Abstract
Musashi1 (Msi1) is an RNA-binding protein that is highly expressed in neural progenitor cells, including neural stem cells. In this study, the RNA-binding sequences for Msi1 were determined by in vitro selection using a pool of degenerate 50-mer sequences. All of the selected RNA species contained repeats of (G/A)UnAGU (n = 1 to 3) sequences which were essential for Msi1 binding. These consensus elements were identified in some neural mRNAs. One of these, mammalian numb (m-numb), which encodes a membrane-associated antagonist of Notch signaling, is a likely target of Msi1. Msi1 protein binds in vitro-transcribed m-numb RNA in its 3′-untranslated region (UTR) and binds endogenous m-numb mRNA in vivo, as shown by affinity precipitation followed by reverse transcription-PCR. Furthermore, adenovirus-induced Msi1 expression resulted in the down-regulation of endogenous m-Numb protein expression. Reporter assays using a chimeric mRNA that combined luciferase and the 3′-UTR of m-numb demonstrated that Msi1 decreased the reporter activity without altering the reporter mRNA level. Thus, our results suggested that Msi1 could regulate the expression of its target gene at the translational level. Furthermore, we found that Notch signaling activity was increased by Msi1 expression in connection with the posttranscriptional down-regulation of the m-numb gene.
Posttranscriptional regulation plays essential roles in the wide variety of events that occur during animal development, including the localization of maternal-effect gene products within oocytes, cell fate determination, and cell polarity formation by controlling alternative splicing, mRNA stability, RNA transport, and/or the translation of existing mRNAs (11, 57, 66). Among developing tissues, the nervous system in particular uses a variety of posttranscriptional means to regulate the vast cellular diversity and synaptic plasticity that is generated. Neural RNA-binding proteins are likely to play essential roles in mediating these processes (44).
Two classes of neural RNA-binding proteins with ribonucleoprotein-type RNA recognition motifs (RRMs), the Elav and Musashi subfamilies, have been proposed (44). The Elav subfamily includes the mammalian Elav homologue, Hu proteins, whose members are expressed in postmitotic neurons and have been proposed to be required for the survival or terminal differentiation of these cells (1, 2, 34, 45, 64, 69). Intensive study in this area revealed that Hu proteins regulate the gene expression involved in neuronal differentiation by controlling RNA stabilization or translation (2, 13, 35, 48, 64). On the other hand, the Musashi subfamily proteins are expressed in neural precursor cells rather than postmitotic neurons (15, 28, 44, 49, 52).
Musashi1 (Msi1) was isolated as a mammalian homologue of Drosophila Musashi (d-Msi), which is required for the asymmetric cell division of sensory neural precursor cells (38). In Drosophila, genes that are responsible for the proper asymmetric cell division of sensory organ precursor cells or central nervous system neuroblasts have been identified and extensively investigated. Interestingly, many of these genes have been shown to be involved in the regulation of Notch signaling, including numb, tramtrack, Notch, and Delta (reviewed by Jan and Jan [23]). Recently, our laboratory showed that d-Msi represses the expression of tramtrack, which encodes a transcriptional repressor, at the level of translation (21, 43). We also identified the mammalian homologue of Drosophila Musashi, Musashi1, which is highly enriched in neural progenitor cells in the developing mouse embryonic central nervous system (28, 52, 53). Msi1 expression is gradually down-regulated during the course of neural differentiation. The Msi1 protein consists of 362 amino acid (aa) residues, and it contains two conserved RRMs in its N-terminal half and a putative nuclear export signal in its C-terminal half, which is consistent with its observed localization of the cytoplasm in embryonic neural progenitor cells (28, 52).
To help elucidate the role of Msi1 protein in neural progenitor cells, we sought to determine (i) the RNA sequences that bind to Msi1, (ii) an in vivo target RNA of Msi1, and (iii) the mechanism of action of Msi1 on the expression of its downstream target genes. To this end, in the present study we identified the RNA sequence for Msi1 and demonstrated putative translational repression of a likely in vivo target gene mammalian numb (m-numb).
MATERIALS AND METHODS
Preparation of Musashi1 fusion protein.
To prepare mouse Musashi1 fusion protein (Msi1-2TR), a plasmid vector, pET21a-msi12TR, was constructed by inserting a part of the coding region (corresponding to aa 7 to 192) of the musashi-1 cDNA into the pET21a expression vector (Novagene). The plasmid was introduced into Escherichia coli strain BL21 (DE3) pLysS. The expression and affinity purification of the fusion protein were performed as described previously (28).
Selection of Musashi-1 RNA ligands.
RNA selection was basically performed according to previously described methods (6, 61). Oligonucleotides harboring a 50-bp random sequence surrounded by primer binding sites (5′-GGGAAGATCTCGACCAGAAG-N50-TATGTGCGTCTACATGGATCCTCA-3′) were synthesized using a DNA synthesizer (Nissinbo). The oligonucleotides were amplified by PCR using a forward primer containing the T7 promoter sequence and a reverse primer (forward primer: 5′-CGGAATTCTAATACGACTCACTATAGGGAAGATCTCGACCAGAAG-3′; reverse primer: 5′-TGAGGATCCATGTAGACGCACATA-3′). The library DNAs were transcribed in vitro with T7 RNA polymerase and [α-32P]UTP (Amersham Pharmacia Biotech). The RNA was applied to a column which was filled with nickel affinity resin preadsorbed with 100 μg of purified histidine-tagged Msi1 fusion protein, in binding buffer (0.5 M LiCl, 20 mM Tris-HCl [pH 7.5], 1 mM MgCl2). The beads were then washed with 10 ml of binding buffer. Bound RNA was eluted from the column in elution buffer (20 mM Tris-HCl [pH 7.5], 1 M imidazol), phenol extracted, and ethanol precipitated. The RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Gibco BRL), cDNA was used for PCR with the forward and reverse primers given above under the following conditions: 15 cycles of 1 min at 94°C, 1 min at 59°C, and 1 min at 72°C. The PCR product was used for the next round in the selection procedure. This process was repeated an additional seven times before the products were subcloned into the pUC119 vector (Clontech). We predicted RNA secondary structure using the commercial sequence analysis software DNASIS (Hitachi Software Engineering Inc.) program based on the Zuker-Stiegler method.
Gel shift assays.
Gel shift assays were performed with various amounts of Msi1 fusion protein in 16 μl of KNET buffer (32). Ten thousand counts per minute (approximately 4 fmol) of selected 32P-labeled RNA ligand (S8-13 and S8-19) was added to the solutions containing Msi1 fusion protein. For the competition experiment, unlabeled RNA was added before the 32P-labeled RNA. Protein and RNA samples were allowed to equilibrate for 30 min at room temperature. After incubation, the mixtures were immediately loaded onto 8 or 15% polyacrylamide gels (0.5× Tris-borate-EDTA buffer, 5% glycerol) and fractionated by electrophoresis. The gels were then dried and exposed to XAR autoradiography film (Kodak).
In vitro binding assay using m-numb 3′ UTR.
[35S]methionine-labeled full-length Msi1 protein was prepared by an in vitro transcription translation system using the pRSETb-msi1 plasmid vector (52), pET21a-msi12TR, pRSETb-C17(C-terminal half), and reticulocyte lysate containing T7 RNA polymerase (Promega). The Msi1 protein was incubated with m-numb RNAs labeled with biotin-14-CTP in binding buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 0.05% NP-40, 0.1% sodium azide) for 30 min. The mixture of Msi1 and m-numb RNA was then added to streptavidin-agarose beads previously resuspended in binding buffer. The beads were then washed with 1 ml of binding buffer five times. The bead pellet was resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, boiled for 5 min, and spun. The supernatant was loaded onto a 15% SDS-polyacrylamide gel and fractionated by electrophoresis. After electrophoresis, the gel was dried and exposed to Fuji RX-U film at −80°C for 1.5 to 8 h.
Cell culture and in vivo binding assay.
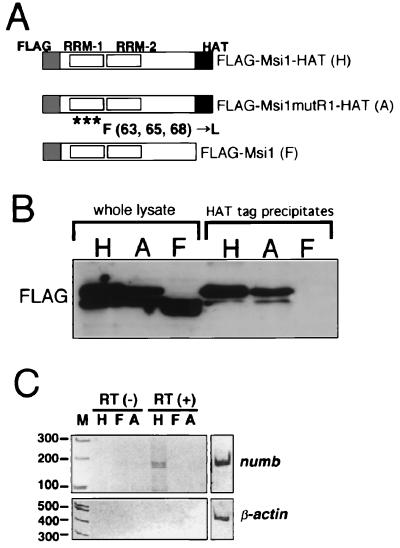
NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium (Nissui) supplemented with 10% calf serum. NIH 3T3 cells were plated onto 60-mm dishes (Falcon) (106 cells/dish). On the following day, cells were transfected with 1 μg of the Msi1 expression constructs (pcDNA3-FLAGMsi1HAT, pcDNA3-FLAGMsi1mutR1HAT, and pcDNA3-FLAGMsi1) shown in Fig. 4A, using the Effective transfection reagent (Qiagen). Two days later, the transfected cells were suspended in 1 ml of NET-Triton buffer (6), homogenized, and spun in a microcentrifuge. In the presence of RNase inhibitor (0.5 U/μl) (Promega), histone affinity tag (HAT)-tagged Msi1-RNA complexes in the supernatants were pulled down by Talon resin (Clontech). Extraction of precipitated RNA, DNase I treatment, and reverse transcription were performed as described previously (6). Subsequently, PCR was performed with m-numb gene-specific primers using 32 cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C or with β-actin-specific primers using 25 cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C. Primers used in the PCR were as follows: the m-numb primer set, 5′-ATGAGCAAGCAGTGTTGTCCTGG-3′ and 5′-CAAGTAGCTGCAACTGGCTGG-3′; and β-actin, 5′-CTTCCTCCCTGGAGAAGAGCTATGAGC-3′ and 5′-GCCTAGAAGCACTTGCGGTGCACG-3′.
FIG. 4.
In vivo binding of Msi1 to m-numb RNA. (A) Schematic diagrams of Msi1 proteins FLAG-Msi1-HAT (H), FLAG-Msi1mutR1-HAT (A), and FLAG-Msi1 (F). The HAT tag at the C-terminal end is an affinity tag for Talon resin (Clontech). FLAG-Msi1mutR1-HAT is a non-RNA-binding form of Msi1 with amino acid replacements in the N-terminal RNA-binding domain. (B) Expression of Msi1 proteins H, A, and F in NIH 3T3 cells and affinity precipitation using HAT-tag were analyzed by immunoblotting with anti-FLAG monoclonal antibody. (C) In vivo RNA-binding assay combining affinity precipitation with RT-PCR. The expression of exogenous Msi1 proteins in NIH 3T3 cells was induced by transient transfection. RNA present in the precipitates with Talon resin was detected by RT-PCR amplification with the indicated primer sets. The RT (−) lanes showing that no amplification was seen without reverse transcriptase activity were controls to ensure that the RT-PCR was RNA dependent. The right panels indicate the amplification control experiment to ensure primer fidelity using the RT product from the initial extract before incubation with affinity resin. Lanes H, FLAG-Msi1-HAT; F, FLAG-Msi1; A, FLAG-Msi1mutR1-HAT.
Reporter assay using luciferase and quantification of reporter mRNA by Northern ELISA system.
NIH 3T3 cells (3 × 105 cells/ml per assay) were transfected with 0.2 μg of firefly luciferase reporter vector, 20 ng of Renilla luciferase control vector pRL-TK (Toyo Ink), and 0.3 μg of pEGFP-N3 vector (Clontech) and with a combination of pcDNA3 vector (Invitrogen) and pCDNA3-T7Msi1 or pCDNA3-T7Msi1mutR1 expression vector (totaling 1.5 μg), using Fugene 6 transfection reagent (Roche). After 2 days of incubation, the cells were lysed with luciferase assay lysis buffer (Toyo Ink). The firefly luciferase (reporter) activities and Renilla luciferase activities (control) were measured with individual reaction substrate mixtures supplied by the manufacturer using a Berthold Lumat LB9507 luminometer. The ratio of reporter luciferase activity in relative light units was divided by the control Renilla luciferase activity to give a normalized reporter luciferase value.
NIH 3T3 cells were transfected and cultured as described above for the reporter assay. Two days after, cells were harvested, and total RNA was extracted from each NIH 3T3 cell with Trizol reagent (Gibco BRL). After DNase I treatment, RNAs (2 μg each) were used for quantification of reporter luciferase RNA and enhanced green fluorescent protein (EGFP) RNA as a control by a Northern enzyme-linked immunosorbent assay (ELISA) system (Rosh Diagnostics). Digoxigenin-labeled detection probes were prepared following PCR amplification using digoxigenin-11-2′-deoxy-uridine-triphosphate as a substrate and 10 ng of plasmid DNA (pGV-P2; Promega, and pEGFP-N3; Clontech) as templates. PCRs were performed using 25 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 30 s, with a final extension phase for 2 min, using Ex Taq DNA polymerase (Takara), luciferase gene-specific primers, and EGFP-specific primers as follows: luciferase forward primer, 5′-GAGGTCCTATGATTATGTCCGG-3′; luciferase reverse primer, 5′-GTTGGAGCAAGATGGATTCC-3′; EGFP forward primer, 5′-CAGAAGAACGGCATCAAGG-3′; and EGFP reverse primer, 5′-TGCTCAGGTAGTGGTTGTCG-3′. The expression levels of luciferase-m-numb 3′-UTR chimeric mRNA versus those of control EGFP mRNA in NIH 3T3 cells were determined from the photometric intensity (measurement of absorbance at 450 nm) with peroxidase and 3,3′,5,5′-tetramethylbenzidine.
Preparation of recombinant adenovirus and infection experiment.
We generated the recombinant adenovirus Adex-FLAGMsi1 using the pAdex1pCAw vector, essentially according to previously described methods (19). The high-titer recombinant adenovirus stock (Adex-FLAGMsi1, 3 × 1010 PFU/ml; Adex- NLLacZ, 3 × 1010 PFU/ml) was obtained and stored at −80°C.
NIH 3T3 cells (2.5 × 106 cells) were infected with 1,000-fold-diluted adenovirus solution in 5 ml of Dulbecco's modified Eagle's medium containing 5% fetal bovine serum. Two days later, the cells were lysed by lysis buffer (6) and used in Western blotting analysis essentially as described previously (28), Northern blot analysis, and sucrose gradient centrifugation analysis. Affinity-purified rabbit polyclonal antibody against chick Numb (65) that recognizes the amino acid sequence which is perfectly conserved between the mouse and chicken proteins as an epitope, anti-FLAG-M2 mouse monoclonal antibody (Sigma), and antitubulin mouse monoclonal antibody (Sigma clone no. 1A2) were used at 1:500, 1:1,000, and 1:1,000 dilutions, respectively, for immunoblots in 3% skim milk phosphate-buffered saline. Each immunoreactivity was detected by diaminobenzidine. Signals were quantified using the NIH Image program version 1.62 (National Institutes of Health).
RNA quantification by Northern blot analysis.
Total RNAs were extracted with Trizol reagent (Gibco BRL) from the NIH 3T3 cells infected with Adex-FLAGMsi1 described above and precipitated with ethanol. The RNAs were loaded onto morpholinepropanesulfonic acid-formaldehyde-agarose gels and then transferred to a Hybond N+ nylon membrane (Amersham Pharmacia Biotech) and probed with 32P-labeled m-numb cDNA and β-actin cDNA. Hybridization signals were detected with XAR autoradiography film (Kodak) and quantified using BAS5000 (Fuji). The ratio of hybridization signals for m-numb mRNA over those for β-actin mRNA yielded normalized quantities of m-numb mRNA level. We performed two independent experiments and calculated the average value.
Sucrose gradient centrifugation.
We performed sucrose gradient centrifugation as described previously (58). NIH 3T3 cells infected with Adex-FLAGMsi1 as described above were harvested by centrifugation, washed with cold phosphate-buffered saline, resuspended in buffer A (10 mM potassium acetate, 2 mM magnesium acetate, 1 mM dithiothreitol, 5 mM HEPES [pH 7.3], 2 μg of leipeptin per ml, 2 μg of pepstatin per ml, and 0.5% aprotinin), incubated on ice for 10 min, and disrupted by passage through needles. Centrifugation at 2,500 × g for 10 min yielded a pellet and a supernatant fraction designated cytoplasmic lysate. The KCl concentration was adjusted to 100 mM at this point. Cytoplasmic lysate was resolved on a linear sucrose gradient (5 to 30%) containing 100 mM KCl, 10 mM potassium acetate, 2 mM magnesium acetate, 1 mM dithiothreitol, 5 mM HEPES [pH 7.3], 2 μg of leipeptin per ml, 2 μg of pepstatin per ml, and 0.5% aprotinin. The gradients were centrifugated at 4°C in a Hitachi P40St1286 rotor at 40,000 rpm for 150 min. Following centrifugation, fractions were collected from the top of gradients (300 μl per fraction) using a Piston gradient fractionator (Biocomp, Inc.). Thirty microliters of each fraction was used for Western blotting. RNA was extracted from the fractions with phenol and precipitated with ethanol, and A254 was measured.
HES1-promoter transactivation assay.
To measure HES-1 promoter activity, NIH 3T3 cells were transfected with 0.2 μg of pHES-1p-luciferase (24) alone, together with 0.025 μg of pEF-BOS-FCDN1 (an expression plasmid for the Notch1 intracellular domain [FCDN1, aa 1747 to 2531]) (41), in combination with various amounts of pcDNA3-T7Msi1 or pEF-BOSneo-R218H (29), or in combination with 1 μg of pCDNA3-HAmNumb; 100 ng of SV40-LacZ construct or 20 ng of the Renilla luciferase control vector pRL-TK (Toyo Ink) was included in each transfection as an internal control. Three independent experiments were carried out. Bars in figures indicate the standard deviations. Luciferase activity was measured 48 h after transfection in a luminometer Lumat LB9507 (Berthold) and normalized according to β-galactosidase activity or Renilla luciferase activity.
RESULTS
In vitro selection of high-affinity RNA ligands for Msi1.
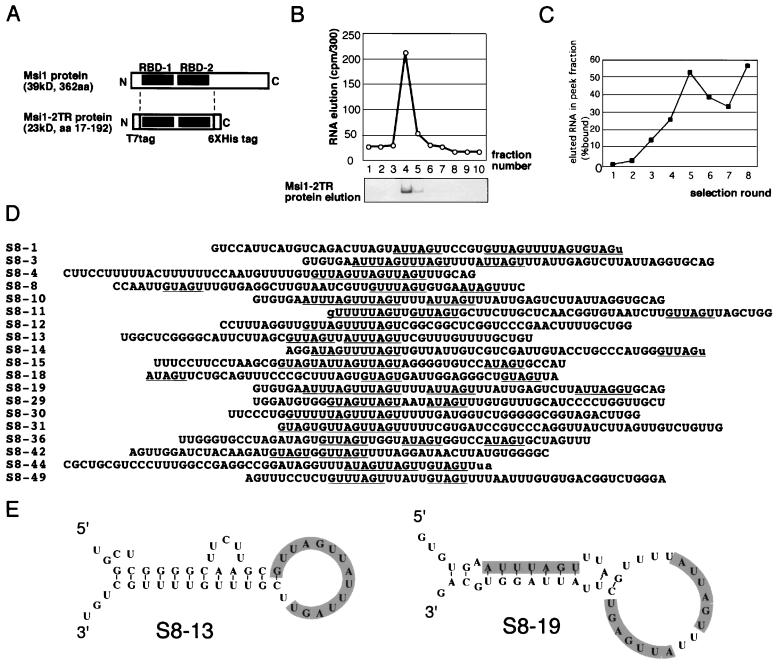
To identify the target RNA sequence of Msi1, we performed an affinity elution-based RNA selection method, SELEX (6, 32, 61). A 32P-labeled RNA pool was synthesized in vitro using a PCR-amplified oligonucleotide library of 50-nucleotide semicomplete random sequences as templates. The synthesized RNA pool was then applied to a nickel affinity column to which Msi1 fusion protein Msi1-2TR had been absorbed previously. Msi1-2TR contained the two tandem RRM-type (8) RNA-binding domains (RBDs) (aa 17 to 192) as well as the histidine tag at its C terminus and a T7 tag at its N terminus (Fig. 1A). After being washed to remove RNAs that did not interact with the Msi1-2TR fusion protein, the Msi1-2TR fusion protein-RNA complexes were eluted in buffer containing 1 M imidazole. The elution profile of the first round of selection is shown in Fig. 1B. We monitored the elution of RNA and protein by counting the radioactivity and by performing SDS-PAGE, respectively (Fig. 1B). After each cycle, the bound RNA was extracted and reverse transcribed to the first strand of cDNA using the SELEX reverse primer (see Materials and Methods for primer sequences), and the cDNAs encoding the selected RNA sequences were amplified by PCR and used again as templates to synthesize RNAs for another cycle of binding and amplification. By repeating this affinity RNA-ligand selection, we found that the fraction of RNA binding to Msi1 increased from 0.2% in the initial RNA pool to 60% after eight selection cycles (Fig. 1C). In this way, we obtained an RNA pool that was enriched in RNA sequences that preferentially bind to Msi1.
FIG. 1.
Selection of RNA ligands of Msi1 from a random RNA library. (A) Schematic representation of the domain structure of the authentic full-length Msi1 protein and the bacterially expressed fusion Msi1 protein, Msi1-2TR, which was used for the in vitro selection. The Msi1-2TR protein contains the two tandem RBDs of authentic Msi1 protein plus a C-terminal polyhistidine tag for affinity elution and an N-terminal T7 tag. (B) Msi1-2TR protein and bound RNAs were coeluted in the first selection column by specifically cluting the histidine-tagged Msi1-2TR fusion protein in 1.0 M imidazole. Eluted protein and RNA were detected by Coomassie brilliant blue staining and radioactivity counting, respectively. (C) Percentage of total radioactivity in loaded RNA represented by bound RNA in each selection is shown. The proportion of bound radioactivity increased to 60% after eight selection cycles. (D) Sequences of Msi1-selected RNAs. Twenty representative PCR clones encoding RNA ligands obtained after eight rounds of RNA selection are shown. The (G/A)UnAGU (n = 1 to 3) consensus sequence motif was often observed to be repeated in each selected clone. (E) Representative secondary structures of the RNA sequences selected by Msi1. Shading indicates the selected sequences. The predictions of RNA secondary structure were based on the Zuker-Stiegler method.
We then sequenced 50 independent cDNA clones that were obtained after the eight selection cycles and used the information to identify the RNA consensus sequence for the binding of Msi1 (Fig. 1D). Twenty representative clones are shown in Fig. 1D, all of which contained short U stretches, 1 to 6 bases long, that were interrupted by A or AG. The other 30 clones that are not shown also contained sequences that matched this consensus, and some were redundant, containing the same RNA sequence as some of the depicted clones in Fig. 1D. In particular, the (G/A)UnAGU motif was seen in most of the selected clones (underlined in Fig. 1D; in many cases, n = 1 to 3). These uridine-rich sequences were often repeated two or three times. The frequencies of the U number (n) were as follows: n = 1, 31%; n = 2, 40%; n = 3, 21%; n = 4, 5%; and n= 5, 2%. Interestingly, in most cases this sequence element was located in the loop region of a stem-loop structure (Fig. 1E), as predicted using commercial sequence analysis software based on the Zuker-Stiegler method (DNasis; Hitachi Software Engineering Inc.).
RNA-protein binding experiments.
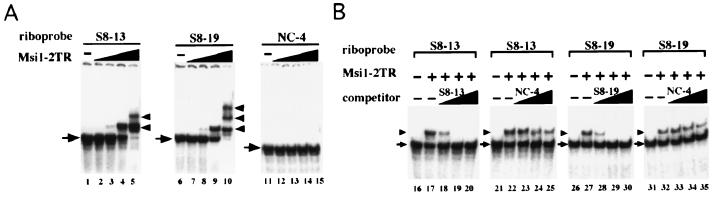
To further investigate whether the repeated (G/A)UnAGU motif is an essential sequence element for the Msi1-RNA interaction, we performed binding assays using the Msi1-2TR fusion protein and RNA sequences derived from the most frequently selected clones, S8-13 and S8-19, which contain, respectively, two and three copies of sequences that match the selected consensus motif (Fig. 2A). Gel shift analysis was performed by incubating 4 fmol of labeled RNAs with various amounts of Msi1-2TR protein. Interestingly, the number of retarded bands in each experiment corresponded to the number of the sequences that matched the consensus sequence motif, (G/A)UnAGU, within a selected clone. S8-13 RNA contains two consensus motifs, and S8-19 RNA has three motifs. Msi1 protein did not recognize the RNA-designated NC-4, which does not contain the selected consensus sequence (Fig. 2A). To examine whether the Msi1 protein bound specifically to the selected RNA, we performed competitive binding assays with unlabeled RNA containing the Msi1 selected-consensus sequence or a nonspecific competitor that did not contain the full consensus sequence (Fig. 2B). Four femtomoles of 32P-labeled RNA (S8-13 or S8-19) was incubated with 100 fmol of Msi1 protein and a 10-, 100-, or 1,000-fold excess of unlabeled cold RNA, followed by gel shift analysis (Fig. 2B, lanes 13 to 15, lanes 18 to 20, lanes 23 to 25, and lanes 28 to 30, respectively). The intensities of the retarded bands representing the protein-RNA complex were decreased by the addition of excess unlabeled RNA containing the Msi1 recognition sequence (i.e., the same sequence as the labeled RNA) as a specific competitor. However, the intensities were not decreased by the addition of RNA that did not contain the Msi1 recognition sequence (NC-4). These observations indicated that the Msi1 protein specifically recognized the RNAs containing the sequence that matched the consensus sequences selected in vitro. The binding affinities of the selected RNA sequences to Msi1 were determined based on the intensity of the retarded band representing the RNA-Msi1 complex in the gel retardation assays. The dissociation constant Kd is equal to the protein concentration at which 50% of RNA is bound. In Fig. 2A, lane 4 and lane 9, 50% of RNA was bound to protein as determined by densitometry evaluation. Kd was calculated to be ∼4 nM for S8-13 and S8-19. Thus, Msi1 was shown to bind to the RNA containing the sequences that match the consensus sequence motifs with high affinity.
FIG. 2.
The sequence motif indicated by the in vitro selection is essential for the specific RNA binding of Msi1. (A) Gel shift assays of Msi1 protein. Msi1-2TR fusion protein binds to selected RNA (S8-13, S8-19; see Fig 1D). Four femtomoles of 32P-labeled RNA was incubated with various amounts of Msi1-2TR protein: 0 fmol (lanes 1, 6, and 11), 1 fmol (lanes 2, 7, and 12), 10 fmol (lanes 3, 8, and 13), 100 fmol (lanes 4, 9, and 14), and 1,000 fmol (lanes 5, 10, and 15), and the complexes were run on nondenaturing polyacrylamide gels. The band representing free RNA is shown with an arrow, and the retarded band is indicated with an arrowhead (also in panel B). Notably, no interaction was detected between the Msi1 fusion protein and NC4 RNA, which lacks the selected consensus sequence motif. (B) Competitive RNA-binding experiments using unlabeled RNAs. Four femtomoles of labeled RNA was incubated without (lanes 16, 21, 26, and 31) or with 10 fmol of Msi1-2TR protein and the following amounts of unlabeled RNA: 0 fmol (lanes 16, 21, 26, and 31), 0 fmol (lanes 17, 22, 27, and 32), 40 fmol (lanes 18, 23, 28, and 33), 400 fmol (lanes 19, 24, 29, and 34), and 4,000 fmol (lanes 20, 25, 30, and 35).
Msi1 binds to m-numb mRNA both in vitro and in vivo.
Candidates for downstream target genes of the Msi1 protein were explored based on the results of the in vitro selection experiments. Since Msi1 is preferentially expressed in undifferentiated neuronal progenitor cells, mRNAs of genes regulating neural differentiation (either positively or negatively) may be possible downstream targets of Msi1. Then, m-numb, which encodes a Notch antagonist (14, 18, 65), is a likely candidate for an Msi1 target gene, based on the following facts. First, the 3′-untranslated region (UTR) of m-numb mRNA contains the consensus sequence motif for Msi1 binding. Second, the region of m-numb gene expression overlaps that of msi1 expression in neuroepithelial cells in the ventricular zone of the neural tube (52, 65, 70, 71). Third, m-numb is involved in the regulation of neuronal differentiation (12, 65, 72; Tokunaga et al., unpublished results).
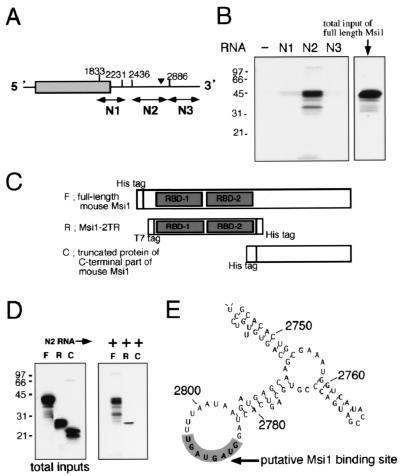
We first examined whether Msi1 binds to the 3′-UTR of m-numb mRNA in vitro. For this purpose, we synthesized various parts of m-numb mRNA in vitro (N1, N2, N3) (Fig. 3A) in the presence of biotin-14 CTP (see Materials and Methods for details). A putative Msi1-binding site exists within N2. The full-length Msi1 protein, a truncated protein containing two tandem RBDs of Msi1 (used for SELEX, Msi1-2TR), and a truncated protein containing the C-terminal portion of Msi1 were examined for their binding abilities to N2 (Fig. 3C and 3D). The full-length Msi1 protein and Msi-2TR were shown to bind to N2 under a moderate ionic strength condition that is close to the physiological condition (150 mM NaCl) (Fig. 3D). [35S]methionine-labeled full-length Msi1 protein coprecipitated with beads conjugated to N2, whereas the other portions of m-numb RNA, N1 and N3, did not interact with the full-length Msi1 protein (Fig. 3B). UV cross-linking experiments showed that Msi-2TR also binds to only N2 (data not shown), indicating that both full-length Msi1 and the truncated protein containing two tandem RBDs of Msi1 (Msi-2TR) preferentially bind to the N2 region within the 3′-UTR of m-numb mRNA in vitro. Thus, m-numb mRNA is a likely in vivo target of the Msi1 protein.
FIG. 3.
Msi1 protein binds to the 3′-UTR of numb mRNA in vitro. (A) Structure of the numb gene according to a previous report (70). Arrows (N1, N2, and N3) indicate the regions that were individually transcribed in vitro. The vertical arrowhead indicates the region containing a putative Msi1-binding sequence (UAGGUAGUAGUUUUA). (B) Binding assays of the Msi1 protein to various transcripts from the 3′-UTR of m-numb mRNA. 35S-labeled full-length Msi1 protein (F) was pulled down with biotin-labeled N2 m-numb RNA-conjugated streptavidin-agarose beads and then the protein-RNA interaction was visualized by autoradiography. The other portions of m-numb RNA, N1 and N3, were not significantly bound by Msi1 protein. In lane −, the absence of biotin-labeled RNA never caused coprecipitation of Msi1. The rectangle on the right shows the amount of total protein included per assay. (C) Schematic representation of Msi1 fusion proteins. F, full-length mouse Msi1; R, truncated protein containing two tandem RBDs (Msi1-2TR); C, truncated protein containing C-terminal potion of mouse Msi1. (D) Both full-length Msi1 protein (F) and truncated Msi1-2TR protein (R, used for SELEX) bind N2 RNA which contains the selected Msi1-binding sequence. The truncated protein containing two RNA-binding domains (R) conserves RNA-recognition specificity similar to that of the full-length Msi1 protein. The C-terminal half of Msi1 protein (C) did not bind N2 RNA. (E) The putative Msi1 binding site in 3′-UTR of m-numb mRNA is located in the loop portion of a predicted stem-loop structure. The predictions of RNA secondary structure were based on the Zuker-Stiegler method.
To determine whether Msi1 binds the 3′-UTR of numb mRNA in vivo, we adopted protocols as described previously (6, 32, 59). We precipitated the Msi1-RNA complex from lysates of NIH 3T3 cells that had been transfected with a series of Msi1-expression vectors (Fig. 4A). In NIH 3T3 cells, the m-numb gene is endogenously transcribed, while Msi1 is not expressed. We thus exogenously induced the expression of HAT-tagged Msi1 protein (Fig. 4B), which binds to Talon metal chelation affinity resin (Clontech) with high specificity in NIH 3T3 cells (Fig. 4B), and examined whether the HAT-tagged Msi1 bound to m-numb mRNA. Cellular lysates from these transfected cells were applied to Talon metal chelation affinity resin (Clontech) to purify the Msi1-RNA complex. RNA that bound to the HAT-tagged Msi1 protein was then phenol extracted, reverse transcribed, and amplified by PCR using primers specific for m-numb or the abundantly expressed β-actin gene (as a negative control). RNA that bound to the HAT-tagged Msi1 protein gave rise to reverse transcription (RT)-PCR product when the m-numb primers, but not the β-actin primers, were used (Fig. 4C, lane H [RT(+)]). To prove the RNA-binding requirement of the Msi1 protein, a mutant Msi1 protein, FLAG-Msi1mutR1HAT (Fig. 4A), in which three aromatic amino acids that are essential for RNA binding had been replaced (63F→L, 65F→L, 68F→L) (8, 31, 36, 43), was also examined for its ability to bind endogenous m-numb RNA. In this case, the mutant Msi1 protein (FLAG-Msi1mutR1-HAT) failed to show binding to m-numb mRNA (Fig. 4C, lane A), indicating that the retention of m-numb RNA on the affinity resin requires the RNA-binding ability of the Msi1 protein. As another control experiment, Msi1 protein without the HAT-affinity tag, FLAG-Msi1 (Fig. 4A), was expressed in NIH 3T3 cells and the same binding assay was performed, which resulted in undetectable retention of m-numb mRNA on this resin (Fig. 4C, lane F). These results demonstrate that Msi1 can interact with the endogenous m-numb RNA in vivo.
Down-regulation of m-numb expression by Msi1: endogenous m-Numb expression and reporter assay.
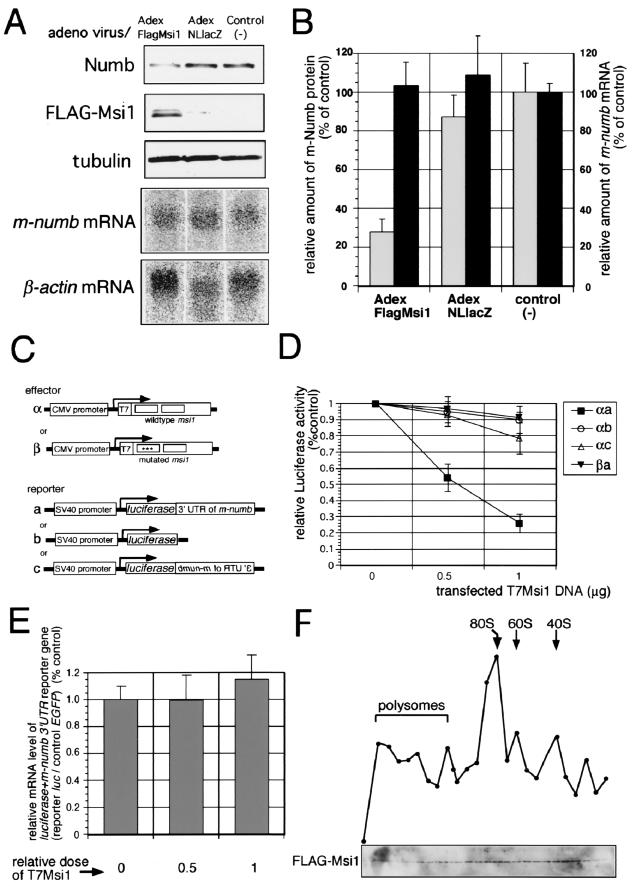
To examine the effect of Msi1 protein on the expression of endogenous m-Numb protein, we misexpressed Msi1 in NIH 3T3 cells using a recombinant adenovirus vector (Fig. 5A and B). NIH 3T3 cells were infected with Adex-FLAGMsi1 or Adex-NLlacZ adenovirus (19) under conditions that are not toxic to the cells (19). Infection with the Adex-FLAGMsi1 vector resulted in the expression of large amounts of FLAG-tagged Msi1 protein under the regulation of the CAG promoter (40), which is a modified chicken β-actin promoter with the cytomegalovirus (CMV) 1E enhancer. Since Msi1 expression did not alter the expression level of tubulin, we used tubulin as an internal control to assess the effect of Msi1 on the expression level of m-Numb protein. Msi1 overexpression decreased the level of the endogenous m-Numb protein to 32% of the level in control cells which expressed LacZ by infection with Adex-NLlacZ (19) (Fig. 5A and B). However, the endogenous m-numb mRNA level remained unchanged in spite of the misexpression of Msi1 and LacZ (Fig. 5A and B). Based on these results, Msi1 protein is likely to be involved in the translational repression of m-Numb protein expression.
FIG. 5.
Posttranscriptional down-regulation of m-numb gene expression by Msi1. (A) Recombinant adenovirus-mediated Msi1-misexpression, immunoblot analyses of m-Numb protein, and Northern blotting analysis. The infection with FLAG-Msi1-expressing adenovirus (Adex-FLAGMsi1 lane) resulted in a decrease of the endogenous expression level of m-Numb protein in NIH 3T3 cells, but the expression of LacZ (Adex NLLacZ lane) (19) and no treatment (control lane) did not. Immunoblot analyses using antitubulin monoclonal antibody and anti-FLAG monoclonal antibody were performed as control experiments. In addition, m-numb and β-actin mRNAs were detected by Northern blotting analysis. (B) Relative amounts of m-Numb protein (blank bar) and m-numb mRNA (filled bar). Msi1 overexpression in NIH 3T3 cells by adenovirus vectors caused a decrease in endogenous m-Numb protein to 32% of the level in control cells expressing LacZ or 28% of the level in untreated cells (control[−]). However, Msi1 overexpression did not affect the m-numb mRNA level in Adex-FLAGMsi1-infected cells compared with that in Adex-NLLacZ-infected cells or untreated cells (control[−]). (C) Schematic representation of Msi1 effector and reporter constructs containing the 3′-UTR of m-numb. α, pcDNA3-T7msi1; β, pcDNA3-T7msi1mutR1 (α and β were under the control of the CMV promoter); a, pGVP2-numb3′-UTR; b, pGV-P2; c, pGVP2-reversednumb3′-UTR (a, b, and c were under the control of the simian virus 40 promoter). (D) Luciferase reporter assay. NIH 3T3 cells were transiently transfected in the indicated combinations with luciferase reporter constructs and increasing concentrations of pcDNA3-T7msi1 or pcDNA3-T7msi1mutR1 effector constructs. The graph depicts the dose-response relationship between exogenously expressed T7Msi1 and the luciferase activity obtained. The results are presented as ratios of firefly luciferase reporter activity over sea pansy (Renilla) luciferase activity, the latter being used as a control (means and standard errors of the means of values from three or four independent experiments). (E) Relative levels of reporter mRNAs quantified by Northern ELISA. The mRNA level of the transcript of EGFP, which does not contain the Msi1-binding site on its mRNA, was used as an internal control. The results were given as the ratios (percentages of control values) of luciferase-numb 3′-UTR chimeric mRNA content to EGFP mRNA content (means and standard errors of the means of results from three independent experiments) and relative doses of T7Msi1 are shown below the panel. The relative amount of luciferase reporter RNA containing the 3′-UTR of m-numb was not affected by increasing doses of exogenous T7Msi1. (F) Sucrose gradient profile of Msi1 protein containing particles in the cytoplasmic fraction of NIH 3T3 cells. Cytoplasmic extracts of NIH 3T3 cells infected with Adex-FLAGMsi1 with Mg2+ were subjected to zone centrifugation through 5 to 30% linear sucrose gradients in a Hitachi P40ST1286 rotor at 40,000 rpm for 2.5 h and fractionated. The curve shows the A254 of each fraction, and the positions of 40S, 60S, and 80S ribosomal particles and polysomes are indicated. The lower panel shows immunodetection analysis of FLAG-Msi1 protein using anti-FLAG monoclonal antibody.
Next, to investigate how the Msi1 protein regulates the expression of its putative targets in vivo, we used a reporter assay system containing heterologous luciferase gene constructs. We transiently cotransfected the firefly luciferase reporter plasmid and the Msi1 expression plasmid into NIH 3T3 cells, in which Msi1 is not expressed endogenously. The luciferase reporter gene combined with the 1.4-kb whole 3′-UTR of the m-numb gene was placed under the control of the simian virus 40 promoter (Fig. 5C). The expression level of the reporter gene was quantified by assaying the luciferase luminescence level. The wild-type msi1 gene and its non-RNA-binding variant (msi1mutR1) were driven under the control of the CMV promoter. As shown in Fig. 5D, the level of luciferase enzymatic activity was reduced in the presence of exogenously expressed wild-type Msi1 in a dose-dependent manner. In contrast, Msi1mutR1, which lacks the RNA-binding activity (31, 36, 43), did not (Fig. 5D). Furthermore, wild-type Msi1 did not decrease the luciferase reporter activity, when the luciferase reporter gene lacked the m-numb 3′-UTR or was combined with the m-numb 3′-UTR in a reversed orientation to eliminate the Msi1-binding site (Fig. 5D). Thus, the repression of the reporter gene expression was shown to be mediated by the RNA-binding activity of Msi1.
Furthermore, Msi1 appeared to translationally repress the expression of the luciferase-m-numb 3′ UTR chimeric reporter gene at the translational level, rather than by regulating the steady-state RNA level. RNA quantification using Northern blotting hybridization showed that increased levels of the msi1 gene product in NIH 3T3 cells did not affect the relative amount of reporter-numb 3′-UTR fusion mRNA in each experiment (Fig. 5E).
To further examine the possibility of translational repression by Msi1 protein, we investigated the subcellular localization of Msi1 protein by the fractionation of the cytoplasmic lysates of NIH 3T3 cells infected with Adex-FLAGMsi1 through the sedimentation on a linear sucrose gradient (5 to 30%) (58). The A254 of each fraction was used to observe ribosomes and ribosomal subunits as size markers. The assignment of ribosomal subunits was confirmed by extracting total RNA from each fraction. The presence of Msi1 protein was determined by Western blotting of each fraction with anti-FLAG monoclonal antibody. In the presence of 2 mM MgCl2, Msi1 protein migrated to the position corresponding to those of polysome, 80S monosome, 60S ribosomal subunit, and 40S ribosomal subunits (Fig. 5F). These findings demonstrate that Msi1 protein is associated with the ribosomes directly or indirectly.
Taken together, these observations indicated that m-numb mRNA is likely to be one of the in vivo targets of Msi1. Msi1 appears to translationally repress the expression of m-Numb protein through direct interaction with the 3′-UTR of m-numb mRNA.
Msi1 potentiates Notch signaling activity.
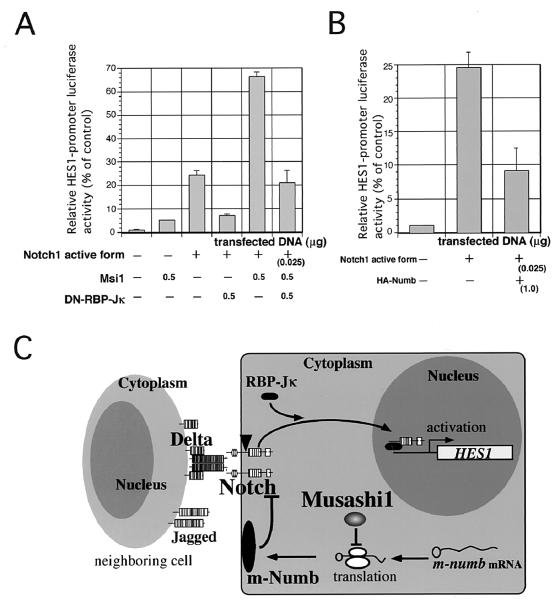
To examine the biological significance of the translational repression of m-numb by the Msi1 protein, we performed a luciferase reporter assay using the HES1 promoter. The minimal HES1 promoter sequence, which has two RBP-Jκ binding sites, is transactivated when Notch signaling is induced (5, 24, 54). Transfection of Msi1 resulted in a slight increase of HES1 promoter activity (5.1-fold activation from the basal level) (Fig. 6A). This slight up-regulation by Msi1 is likely to be attributed to the activation of endogenous Notch. We examined how the transactivating activity of the Notch1 intracellular domain (FCDN1), which is a dominant active form of Notch1 (41), was modified by transfecting exogenous Msi1 into NIH 3T3 cells. Expression of the Notch1 active form alone resulted in a 24.5-fold activation of the HES1 promoter from the basal level (Fig. 6 A). This activation is inhibited by the expression of RBP-Jκ dominant negative form (R218H, designated DN-RBP-Jκ in Fig. 6), which has no binding site to the target DNA and blocks the activation of Notch signal (10, 29). Furthermore, when exogenous Msi1 expression was induced together with the Notch1 dominant active form, the HES1 promoter activity was up-regulated another 2.7-fold over the activation caused by the dominant active Notch1 alone (66-fold activation from the basal level) (Fig. 6A). We found that the expression of Msi1 potentiated the HES1 promoter activity synergistically with that of dominant active Notch1 (Fig. 6A). The potentiation of HES1 promoter luciferase reporter activity by Msi1 is also suppressed by the expression of DN-RBP-Jκ (Fig. 6A). Consequently, the induction of the HES1 promoter by Msi1 is likely to be due to the activation of Notch signaling through the RBP-Jκ-dependent pathway. On the other hand, we found that transactivation of the HES1 promoter by Notch1 is inhibited by misexpression of m-Numb protein (Fig. 6B). Taken together, we demonstrated that misexpression of Msi1 in NIH 3T3 cells decreased the endogenous m-Numb protein level without affecting the mRNA level (Fig. 5A and B) and that m-Numb acts as an antagonist of Notch signaling in NIH 3T3 cells (Fig. 6B). Thus, Msi1 is likely to be involved in activation of Notch-signaling through the RBP-Jκ-dependent pathway by the translational repression of m-Numb.
FIG. 6.
Msi1 expression potentiates Notch1-mediated activation of the HES1 promoter. (A) Transactivation of pHES1-Luc in NIH 3T3 cells. NIH 3T3 cells were transfected with HES1 luciferase reporter plasmids as indicated. The firefly luciferase activities, which represent the transactivation of the HES1 promoter, were determined and normalized against Renilla luciferase activity measurements (means and standard errors of the means of results of three independent experiments). (B) Expression of m-Numb protein inhibits the transcriptional activity of the HES1 promoter which is potentiated by the constitutive active form of Notch1. When the Notch1 active form is expressed in NIH 3T3 cells, the HES1 promoter is activated 24.6-fold from basal level. Cotransfection of HA-tagged m-Numb with the active form of Notch1 caused a decrease in HES1 promoter activity to 37% of the level in cells expressing the active form of Notch1 alone. Therefore, m-Numb antagonistically acts against Notch1 in NIH 3T3 cells. The firefly luciferase activities, which represent the transactivation of the HES1 promoter, were determined and normalized against Renilla luciferase activity measurements (means and standard errors of the means of results of three independent experiments). (C) Model of Msi1 function in the regulation of the Notch signal. Msi1 translationally regulates the m-numb gene expression. Since m-Numb blocks the Notch signal activation (cleavage and/or nuclear translocation with RBP-Jκ, etc.) induced by Notch ligands (Delta, Jagged) which are expressed in neighboring cells, translational repression of m-Numb by Msi1 stimulates Notch signaling through the Notch1, RBP-Jκ, and HES1 pathways. This potentiation of the Notch signal by Msi1 should maintain the immature proliferating status of cells expressing Msi1. The vertical arrowhead shows the Notch1 intracellular cleavage site.
DISCUSSION
Msi1 is a sequence-specific RNA-binding protein.
To investigate the functions of RNA-binding proteins, it is important to identify the target sequences (or target genes). In this study, we used an in vitro selection method (SELEX) to identify high-affinity binding sequences for Msi1 from a random RNA pool. We successfully identified sequence-specific uridine-rich RNAs that bind Msi1. All the selected RNAs contained the (G/A)UnAGU sequence motif (n = 1 to 3), and these motifs frequently appeared as two or three tandem repeats. The RNA-binding specificities of other RNA-binding proteins, including those of the RRM type, have been determined using SELEX or other in vitro selection approaches. hnRNP A1 specifically binds to RNA containing the sequence UAGGGA/U (7). hnRNP D binds to RNA containing UUAG (27). The recognition sequence for HuB (Hel-N1) contains short uridine-rich stretches, like AUUUA, GUUUA, and CUUUA (32). The KH-type RNA-binding protein, Nova-1, interacts with RNA containing three repeats of the sequence UCAU(N) (6). The selected consensus sequence motif for Msi1 is similar to those of hnRNP A1 and HuB in its inclusion of UAG and a short uridylate stretch, respectively. Interestingly, the uridine-rich Msi1-binding site is likely to be recognized by Hu proteins also, including HuB. The selected consensus sequences for Msi1, however, are distinguishable from any that have been previously reported for other RNA-binding proteins.
RNA-binding experiments showed that some of these RNAs were bound by Msi1-2TR with affinity values for Kd as high as ∼4 nM. Kd values for the binding of other RNA by other RRM-type RNA-binding proteins were previously reported. hnRNP A1 binds its target RNA selected by SELEX with affinity values for Kd as high as 1 to 3 nM (7). Full-length hnRNP D (C7) and the truncated protein containing two RBDs (D12L) bind to the target RNA (rH4) at Kd of 34 nM and 490 nM, respectively (27). When comparing Kd values of RRM-type RNA-binding proteins, the binding affinity of the Msi1-2TR protein is found to be one of the highest values. We found that the affinities of the Msi1-2TR protein for oligo-RNAs encoding only two or three copies of the selected sequence motif, however, were comparatively lower than those for the RNAs obtained in the in vitro selection experiment (Fig. 2) (Imai et al., unpublished results). This difference in affinities suggests that not only the consensus sequence motif but also the flanking region might be required for the higher affinity interaction between Msi1 and RNA. The regions flanking the consensus sequence motif are likely to be essential for secondary structure formation, such as stem-loop structures. Interestingly, the sequence motifs recognized by Msi1 were often located in the loops of putative stem-loop structures (Fig. 1E and 3E). Thus, it is possible that the formation of a stem-loop structure surrounding the Msi1-recognition sequence facilitated the Msi1-RNA interaction. A number of RNA-binding proteins have been shown to bind to stem-loop structures both in vitro and in vivo, including Rev (20), U1-A (46) and Nova-1 (6, 25, 33). Thus, the Msi1 protein may also recognize RNA sequences on stem-loop structures.
Taken together, these observations lead us to conclude that Msi1 is a sequence-specific RNA-binding protein.
Putative target gene of the Msi1 protein.
We proposed previously that Msi1 is involved in maintaining the undifferentiated state of neural stem cells through the posttranscriptional control of downstream genes (43, 52). To identify possible target genes of the Msi1 protein, we explored a number of genes responsible for the generation of cell diversity, neuronal cell differentiation, and other neural stem cell characteristics. One of the most likely candidate genes appeared to be m-numb (70). Its Drosophila homologue, numb (62), is required for the asymmetric cell division of the neural precursor cells of the external sensory organ and acts through the inhibition of Notch signaling (18). Recently, we and others have performed functional analysis of the vertebrate Numb protein family. m-Numb binds the post-CDC10/ankyrin repeat region of Notch (70). Chicken Numb protein binds the cytoplasmic region (C-terminal PEST region) of the Notch1 protein and blocks the nuclear transport of a truncated form of Notch1 (65). The findings from both groups suggested that the physical interaction between Numb and Notch1 may reduce the Notch1 activity (65, 70, 71). Correspondingly, we could demonstrate that m-Numb overexpression antagonized the dominant active Notch1-induced HES1 promoter transactivation (Fig. 6B). Furthermore, vertebrate Numb has been shown to play important roles in neural differentiation. Targeted disruption of the m-numb gene in mice results in severe defects in cortical neurogenesis (70). Overexpression of Numb protein promoted the differentiation of the MNS70 cell line of rat neural progenitor cells (37) (mouse Numb) (Tokunaga et al., unpublished results) and of chick neural progenitor cells (chick Numb) (65). Thus, it was expected that proper regulation of vertebrate Numb gene expression would be very important in the control of normal neural differentiation.
In the present study, we demonstrated that the Msi1 protein interacts with m-numb mRNA in vivo, using methods of coaffinity precipitation and RT-PCR analysis. The binding of Msi1 to the 3′-UTR of m-numb demonstrated here may contribute to the regulation of m-numb gene expression. Interestingly, we found the phylogenetic conservation of the Msi1-binding consensus in the 3′-UTR of vertebrate Numb mRNAs (including chick [Y. Wakamatsu, personal communication], rat [63] and mouse [70]), indicating the functional importance of posttranscriptional regulation of vertebrate Numb gene expression by Msi1.
Msi1 inhibits m-numb expression at the translational level.
We next examined the physiological role of the Msi1-numb mRNA interaction in regulating gene expression. Since Msi1 is predominantly and uniformly localized in the cytoplasm of immature neural cells (28, 52), not in their nuclei, Msi1 is not likely to participate in pre-mRNA splicing. The present results indicated that exogenously expressed Msi1 repressed the expression of endogenous m-Numb protein without altering its mRNA level (Fig. 5A and B). Msi1 also repressed the expression of a luciferase reporter containing the 3′-UTR of m-numb cDNA without affecting the reporter's steady-state RNA level (Fig. 5E). Furthermore, Msi1 comigrated with polysomes and monoribosomes and ribosome subunits through sucrose gradient centrifugation using lysate of NIH 3T3 cells infected with Adex-FLAGMsi (Fig. 5F) and localized to free or membrane-associated ribosomes in the cytoplasm of neural progenitor cells as viewed by electron microscopy (unpublished results). These subcellular localization studies on Msi1 suggest that Msi1 is likely to associate with ribosomes and regulate gene expression of downstream targets translationally. Taken together, these observations suggested that Msi1 is likely to repress the expression of the m-Numb protein at the level of translation rather than by reducing the RNA stability mediating the 3′-UTR of m-numb RNA.
The translational control of gene expression often relies on the 5′- or 3′-UTR of the mRNA as cis-elements in eukaryotes (16, 17). ferritin gene expression is regulated at the level of translational initiation by the activation of a trans-acting protein (iron response element binding protein) which binds to a secondary structure on the 5′-UTR of the ferritin mRNA (30). In Drosophila germ line development, translational down-regulation of maternal hunchback (hb) mRNA is essential for the posterior patterning of the Drosophila embryo, which is mediated by a cis-acting element in the 3′-UTR of hb mRNA (the Nanos response elements) as well as two trans-acting factors, Nanos and Pumilio (reviewed by Paris and Lin [47]). Another example of a posttranscriptional trans-acting factor is the Elav-like neural RNA-binding protein family, Hu, which regulates RNA stabilization or translation through binding to an AU-rich element in the 3′-UTR of its target mRNAs (e.g., c-myc, c-fos, neurofilament M, p21) (2, 13, 17, 26, 32, 35, 48, 55, 68).
The molecular mechanisms by which Msi1 regulates the translation of m-numb RNA are still elusive. The possibility that Msi1 induces other RNA-binding proteins that in turn regulate m-Numb expression cannot be excluded. Based on analogy to the work of Nanos and Pumilio (47), a cofactor of Msi1 may be required to regulate the translation of m-numb RNA. Such a cofactor may be identified by yeast two-hybrid screening or other methods in future experiments.
HES1 promoter transactivation by Msi1 in nonneuronal cell line NIH 3T3 cells.
NIH 3T3 fibroblast cells are widely used together with C2C12 cells to determine whether unknown factors, which are expressed in hematopoietic tissue or other tissues, can modulate Notch signaling (9, 41, 42, 50, 51, 67). One of the advantages of NIH 3T3 cells is the endogenous expressions of essential members of the Notch signaling pathway (Notch receptors, RBP-Jκ, Deltex, m-Numb, etc.) at moderate levels. For example, RBP-Jκ, which cotranslocates with the activated form of Notch1 and stimulates the transcriptional activity of HES1 promoter, is endogenously expressed in NIH 3T3 cells at a level high enough to activate the HES1 promoter upon the introduction of the dominant active form of Notch1. Thus, functions of a certain gene product can be investigated by assaying the level of HES1 promoter transactivation when it is coexpressed with the dominant active form of Notch1.
The following points are further advantages of NIH 3T3 cells for the analysis of the functions of Msi1 in the regulation of Notch signaling. First, Msi1 is not endogenously expressed in NIH 3T3 cells, thus providing a highly sensitive assay system for the exogenously expressed Msi1. We found that high levels of endogenous Msi1 expression in neural cells (e.g., rat neural stem cell line MNS-70 cells) sometimes hindered our analysis of the effects of the exogenously expressed Msi1 (Tokunaga et al., unpublished results). Second, NIH 3T3 cells endogenously expressed m-Numb (Fig. 5A), the target of Msi1, thus providing an excellent assay system for the exogenously expressed Msi1. Third, since m-Numb was shown to act as a Notch1 antagonist in NIH 3T3 cells (Fig. 6B), it was expected that exogenously expressed Msi1 would be able to modulate Notch signaling by regulating the expression of m-Numb.
In fact, we found that the misexpression of Msi1 potentiated the HES1 promoter activity synergistically with that of dominant active Notch1 in NIH 3T3 cells (Fig. 6A). Furthermore, Msi1-induced transactivation of the HES1 promoter was RBP-Jκ dependent. We believe that the observed Msi1-induced activation of Notch signaling is mediated by its posttranscriptional regulation of m-Numb expression (Fig. 6C). The expression level of m-Numb was shown to be closely related to the extent of HES1 promoter transactivation by Notch1 activation (Fig. 6B), consistent with the previous reports showing that chicken Numb inhibits the nuclear translocation of the dominant active form of chick Notch1 (65). In the present study, Msi1-misexpression in NIH 3T3 cells induced the down-regulation of endogenous m-Numb protein level translationally, which is likely to have initiated the gene cascade leading to the activation of Notch signaling (Fig. 6C). Since Msi1 and Notch1 are strongly expressed in neural stem cells, it is highly possible that the above described translational repression of m-numb by Msi1 is conserved in neural stem cells in a manner similar to that of NIH 3T3 cells.
Biological significance of m-Numb down-regulation by Musashi1: self-renewing activity and survival of neural stem cells and possible involvement of Msi1 in diseases.
What is the biological significance of the putative translational repression of the m-numb gene by Msi1? Previously, we demonstrated using neurospheres that mammalian Notch/HES1 signaling is essential for the self-renewing activity of neural stem cells and for the repression of their commitment to neuronal lineage (39). Furthermore, Notch signaling, which usually directs cells toward a nonneuronal fate, also has an antiapoptotic function in various cell types (3, 4). Consistent with the fact that Numb family proteins inhibit Notch signaling (14, 18, 65, 71), we and others have observed that overexpression of the vertebrate Numb protein in neural progenitor cells causes cell death (in the absence of caspase inhibitor) or neuronal differentiation (in the presence of caspase inhibitor) (65; Tokunaga et al., unpublished results). Thus, the translational repression of m-Numb expression by Msi1 could promote the self-renewing activity and/or survival of neural stem cells through the modulation of Notch signaling. To test the roles of Msi1 in the maintenance and differentiation of neural stem cells, loss-of-function studies are currently being performed.
In addition to its involvement in such normal development events, Msi1 could also be involved in the pathogenesis of various human diseases through its posttranscriptional regulation of downstream target genes, including m-numb. Many lines of evidence indicate that aberrant posttranscriptional gene regulation causes various hereditary human neurological diseases and neoplasms, i.e., fragile X mental retardation syndrome (56), frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17) (22), myotonic dystrophy, Fukuyama-type congenital muscular dystrophy and neuroblastoma (reviewed by Conne et al. [11]). We have recently obtained evidence that Msi1 is strongly expressed in particular types of human brain tumors, including medulloblastomas, astrocytomas, and glioblastomas (60). Interestingly, the Msi1 expression level was positively correlated with the malignancy and proliferative activities of these tumors. Thus, we proposed that a high level of Msi1 expression leads to the clonal expansion of the above-mentioned tumor cells, mediated by the activation of Notch signaling, presumably through the translational inhibition of m-Numb (Kanemura et al., submitted for publication).
An evaluation of all of these results leads us to propose that Msi1 plays an important role in the self-renewing activity and survival of neural stem cells by regulating Notch signaling activity through posttranscriptional gene regulation. However, the detailed molecular mechanism underlying the potentiation of the Notch signal by Msi1 remains to be elucidated. Furthermore, it is possible that Msi1 regulates the expression of genes other than m-numb and is involved in the pathogenesis of various human diseases (Cuadrado et al., submitted for publication). To elucidate the in vivo functions of Msi1 under normal and pathogenic conditions, functional ablation, including targeted gene disruption, would be essential. Such experiments are currently in progress in our laboratory.
ACKNOWLEDGMENTS
We are grateful to Mikiko Siomi and Haruhiko Siomi for kind instructions for subcellular fractionation using sucrose density gradients and to Yuh Nung Jan for providing us with the m-numb cDNA. We thank Keiko Nakao-Sawai, Wado Akamatsu, Shinichi Sakakibara, and Hirotaka James Okano for valuable discussions and comments and Yoshio Wakamatsu for providing us with the anti-chicken-Numb antiserum and for the valuable suggestion regarding the 3′-UTR sequence of chick and human numb.
This work was supported by grants to H.O. from the Japanese Ministry of Education, Science and Culture and from CREST, Japan and Technology Corporation. T.I. was a research fellow of the Japan Society for the Promotion of Science. This work was also supported by grants from the Human Frontier Science Program to H.O.
REFERENCES
- 1.Akamatsu W, Okano H J, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell R B, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and peripheral nervous systems. Proc Natl Acad Sci USA. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antic D, Lu N, Keene J D. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas S, Matsuno K, Fortini M F. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 4.Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 5.Beatus P, Lundkvist J, Oberg C, Lendahl U. The Notch3 intracellular domain represses Notch1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development. 1999;126:3925–3935. doi: 10.1242/dev.126.17.3925. [DOI] [PubMed] [Google Scholar]
- 6.Buckanovich R J, Darnell R B. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burd C G, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding protein. Science. 1994;263:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 9.Carlesso N, Aster J C, Sklar J, Scadden D T. Notch1-induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood. 1999;93:838–848. [PubMed] [Google Scholar]
- 10.Chung C N, Hamaguchi Y, Honjo T, Kawaichi M. Site-directed mutagenesis study on DNA binding regions of the mouse homologue of Suppressor of Hairless, RBP-J kappa. Nucleic Acids Res. 1994;22:2938–2944. doi: 10.1093/nar/22.15.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conne B, Stutz A, Vassalli J D. The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 12.Dho S E, French M B, Woods S T, McGlade S J. Characterization of four mammalian Numb protein isoforms. J Biol Chem. 1999;274:33097–33104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- 13.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frise E, Knoblich J A, Younger-Shepherd S, Jan L Y, Jan Y N. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc Natl Acad Sci USA. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Good P J, Akinori Y, Sakakibara S, Yamamoto A, Imai T, Sawa H, Ikeuti T, Tsuji S, Satoh H, Okano H. The human Musashi homolog1 (MSI1) encoding the homologue of Musashi/Nrp-1, a neural RNA-binding protein putatively expressed in CNS stem cell and neural progenitor cells. Genomics. 1998;52:382–384. doi: 10.1006/geno.1998.5456. [DOI] [PubMed] [Google Scholar]
- 16.Gray N K, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 17.Grosset C, Chen C Y, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu A B. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 18.Guo M, Jan L Y, Jan Y N. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto M, Aruga J, Hosoya Y, Kanegae Y, Saito I, Mikoshiba K. A neural cell-type-specific expression system using recombinant adenovirus vectors. Hum Gene Ther. 1996;7:149–158. doi: 10.1089/hum.1996.7.2-149. [DOI] [PubMed] [Google Scholar]
- 20.Heaphy S, Dingwall C, Ernberg I, Gait M J, Green S M, Karn J, Lowe A D, Singh M, Skinner M A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990;60:685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- 21.Hirota Y, Okabe M, Imai T, Kurusu M, Yamamoto A, Miyao S, Nakamura M, Sawamoto K, Okano H. Musashi and seven in absentia downregulate tramtrack through distinct mechanisms in Drosophila eye development. Mech Dev. 1999;87:93–101. doi: 10.1016/s0925-4773(99)00143-4. [DOI] [PubMed] [Google Scholar]
- 22.Hutton M, Lendon C L, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 23.Jan Y N, Jan L Y. Asymmetric cell division. Nature. 1998;392:775–778. doi: 10.1038/33854. [DOI] [PubMed] [Google Scholar]
- 24.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 25.Jensen K B, Dredge B K, Stetani G, Zhong R, Buckanovich R J, Okano H J, Yang Y Y, Daenell R B. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 26.Joseph B, Orlian M, Furneaux H. p21(waf1) mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J Biol Chem. 1998;273:20511–20516. doi: 10.1074/jbc.273.32.20511. [DOI] [PubMed] [Google Scholar]
- 27.Kajita Y, Nakayama J, Aizawa M, Ishikawa F. The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0. J Biol Chem. 1995;270:22168–22175. doi: 10.1074/jbc.270.38.22167. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, Ogawa Y, Toyama T, Okano H. Musashi1: evolutionarily conserved markers for CNS progenitor cells including neural stem cells. Dev Neurosci. 2000;22:138–152. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- 29.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 30.Klausner R D, Harford J B. cis-trans models for post-transcriptional gene regulation. Science. 1989;246:870–872. doi: 10.1126/science.2683086. [DOI] [PubMed] [Google Scholar]
- 31.Kurihara Y, Nagata Y, Imai T, Hiwatasi A, Horiuchi M, Sakakibara S, Katahira M, Okano H, Uesugi S. Structural properties and RNA-binding activities of two RNA recognition motifs of a mouse neural RNA-binding protein, mouse-Musashi-1. Gene. 1997;186:21–27. doi: 10.1016/s0378-1119(96)00673-7. [DOI] [PubMed] [Google Scholar]
- 32.Levine T D, Gao F, King P H, Andrews L G, Keene J D. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis H A, Musunuru K, Jensen K B, Edo C, Chen H, Darnell R B, Burley S R. Sequence-specific RNA binding by Nova KH domain: implications for paraneoplastic disease and the fragile X. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Dalmau J, Szabo A, Rosenfeld M, Huber J, Furneaux H. Paraneoplastic encephalomyelitis antigens bind to the AU-rich elements of mRNA. Neurology. 1995;45:544–550. doi: 10.1212/wnl.45.3.544. [DOI] [PubMed] [Google Scholar]
- 35.Myer V E, Fan X C, Steitz J A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagata T, Kanno R, Kurihara Y, Uesugi S, Imai T, Sakakibara S, Okano H, Katahira M. Structural, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1. J Mol Biol. 1999;287:315–330. doi: 10.1006/jmbi.1999.2596. [DOI] [PubMed] [Google Scholar]
- 37.Nakafuku M, Nakamura S. Establishment and characterization of a multipotential neural cell line that can conditionally generate neurons, astrocytes, and oligodendrocytes in vitro. J Neurosci Res. 1995;41:153–168. doi: 10.1002/jnr.490410203. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura M, Okano H, Blendy J, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:68–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura Y, Sakakibara S, Miyata T, Ogawa M, Shimazaki T, Weiss S, Kageyama R, Okano H. The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J Neurosci. 2000;20:283–293. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 41.Nofziger D, Miyamoto A, Lyons K M, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 42.Ordentlich P, Lin A, Shen C P, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, Kadesch T. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol Cell Biol. 1998;18:2230–2239. doi: 10.1128/mcb.18.4.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okabe, M., T. Imai, M. Kurusu, Y. Hiromi, and H. Okano. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature, in press. [DOI] [PubMed]
- 44.Okano H. Two major mechanisms regulating cell-fate decisions in the developing nervous system. Dev Growth Differ. 1995;37:619–629. doi: 10.1046/j.1440-169X.1995.t01-5-00001.x. [DOI] [PubMed] [Google Scholar]
- 45.Okano H J, Darnell R B. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oubridge C, Ito N, Evans P R, Teo C H, Nagai K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 47.Paris M, Lin H. Translational repression: a duet of Nanos and Pumilio. Curr Biol. 2000;10:R81–R83. doi: 10.1016/s0960-9822(00)00283-9. [DOI] [PubMed] [Google Scholar]
- 48.Pengs, S. S., C. Y. Chen, N. Xu, and A. B. Shyu. RNA stabilization by AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17:3461–3470. [DOI] [PMC free article] [PubMed]
- 49.Pincus D W, Keyoung H, Restelli C, Goodman R R, Fraser R A R, Edgar M, Sakakibara S, Okano H, Nedergaad M, Goldman S A. FGF2/BDNF-responsive neuronal progenitor cells in the adult human subependyma. Ann Neurol. 1998;43:576–585. doi: 10.1002/ana.410430505. [DOI] [PubMed] [Google Scholar]
- 50.Rand M D, Grimm L M, Artavanis-Tsakonas S, Patriub V, Blacklow S C, Sklar J, Aster J C. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao P K, Dorsch M, Chickering T, Zheng G, Jiang C, Goodearl A, Kadesch T, McCarthy S. Isolation and characterization of the notch ligand delta4. Exp Cell Res. 2000;260:379–386. doi: 10.1006/excr.2000.5034. [DOI] [PubMed] [Google Scholar]
- 52.Sakakibara S, Imai T, Hamaguchi K, Okabe M, Aruga J, Nakajima K, Yasutomi D, Nagata T, Kurihara Y, Uesugi S, Miyata T, Ogawa M, Mikoshiba K, Okano H. Mouse-musashi-1, neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- 53.Sakakibara S, Okano H. Expression of neural RNA-binding proteins in the postnatal CNS: implication of their roles in neural and glial cell development. J Neurosci. 1997;17:8300–8312. doi: 10.1523/JNEUROSCI.17-21-08300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 55.Shyu A B, Wilkinson M F. The double lives of shuttling mRNA binding protein. Cell. 2000;102:135–138. doi: 10.1016/s0092-8674(00)00018-0. [DOI] [PubMed] [Google Scholar]
- 56.Siomi H, Siomi M C, Nussbaum R L, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 57.Siomi H, Dreyfuss G. RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- 58.Siomi M C, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steitz J A. Immunoprecipitation of ribonucleoproteins using autoantibodies. Methods Enzymol. 1989;180:468–481. doi: 10.1016/0076-6879(89)80118-1. [DOI] [PubMed] [Google Scholar]
- 60.Toda M, Iizuka Y, Yu W-J, Ikeda E, Yoshida K, Imai T, Kawase T, Kawakami Y, Okano H, Uyemura K. Expression of the neural RNA-binding protein Musashil in human gliomas. Glia. 2001;34:1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- 61.Tsai D E, Harper D S, Keene J D. U1-snRNP-A protein selects a ten nucleotide consensus sequence from a degenerate RNA pool presented in various structural contexts. Nucleic Acids Res. 1991;19:4931–4936. doi: 10.1093/nar/19.18.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uemura T, Shepherd S, Ackerman L, Jan L Y, Jan Y N. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 63.Verdi J M, Schmandt R, Bashirullah A, Jacob S, Salvino R, Craig C G, Program A E, Lipshitz H D, McGlade C J. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr Biol. 1996;6:1134–1145. doi: 10.1016/s0960-9822(02)70680-5. [DOI] [PubMed] [Google Scholar]
- 64.Wakamatsu Y, Weston J A. Sequential expression and role of Hu RNA-binding proteins during neurogenesis. Development. 1997;124:3449–3460. doi: 10.1242/dev.124.17.3449. [DOI] [PubMed] [Google Scholar]
- 65.Wakamatsu Y, Maynard T M, Jones S U, Weston J A. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron. 1999;23:71–81. doi: 10.1016/s0896-6273(00)80754-0. [DOI] [PubMed] [Google Scholar]
- 66.Wickens M, Kimble J, Strickland S. Translational control of developmental decisions. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Vol. 30. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 411–450. [Google Scholar]
- 67.Wong M K, Prudovsky L, Vary C, Booth C, Liaw L, Mousa S, Small D, Maciag T. A non-transmembrane form of Jagged-1 regulates the formation of matrix-dependent chord-like structures. Biochem Biophys Res Commun. 2000;268:853–859. doi: 10.1006/bbrc.2000.2173. [DOI] [PubMed] [Google Scholar]
- 68.Xu N, Chen C Y, Shyu A B. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao K M, Samson M L, Reeves R, White K. Gene elav of Drosophila melanogaster: a prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J Neurobiol. 1993;24:723–739. doi: 10.1002/neu.480240604. [DOI] [PubMed] [Google Scholar]
- 70.Zhong W, Feder J H, Jiang M M, Jan L Y, Jan Y N. Asymmetric localization of a mammalian Numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 71.Zhong W, Jiang M M, Weinmaster G, Jan L Y, Jan Y N. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development. 1997;124:1887–1897. doi: 10.1242/dev.124.10.1887. [DOI] [PubMed] [Google Scholar]
- 72.Zhong W, Jiang M M, Schonemann M D, Meneses J J, Pedersen R A, Jan L Y, Jan Y N. Mouse numb is an essential gene involved in cortical neurogenesis. Proc Natl Acad Sci USA. 2000;97:6844–6849. doi: 10.1073/pnas.97.12.6844. [DOI] [PMC free article] [PubMed] [Google Scholar]