Fig. 9 B). Moreover, the overall number and extent of these contacts were significantly greater in SNX13-overexpressing cells (Fig. 9, C and D). In summary, these data demonstrate that SNX13 functions as an ER–EL tether (Fig. 9 E).
Discussion
We have presented here parallel screens to investigate the pathways that regulate cholesterol and BMP in cells. Our screens generated a comprehensive dataset that not only recapitulates many known trafficking, metabolic, and transcriptional pathways and proteins that regulate intracellular cholesterol and BMP physiology but also, importantly, revealed previously unrecognized roles for many genes and protein complexes that deserve further validation and characterization, opening new avenues for future investigation. This study also revealed a role for ER-localized SNX13 as a membrane contact site protein that controls triacylglyceride content and LD numbers, the distribution of accessible cholesterol in lysosomes, and the levels of lysosomal BMP.
Our findings complement other previous screens related to cholesterol trafficking and highlight the strong, coordinate regulation of PFO*-labeled cholesterol and antibody-stained BMP. Despite the importance of BMP in cholesterol transport and other lipid degradative processes, little is known about how this lipid is synthesized or degraded. Strikingly, no single specific metabolic enzyme revealed itself as a strong candidate for BMP biosynthesis; rather, knockout of the mannose 6-phosphate receptor yielded the strongest phenotype. The simplest explanation is that BMP is up-regulated in response to a severe defect in lysosomal enzyme delivery; alternatively, BMP degradation may be blocked.
Our screen relied on two protein reagents, PFO* and anti-BMP antibodies, to monitor cholesterol and BMP changes in millions of cells. These reagents are not able to detect the entire pool of either lipid, as total lipid determinations using biochemical assays and MS of SNX13-depleted cell extracts, for example, did not match changes seen with either PFO* or anti-BMP antibody staining: PFO* staining decreased in SNX13-depleted, U18666A-treated cells (Fig. 5) under conditions in which total cellular cholesterol determined biochemically did not (Fig. 8 C). Similarly, treatment of control cells with U18666A led to increased anti-BMP staining (Fig. 8 B), while the major species of BMP measured by MS were essentially unchanged upon U18666A administration (Fig. 8). Thus, while PFO* and anti-BMP antibodies are valuable reagents without which these screens could not have been completed, additional assays are needed to fully understand the phenotypes obtained. We think of PFO* and anti-BMP as detecting pools of “accessible” lipids, the precise nature of which will require further work.
Recent interest has turned to BMP in relation to multiple disease processes. In addition to NPC disease, BMP is currently a promising biomarker for Parkinson’s disease (compare Alcalay et al., 2020). In models of frontotemporal dementia due to loss of progranulin, BMP levels are altered (granulin [GRN]; Logan et al., 2021). Progranulin is targeted to lysosomes, where it is cleaved to yield several granulin peptides that contribute to endolysosomal homeostasis (Logan et al., 2021). In this study, GRN deletion increased the appearance of accessible BMP but not accessible cholesterol (Fig. S2, far right). On the other hand, prosaposin that heterodimerizes with GRN and is also cleaved in the lysosome (Zhou et al., 2015) increased both accessible cholesterol and accessible BMP, consistent with its role in providing lipid substrates to lysosomal degradative enzymes (Fig. S2).
Our screens revealed a number of unexpected consequences of deleting well-characterized players. For example, why does deletion of any of the endosomal sorting complex required for transport (ESCRT) subunits consistently increase cholesterol under NPC1 inhibition conditions (Fig. S2)? Depletion of ESCRT complexes decreases, but does not abolish, the number of intralumenal vesicles in MVEs (Stuffers et al., 2009). In other work, depletion of the ESCRT-0 subunit HRS, but not other ESCRT subunits, caused an NPC-like phenotype (Du et al., 2012). It is possible that this phenotype is only seen upon full knockout of ESCRTs performed here. In the absence of ESCRT function, the alternative, ceramide-dependent intralumenal vesicle biogenesis pathway may be up-regulated (Stuffers et al., 2009; Trajkovic et al., 2008), perhaps generating intralumenal vesicles with higher cholesterol content. Further work is needed to resolve this question. Another example is the discovery of RP11-512M8.5, which encodes a protein of unknown function that shows high homology (>90%) to VPS33A, a HOPS/CORVET (complete information for VPS8 gene) subunit (Figs. S1 and S2). RP11-512M8.5 showed the same profile as other known HOPS/CORVET core subunits: increasing or decreasing cholesterol/BMP in control or U18666A conditions, respectively (Figs. S1 and S2). The contribution of this novel subunit to HOPS/CORVET function will be of interest for future study.
SNX13 bypass of NPC1 function
SNX13 was an especially interesting hit that, when knocked out, decreased accessible lysosomal cholesterol accumulation in cells lacking NPC1 function, triggered either via NPC1 knockout or U18666A inhibition. Moreover, under these conditions, accessible cholesterol was detected at the cell surface compared with control cells, despite lack of NPC1 function. How might NPC1 function be bypassed? NPC1 bypass can be achieved by treatment of cells with cyclodextrin (Abi-Mosleh et al., 2009; Rosenbaum et al., 2010); in this case, it is likely that endocytosed cyclodextrin enables hydrophobic cholesterol to permeate the glycocalyx and gain access to the limiting lysosome membrane for eventual transport to the ER. Cyclodextrin may also trigger MVE fusion with the cell surface (Llorente et al., 2007), triggering cholesterol release.
Another example of NPC1 bypass was recently reported by Spiegel and colleagues (Newton et al., 2020). Remarkably, activation of sphingosine kinase was sufficient to drive cholesterol export from NPC1-deficient lysosomes. Sphingolipids are normally degraded by lysosomal sphingomyelinase, yielding sphingosine that is transported into the cytoplasm. There, phosphorylation by sphingosine kinase pulls overall sphingosine export by creating a substrate for a cytoplasmic degradative lyase; alternatively, sphingosine can be reused for ceramide synthesis in the ER. There are several possible explanations for this NPC1 bypass. First, increased sphingosine kinase could increase membrane contact sites, thereby providing access of accumulated lysosomal cholesterol to an alternative cholesterol export route (Höglinger et al., 2019; Meneses-Salas et al., 2020). It is also possible that NPC2 could deliver cholesterol directly to lumenal membranes that contain a less dense glycocalyx, such as those of late endosomes (Cheruku et al., 2006; McCauliff et al., 2019), where LAMP proteins that contribute to the glycocalyx are less abundant (Li et al., 2015). In this case, enhanced sphingosine kinase could somehow enhance late endosome–lysosome fusion to permit such egress. Alternatively, sphingosine kinase activation could enhance MVE exocytosis (Kajimoto et al., 2013), releasing accumulated cholesterol into the extracellular space. All of these scenarios may equally well explain the effect of SNX13 depletion studied here.
In summary, SNX13 appears to be a negative regulator of cholesterol egress from lysosomes. In its absence, PM-localized, accessible cholesterol pools are replenished, even in the absence of NPC1 function (Fig. 9 E). We have shown that SNX13 normally functions as an interorganelle tether, mediating membrane contacts between the ER and LDs and endolysosomes, analogously to its yeast orthologue (Henne et al., 2015). SNX13 may coordinate the association of membrane-bound organelles to maintain appropriate and tightly regulated cholesterol levels in distinct cellular compartments, particularly in the ER. Further work will be needed to parse the differences in phenotypes observed for the related but distinct SNX13 and SNX14 proteins that represent important regulators of both cholesterol and neutral lipid accumulation (Bryant et al., 2018; Datta et al., 2019, 2020).
Materials and methods
Cell culture, antibodies, and other reagents
U2OS, HeLa, and RPE cells were obtained from American Type Culture Collection and grown in DMEM containing 10% FBS, 2 mM L-glutamine, and penicillin (100 U/ml)/streptomycin (100 µg/ml). Cas9-expressing K562 cells (Lu et al., 2018) were grown in RPMI 1640 medium supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, and penicillin (100 U/ml)/streptomycin (100 µg/ml). All cell lines were cultured at 37°C with 5% CO2. The NPC1 CRISPR knockout HeLa cell line was previously generated (Saha et al., 2020). Primary antibodies diluted in PBS with 1% BSA (for immunofluorescence) or 5% skim milk (for immunoblotting) were monoclonal anti–lysobisphosphatidic acid (anti-BMP) clone 6C4 1:1,000 (EMD Millipore; catalog no. MABT837), rabbit polyclonal anti-LAMP2 1:500 (Invitrogen; catalog no. PA1-655), mouse monoclonal anti-GST 1:1,000 (Cell Signaling Technology; catalog no. 2624S), rabbit polyclonal anti-SNX13 1:1,000 (Abcepta; catalog no. AP12244b), mouse monoclonal anti-tubulin 1:10,000 (Sigma-Aldrich; catalog no. T5168), and rabbit monoclonal anti-HA tag (Cell Signaling Technology; catalog no. 3724S). U18666A (Sigma-Aldrich; catalog no. U3633) was used at 1 µM for 16 h. Oleic acid (Sigma-Aldrich; catalog no. O1008) was conjugated with fatty acid–free BSA (Sigma-Aldrich; A8806) at a 6:1 molar ratio and used at 0.5 mM for 16 h. The ACAT pharmacological inhibitor Sandoz 58-035 (Sigma-Aldrich; S9318) was used at 20 µg/ml for 16 h.
Plasmids and transfections
Plasmids encoding SNX13-GFP and SNX14-GFP were a gift from Mike Henne (University of Texas Southwestern, Dallas, TX). The CFP-VAPA construct was a gift from Clare Futter (University College London, London, UK). Plasmids encoding GFP-HNeu and OFP-HPos (Kassan et al., 2013) were a gift from Albert Pol (Institut d'Investigacions Biomèdiques August Pi i Sunyer, University of Barcelona, Barcelona, Spain). The WT-SNX13-2XHA plasmid was prepared as follows. First, the BioID2 insert from a target MCS-BioID2-HA plasmid (Addgene; catalog no. 74224) was removed by restriction digestion at BspEI and HindIII sites and replaced with a 2×HA oligo sequence flanked by the same restriction sites. Next, the WT-SNX13 ORF sequence from the SNX13-GFP plasmid was shuttled into the NheI and BspEI restriction sites of the previously modified MCS-BioID2-HA. The SNX13 truncated constructs were cloned by Gibson assembly using WT-SNX13-2XHA as a template (see primers in Table S1). Transfections of U2OS cells plated on coverslips were performed using GenJet Plus Reagent (SigmaGen Laboratories; catalog no. SL10050) according to the manufacturer’s instructions. For siRNA-mediated knockdown experiments, cells were transfected with siRNAs targeting human SNX13 (5′-CAGAAAGGCUCAACAGAAAUU-3′) or SNX14 (5′-GGAUGAAAGUAUUGACAAAUU-3′) using Lipofectamine RNAiMax (Invitrogen; catalog no. 13778-075) according to the manufacturer’s instructions. Studies were conducted 48–72 h after siRNA transfection. A scrambled siRNA was used as a negative control (Ambion; catalog no. AM4635). For siRNA rescue experiments, human SNX13 was targeted with an siRNA (same sequence as above) labeled with the DY547 fluorophore. The siRNA-resistant WT SNX13-2XHA construct used for these experiments was generated by site-directed mutagenesis.
Quantitative real-time PCR
Total RNA from SNX14 siRNA- or control siRNA–treated cells was extracted using QIAzol (Qiagen; catalog no. 79306) as per the manufacturer. 1 µg of isolated RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems; catalog no. 4368814). Quantitative real-time PCR was performed using SYBR Green–based technology and GoTaq qPCR Master Mix (Promega; catalog no. A600A). Specific SNX14 primers and HPRT1 primers (reference gene) were used (see Table S1). PCRs were run in a LightCycler 96 Real-Time PCR system (Roche). Transcript levels relative to HPRT1 were calculated using the ΔCt method.
Flow cytometric analysis of cholesterol and BMP
K562 cells were treated with or without 1 µM U18666A. After 16 h, cells were harvested and washed before fixation in 2% PFA, then permeabilized in 0.1% saponin and stained with PFO*–Alexa Fluor 647 or anti-BMP and Alexa Fluor 647 goat anti-mouse antibodies (Invitrogen). Quantification of cholesterol and BMP total fluorescence was performed using a BD Accuri C6 flow cytometer, and data were analyzed using FlowJo software (BD Biosciences).
PFO* purification
Recombinant PFO* was purified as previously described (Li et al., 2017). Briefly, expression was induced in Rosetta 2 cells with 1 mM isopropyl β-p-thiogalactopyranoside for 4 h at 37°C. Cells were resuspended in buffer A (PBS, 10% [vol/vol] glycerol, protease inhibitors) and lysed using an Emulsiflex C-5 homogenizer (Avestin). Clarified lysate was incubated with Ni–nitrilotriacetic acid agarose (Qiagen; catalog no. 30210) for 1 h at 4°C, and, after washing with buffer A + 50 mM imidazole, bound protein was eluted with buffer A + 300 mM imidazole. The eluate was concentrated with an Amicon Ultra 10 kD cutoff centrifugal filter (EMD Millipore; catalog no. UFC901024) and then exchanged into buffer A + 1 mm EDTA. PFO* was directly conjugated to N-hydroxysuccinimide ester Alexa Fluor 647 dye as per the manufacturer’s instructions (Life Technologies). For immunofluorescence analysis of cholesterol, GST-PFO* was used (Meneses-Salas et al., 2020).
CRISPR/Cas9 screens in K562 cells
The genome-wide K562 CRISPR knockout library line was generated as previously described (Morgens et al., 2017). Briefly, a whole-genome library of exon-targeting sgRNAs (10 sgRNA per gene; Morgens et al., 2017) was synthesized and cloned into a lentiviral vector (Addgene; catalog no. 89359), which, together with third-generation lentiviral packaging plasmids (pVSVG, pRSV, and pMDL), was transfected into HEK293T cells to generate lentiviral particles. Then ∼300 million Cas9-expressing K562 cells were infected at low multiplicity of infection (<1). Transduced cells were selected and expanded in puromycin-supplemented media over 5–7 d before experiments were conducted 10 d after infection. All screens were performed as independent replicates. Two independent screens were performed for each lipid (cholesterol and BMP) and condition (with or without U18666A). For each screen, 600 million cells were stained. 16 h before staining, cells were treated with 1 µM U18666A or vehicle (control). The next day, cells were first pelleted and washed twice in cold PBS, followed by fixation in 2% PFA-PBS for 30 min at 4°C. Cells were then washed twice in PBS and permeabilized/blocked in 0.1% saponin–1% BSA-PBS for 10 min. Cells were then incubated with 10 µg/ml Alexa 647–labeled PFO* (Li et al., 2017) for 45 min in the cold. Alternatively, for BMP screens, staining was performed with mouse anti-BMP antibody for 1 h at 4°C, followed by 1-h incubation with Alexa Fluor 647–conjugated secondary antibody (Life Technologies) used at 1:2,000. Finally, cells were washed once in cold PBS and then kept in 15 ml PBS–0.5% BSA at 4°C for 16 h before sorting. The next day, cells were separated into 10% high or 10% low PFO/BMP fluorescence populations by sorting on a BD FACSAria II. Approximately 20 million cells were recovered from each gated population. Sorted cells were then sedimented by centrifugation, and the cell pellet was frozen at −80°C before genomic DNA isolation. Approximately 200 million unsorted cells (1,000× coverage per library element) were saved for screen data normalization. Genomic DNA was extracted using Qiagen Blood Midi or Maxi kits (Qiagen; catalog nos. 51183 and 51194) for sorted or unsorted cells, respectively, as per the manufacturer. To prepare sequencing libraries, the sgRNA sequences were PCR amplified from genomic DNA, and the number of PCRs was scaled to use 40–60 µg isolated genomic DNA. Each of these first PCRs contained 10 µg of genomic DNA, 2 µl Herculase II Fusion DNA polymerase (Agilent Technologies; catalog no. 600677), 20 µl 5× Herculase buffer, 1 µl 100 mM deoxynucleoside triphosphates, 1 µl 100 µM oMCB 1562 (forward primer), 1 µl 100 µM oMCB 1563 (reverse primer), and water to adjust the volume to 100 µl. PCR amplification was conducted as follows: 1× 98°C/2 min, 18× (98°C/30 s, 59.1°C/30 s, 72°C/45 s), and 1× 72°C/3 min first PCRs from each sample were pooled. Then second PCRs were set up for each sample as follows: 5 µl from first PCR pooled amplicons, 2 µl Herculase II Fusion DNA polymerase, 20 µl 5× Herculase buffer, 2 µl 100 mM deoxynucleoside triphosphates, 0.8 µl 100 µM oMCB 1439 (forward primer), 0.8 µl 100 µM of barcoded CRISPR knockout reverse primer, and 69.4 µl H2O. The PCR protocol for this second PCR was as follows: 1× 98°C/2 min, 20× (98°C/30 s, 59.1°C/30 s, 72°C/45 s), and 1× 72°C/3 min. Finally, 50 µl of second PCR was separated by running on a 2% Tris-borate-EDTA agarose gel. The PCR products were excised and purified using a QIAquick Gel Extraction Kit (Qiagen; catalog no. 28704) according to the manufacturer’s instructions. The sgRNA libraries were analyzed by deep sequencing on an Illumina NextSeq 500 using a custom sequencing primer (oMCB 1672) with ∼40 million reads per condition (∼200× coverage per library element). Computational analysis and comparison of sgRNA composition of sorted versus unsorted cells were performed using casTLE version 1.0 (https://bitbucket.org/dmorgens/castle) as previously described (Lu et al., 2018; Morgens et al., 2016). Briefly, sgRNA distribution was compared between the sorted and unsorted cell samples, and sgRNA enrichments were calculated as logarithmic ratios between sorted and unsorted cells. A maximum likelihood estimator was used to estimate the phenotypic effect size for each gene and the log-likelihood ratio (confidence score) by comparing the distribution of the 10 different sgRNAs targeting each gene with the distribution of negative control sgRNAs. P values were determined by permuting the gene-targeting sgRNAs in the screen and comparing them with the distribution of negative controls using casTLE. For genome-wide cholesterol screens, we used a threshold of 5% false discovery rate (calculated using the Benjamini-Hochberg procedure) to define hits. For the BMP screens, the 100 top-ranked genes in the analysis were considered as hits. Because cholesterol is functionally linked to BMP (Chevallier et al., 2008), despite lower statistical significance for some of the BMP screen hits, we found that a significant number of genes that passed this cutoff overlapped with those identified as hits in the cholesterol screens. See Table S1 for complete genome-wide screen datasets.
Immunofluorescence
To visualize endolysosomes in suspension K562 cells (Fig. 1, B–D), cells were attached to glass coverslips using a cytospin (Shandon) at 800 rpm for 5 min. Cells were fixed with 3.7% (vol/vol) PFA for 15 min, washed, and permeabilized/blocked with 0.1% saponin/1% BSA-PBS, except for cells labeled with GST-PFO*, which were permeabilized for 3 min in 0.1% Triton X-100 and blocked with 1% BSA in PBS. Primary antibodies were diluted in PBS–1% BSA and incubated for 1 h at RT. Highly cross-adsorbed H+L secondary antibodies (Life Technologies) conjugated to Alexa Fluor 488, 568, or 647 were used at 1:2,000 in PBS–1% BSA and incubated at RT for 45 min. Nuclei were stained using 0.1 µg/ml DAPI (Sigma-Aldrich; catalog no. D9542), and coverslips were mounted on glass slides with Mowiol. Microscopy images were acquired using a Zeiss LSM880 laser scanning spectral confocal microscope (Carl Zeiss) equipped with an Axio Observer 7 inverted microscope, a blue diode (405 nm), argon (488 nm), diode-pumped solid-state (561 nm), and HeNe (633 nm) lasers, and a Plan Apochromat 63× /NA1.4 oil-immersion objective lens. DAPI, Alexa Fluor 488, Alexa Fluor 555, and Alexa Fluor 647 images were acquired sequentially using 405-, 488-, 561-, and 633-nm laser lines; Acousto-Optical Beam Splitter as a beam splitter; and emission detection ranges 415–480 nm, 500–550 nm, 571–625 nm, and 643–680 nm, respectively. The confocal pinhole was set at 1 Airy unit. All images were acquired in a 1,024 × 1,024–pixel format. In some experiments (Fig. 1, B–D; Fig. S4 B), images were obtained using MetaMorph software with a spinning-disk confocal microscope (Yokogawa) with an electron-multiplying charge-coupled device camera (Andor) and a 100×/1.4 NA oil-immersion objective. Typical exposure times of 100–300 ms were used. 3D-rendered images (Fig. S4 E) were generated using IMARIS software (Bitplane AG). All image quantifications were performed using CellProfiler (Carpenter et al., 2006).
Live-cell microscopy analysis of ER–EL/LD contacts
U2OS cells were plated in 25-mm coverslips and transfected with SNX13-GFP or a GFP-ER probe (GFP-HNeu; Kassan et al., 2013) used as a control. 24 h after transfection, cells were pulsed for 2 h with 0.1 mg/ml TRITC-dextran, followed by extensive PBS washes and a chase of 2 h in full media to label ELs before imaging. For monitorization of ER–LD contacts, cells were cotransfected with SNX13-GFP and an OFP-tagged LD probe (OFP-HPos; Kassan et al., 2013). Coverslips were mounted on an Attofluor chamber (Molecular Probes, Thermo Fisher Scientific; catalog no. A7816). Automated multiposition live-cell imaging was performed using a Zeiss LSM 880 confocal microscope (Carl Zeiss) equipped with Definite Focus control to keep the specimen in focus and an incubation system with temperature (37°C), humidity, and CO2 (5%) control. 10 min after swelling cells in hypotonic media (5% DMEM), GFP (ER) and TRITC (ELs) signals were acquired sequentially line by line using 488- and 561-nm laser lines and detection ranges at 500–550 nm and 570–650 nm, respectively. Single-section images were acquired every 15 s during ∼10 min using a 63× oil immersion objective lens (NA 1.4) and a pinhole set at 1 Airy unit.
Image analysis was performed using FIJI-ImageJ (Wayne Rasband, National Institutes of Health). The following workflow was automated in an ImageJ macro script (TrackTheContacts) to track contacts between the ER and ELs in time-lapse experiments, based on the previously described macro code ContactJ (Bosch et al., 2020; Martín et al., 2021). Time-lapse videos were first recorded using the StackReg plugin, and target cells were segmented manually and analyzed individually. First, by adapting the previous ContactJ macro code, ER–EL contact regions were initially obtained using the Colocalization Highlighter plugin and then converted into a contour line section by skeletonization. Next, binary individually segmented ELs were tracked using the TrackMate plugin and coded with their corresponding track identity value. Finally, the contact length from each track-coded vesicle was measured frame by frame and stored in a table together with its corresponding track identity, time frame, and vesicle area. For contact persistence quantitation (Fig. 9 B), only contacts that could be tracked for at least three consecutive frames (45 s) were measured. The average ER–EL contact number (Fig. 9 C) or length (Fig. 9 D) per frame was calculated as the total number of contacts or as the sum of all contact lengths, divided by the total number of ELs in each frame, respectively.
Immunoblotting
Cells were lysed in lysis buffer (50 mM Hepes, 150 mM KCl, 1% Triton X-100, 5 mM MgCl2, pH 7.4) supplemented with a protease/phosphatase inhibitor cocktail (1 mM Na3VO4, 10 mM NaF, 1 mM PMSF, 10 µg/ml leupeptin, and 10 µg/ml aprotinin). Lysates were boiled in 1× sample buffer, resolved on SDS-PAGE gel, and transferred onto nitrocellulose membranes (Bio-Rad Laboratories; catalog no. 1620115) using a Bio-Rad Trans-Blot system. Membranes were blocked with 5% skim milk in TBS with Tween-20 for 60 min at RT. Primary antibodies were diluted in blocking buffer and incubated either for 1 h at RTor overnight at 4°C. HRP-conjugated secondary antibodies (Bio-Rad Laboratories or Abcam) diluted in blocking buffer at 1:5,000 were incubated for 60 min at RT and developed using EZ-ECL (Biological Industries). Blots were imaged using an ImageQuant LAS 4000 system (GE Healthcare) and quantified using ImageJ software.
TLC
Total lipids were extracted from control or SNX13-depleted cells as follows. Briefly, cells were washed with PBS and resuspended in 1 vol of methanol:chloroform (1:2). Next, one-half volume of chloroform and one-half volume of H2O were added, and tubes were centrifuged at 5,000 rpm for 5 min. The organic phase was collected, dried, resuspended with chloroform, spotted on TLC silica gel 60 plates (EMD Millipore; catalog no. 1055530001), and dried for several minutes. Plates were run in a solvent system of hexane/diethyl ether/acetic acid (70:30:1) until three-fourths of the total length of the plate was reached. Lipids were stained using a phosphomolybdic acid (ACROS Organics; catalog no. 206385000) ethanol solution.
BMP lipidomics
Targeted high-resolution ultraperformance liquid chromatography MS/MS was used to accurately quantitate the three geometrical isoforms (2,2′-, 2,3′-, and 3,3′-) of di-22:6-BMP and di-18:1-BMP in control or SNX13 siRNA-treated cells with or without U18666A. Lipidomic analyses were conducted by Nextcea, Inc. as previously described (Liu et al., 2014) using a SCIEX TripleTOF 6600 mass spectrometer equipped with an IonDrive Turbo V source (SCIEX). Standard curves were prepared using authentic BMP reference standards. Protein was determined by bicinchoninic acid protein assay.
Biochemical cholesterol determination
Total cell cholesterol content of control or SNX13 siRNA-treated U2OS cells was determined using the Amplex Red Cholesterol Assay kit (Molecular Probes; catalog no. A12216) according to the manufacturer’s instructions. Samples were treated with cholesterol esterase before the assay to cleave cholesterol esters. Protein was determined by Bio-Rad Laboratories protein assay.
Statistics
Results are expressed as the mean ± SEM unless otherwise specified. Means were compared using Student’s t test when two experimental conditions were compared. When three or more experimental conditions were compared, statistical significance was assessed via multiple t tests by the Holm-Sidak method with α = 0.05 or two-way ANOVA with Tukey’s post hoc test using GraphPad Prism version 9, unless otherwise specified. Two-tailed P values <0.05 were considered statistically significant (Lord et al., 2020).
Online supplemental material
Fig. S1 shows hierarchical clustering analysis comparing 10% low and 10% high populations. Fig. S2 shows subcellular localization of screen hits and their phenotypes. Fig. S3 shows a comparison of hits from four independent studies analyzing cholesterol homeostasis. Fig. S4 shows how SNX13 depletion redistributes cholesterol to the cell surface in the absence of NPC1 function. Video 1 shows live-cell visualization of SNX13-positive ER domains in contact with LDs. Video 2 shows live imaging of ER–EL contacts in U2OS cells overexpressing the GFP-ER probe 10 min after incubation in hypotonic media. Video 3 shows live imaging of ER–endolysosome contacts in U2OS cells overexpressing SNX13-GFP 10 min after incubation in hypotonic media. Table S1 lists oligonucleotide sequences used in this study and all screen results in an Excel file.
Supplementary Material
lists oligonucleotide sequences used in this study and all screen results in an Excel file.
Acknowledgments
We are grateful to Dr. Mike Bassik (Stanford University School of Medicine, Stanford, CA) for sharing Cas9-positive K562 cells infected with the sgRNA-CRISPR knockout lentivirus library and running sequencing experiments. We also thank Maria Calvo and Gemma Martín for help with automated membrane contact microscopy analysis.
This research was funded by grants to S.R. Pfeffer from the National Heart, Lung, and Blood Institute (5R01HL134991-04) and the Ara Parseghian Medical Research Foundation/University of Notre Dame. C. Enrich was funded by the Spanish Ministerio de Economía y Competitividad (grant BFU2015-66785-P) and the Universitat de Barcelona (grant AR0RM005).
The authors declare no competing financial interests.
Author contributions: A. Lu and S.R. Pfeffer conceived the project. All experiments were carried out by A. Lu except for biochemical determination of cellular cholesterol content (S.R. Pfeffer, B.R. Sharma, and S.R. Vaughn) and mass spectrometric analysis of BMP (F. Hsieh). C. Enrich provided materials and advice, and S.R. Pfeffer oversaw the project and obtained research funding. A. Lu and S.R. Pfeffer wrote and edited the manuscript.
Data availability
The adapted ContactJ Fiji macro (Martín et al., 2021) used for ER–EL membrane contact image analysis is available at https://zenodo.org/record/5639498#.YYF4fBDMLOQ.
References
- Abi-Mosleh, L., Infante R.E., Radhakrishnan A., Goldstein J.L., and Brown M.S.. 2009. Cyclodextrin overcomes deficient lysosome-to-endoplasmic reticulum transport of cholesterol in Niemann-Pick type C cells. Proc. Natl. Acad. Sci. USA. 106:19316–19321. 10.1073/pnas.0910916106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi, S., Homoto M., Tanaka R., Hioki Y., Murakami H., Suga H., Matsumoto M., Nakayama K.I., Hatta T., Iemura S., and Natsume T.. 2014. ZFP36L1 and ZFP36L2 control LDLR mRNA stability via the ERK-RSK pathway. Nucleic Acids Res. 42:10037–10049. 10.1093/nar/gku652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcalay, R.N., Hsieh F., Tengstrand E., Padmanabhan S., Baptista M., Kehoe C., Narayan S., Boehme A.K., and Merchant K.. 2020. Higher urine bis(monoacylglycerol)phosphate levels in LRRK2 G2019S mutation carriers: implications for therapeutic development. Mov. Disord. 35:134–141. 10.1002/mds.27818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aregger, M., Lawson K.A., Billmann M., Costanzo M., Tong A.H.Y., Chan K., Rahman M., Brown K.R., Ross C., Usaj M., et al. 2020. Systematic mapping of genetic interactions for de novo fatty acid synthesis identifies C12orf49 as a regulator of lipid metabolism. Nat. Metab. 2:499–513. 10.1038/s42255-020-0211-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio, A., and Bonifacino J.S.. 2020. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21:101–118. 10.1038/s41580-019-0185-4 [DOI] [PubMed] [Google Scholar]
- Bayraktar, E.C., La K., Karpman K., Unlu G., Ozerdem C., Ritter D.J., Alwaseem H., Molina H., Hoffmann H.H., Millner A., et al. 2020. Metabolic coessentiality mapping identifies C12orf49 as a regulator of SREBP processing and cholesterol metabolism. Nat. Metab. 2:487–498. 10.1038/s42255-020-0206-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, M., Sánchez-Álvarez M., Fajardo A., Kapetanovic R., Steiner B., Dutra F., Moreira L., López J.A., Campo R., Marí M., et al. 2020. Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science. 370:eaay8085. 10.1126/science.aay8085 [DOI] [PubMed] [Google Scholar]
- Bryant, D., Liu Y., Datta S., Hariri H., Seda M., Anderson G., Peskett E., Demetriou C., Sousa S., Jenkins D., et al. 2018. SNX14 mutations affect endoplasmic reticulum-associated neutral lipid metabolism in autosomal recessive spinocerebellar ataxia 20. Hum. Mol. Genet. 27:1927–1940. 10.1093/hmg/ddy101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100. 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova, J.E., and Winckler B.. 2017. A new Rab7 effector controls phosphoinositide conversion in endosome maturation. J. Cell Biol. 216:2995–2997. 10.1083/jcb.201709034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerikan, B., Shaheen R., Colo G.P., Gläßer C., Hata S., Knobeloch K.P., Alkuraya F.S., Fässler R., and Schiebel E.. 2016. Cell-intrinsic adaptation arising from chronic ablation of a key Rho GTPase regulator. Dev. Cell. 39:28–43. 10.1016/j.devcel.2016.08.020 [DOI] [PubMed] [Google Scholar]
- Cheruku, S.R., Xu Z., Dutia R., Lobel P., and Storch J.. 2006. Mechanism of cholesterol transfer from the Niemann-Pick type C2 protein to model membranes supports a role in lysosomal cholesterol transport. J. Biol. Chem. 281:31594–31604. 10.1074/jbc.M602765200 [DOI] [PubMed] [Google Scholar]
- Chevallier, J., Chamoun Z., Jiang G., Prestwich G., Sakai N., Matile S., Parton R.G., and Gruenberg J.. 2008. Lysobisphosphatidic acid controls endosomal cholesterol levels. J. Biol. Chem. 283:27871–27880. 10.1074/jbc.M801463200 [DOI] [PubMed] [Google Scholar]
- Chu, B.B., Liao Y.C., Qi W., Xie C., Du X., Wang J., Yang H., Miao H.H., Li B.L., and Song B.L.. 2015. Cholesterol transport through lysosome-peroxisome membrane contacts. Cell. 161:291–306. 10.1016/j.cell.2015.02.019 [DOI] [PubMed] [Google Scholar]
- Das, A., Goldstein J.L., Anderson D.D., Brown M.S., and Radhakrishnan A.. 2013. Use of mutant 125I-perfringolysin O to probe transport and organization of cholesterol in membranes of animal cells. Proc. Natl. Acad. Sci. USA. 110:10580–10585. 10.1073/pnas.1309273110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A., Brown M.S., Anderson D.D., Goldstein J.L., and Radhakrishnan A.. 2014. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife. 3:e02882. 10.7554/eLife.02882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, S., Liu Y., Hariri H., Bowerman J., and Henne W.M.. 2019. Cerebellar ataxia disease-associated Snx14 promotes lipid droplet growth at ER-droplet contacts. J. Cell Biol. 218:1335–1351. 10.1083/jcb.201808133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, S., Bowerman J., Hariri H., Ugrankar R., Eckert K.M., Corley C., Vale G., McDonald J.G., and Henne W.M.. 2020. Snx14 proximity labeling reveals a role in saturated fatty acid metabolism and ER homeostasis defective in SCAR20 disease. Proc. Natl. Acad. Sci. USA. 117:33282–33294. 10.1073/pnas.2011124117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, O.B., Shin H.R., Lim C.Y., Wu E.Y., Kukurugya M., Maher C.F., Perera R.M., Ordonez M.P., and Zoncu R.. 2021. NPC1-mTORC1 signaling couples cholesterol sensing to organelle homeostasis and is a targetable pathway in Niemann-Pick type C. Dev. Cell. 56:260–276.e7. 10.1016/j.devcel.2020.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diofano, F., Weinmann K., Schneider I., Thiessen K.D., Rottbauer W., and Just S.. 2020. Genetic compensation prevents myopathy and heart failure in an in vivo model of Bag3 deficiency. PLoS Genet. 16:e1009088. 10.1371/journal.pgen.1009088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, X., Kazim A.S., Brown A.J., and Yang H.. 2012. An essential role of Hrs/Vps27 in endosomal cholesterol trafficking. Cell Rep. 1:29–35. 10.1016/j.celrep.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Gruenberg, J. 2020. Life in the lumen: The multivesicular endosome. Traffic. 21:76–93. 10.1111/tra.12715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne, W.M., Zhu L., Balogi Z., Stefan C., Pleiss J.A., and Emr S.D.. 2015. Mdm1/Snx13 is a novel ER-endolysosomal interorganelle tethering protein. J. Cell Biol. 210:541–551. 10.1083/jcb.201503088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höglinger, D., Burgoyne T., Sanchez-Heras E., Hartwig P., Colaco A., Newton J., Futter C.E., Spiegel S., Platt F.M., and Eden E.R.. 2019. NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress. Nat. Commun. 10:4276. 10.1038/s41467-019-12152-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante, R.E., and Radhakrishnan A.. 2017. Continuous transport of a small fraction of plasma membrane cholesterol to endoplasmic reticulum regulates total cellular cholesterol. eLife. 6:e25466. 10.7554/eLife.25466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimoto, T., Okada T., Miya S., Zhang L., and Nakamura S.. 2013. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat. Commun. 4:2712. 10.1038/ncomms3712 [DOI] [PubMed] [Google Scholar]
- Kassan, A., Herms A., Fernández-Vidal A., Bosch M., Schieber N.L., Reddy B.J., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203:985–1001. 10.1083/jcb.201305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, C., Sengupta P., Seo A.Y., and Lippincott-Schwartz J.. 2020. ER membranes exhibit phase behavior at sites of organelle contact. Proc. Natl. Acad. Sci. USA. 117:7225–7235. 10.1073/pnas.1910854117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter, T., and Sandhoff K.. 2005. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu. Rev. Cell Dev. Biol. 21:81–103. 10.1146/annurev.cellbio.21.122303.120013 [DOI] [PubMed] [Google Scholar]
- Li, J., Deffieu M.S., Lee P.L., Saha P., and Pfeffer S.R.. 2015. Glycosylation inhibition reduces cholesterol accumulation in NPC1 protein-deficient cells. Proc. Natl. Acad. Sci. USA. 112:14876–14881. 10.1073/pnas.1520490112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Lee P.L., and Pfeffer S.R.. 2017. Quantitative measurement of cholesterol in cell populations using flow cytometry and fluorescent perfringolysin O. Methods Mol. Biol. 1583:85–95. 10.1007/978-1-4939-6875-6_8 [DOI] [PubMed] [Google Scholar]
- Lim, C.Y., Davis O.B., Shin H.R., Zhang J., Berdan C.A., Jiang X., Counihan J.L., Ory D.S., Nomura D.K., and Zoncu R.. 2019. ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C. Nat. Cell Biol. 21:1206–1218. 10.1038/s41556-019-0391-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N., Tengstrand E.A., Chourb L., and Hsieh F.Y.. 2014. Di-22:6-bis(monoacylglycerol)phosphate: A clinical biomarker of drug-induced phospholipidosis for drug development and safety assessment. Toxicol. Appl. Pharmacol. 279:467–476. 10.1016/j.taap.2014.06.014 [DOI] [PubMed] [Google Scholar]
- Llorente, A., van Deurs B., and Sandvig K.. 2007. Cholesterol regulates prostasome release from secretory lysosomes in PC-3 human prostate cancer cells. Eur. J. Cell Biol. 86:405–415. 10.1016/j.ejcb.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Logan, T., Simon M.J., Rana A., Cherf G.M., Srivastava A., Davis S.S., Low R.L.Y., Chiu C.L., Fang M., Huang F., et al. 2021. Rescue of a lysosomal storage disorder caused by Grn loss of function with a brain penetrant progranulin biologic. Cell. 184:4651–4668.e25. 10.1016/j.cell.2021.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, S.J., Velle K.B., Mullins R.D., and Fritz-Laylin L.K.. 2020. SuperPlots: Communicating reproducibility and variability in cell biology. J. Cell Biol. 219:e202001064. 10.1083/jcb.202001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loregger, A., Raaben M., Nieuwenhuis J., Tan J.M.E., Jae L.T., van den Hengel L.G., Hendrix S., van den Berg M., Scheij S., Song J.Y., et al. 2020. Haploid genetic screens identify SPRING/C12ORF49 as a determinant of SREBP signaling and cholesterol metabolism. Nat. Commun. 11:1128. 10.1038/s41467-020-14811-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, F., Liang Q., Abi-Mosleh L., Das A., De Brabander J.K., Goldstein J.L., and Brown M.S.. 2015. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife. 4:e12177. 10.7554/eLife.12177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, A., Wawro P., Morgens D.W., Portela F., Bassik M.C., and Pfeffer S.R.. 2018. Genome-wide interrogation of extracellular vesicle biology using barcoded miRNAs. eLife. 7:e41460. 10.7554/eLife.41460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín, G., Bosch M., Coll E., Parton R.G., Pol A., and Calvo M.. 2021. ContactJ: lipid droplets-mitochondria contacts characterization through fluorescence microscopy and image analysis [version 1; peer review: 2 approved with reservations]. F1000Res. 10:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauliff, L.A., Langan A., Li R., Ilnytska O., Bose D., Waghalter M., Lai K., Kahn P.C., and Storch J.. 2019. Intracellular cholesterol trafficking is dependent upon NPC2 interaction with lysobisphosphatidic acid. eLife. 8:e50832. 10.7554/eLife.50832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneses-Salas, E., García-Melero A., Kanerva K., Blanco-Muñoz P., Morales-Paytuvi F., Bonjoch J., Casas J., Egert A., Beevi S.S., Jose J., et al. 2020. Annexin A6 modulates TBC1D15/Rab7/StARD3 axis to control endosomal cholesterol export in NPC1 cells. Cell. Mol. Life Sci. 77:2839–2857. 10.1007/s00018-019-03330-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens, D.W., Deans R.M., Li A., and Bassik M.C.. 2016. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 34:634–636. 10.1038/nbt.3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens, D.W., Wainberg M., Boyle E.A., Ursu O., Araya C.L., Tsui C.K., Haney M.S., Hess G.T., Han K., Jeng E.E., et al. 2017. Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat. Commun. 8:15178. 10.1038/ncomms15178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, J., Palladino E.N.D., Weigel C., Maceyka M., Gräler M.H., Senkal C.E., Enriz R.D., Marvanova P., Jampilek J., Lima S., et al. 2020. Targeting defective sphingosine kinase 1 in Niemann-Pick type C disease with an activator mitigates cholesterol accumulation. J. Biol. Chem. 295:9121–9133. 10.1074/jbc.RA120.012659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentchev, P.G. 2004. Niemann-Pick C research from mouse to gene. Biochim. Biophys. Acta. 1685:3–7. 10.1016/j.bbalip.2004.08.005 [DOI] [PubMed] [Google Scholar]
- Pfeffer, S.R. 2019. NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes. J. Biol. Chem. 294:1706–1709. 10.1074/jbc.TM118.004165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, A.I., Zhang G., Warren J.D., and Maxfield F.R.. 2010. Endocytosis of β-cyclodextrins is responsible for cholesterol reduction in Niemann-Pick type C mutant cells. Proc. Natl. Acad. Sci. USA. 107:5477–5482. 10.1073/pnas.0914309107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, P., Shumate J.L., Caldwell J.G., Elghobashi-Meinhardt N., Lu A., Zhang L., Olsson N.E., Elias J.E., and Pfeffer S.R.. 2020. Inter-domain dynamics drive cholesterol transport by NPC1 and NPC1L1 proteins. eLife. 9:e57089. 10.7554/eLife.57089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamaki, J.I., Wilkinson S., Hahn M., Tasdemir N., O’Prey J., Clark W., Hedley A., Nixon C., Long J.S., New M., et al. 2017. Bromodomain protein BRD4 is a transcriptional repressor of autophagy and lysosomal function. Mol. Cell. 66:517–532.e9. 10.1016/j.molcel.2017.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanga, C.M., and Salanga M.C.. 2021. Genotype to phenotype: CRISPR gene editing reveals genetic compensation as a mechanism for phenotypic disjunction of morphants and mutants. Int. J. Mol. Sci. 22:3472. 10.3390/ijms22073472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saric, A., Freeman S.A., Williamson C.D., Jarnik M., Guardia C.M., Fernandopulle M.S., Gershlick D.C., and Bonifacino J.S.. 2021. SNX19 restricts endolysosome motility through contacts with the endoplasmic reticulum. Nat. Commun. 12:4552. 10.1038/s41467-021-24709-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, C.C., Vossio S., Vacca F., Snijder B., Larios J., Schaad O., Guex N., Kuznetsov D., Martin O., Chambon M., et al. 2015. Wnt directs the endosomal flux of LDL-derived cholesterol and lipid droplet homeostasis. EMBO Rep. 16:741–752. 10.15252/embr.201540081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, L.A., Podder S., He J., Jackson-Chornenki N.L., Gibson K., Ziliotto R.G., Rhee J., and DeKoter R.P.. 2017. Coordination of myeloid differentiation with reduced cell cycle progression by PU.1 induction of microRNAs targeting cell cycle regulators and lipid anabolism. Mol. Cell. Biol. 37:e00013–e00017. 10.1128/MCB.00013-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, K., Goebel C., Runz H., Möbius W., Weiss S., Feussner I., Simons M., and Schneider A.. 2010. Exosome secretion ameliorates lysosomal storage of cholesterol in Niemann-Pick type C disease. J. Biol. Chem. 285:26279–26288. 10.1074/jbc.M110.134775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuffers, S., Sem Wegner C., Stenmark H., and Brech A.. 2009. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic. 10:925–937. 10.1111/j.1600-0854.2009.00920.x [DOI] [PubMed] [Google Scholar]
- Tan, J.M.E., Cook E.C.L., van den Berg M., Scheij S., Zelcer N., and Loregger A.. 2019. Differential use of E2 ubiquitin conjugating enzymes for regulated degradation of the rate-limiting enzymes HMGCR and SQLE in cholesterol biosynthesis. Atherosclerosis. 281:137–142. 10.1016/j.atherosclerosis.2018.12.008 [DOI] [PubMed] [Google Scholar]
- Thelen, A.M., and Zoncu R.. 2017. Emerging roles for the lysosome in lipid metabolism. Trends Cell Biol. 27:833–850. 10.1016/j.tcb.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic, K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., and Simons M.. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 319:1244–1247. 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- Trinh, M.N., Brown M.S., Goldstein J.L., Han J., Vale G., McDonald J.G., Seemann J., Mendell J.T., and Lu F.. 2020. Last step in the path of LDL cholesterol from lysosome to plasma membrane to ER is governed by phosphatidylserine. Proc. Natl. Acad. Sci. USA. 117:18521–18529. 10.1073/pnas.2010682117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugrankar, R., Bowerman J., Hariri H., Chandra M., Chen K., Bossanyi M.F., Datta S., Rogers S., Eckert K.M., Vale G., et al. 2019. Drosophila Snazarus Regulates a Lipid Droplet Population at Plasma Membrane-Droplet Contacts in Adipocytes. Dev. Cell. 50:557–572.e5. 10.1016/j.devcel.2019.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boomen, D.J.H., Sienkiewicz A., Berlin I., Jongsma M.L.M., van Elsland D.M., Luzio J.P., Neefjes J.J.C., and Lehner P.J.. 2020. A trimeric Rab7 GEF controls NPC1-dependent lysosomal cholesterol export. Nat. Commun. 11:5559. 10.1038/s41467-020-19032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B., and Tontonoz P.. 2018. Liver X receptors in lipid signalling and membrane homeostasis. Nat. Rev. Endocrinol. 14:452–463. 10.1038/s41574-018-0037-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka, T., Tosaki A., Kurosawa M., Shimogori T., Hattori N., and Nukina N.. 2016. Genome-wide analyses in neuronal cells reveal that upstream transcription factors regulate lysosomal gene expression. FEBS J. 283:1077–1087. 10.1111/febs.13650 [DOI] [PubMed] [Google Scholar]
- Youn, D.Y., Xiaoli A.M., Pessin J.E., and Yang F.. 2016. Regulation of metabolism by the Mediator complex. Biophys. Rep. 2:69–77. 10.1007/s41048-016-0031-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., Sun L., Bastos de Oliveira F., Qi X., Brown W.J., Smolka M.B., Sun Y., and Hu F.. 2015. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J. Cell Biol. 210:991–1002. 10.1083/jcb.201502029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists oligonucleotide sequences used in this study and all screen results in an Excel file.
Data Availability Statement
The adapted ContactJ Fiji macro (Martín et al., 2021) used for ER–EL membrane contact image analysis is available at https://zenodo.org/record/5639498#.YYF4fBDMLOQ.