Abstract
The nature of the interaction of coactivator proteins with transcriptionally active promoters in chromatin is a fundamental question in transcriptional regulation by RNA polymerase II. In this study, we used a biochemical approach to examine the functional association of the coactivator p300 with chromatin templates. Using in vitro transcription template competition assays, we observed that p300 forms a stable, template-committed complex with chromatin during the transcription process. The template commitment is dependent on the time of incubation of p300 with the chromatin template and occurs independently of the presence of a transcriptional activator protein. In studies examining interactions between p300 and chromatin, we found that p300 binds directly to chromatin and that the binding requires the p300 bromodomain, a conserved 110-amino-acid sequence found in many chromatin-associated proteins. Furthermore, we observed that the isolated p300 bromodomain binds directly to histones, preferentially to histone H3. However, the isolated p300 bromodomain does not bind to nucleosomal histones under the same assay conditions, suggesting that free histones and nucleosomal histones are not equivalent as binding substrates. Collectively, our results suggest that the stable association of p300 with chromatin is mediated, at least in part, by the bromodomain and is critically important for p300 function. Furthermore, our results suggest a model for p300 function that involves distinct activator-dependent targeting and activator-independent chromatin binding activities.
Activated transcription of genes by RNA polymerase II (RNA Pol II) requires the concerted actions of sequence-specific DNA-binding transcriptional activators, chromatin remodeling complexes, histone acetyltransferases (HATs), and coactivators. Together, these factors function to overcome the transcriptional repression caused by the packaging of genes into chromatin and to stimulate the activity of RNA Pol II. The initiating event in the process of activated transcription is the binding of transcriptional activator proteins to binding sites in the promoters of target genes. In many cases, the activities of the activator proteins are regulated by input from cellular signaling pathways (e.g., steroid hormones and mitogen-activated protein kinase pathways). Once bound to DNA, activators can recruit chromatin remodeling complexes, HATs, and coactivators to the promoter, leading to the formation of a stable RNA Pol II preinitiation complex and, subsequently, transcription initiation (reviewed in references 3, 19, 23, 30, 34, 50, and 67).
Transcriptional coactivators are a diverse group of factors and multipolypeptide complexes, some of which possess intrinsic HAT activity (16, 42, 43, 45, 63, 68). Coactivators with HAT activity include p300 and the closely related CREB binding protein (CBP; often referred to with p300 as p300/CBP) (4, 20), as well as the p300/CBP-associated factor (PCAF), which functions as part of a multipolypeptide complex (54, 69). In general, coactivators play one or more of the following roles: (i) they function as bridging factors to recruit other coactivators to the DNA bound transcriptional activator (e.g., members of the p160/steroid receptor coactivator [SRC] family of proteins which recruit p300/CBP to ligand-activated steroid hormone receptors) (reviewed in reference 43); (ii) they acetylate nucleosomal histones in and around the promoters of activated genes (e.g., the PCAF complex which acetylates nucleosomal histones H3 and H4) (58); (iii) they alter the activities of transcription factors and chromatin-associated proteins by acetylating them (e.g., acetylation of p53 by p300/CBP increases the sequence-specific DNA binding of p53) (22; see references 9, 25, 49, 61 for additional examples); and (iv) they make contacts with the basal transcriptional machinery to stimulate the recruitment of RNA Pol II and the formation of stable transcription preinitiation complexes (e.g., the multipolypeptide TRAP-DRIP-ARC complex, which can enhance the transcriptional activity of nuclear receptors and other activators) (15, 23, 42, 45).
p300 and CBP are large multifunctional coactivators that participate in all four of these processes. In addition to their intrinsic HAT activity, p300 and CBP contain distinct domains for binding to transcriptional activator proteins, other coactivators, and components of the basal transcriptional machinery (4, 20). They also contain a single bromodomain, a conserved 110-amino-acid sequence found in many chromatin-associated proteins, including almost all nuclear HATs (reviewed in references 28 and 66). The bromodomain is thought to function as a histone binding motif (28, 66). Recent studies have shown the bromodomain to have a conserved structure consisting of four antiparallel alpha helices (a four-helix bundle) with a left-handed twist (12, 27, 57). The multiple distinct domains and functions of p300/CBP are differentially required for coactivator function with different classes of transcriptional activator proteins (35, 38, 39).
Although activator-mediated recruitment of coactivators has been well established, the events occurring before and after recruitment are less clear. Many fundamental questions regarding multifunctional coactivators such as p300/CBP relate to how their multiple functional activities are coordinated at the promoter during the course of events leading to transcription initiation. Do coactivators bind to chromatin prior to targeting to specific promoters by DNA-bound activators? Do coactivators remain stably associated with a promoter once recruited? If so, are contacts with a transcriptional activator sufficient to keep the coactivators associated with the promoter, or are other protein-protein contacts also required? In this study, we have used a biochemical approach to address these questions using p300 as a model multifunctional coactivator. Our results indicate that p300 forms a stable, template-committed complex with chromatin. We have also found that p300 binds directly to chromatin and that the binding requires the bromodomain. Taken together, our results suggest a model for p300 function that involves distinct activator-dependent targeting and activator-independent chromatin binding activities.
MATERIALS AND METHODS
Synthesis and purification of recombinant proteins.
Wild-type and bromodomain-deletion-containing His6-tagged human p300 proteins were synthesized in Sf9 cells using a baculovirus expression system and purified as described previously (35–38). His6-tagged NF-κB p65 subunit (44) was synthesized in Sf9 cells by using a recombinant baculovirus kindly provided by J. Hiscott (Lady Davis Institute, Montreal, Canada) and purified essentially as described for His6-tagged p300 (36). Purified Sp1 was obtained from Promega (Madison, Wis.). FLAG-tagged human PCAF was synthesized in Sf9 cells by using a recombinant baculovirus kindly provided by Y. Nakatani (Dana-Farber Cancer Institute) and purified as described for FLAG-tagged human estrogen receptor (36).
DNA templates.
The wild-type human interferon regulatory factor 1 (IRF-1) template in pUC118 contains 1.3 kb of the native gene, which includes the core promoter and the upstream regulatory region with the NF-κB and Sp1 binding sites (6, 60). The IRF+30 construct contains a 30-bp insert in the IRF-1 promoter located 44 bp downstream of the transcription start site and is otherwise identical to the wild-type IRF-1 construct.
Chromatin assembly and in vitro transcription reactions.
Chromatin assembly reactions were performed with a chromatin assembly extract derived from Drosophila embryos (S190) (5, 31, 36–38). The transcriptional activator proteins NF-κB p65 and Sp1 were added during the chromatin assembly reactions at concentrations of 200 and 30 nM, respectively. p300 was added at a concentration of 50 nM to the chromatin templates, where indicated, after the assembly reactions were complete. The reaction mixtures were incubated for an additional 30 min at 27°C after the p300 was added to allow the interaction of the p300 with the chromatin templates prior to the addition of the HeLa cell nuclear extracts for transcription.
In vitro transcription reactions were performed with HeLa cell nuclear extracts that were prepared essentially by the method of Dignam et al. (13) with slight modification (37). The template competition experiments were set up as indicated in Fig. 1 and described below and were analyzed in single-round transcription reactions as described previously (36), except that transcription preinitiation complexes were formed for 45 min at 27°C after the addition of the HeLa nuclear extract. The RNA products from the in vitro transcription reactions were analyzed by primer extension analysis (36). All reactions were performed in duplicate, and each experiment was performed a minimum of three separate times to ensure reproducibility. The data were analyzed and quantified with a PhosphorImager (Molecular Dynamics).
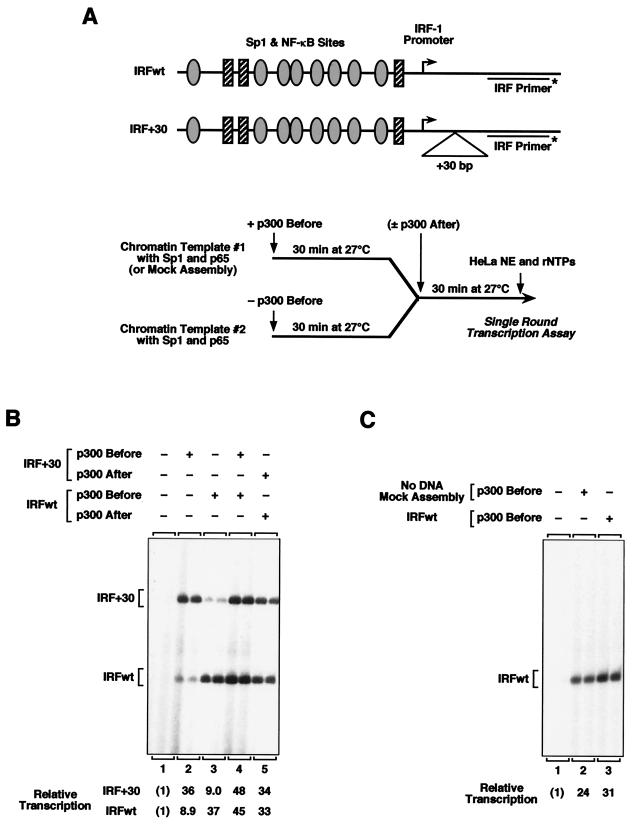
FIG. 1.
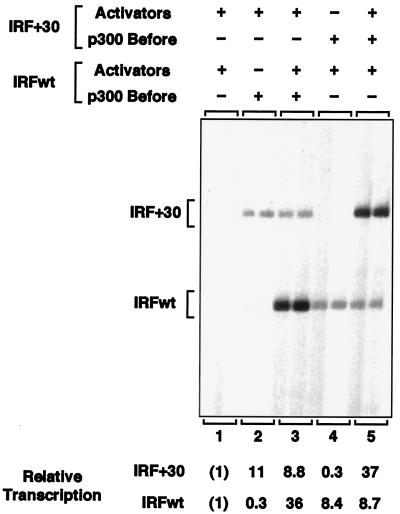
p300 forms a stable, template-committed complex with chromatin. (A) (Top) Schematic diagram of the templates IRFwt and IRF+30. Ovals and rectangles represent binding sites for Sp1 and NF-κB, respectively, and are not drawn to scale. (Bottom) Outline of the template competition experiments, which are described in the text. p300 was added to the chromatin templates at a concentration of 50 nM, which corresponds to approximately one p300 polypeptide per nucleosome prior to the addition of the challenge template or one p300 polypeptide per two nucleosomes after the addition of the challenge template. (B) p300 forms a template-committed complex with chromatin. Template competition experiments were carried out as shown in panel A (bottom). Briefly, the two templates were separately assembled into chromatin in the presence of NF-κB p65 and Sp1. After completion of chromatin assembly, purified p300 was added to one template (p300 Before) but not the other, and the samples were incubated for 30 min to allow the interaction of p300 with the template. Next, the two templates were mixed and incubated for an additional 30 min to allow the potential exchange of p300 from one template to the other. In some instances, p300 was added at the time of template mixing as a control or reference (p300 After). The amount of transcription from each template was analyzed in single-round transcription assays with a HeLa cell nuclear extract. The RNA products were analyzed by primer extension analysis with the radiolabeled IRF primer shown in panel A (top) (IRF Primer*). (C) p300 remains functionally active throughout the template competition experiments. Template competition experiments were carried out as described for panel B, except that one chromatin assembly reaction mixture lacked template DNA (mock assembly). The transcription products from single-round transcription assays were analyzed by primer extension analysis.
p300-chromatin interaction assays.
One 15-cm diameter dish of Sf9 cells was infected with recombinant baculovirus for the expression of wild-type or bromodomain-deletion-containing His6-tagged human p300 protein (38). After 3 days of infection, the p300 proteins were purified as described previously (36) but were left bound to the nickel-nitrilotriacetic acid (NTA) resin. The p300-bound resin, or resin similarly treated with uninfected Sf9 cell extract, was mixed with 200 μl of chromatin prepared using S190 (consisting of 1 μg of pUC118 plasmid DNA and 1.4 μg of core histones) and 300 μl of buffer R (10 mM HEPES [pH 7.6], 10 mM KCl, 1.5 mM MgCl2, 10% glycerol, 2 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT]) containing 0.5% NP-40 and 10 mM imidazole. The resin was then incubated with the chromatin for 2 h at 4°C with gentle mixing. After the incubation, the resin was washed twice with 1 ml of buffer R containing 0.5% NP-40 and 10 mM imidazole. The material bound to the p300 was eluted in successive 100-μl volumes of buffer R containing 0.5% NP-40, 10 mM imidazole, and NaCl (0.2, 0.4, 0.6, or 0.8 M). The final elution, which contained 0.25 M imidazole and no NaCl, was used to elute the p300 itself from the resin. The various fractions were deproteinized with proteinase K, extracted with phenol-chloroform, ethanol precipitated, and subjected to 1% agarose gel electrophoresis in 1× Tris-borate-EDTA with ethidium bromide staining for DNA analysis.
Glycerol gradient analyses.
Chromatin was assembled using the S190 extract as described above or by salt dialysis essentially as described previously (29). Prior to use in the glycerol gradient analyses, the salt-dialyzed chromatin was run on a linear 15-to-40% glycerol gradient to purify the slower-migrating chromatin, which has a lower density of nucleosomes (i.e., not closely packed) relative to the faster-migrating chromatin (26). For the experiments whose results are shown in Fig. 4B, a volume of S190-assembled chromatin containing 2.0 μg of plasmid DNA (pGIE0) and 2.8 μg of purified Drosophila core histones was incubated with 2.0 μg of purified p300 for 1 h at 25°C. A corresponding control sample that lacked the chromatin but contained the p300 was set up in buffer R. For the experiments whose results are shown in Fig. 5, a volume of purified salt-dialyzed chromatin containing 1.6 μg of plasmid DNA (pGIE0) and 1.0 μg of purified Drosophila core histones (or an equivalent amount of the DNA alone) was incubated in various combinations with 0.3 μg of purified p300 in 200 μl of buffer G (20 mM HEPES [pH 7.6], 75 mM KCl, 0.1 mM EDTA, 10% glycerol, 0.01% NP-40, 2 mM PMSF, 1 mM DTT) for 1 h at 25°C. In both cases, after the incubation, the reaction products were applied to 4-ml linear 15-to-40% glycerol gradients prepared in buffer G. The gradients were centrifuged at 60,000 rpm in a Beckman SW60 rotor for 4 h at 4°C. When the run was complete, 350-μl (for Fig. 4B) or 400-μL (for Fig. 5) fractions were removed from the top down for each gradient. Fifty microliters of each fraction was extracted with phenol-chloroform, ethanol precipitated, and subjected to 1% agarose gel electrophoresis in 1× Tris-borate-EDTA with ethidium bromide staining for DNA analysis. The remaining portion of each fraction was precipitated with 25% trichloroacetic acid, run on 6 or 18% acrylamide–sodium dodecyl sulfate (SDS) gels for p300 and core histones, respectively, transferred to nitrocellulose, and analyzed by Western blotting using an alkaline phosphatase detection system (for Fig. 4B) or a 125I-labeled-protein A detection system (for Fig. 5). The 125I-protein A blots were analyzed with a phosphorimager (Fuji).
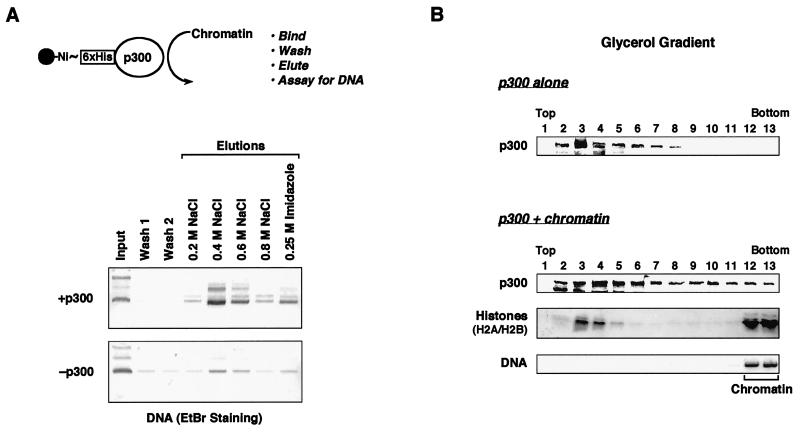
FIG. 4.
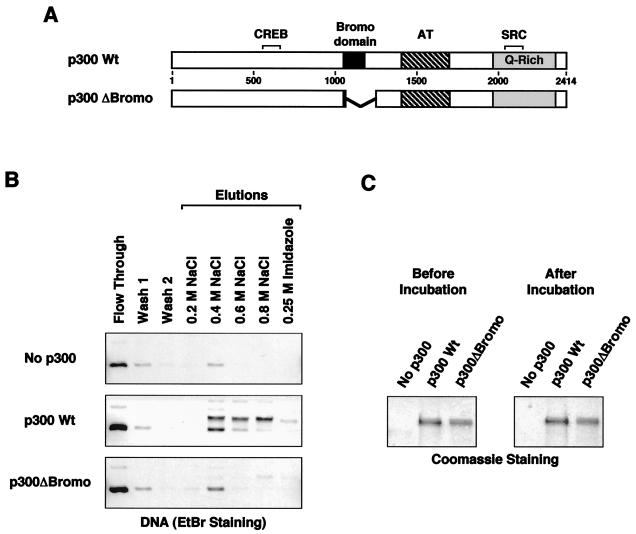
Full-length p300 binds to chromatin. (A) Immobilized p300 binds to S190-assembled chromatin. (Top) Schematic representation of the p300-chromatin interaction assay. (Bottom) Results of a representative assay performed as follows. Full-length, wild-type, His6-tagged p300 was immobilized on nickel-NTA resin and incubated with chromatin assembled using S190 as described in Materials and Methods. After the incubation, the resin was washed and the bound material was eluted using buffers containing increasing amounts of NaCl. The final elution contained 0.25 M imidazole to elute the p300 itself from the resin. The various fractions were deproteinized and subjected to agarose gel electrophoresis with ethidium bromide (EtBr) staining for DNA analysis. Twenty percent of the input from each experiment was loaded. (B) p300 cofractionates with S190-assembled chromatin on glycerol gradients. Purified p300 was incubated with or without S190-assembled chromatin for 1 h at 25°C. After the incubation, each reaction was run on a linear 15-to-40% glycerol gradient. Fractions from the gradients were analyzed for p300 and core histones by Western blotting and for DNA by agarose gel electrophoresis with ethidium bromide staining. Note that the antihistone antiserum used in this experiment preferentially detects H2A and H2B.
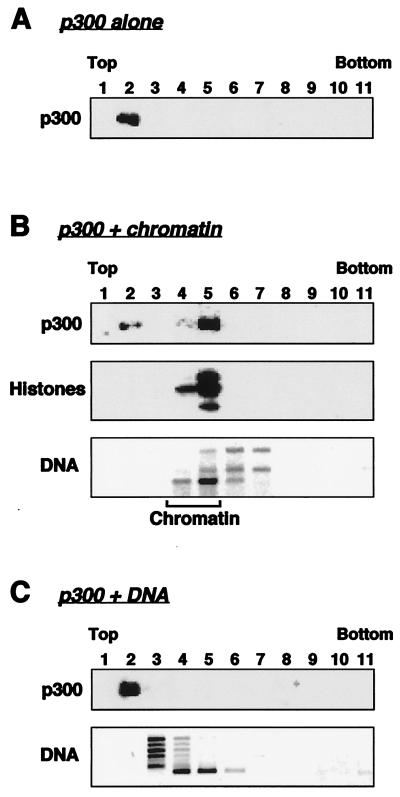
FIG. 5.
p300 cofractionates with salt-dialyzed chromatin on glycerol gradients. Salt-dialyzed chromatin or an equivalent amount of DNA alone was incubated with purified p300 for 1 h at 25°C. After the incubation, each reaction was run on a linear 15-to-40% glycerol gradient. Fractions from the gradients were analyzed for p300 and core histones by Western blotting and for DNA by agarose gel electrophoresis with ethidium bromide staining.
Histone acetylation reactions and assays.
Large-scale reactions to generate acetylated histones for the binding assays and for assembly into chromatin by salt dialysis were set up using core histones purified from 0- to 12-h Drosophila embryos. Note that these histones are hypoacetylated (essentially unacetylated), as determined by Triton-acid-urea gels (M. Levenstein and J. Kadonaga, unpublished data). About 110 μg of core histones was incubated with 1 μg of purified recombinant p300 or PCAF in 1× HAT buffer (50 mM Tris · HCl [pH 8], 0.1 mM EDTA, 150 mM NaCl, 0.2 mM PMSF, 0.5 mM DTT) in the presence of 1 mM acetyl coenzyme A (acetyl-CoA) for 30 min at 30°C. The final concentrations of histones, p300, and PCAF in the reaction mixtures were about 50 μM, 25 nM, and 100 nM, respectively.
To test the extent of histone acetylation, parallel reactions were performed under similar conditions, replacing 10% of the cold acetyl-CoA with [3H]acetyl-CoA (New England Nuclear). The test samples were divided into three portions. The first portion was used to determine the total counts per minute in the reaction mixture by liquid scintillation counting. The second was used to analyze the incorporation of [3H]acetyl-CoA into the histones by trichloroacetic acid precipitation, coupled to a filter binding assay and liquid scintillation counting. The third was analyzed on 18% acrylamide–SDS gels stained using Coomassie brilliant blue R-250, with subsequent fluorography. After fluorography, the individual histone bands were excised and subjected to liquid scintillation counting. Using the information from these assays, as well as the known total concentration of acetyl-CoA in the reactions, the number of acetyl groups added per histone was calculated.
p300 bromodomain-histone interaction assays.
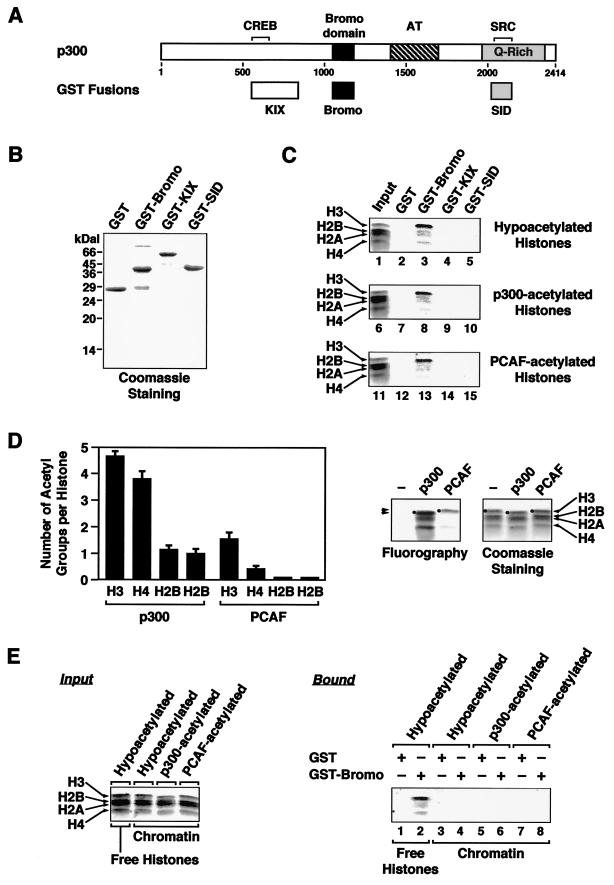
Fusions of glutathione-S-transferase (GST) with fragments of p300 were expressed in Escherichia coli and purified using standard glutathione-agarose chromatography. The fragments of p300 contained the CREB interaction domain (KIX; residues 568 to 828) (10), the bromodomain (residues 1047 to 1157) (28), and the SRC interaction domain (SID; residues 2021 to 2156) (32). The purified proteins were left bound to the resin, adjusted to equal resin and protein amounts per unit of volume, and stored at 4°C for subsequent in vitro binding assays. The binding assays were performed using the hypo- or hyperacetylated Drosophila core histones described above or the same histones assembled into chromatin by salt dialysis (29).
For the binding assays, equal volumes of resin (15 μl) containing about 1.5 μg of GST fusion protein were incubated with 2.8 μg of hypo- or hyperacetylated Drosophila core histones (or with salt-dialyzed chromatin containing the same amount of histones) in 400 μl of binding buffer (10 mM HEPES [pH 7.6], 200 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 10% glycerol, 0.05% NP-40, 2 mM PMSF, 1 mM DTT) for 2 h at 4°C with gentle mixing. After the incubation, the resin was washed three times in ice-cold wash buffer (10 mM HEPES [pH 7.6], 400 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 10% glycerol, 0.1% NP-40, 2 mM PMSF, 1 mM DTT). After the last wash, the wash buffer was aspirated completely from the resin and the resin was resuspended in 20 μl of 1× SDS gel loading solution. The samples were boiled, and 10-μl aliquots were run on an 18% polyacrylamide–SDS gel stained with Coomassie brilliant blue R-250.
RESULTS
p300 forms a stable, template-committed complex with chromatin.
In previous studies, we observed that p300 acts effectively as a transcriptional coactivator in vitro with chromatin templates but not with nonchromatin templates (37, 38). To investigate further the basis for this effect, we examined whether p300 interacts or associates with chromatin in a functionally important manner. To this end, we carried out template competition experiments (Fig. 1A). In these studies, we used two nearly identical DNA templates which are responsive to the transcriptional activator proteins NF-κB and Sp1, as well as the coactivator p300. They are IRFwt, which is the wild-type version of the IRF-1 promoter (from −1312 to +38 relative to the transcription start site), and IRF+30, which is identical to IRFwt except that it has a 30-bp insert downstream of the core promoter (Fig. 1A, top). The transcript derived from IRF+30 is 30 nt longer than the transcript produced from IRFwt, and thus, the two transcripts can be clearly distinguished by primer extension analysis.
In the template competition experiments (Fig. 1A, bottom), IRFwt and IRF+30 were separately assembled into chromatin using a Drosophila embryo-derived chromatin assembly extract (S190) in the presence of the NF-κB p65 subunit and Sp1. After the completion of chromatin assembly, purified recombinant human p300 was added to one template (template 1) but not to the other (template 2), and the samples were incubated for 30 min to allow the interaction of p300 with template 1. Next, the two templates were mixed and incubated for an additional 30 min to allow the possible exchange of p300 from one template to the other. In some cases, as a control, p300 was added after the mixing of the two templates. The amount of transcription from each template was analyzed in single-round transcription assays with a HeLa cell nuclear extract.
The results of template competition experiments with IRFwt and IRF+30 are shown in Fig. 1B. When IRF+30 was used as template 1 and IRFwt was template 2, commitment of p300 to IRF+30 was seen (i.e., p300 remained associated with IRF+30 and activated its transcription fourfold more strongly than IRFwt [Fig. 1B, lane 2]). Alternatively, when IRFwt was template 1 and IRF+30 was template 2, commitment of p300 to IRFwt was observed (Fig. 1B, lane 3). Control experiments confirmed that transcription from either IRFwt or IRF+30 was dependent on exogenously added, purified p300 (Fig. 1B, compare lane 1 with lanes 4 and 5). Since it was possible that the apparent template commitment was due to the loss of p300 activity during the first 30 min incubation prior to the addition of template 2, we carried out an additional control experiment in which a mock chromatin assembly reaction mixture (i.e., complete chromatin assembly reaction mixture minus DNA) was used instead of template 1. This experiment revealed that more than 75% of p300 coactivator activity remained after the first 30 min of incubation (Fig. 1C, compare lanes 2 and 3). Together, these results suggest that p300 can form a template-committed complex with chromatin during transcriptional activation.
Time course and activator independence of p300 template commitment.
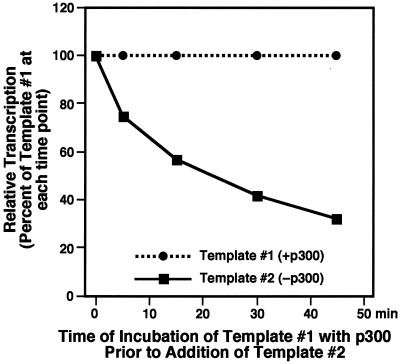
To investigate further the template commitment by p300, we examined the time course for the stable association of p300 with the chromatin templates. As shown in Fig. 2, the extent of commitment of p300 to template 1 (i.e., the template to which p300 was first added) increased as the amount of time before the addition of template 2 increased. For example, when the preincubation time was 5 min, template 2 was activated to about 75% of the level of template 1. When the preincubation time was increased to 45 min, template 2 was activated to about 30% of the level of template 1, indicating a greater commitment of p300 to template 1 with longer preincubation time. These results illustrate the time-dependent nature of the association and commitment of p300 with chromatin templates.
FIG. 2.
Time-dependent commitment of p300 with chromatin. A time course of p300 commitment with chromatin templates is shown. IRF+30 (template 1) and IRFwt (template 2) were assembled into chromatin in the presence of NF-κB p65 and Sp1. The amount of transcription from each template was analyzed in single-round transcription assays with a HeLa cell nuclear extract as described for Fig. 1. The RNA products were analyzed by primer extension analysis. Each point is the mean from two independent determinations. Similar results were obtained when IRFwt was used as template 1 and IRF+30 was used as template 2.
Activator-dependent targeting is one mechanism thought to be involved in directing coactivators to specific promoters. Thus, we tested whether the p300 template commitment required the presence of the activator proteins (i.e., NF-κB p65 and Sp1). In control reactions, no transcription was observed when the activator proteins were left out of the reaction mixtures, even in the presence of exogenously added p300 (Fig. 3, compare lanes 2 and 3 for IRFwt and lanes 4 and 5 for IRF+30). Thus, the transcriptional activity of the test promoters was dependent on both the activators and p300. Surprisingly, when template 1 lacked activator proteins, commitment of p300 to that template was still observed, as indicated by a failure of p300 to fully activate template 2 even in the absence of transcription from template 1 (Fig. 3, lanes 2 and 4). For example, even though no transcription from IRFwt was observed in the absence of activators when p300 was added, commitment of p300 to the IRFwt template was still observed, as indicated by the failure of p300 to substantially activate the IRF+30 challenge template which contained activator proteins (Fig. 3, lane 2). These results indicate that p300 can commit to a chromatin template even in the absence of transcriptional activator proteins. Furthermore, they suggest that activator-dependent recruitment and template commitment are distinct activities of p300. Collectively, the results from the template competition experiments indicate that there is a stable, activator-independent association of p300 with chromatin.
FIG. 3.
The formation of a template-committed complex by p300 does not require the presence of a transcriptional activator. p300 template competition experiments were carried out in the presence or absence of transcriptional activators (NF-κB p65 and Sp1) using the experimental scheme described for Fig. 1. The amount of transcription from each template was analyzed in single-round transcription assays with a HeLa cell nuclear extract, and the RNA products were analyzed by primer extension analysis.
p300 binds directly and stably to chromatin.
Three lines of evidence suggested to us that p300 might bind to chromatin. First, the results from our template competition experiments are consistent with a binding process. Second, p300 has HAT activity, which presumably requires the binding of p300 to histones. Last, p300 contains a bromodomain, a protein-protein interaction motif thought to be important for binding to the amino-terminal tails of histones (reviewed in reference 66). To determine if p300 can bind to chromatin, we performed the assay shown in Fig. 4A. In the assay, full-length His6-tagged p300 was immobilized on nickel-NTA resin and incubated with chromatin assembled using the S190 extract (Fig. 4A, top). The chromatin was not purified, and thus the binding reaction mixtures contained all of the S190 extract proteins. The resin was washed and elutions were performed using increasing amounts of NaCl. The binding of chromatin to the immobilized p300 was assessed by assaying for the presence of DNA in the elutions. As shown in Fig. 4A, a considerable amount of chromatin binding to the p300-containing resin was observed, in contrast to the control resin lacking p300, with the peak occurring in the 0.4 and 0.6 M NaCl fractions.
To confirm this result using a different approach, we assayed the ability of p300 to bind to and cofractionate with S190-assembled chromatin on a linear 15-to-40% glycerol gradient. After centrifugation, fractions were removed and analyzed for p300 and core histones by Western blotting with appropriate antibodies and for DNA by agarose gel electrophoresis with ethidium bromide staining. As shown in Fig. 4B (top), p300 sedimented in the top half of the gradient (fractions 2 to 8) when analyzed in the absence of chromatin. However, when preincubated with S190-assembled chromatin, a portion of the p300 (about 10%) cofractionated with the chromatin (i.e., histones and DNA) near the bottom of the gradient (fractions 12 and 13). Together, the results in Fig. 4 indicated that p300 can bind to chromatin, but due to the use of unpurified chromatin, we could not determine if the binding was direct or indirect.
To determine if p300 can bind directly to chromatin, we assayed the ability of purified p300 to cofractionate with salt-dialyzed chromatin, prepared using core histones and plasmid DNA only, on a glycerol gradient (Fig. 5). p300 was incubated with salt-dialyzed chromatin or an equivalent amount of plasmid DNA alone and was then subjected to centrifugation on a glycerol gradient with subsequent analysis for p300, histones, and DNA as described above. When loaded on the gradient alone, p300 was found as a peak in fraction 2 (Fig. 5A). In contrast, when p300 was preincubated with salt-dialyzed chromatin, the peak of p300 appeared in fraction 5, coincident with the chromatin (i.e., histones and DNA) (Fig. 5B), indicating a direct interaction between p300 and the chromatin. No interactions between p300 and DNA alone were observed in this assay (Fig. 5C). (Note that the differences between the sedimentation profiles in Fig. 4B and 5 are due to the use of crude versus purified chromatin, as well as differences in the amounts of p300 in the reaction mixtures; see Materials and Methods.) Together, the results from Fig. 4 and 5 indicate that p300 can bind directly and stably to chromatin.
The p300 bromodomain is required for the interaction of p300 with chromatin.
Previous reports have suggested that the bromodomain is important for the interaction of HAT proteins (e.g., Gcn5, PCAF, and TAFII250) with the amino-terminal tails of histones (12, 27, 56, 57). To determine if the p300 bromodomain is required for the binding of p300 to chromatin, we performed an assay similar to the one shown in Fig. 4A. In this case, we compared the chromatin-binding ability of wild-type p300 with that of a mutant p300 lacking the bromodomain (Fig. 6A). As shown in Fig. 6B, deletion of the bromodomain dramatically reduced the ability of p300 to bind chromatin. This was not due simply to instability of the mutant p300 protein as indicated by SDS-polyacrylamide gel electrophoresis (PAGE) with samples of the p300 resins pre- and postincubation (Fig. 6C). These results indicate that an intact bromodomain is required for the binding of p300 to chromatin.
FIG. 6.
The p300 bromodomain is required for the interaction of p300 with chromatin. (A) Schematic diagrams of wild-type (Wt) p300 and a bromodomain-deletion-containing p300 mutant (p300 ΔBromo). The locations of the CREB binding site, the bromodomain, the acetyltransferase domain (AT), the glutamine-rich region (Q-rich), and the SRC binding site of p300 are shown. (B) The p300 bromodomain is required for the binding of p300 to chromatin. The interaction of chromatin with immobilized wild-type or bromodomain-deletion-containing p300 was assayed as described for Fig. 4A. The DNA component of the bound chromatin was resolved on agarose gels with ethidium bromide (EtBr) staining. Twenty percent of the input from each experiment was loaded on the gel. (C) The wild-type and p300 ΔBromo proteins are stable during the interaction assay. Portions of the p300 proteins immobilized on the resin were analyzed before and after the incubation with chromatin by SDS-PAGE analysis with subsequent staining using Coomassie brilliant blue R-250.
The isolated p300 bromodomain can bind to free histones but not to nucleosomal histones.
To determine if the isolated p300 bromodomain can bind to histones as reported previously for other bromodomain-containing proteins (12, 27, 56, 57), we performed a series of in vitro interaction assays. GST fusions with fragments of p300 (the bromodomain, the CREB interaction domain [KIX], or the SID) (Fig. 7A) were expressed in E. coli and purified using glutathione-agarose resin (Fig. 7B). The GST resins were incubated with native Drosophila core histones (which are hypoacetylated as determined by Triton-acid-urea gel analysis [Levenstein and Kadonaga, unpublished]), and the specifically bound material was assayed by acrylamide-SDS gel analysis with subsequent staining using Coomassie blue. In this assay, the binding of all four core histones, but preferentially histone H3, to the GST-p300 bromodomain fusion was observed (Fig. 7C, top, lane 3). In contrast, no binding of the histones to GST alone or to the GST fusions containing the p300 KIX or SID fragments was observed (Fig. 7C, top, lanes 2, 4, and 5). These results indicate that the isolated p300 bromodomain can bind to core histones and that it preferentially binds to histone H3. However, our results cannot distinguish between the weak binding of histones H2A, H2B, and H4 directly to the GST-p300 bromodomain fusion and their weak association indirectly via interactions with histone H3 (Fig. 7C, top, lane 3).
FIG. 7.
The p300 bromodomain preferentially binds to histone H3 as free histones, but it does not bind to nucleosomal histones. (A) Schematic diagrams of wild-type p300 and fragments of p300 used as GST fusions. The locations of the CREB binding site, the bromodomain, the acetyltransferase domain (AT), the glutamine-rich region (Q-rich), and the SRC binding site of p300 are shown. (B) Purification of GST fusion proteins. Fusions of GST with the fragments of p300 indicated in panel A were expressed in E. coli, purified using standard glutathione-agarose chromatography, and left bound to the resin. Aliquots of each resin were analyzed by SDS-PAGE with subsequent staining using Coomassie brilliant blue R-250. (C) The p300 bromodomain preferentially binds to histone H3 as free histones. Equal volumes of resin containing equal amounts of GST fusion protein were incubated with purified native Drosophila core histones (which are hypoacetylated; see Materials and Methods) or the same histones acetylated with purified recombinant p300 or PCAF. After the incubation, the resins were washed three times and the bound proteins were analyzed by SDS-PAGE with subsequent staining using Coomassie brilliant blue R-250. Twenty percent of the input from each experiment was loaded. (D) Control experiment showing the extent of acetylation of the Drosophila core histones used for panels C and E. Acetylation reactions similar to those used to generate the acetylated histones for panels C and E were performed in the presence of a known concentration of [3H]acetyl-CoA. The reaction products were analyzed by filter binding assays (not shown), as well as by SDS-PAGE with subsequent fluorography or staining using Coomassie brilliant blue R-250 (right panel). The filter binding assays and the fluorography were used to calculate the number of acetyl groups incorporated per histone, plotted as the means plus the range for two separate determinations (left panel). Note the observable shift in the mobility of the hyperacetylated histones (most evident with histone H3 plus p300) to a faster-migrating species on both the fluorogram and the Coomassie-stained gel (marked by dots and arrows). (E) The isolated p300 bromodomain does not bind to nucleosomal histones. Binding assays were performed as described for panel C using salt-dialyzed chromatin in place of the free histones for lanes 3 through 8. The salt-dialyzed chromatin was assembled from purified native Drosophila core histones or the same histones acetylated with purified recombinant p300 or PCAF before chromatin assembly. Note that the salt-dialyzed chromatin input contained the same amount of histones as used for panel C.
Previous studies have suggested a role for acetylation in the binding of histones to bromodomains (12, 27, 57). Thus, we analyzed whether preacetylating the histones with p300 or PCAF would affect their binding to the p300 bromodomain. Large-scale histone acetylation reactions were set up using native Drosophila core histones to generate material for the binding assays. Smaller-scale reactions were performed in parallel in the presence of [3H]acetyl-CoA under similar conditions to monitor the extent of acetylation by filter binding assays (data not shown) and fluorography (Fig. 7D, right). p300 added about four acetyl groups per molecule of histone H3 and H4 and about one acetyl group per molecule of histone H2A and H2B, while PCAF added about one acetyl group per molecule of histone H3 and less than one acetyl group per molecule of H2A, H2B, and H4 (Fig. 7D, left). These results are in good agreement with the results of a previous study showing that, with free histones, recombinant p300 preferentially acetylates H3 and H4 at multiple sites, while recombinant PCAF preferentially acetylates H3 at a single site (58). In the in vitro binding assays, the acetylated histones (preferentially H3) bound to the isolated p300 bromodomain (Fig. 7C, lanes 8 and 13) but did not do so more strongly than the hypoacetylated histones (Fig. 7C, compare lanes 8 and 13 with lane 3). Thus, two different histone acetylation patterns, namely, those generated by p300 and PCAF, do not affect the binding of histones to the p300 bromodomain.
Finally, we examined the ability of the isolated p300 bromodomain to bind directly to chromatin. Preparations of histones similar to those used for the binding experiments in Fig. 7C were assembled into chromatin by salt dialysis. The binding of the salt-dialyzed chromatin to the GST-p300 bromodomain fusion was assayed under the same conditions used for the free histones in Fig. 7C (i.e., the same buffer, salt, and detergent conditions, as well as the same total amount of histones in the reaction mixtures). Under these conditions, we observed no binding of chromatin to the isolated p300 bromodomain (Fig. 7E, compare lanes 2 and 4), even when the histones used to assemble the chromatin were preacetylated by p300 or PCAF (lanes 6 and 8). Similar results were obtained using S190-assembled chromatin or mononucleosomes (data not shown). Together, the results in Fig. 6B and 7E suggest that the p300 bromodomain is necessary, but not sufficient, for the binding of p300 to chromatin.
DISCUSSION
In this study, we examined the functional association of the coactivator p300 with chromatin templates. We observed that p300 forms a stable, template-committed complex with chromatin during transcriptional activation. These results led us to examine the binding of p300 to chromatin. In doing so, we found that p300 binds directly to chromatin and that the binding requires the bromodomain. In addition, we observed that the isolated p300 bromodomain binds directly to histones, preferentially to histone H3, but does not bind to chromatin. Thus, the bromodomain is necessary, but not sufficient, for the binding of p300 to chromatin. Considering these results together with our previous results showing that the bromodomain is functionally important for p300 HAT and coactivator activities (38), we conclude that the stable association of p300 with chromatin is mediated, at least in part, by the bromodomain and is critical for p300 function.
Functional significance of p300-chromatin interactions and template commitment.
Current models of coactivator function suggest that sequence-specific DNA-binding transcriptional activator proteins, such as NF-κB, target coactivators, such as p300/CBP, to promoters via direct protein-protein interactions. Indeed, the p65 subunit of NF-κB has been found to bind directly to p300/CBP, and this interaction has been shown to be critical for the enhancement of NF-κB activity by p300 (47, 71). In Fig. 3, we have shown that the formation of a stable, template-committed complex of p300 with chromatin templates does not require the presence of a transcriptional activator protein. Transcriptional activation by p300, however, does require an activator. Thus, activator-mediated targeting and template commitment are distinct activities that contribute to transcriptional activation.
What might be the role of p300-chromatin interactions and template commitment during transcriptional activation? Interactions between p300 and chromatin prior to activator-mediated targeting might represent the initial association of p300 with chromatin. Such interactions would allow DNA-bound activators to recruit p300 from chromatin, rather than from solution, and target it to specific promoters. Alternatively, the initial interactions could allow p300 to preacetylate the chromatin template, allowing more efficient binding of the transcriptional activator to the chromatin. These distinct possibilities will need to be explored in more detail.
Interactions between p300 and chromatin after activator-mediated targeting could stabilize the association of p300 at transcriptionally active promoters. For example, in a previous study using the estrogen receptor as a transcriptional activator, we showed that although p300 does not function to enhance transcription reinitiation, it does act cooperatively with activators to enhance initiation in each round of transcription through multiple successive rounds (37). The formation of a stable, template-committed complex of p300 with chromatin at the promoter through multiple rounds of transcription could facilitate this activity. Indeed, we have observed p300 template commitment through three rounds of NF-κB–Sp1-mediated transcription in multiple-round transcription experiments (i.e., without Sarkosyl [data not shown]). Additionally, interactions between p300 and chromatin after activator-mediated targeting could allow the continued presence of p300 at promoters in the absence of ongoing DNA binding by a transcriptional activator. Such a result has been observed at the Saccharomyces cerevisiae HO promoter using chromatin immunoprecipitation assays (11). Components of the SWI/SNF chromatin remodeling complex and the Spt-Ada-GcnS acetyltransferase (SAGA) coactivator complex, both of which have bromodomain-containing subunits (28), were found to associate with the HO promoter even after a transient association of the Swi5 transcriptional activator protein subsided (11). Taken together, our results suggest that distinct activator-dependent targeting and activator-independent chromatin binding activities can contribute to the function of p300 as a coactivator at promoters in chromatin.
Role of the bromodomain in the function of p300 and other chromatin-associated factors.
We have shown previously that the bromodomain is critical for at least two aspects of p300 activity, namely, the efficient acetylation of nucleosomal histones and the activation of transcription (38). With regard to p300 HAT activity, it is likely that the bromodomain participates in the recognition and binding of the nucleosomal substrate. However, although the bromodomain is required for the binding of p300 to chromatin, it is not sufficient (Fig. 6B and 7E). Thus, other domains of p300 must also contribute to this activity. The HAT domain is a likely candidate. This is supported by previous studies demonstrating that fragments of p300/CBP containing the HAT domain retain at least some ability to acetylate nucleosomal histones (1, 55), an activity that requires binding of the substrate.
With regard to p300 transcriptional activity, the bromodomain is likely to have two roles, both involving the binding of nucleosomal histones. First, as described above, the bromodomain is required for efficient p300 HAT activity with nucleosomal substrates. Since the p300 HAT activity is required for full transcriptional activity (38), the bromodomain contributes to p300 transcriptional activity, at least in part, by supporting p300 HAT activity. In fact, we have found that the transcriptional phenotype of the p300 bromodomain mutant shown in Fig. 6 is similar to that of a p300 HAT mutant (38). Second, the bromodomain is important for the stable interaction of p300 with chromatin, thus contributing to its ability to form a template-committed complex with chromatin. As described above, this could contribute to p300's ability to associate with chromatin prior to activator-mediated targeting and to enhance transcription initiation by stable association with the promoter through multiple rounds of transcription.
Bromodomains have also been identified in numerous other chromatin-associated factors, including S. cerevisiae Gcn5 (yGcn5), a transcription-related HAT protein. Interestingly, the bromodomain of yGcn5 plays roles in the activity of the yeast SAGA complex, a Gcn5-containing coactivator complex (21), similar to those of the bromodomain in p300. For example, a recent study showed that purified SAGA complex containing wild-type yGcn5, but not a bromodomain deletion mutant of yGcn5, was able to acetylate nucleosomal histones in vitro (62). Interestingly, the HAT activity of another S. cerevisiae Gcn5-containing complex, Ada (21), was found to be unaffected by deletion of the yGcn5 bromodomain (62). Further analysis revealed that a specific transcriptional defect for the HIS3 gene was similar in yeast strains harboring mutations in the yGcn5 bromodomain or HAT domain, although the transcriptional defect was not as severe as we have observed for the p300 bromodomain mutant in our studies (62). Other studies with S. cerevisiae have shown that deletion of the yGcn5 bromodomain results in minor growth defects and reduced activation by weak transcriptional activators (8, 18, 46). Thus, with respect to histone acetylation and transcriptional activation, the bromodomains of human p300 and yGcn5 appear to have similar roles, suggesting an evolutionarily conserved function for this motif. The role of the bromodomain in the activities of other chromatin-associated proteins seems to vary depending on the protein. In many cases, bromodomains are required for full activity of the proteins that contain them (2, 7, 14, 48, 65) and in other cases they are not (15, 17, 41, 62). Together, the available data suggest that bromodomains play essential roles in the activities of many, but not all, of the proteins that contain them.
The bromodomain as a histone-interacting domain.
The biochemical function of the bromodomain had remained elusive since it was first identified as a conserved sequence motif found in human, Drosophila, and yeast proteins (24). However, recent studies have begun to elucidate the biochemical function of the bromodomain as a histone-interacting domain. Results from a number of recent studies suggest that the bromodomains from various factors (e.g., Gcn5, PCAF, and TAFII250) can bind directly to histones (12, 27, 56, 57). Those studies were performed using short polypeptides representing sequences from the amino-terminal tails of the histones. Here we show that the isolated p300 bromodomain can bind directly to purified, native, full-length histones (Fig. 7C). Furthermore, we have demonstrated a critical role for the bromodomain in the binding of p300 to chromatin (Fig. 6B), although the isolated p300 bromodomain is unable to bind to nucleosomes under our assay conditions (Fig. 7E). Thus, bromodomains likely serve as histone-interacting domains, contributing to chromatin binding, in most of the proteins in which they are found.
In our studies (Fig. 7C), we observed no effect of histone acetylation on the binding of the p300 bromodomain to free or nucleosomal histones. These results are in contrast to the results of three recent reports suggesting that histone acetylation plays an important role in the binding of the amino-terminal tail of histone H4 to the bromodomains of human PCAF (12), human TAFII250 (27), and yGcn5 (57). These contrasting results may reflect differences in the patterns of acetylation, intrinsic differences among the four different bromodomains used in the studies, the use of full-length histones versus short polypeptides from the histone tails, or differences in the methodologies used. With respect to the first possibility, we tested histones acetylated with only two different HATs, namely p300 and PCAF. It is possible that a particular pattern of acetylation is required to enhance the binding of histones to a bromodomain and that the patterns of acetylation generated by p300 or PCAF are not recognized by the p300 bromodomain. Alternatively, our results might indicate that the p300 bromodomain binds to a region other than the amino-terminal tails of the histones. Additional studies will be required to explore these possibilities.
Multiple p300 interactions at transcriptionally active promoters.
Our results indicate that the binding of p300/CBP to chromatin via its bromodomain is critically important for directing the stable association of p300 with transcriptionally active promoters in chromatin, as well as for p300 HAT and coactivator functions. However, other interactions involving p300 also seem to play a role. They include stable binding to transcriptional activators, contacts with components of the basal transcriptional machinery, and indirect association with chromatin via histone-binding proteins. For example, p300/CBP makes multiple contacts within the beta interferon enhanceosome (33, 47, 71). In addition, p300/CBP has been shown to interact with components of the basal transcriptional machinery, including RNA polymerase-containing complexes (51, 52), TFIIB (40, 53), and TBP (64). Furthermore, recent studies have indicated that p300/CBP can interact with histone binding proteins, such as RbAp48 and nucleosome assembly protein 1 (59, 70). It is likely that multiple distinct interactions such as those listed above serve to stabilize the association of p300/CBP with transcriptionally active promoters in vivo after activator-dependent targeting and activator-independent chromatin binding have occurred. Such interactions could also contribute to the stable commitment of p300 to a chromatin template and enhanced transcription initiation through multiple rounds of transcription.
ACKNOWLEDGMENTS
This work was supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and a grant from the National Institutes of Health (DK58110) to W.L.K, a grant from the National Institutes of Health (GM46995) to J.T.K, and a grant from the Japanese Society for the Promotion of Science (JSPS) Research for the Future Program to T.I.
We thank Kathy Lee, Edwin Cheung, and Erik Andrulis for critical reading of the manuscript. We are grateful to John Hiscott for the p65 baculovirus; Y. Cha Henderson, A. Deisseroth, and T. Burke for the hIRF-1 constructs; and Pat Nakatani for the PCAF baculovirus.
REFERENCES
- 1.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 2.Barlev N A, Poltoratsky V, Owen-Hughes T, Ying C, Liu L, Workman J L, Berger S L. Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol Cell Biol. 1998;18:1349–1358. doi: 10.1128/mcb.18.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berk A J. Activation of RNA polymerase II transcription. Curr Opin Cell Biol. 1999;11:330–335. doi: 10.1016/S0955-0674(99)80045-3. [DOI] [PubMed] [Google Scholar]
- 4.Blobel G A. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- 5.Bulger M, Kadonaga J T. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol Genet. 1994;5:241–262. [Google Scholar]
- 6.Burke T W, Kadonaga J T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns B R, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg R D, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 8.Candau R, Zhou J X, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 10.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 11.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 12.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M-M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 13.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du J, Nasir I, Benton B K, Kladde M P, Laurent B C. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elfring L K, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek S J, Waldrip W R, Daubresse G, DePace A, Kennison J A, Tamkun J W. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics. 1998;148:251–265. doi: 10.1093/genetics/148.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freedman L P. Multimeric coactivator complexes for steroid/nuclear receptors. Trends Endocrinol Metab. 1999;10:403–407. doi: 10.1016/s1043-2760(99)00208-8. [DOI] [PubMed] [Google Scholar]
- 17.Gansheroff L J, Dollard C, Tan P, Winston F. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgakopoulos T, Gounalaki N, Thireos G. Genetic evidence for the interaction of the yeast transcriptional co-activator proteins GCN5 and ADA2. Mol Gen Genet. 1995;246:723–728. doi: 10.1007/BF00290718. [DOI] [PubMed] [Google Scholar]
- 19.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 20.Goodman R H, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 21.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 22.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 23.Hampsey M, Reinberg D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 24.Haynes S R, Dollard C, Winston F, Beck S, Trowsdale J, Dawid I B. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrera J E, Sakaguchi K, Bergel M, Trieschmann L, Nakatani Y, Bustin M. Specific acetylation of chromosomal protein HMG-17 by PCAF alters its interaction with nucleosomes. Mol Cell Biol. 1999;19:3466–3473. doi: 10.1128/mcb.19.5.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 27.Jacobson R H, Ladurner A G, King D S, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 28.Jeanmougin F, Wurtz J M, Le Douarin B P, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 29.Jeong S W, Lauderdale J D, Stein A. Chromatin assembly on plasmid DNA in vitro. Apparent spreading of nucleosome alignment from one region of pBR327 by histone H5. J Mol Biol. 1991;222:1131–1147. doi: 10.1016/0022-2836(91)90597-y. [DOI] [PubMed] [Google Scholar]
- 30.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 31.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 32.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 33.Kim T K, Kim T H, Maniatis T. Efficient recruitment of TFIIB and CBP-RNA polymerase II holoenzyme by an interferon-beta enhanceosome in vitro. Proc Natl Acad Sci USA. 1998;95:12191–12196. doi: 10.1073/pnas.95.21.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornberg R D, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 35.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 36.Kraus W L, Kadonaga J T. Ligand- and cofactor-regulated transcription with chromatin templates. In: Picard D, editor. Steroid/nuclear receptor superfamily: a practical approach. Oxford, England: Oxford University Press; 1999. pp. 167–189. [Google Scholar]
- 37.Kraus W L, Kadonaga J T. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraus W L, Manning E T, Kadonaga J T. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurokawa R, Kalafus D, Ogliastro M H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 40.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 41.Laurent B C, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 42.Lemon B D, Freedman L P. Nuclear receptor cofactors as chromatin remodelers. Curr Opin Genet Dev. 1999;9:499–504. doi: 10.1016/s0959-437x(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 43.Leo C, Chen J D. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 44.Lin R, Gewert D, Hiscott J. Differential transcriptional activation in vitro by NF-kappa B/Rel proteins. J Biol Chem. 1995;270:3123–3131. doi: 10.1074/jbc.270.7.3123. [DOI] [PubMed] [Google Scholar]
- 45.Malik S, Roeder R G. Transcriptional regulation through mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 46.Marcus G A, Silverman N, Berger S L, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 48.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 50.Myer V E, Young R A. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 51.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 53.O'Connor M J, Zimmermann H, Nielsen S, Bernard H U, Kouzarides T. Characterization of an E1A-CBP interaction defines a novel transcriptional adapter motif (TRAM) in CBP/p300. J Virol. 1999;73:3574–3581. doi: 10.1128/jvi.73.5.3574-3581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogryzko V V, Kotani T, Zhang X, Schlitz R L, Howard T, Yang X J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 55.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 56.Ornaghi P, Ballario P, Lena A M, Gonzalez A, Filetici P. The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J Mol Biol. 1999;287:1–7. doi: 10.1006/jmbi.1999.2577. [DOI] [PubMed] [Google Scholar]
- 57.Owen D J, Ornaghi P, Yang J-C, Lowe N, Evans P R, Ballario P, Neuhaus D, Filetici P, Travers A A. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schiltz R L, Mizzen C A, Vassilev A, Cook R G, Allis C D, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 59.Shikama N, Chan H M, Krstic-Demonacos M, Smith L, Lee C-W, Cairns W, La Thangue N B. Functional interaction between nucleosome assembly proteins and p300/CREB-binding family coactivators. Mol Cell Biol. 2000;20:8933–8943. doi: 10.1128/mcb.20.23.8933-8943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sims S H, Cha Y, Romine M F, Gao P Q, Gottlieb K, Deisseroth A B. A novel interferon-inducible domain: structural and functional analysis of the human interferon regulatory factor 1 gene promoter. Mol Cell Biol. 1993;13:690–702. doi: 10.1128/mcb.13.1.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 62.Sterner D E, Grant P A, Roberts S M, Duggan L J, Belotserkovskaya R, Pacella L A, Winston F, Workman J L, Berger S L. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Struhl K, Moqtaderi Z. The TAFs in the HAT. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- 64.Swope D L, Mueller C L, Chrivia J C. CREB-binding protein activates transcription through multiple domains. J Biol Chem. 1996;271:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]
- 65.Syntichaki P, Topalidou I, Thireos G. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature. 2000;404:414–417. doi: 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- 66.Winston F, Allis C D. The bromodomain: a chromatin-targeting module? Nat Struct Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 67.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 68.Xu L, Glass C K, Rosenfeld M G. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 69.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Q, Vo N, Goodman R H. Histone binding protein RbAp48 interacts with a complex of CREB binding protein and phosphorylated CREB. Mol Cell Biol. 2000;20:4970–4978. doi: 10.1128/mcb.20.14.4970-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhong H, Voll R E, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]