Abstract
Transforming growth factor β (TGFβ) signaling is transduced via Smad2–Smad4–DNA-binding protein complexes which bind to responsive elements in the promoters of target genes. However, the mechanism of how the complexes activate the target genes is unclear. Here we identify Xenopus Swift, a novel nuclear BRCT (BRCA1 C-terminal) domain protein that physically interacts with Smad2 via its BRCT domains. We examine the activity of Swift in relation to gene activation in Xenopus embryos. Swift mRNA has an expression pattern similar to that of Smad2. Swift has intrinsic transactivation activity and activates target gene transcription in a TGFβ-Smad2-dependent manner. Inhibition of Swift activity results in the suppression of TGFβ-induced gene transcription and defective mesendoderm development. Blocking Swift function affects neither bone morphogenic protein nor fibroblast growth factor signaling during early development. We conclude that Swift is a novel coactivator of Smad2 and that Swift has a critical role in embryonic TGFβ-induced gene transcription. Our results suggest that Swift may be a general component of TGFβ signaling.
Transforming growth factor β (TGFβ) superfamily signaling mediates a very diverse array of biological processes, including immune function, growth control, cell differentiation, patterning the embryonic body, sexual reproduction, and skeletal formation (reviewed in reference 29). The TGFβ family member activin elicits several different gene expression and cell fate pathways in a concentration-dependent way in early Xenopus development (15–17). The study of activin response genes in early Xenopus development has provided an excellent tool to elucidate the general mechanism of the TGFβ signaling pathway.
TGFβ-activin signals through serine-threonine kinase receptors I and II at the cell surface and initiates the following sequence of events (reviewed in reference 30). Binding of activin induces the formation of heteromeric complexes of these receptors, and signaling is initiated when receptor I is phosphorylated and activated by receptor II. Activated receptor I phosphorylates Smad2. Activated Smad2 forms a complex with Smad4 (Smad2/4 complex) in the cytoplasm and then enters the nucleus. In the nucleus, the Smad2/4 complex interacts with specific DNA-binding cofactors that help to select target genes. In Xenopus development, FAST, Mixer, and Milk have been identified as Smad2-recruiting DNA-binding cofactors (4, 13). FAST, which contains a winged helix DNA-binding domain, binds to an activin-responsive element and is required for activation of Mix.2 gene in response to activin. FAST bound to DNA alone does not appear to activate transcription (41). However, recruitment of an activated Smad2/4 complex to the activin-responsive element by FAST results in activation of Mix.2 gene expression (5). Mixer and Milk are paired-like homeodomain transcription factors of the Mix family (7, 19). In a mechanism similar to FAST, they bind to the activin-inducible distal element of the Xgsc promoter, recruit Smad2, and initiate transcription of Xgsc (13).
Recently, it has been demonstrated that the Smad2/4 complex recruits the transcriptional coactivators p300 and CREB-binding protein (CBP) (10, 21, 32). However, little is known about how the Smad2/4 complex with DNA-binding proteins activates gene expression. Here we describe the identification and function of a novel Xenopus Smad2 coactivator which we call Swift. Interestingly, Swift contains six BRCT (BRCA1 C-terminal) domains, first described in the breast cancer suppressor protein BRCA1. Swift binds to Smad2 via its BRCT domains. We show that Swift mRNA is maternally expressed and that its expression pattern during early development is similar to that of Smad2. Swift synergistically activates gene expression with Smad2 in an activin signaling-dependent manner. Swift has intrinsic transactivation activity that is enhanced by activin signaling. Using two different dominant interfering approaches, we show that Swift function is required for TGFβ-activin-induced gene expression in vivo and for normal mesendoderm formation. Our results suggest that Swift is a necessary component of the embryonic TGFβ signaling pathway. Swift may have a role in many instances of TGFβ signaling.
MATERIALS AND METHODS
Isolation of Xenopus Swift.
For the yeast two-hybrid screening, the linker region and a part of the MH2 domain of Smad2 (amino acids 180 to 432) was subcloned into a pBTM116 bait vector; the expressed fusion protein contains a LexA DNA-binding domain (40). The cDNA library, consisting of poly(A) mRNA and poly(A)+ mRNA random primed from Xenopus eggs, was cloned into the EcoRI site of the pACTII library vector that contains a GAL4 transactivation domain. Yeast two-hybrid screening was performed using Saccharomyes cerevisiae strain L40 as described previously (40). A screen of 107 clones yielded 31 positive clones, 6 of which contained identical 2.8-kb cDNAs. A full-length Swift was obtained by a 5′ primer extension on a Xenopus egg cDNA library (33).
RNA expression constructs.
For Swift expression constructs, the open reading frame was amplified by PCR using Pfu Turbo DNA polymerase (Stratagene) and subcloned into expression vectors. The capped mRNAs were synthesized in vitro (33). pT7TSHA-HA (hemagglutinin)-Swift was constructed by inserting full-length Swift cDNA into pT7TSHA-HA (36), linearized by XbaI, and transcribed with T7. pT7TSHA-HA-EnR-Swift ΔN was constructed by inserting a DNA fragment encoding the Engrailed sequence into the C terminus of Swift ΔN (amino acids 567 to 1256) in pT7TSHA-HA-Swift ΔN, linearized by XbaI, and transcribed with T7. pActRIB* was linearized by HindIII and transcribed with T7. pT7TSHA-HA ActRIB* was constructed by inserting an ActRIB* cDNA into pT7TSHA-HA, linearized by XbaI, and transcribed with T7. pT7TS-VP16A-HA-HA was constructed by inserting a DNA fragment encoding a VP16 transactivation domain into pT7TSHA-HA. pT7TS-VP16A-HA-HA-Swift and -Smad2 were constructed by inserting full-length Swift and Smad2 cDNAs, respectively, into pT7TS-VP16A-HA-HA, linearized by XbaI, and transcribed with T7. pT7TS-GAL-HA was constructed by inserting a DNA fragment encoding a GAL4 DNA-binding domain into pT7TSHA. pT7TS-GAL-HA-Swift was constructed by inserting a Swift cDNA into pT7TS-GAL-HA, linearized by XbaI, and transcribed with T7. pT7TS-GST-Swift B3-6 was constructed by inserting a DNA fragment encoding glutathione S-transferase (GST) into pT7TS-Swift B3-6, linearized by XbaI, and transcribed with T7. HA-Smad2 and activin mRNAs were prepared as described elsewhere (33, 36). pSP64-Smad1, pSP64-Smad2, and pdn-BRII were gifts from D. A. Melton. The Engrailed repressor construct and pActRIB* (pALK4*) were gifts from J. Smith.
Oocyte synthesis of activin.
The procedure for oocyte synthesis of activin was as described elsewhere (31).
In vitro binding assays.
35S-labeled Swift was in vitro translated using the TNT coupled reticulocyte lysate system (Promega). GST, GST-Smad1, and GST-Smad2 were purified from overexpressing Escherichia coli (37). One microgram of GST, GST-Smad1, or GST-Smad2 was incubated with 0.5 μl of 35S-labeled Swift in 0.1 ml of buffer A (20 mM Tris-Cl [pH 8.0], 0.5 mM EDTA, 1 mM dithiothreitol, 1% NP-40, 0.2 M NaCl) at 4°C for 1 h; then 10 μl of glutathione-Sepharose beads was added. After 1 h of incubation, the beads were washed with buffer A four times. Bound proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Coomassie brilliant blue staining and autoradiography.
In vivo coprecipitation.
Each embryo was injected with various combinations of 2 ng of GST-Swift B3-6, 2 ng of HA-Smad2, and 1 ng of activin mRNAs into an animal pole of the one-cell stage embryo and cultured to stage 9. Forty embryos were homogenized in 0.4 ml of buffer B (50 mM Tris-Cl [pH 7.5], 50 mM KCI, 25% glycerol, 25 mM β-glycerophosphate, phosphatase inhibitors [Sigma], protease inhibitors [Roche], 2 mM Na3VO4, 0.2 mM NaF, 1 mM dithiothreitol) and cleared by centrifugation at 4°C; then 20 μl of glutathione-Sepharose beads was added. After 2 h of incubation, the beads were washed four times with buffer B containing 0.25% NP-40 and 0.2 M NaCl. Bound proteins were resolved by SDS-PAGE and analyzed by Western blotting.
Northern blot, in situ, and RNase protection assays.
Northern blot, in situ, and RNase protection assays were performed as described elsewhere (33), and experiments were repeated at least three times. The RNase protection probe template for Swift was constructed as follows: pSwiftΔN(NcoI), which consisted of Swift nucleotides 1 to 2472, was linearized at the PstI site (nucleotide 2231), and an antisense probe was synthesized using SP6 RNA polymerase.
Subcellular localization by immunofluorescence microscopy.
U20S human osteosarcoma cells were maintained in Dulbecco Modified Eagle medium containing 10% fetal bovine serum under 5% CO2, at 37°C. Cultured cells were transfected with pcDNAmyc-Swift (full length) using FuGENETM6 transfection reagent (Roche) and cultured for 24 h. Immunostaining was performed as described elsewhere (39). Cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.5% Triton X-100 for 5 min. After being washed with phosphate-buffered saline, cells were incubated with an anti-Myc antibody (9E10) (Roche) for 1 h. Fluorescein-conjugated rabbit anti-mouse immunoglobulin (Sigma) was used as the second antibody. Cells were observed and photographed under fluorescein or UV illumination using immunofluorescence microscopy.
Luciferase assays.
Each embryo was coinjected in the animal pole at stage 1 with mRNA (0.5 ng) encoding a GAL fusion construct with 5xGAL4-luciferase reporter plasmid DNA (0.3 ng) (46) or 5xGAL4-TK-luciferase reporter plasmid DNA (0.1 ng) (12) with or without ActRIB* (0.2 ng) or VP16A-Smad2 (3 ng) mRNA. Embryos were cultured to stage 10. Five embryos at stage 10 were homogenized in 0.1 ml of buffer C (50 mM Tris-Cl [pH 7.5], protease inhibitors [Roche]) and cleared by centrifugation at 4°C. One to five microliters of the supernatant was assayed for luciferase activity in a Monolight 2010 luminometer (Analytical Luminescence Laboratory).
Nucleotide sequence accession number.
The cDNA sequence has been deposited in GenBank under accession number AF172855.
RESULTS
Identification of Swift.
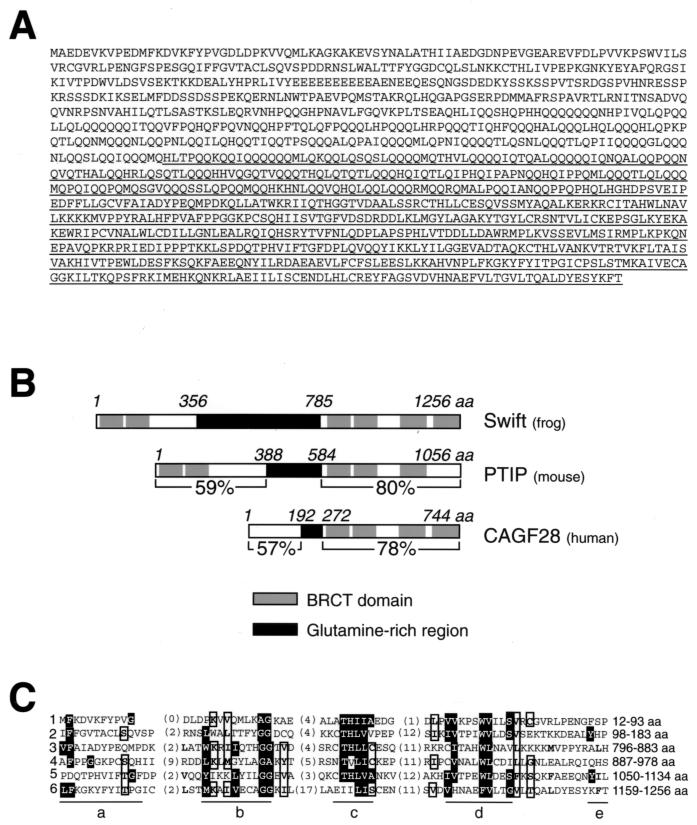
Smad2 is a critical intracellular mediator of the TGFβ signaling pathway during early Xenopus development (14). To identify Smad2-binding proteins that participate in TGFβ signaling, we used a two-hybrid screen of a Xenopus egg cDNA library with pLexA-Smad2. This LexA fusion construct encoded the linker region and a part of the MH2 domain of Smad2. Of 31 positive clones isolated, six were identical and encoded a novel amino acid sequence. Because all isolated cDNAs were partial clones, we performed a 5′ primer extension on a Xenopus egg cDNA library to obtain the entire open reading frames. The complete protein consists of 1,256 amino acids (Fig. 1A). Based on our subsequent functional analysis, we name this protein Swift, for Xenopus Smad wing for transcriptional activation.
FIG. 1.
Cloning of Swift. (A) Deduced amino acid sequence of Swift. The cDNA fragment isolated from the yeast two-hybrid screen is underlined. (B) Comparison of the primary structure of Swift with those of PTIP and CAGF28. The percentage values in the N-terminal regions of PTIP (amino acids 1 to 388) and CAGF28 (amino acids 1 to 192) show amino acid sequence identity with that of Swift (amino acids 1 to 356 and 207 to 356 respectively). The percentage values in the C-terminal regions of PTIP (amino acids 584 to 1056) and CAGF28 (amino acids 272 to 744) show amino acid sequence identity with that of Swift (amino acids 785 to 1256). (C) Consensus sequence for the six BRCT domains of Swift. Five aligned blocks (designated a to e) constituting the five conserved regions of the domain are separated from each other as described elsewhere (3). Matches to the consensus are in black boxes. Bulky hydrophobic residues (I, L, V, M, F, Y, W) and small residues (G, A, S, T, C) are grouped; residues conserved in four out of six sequences are in white boxes (24).
Sequence analysis reveals that Swift has six BRCT domains, first identified in the C-terminal region of the breast cancer suppressor protein BRCA1 (24) (Fig. 1B and C). The BRCT domain is defined by distinct hydrophobic clusters of amino acids and is believed to occur as an autonomous folding unit of ∼95 amino acids that is implicated in protein-protein interactions (3). Swift also contains a glutamine-rich region and a putative nuclear localization signal (amino acids 857 to 874). GenBank searches reveal a mouse homologue named PTIP (Pax transcription activation domain-interacting protein) (27) and a human homologue named CAGF28 (28) (GenBank accession numbers AF104261 and U80735, respectively). PTIP is a nuclear protein with five BRCT domains that interacts with the Pax2 DNA-binding protein (27). However, these two genes were not previously implicated in TGFβ signaling. Their C-terminal domains share about 80% amino acid sequence identity with that of Swift and contain BRCT domains (Fig. 1B). These results suggest that Swift is conserved at least from Xenopus to humans.
Expression pattern and nuclear localization of Swift.
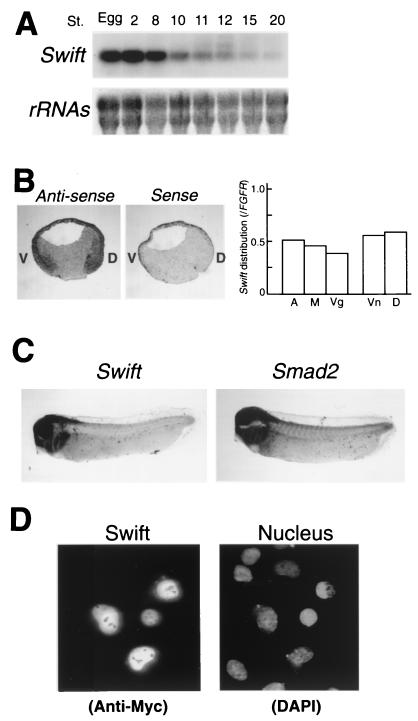
Northern blot analysis of the developmental expression pattern of Swift reveals that Swift mRNA is maternally expressed and that the level of its mRNA declines during gastrulation and continues at a lower level until late stages (Fig. 2A). In situ hybridization analysis reveals that Swift mRNA is ubiquitously expressed at the gastrula stage, and its expression pattern is similar to that of Smad2 mRNA (14) (Fig. 2B, left). This result is confirmed by RNase protection assays (Fig. 2B, right). Later in development, Swift mRNA becomes enriched in the head and brain as does Smad2 mRNA (Fig. 2C). The coincident expression pattern of Swift and Smad2 mRNAs supports the possibility that they interact functionally during early development. A Myc-tagged Swift expressed in U20S human osteosarcoma cells is mostly localized in the nucleus (Fig. 2D), indicating that Swift is a nuclear protein.
FIG. 2.
Developmental expression of Swift. (A) Temporal expression of Swift mRNA during Xenopus development. Northern blots were probed with random-prime-labeled Swift cDNA (nucleotides 2991 to 3768). The bottom panel shows ethidium bromide staining of rRNAs. (B) Distribution of Swift mRNA at the gastrula stage. Left, in situ hybridization to sections cut vertically through the dorsoventral axis of stage 11 embryos, using full-length antisense and sense Swift mRNA probes. Ventral (V) and dorsal (D) are marked. Grey staining shows the expression of Swift mRNA. Right, quantitation of Swift mRNA. Stage 11 embryos were dissected into roughly thirds (animal [A], marginal [M], or vegetal [Vg]) or halves (ventral [Vn] or dorsal [D]), and total RNA was harvested. The level of Swift mRNA was quantitated by RNase protection assays, and FGFR was used as a loading control. (C) Whole-mount hybridization with full-length Swift and Smad2 mRNA probes, using Xenopus embryos at stage 28. (D) Swift is a nuclear protein. U20S human osteosarcoma cells were transiently transfected with pcDNAmyc-Swift (full length). Cells expressing Myc-tagged Swift were analyzed by indirect immunofluorescence using an anti-Myc monoclonal antibody and fluorescein-conjugated rabbit anti-mouse immunoglobulin. 4′,6-Diamidino-2-phenylindole (DAPI) staining of DNA is shown for comparison. Myc-tagged cyclin B1 is cytoplasmic, whereas an nuclear localization site-tagged cyclin B1 enters the nucleus; therefore the Myc tag does not contain a nuclear localization site (personal communication from J. Pines).
Interaction of Swift with Smad2.
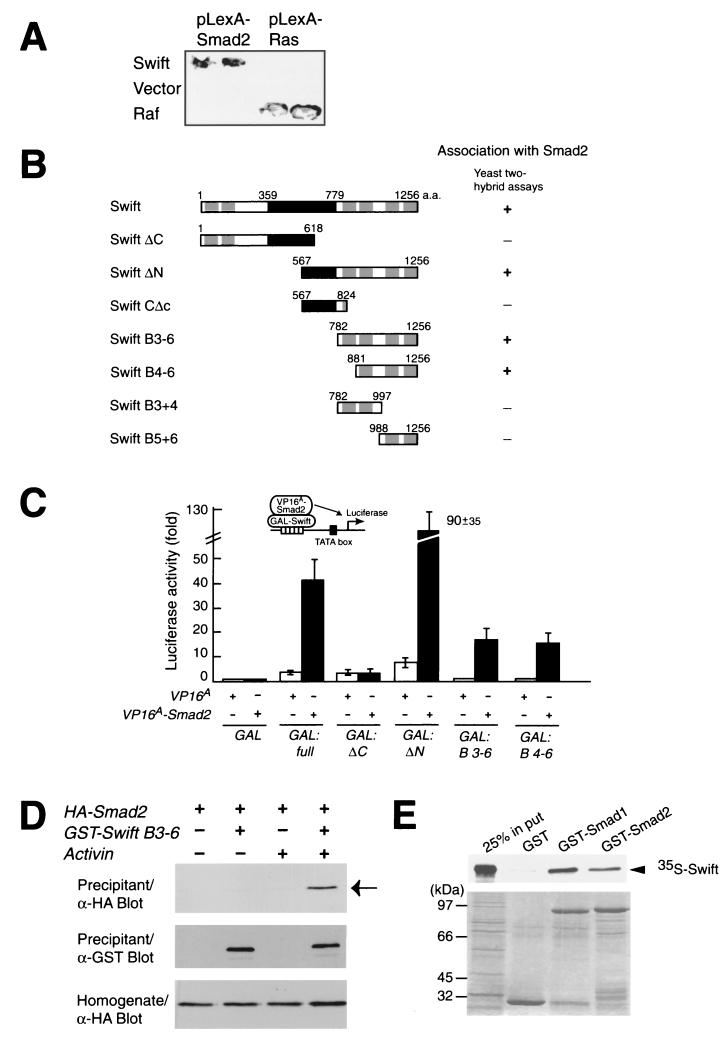
To confirm that full-length Swift interacts with Smad2, we transformed pLexA-Smad2 with pACTIIHK-Swift in yeast, where Swift binds to Smad2 but not to the negative control bait (Fig. 3A). To define the domain of Swift required for Smad2 binding, various fragments of Swift were examined by yeast two-hybrid assays. We tested the fragments encoding full-length Swift, the N-terminal fragment (ΔC), the C-terminal fragment (ΔN), the C-terminal fragment lacking BRCT domains (CΔc), the last four BRCT domains (B3−6), the last three BRCT domains (B4−6), the two BRCT domains (B3+4), and the last two BRCT domains (B5+6). Smad2 binds to full-length Swift, ΔN, B3−6, and B4−6 but not to ΔC, CΔc, B3+4, or B5+6 (Fig. 3B). Then, using luciferase assays, we confirmed the Smad2-binding region of Swift in Xenopus embryos. We coinjected mRNA encoding Swift fused to a GAL4 DNA-binding domain and GAL4-luciferase reporter plasmid DNA with VP16A or VP16A-Smad2 mRNA at stage 1 and measured the luciferase activity at stage 10. VP16A Smad2 enhances the transcriptional activity of full-length, ΔN, B3−6, and B4−6 but not that of ΔC fused to a GAL4 DNA-binding domain (Fig. 3C). This result indicates that Smad2 binds to full-length Swift, ΔN, B3−6, and B4−6 but not ΔC in embryos. In summary, the last three BRCT domains, B4−6, are necessary and sufficient for Smad2 interaction in yeast and embryos.
FIG. 3.
Interaction of Swift with Smad2. (A) Interaction of Swift with Smad2 in yeast two-hybrid assays. Strain L40 was transformed with pLexA-Smad2 (amino acids 180 to 432) or pLexA-Ras (G12V) with pACTIIHK-Swift (full length), pACTIIHK, or pVP-Raf. Interaction of Swift with Smad2 was examined by qualitative assays for β-galactosidase activity, and pLexA-Ras and pVP-Raf were used as positive controls (40). (B) Determination of the Smad2-interacting region of Swift by yeast two-hybrid assays. The indicated fragments of Swift were tested for their interaction with Smad2 in yeast two-hybrid assays. Interaction of Swift with Smad2 in yeast was examined by qualitative assays for β-galactosidase activity. (C) Determination of the Smad2-interacting region of Swift in embryos by luciferase assays. Each embryo was coinjected in the animal pole of the one-cell-stage embryo with mRNA encoding a GAL fusion construct and 5xGAL4-luciferase reporter plasmid DNA with VP16A or VP16A-Smad2 mRNA. Interaction of Swift with Smad2 in embryos was examined by luciferase assays for luciferase activity. The values are means ± standard error of three independent experiments. Closed and open bars show luciferase activities with VP16A-Smad2 and VP16A mRNAs, respectively. (D) In vivo coprecipitation of Smad2 with Swift in embryos. Embryos were coinjected with various combinations of the indicated mRNAs at stage 1; in vivo coprecipitation assays were performed at stage 9, followed by Western blotting using the indicated antibodies. The arrow indicates HA-Smad2 coprecipitated with GST-Swift B3-6. (E) Direct interaction of Swift with Smads using in vitro binding assays. GST, GST-Smad1, or GST-Smad2 was incubated with 35S-labeled full-length Swift, and glutathione-Sepharose beads were then added. Bound proteins were resolved on SDS-PAGE and visualized by autoradiography (top) and Coomassie brilliant blue staining (bottom).
We then asked if Swift interacts with Smad2 in an activin signal-dependent manner in embryos. We injected HA-Smad2 (full-length) mRNA with or without GST-Swift B3−6 and activin mRNAs at stage 1 and performed in vivo coprecipitation assays at stage 9. GST-Swift B3−6 interacts with HA-Smad2 in the presence but not in the absence of activin signaling (Fig. 3D). We conclude that Swift interacts with Smad2 in an activin signal-dependent manner in embryos.
To test if Swift directly binds to Smad2, we prepared 35S-labeled Swift protein. 35S-Swift directly binds to GST-Smad2 but not to GST alone (Fig. 3E). Swift also binds to Smad1 as well as Smad2 in in vitro binding assays. First, we focused on the analysis of Swift function for Smad2 in early embryos, because Smad2, but not Smad1, is a downstream component of TGFβ signaling. Then, we tested if Swift functioned with Smad1 in early embryos (see Fig. 7A). We conclude that Swift physically interacts with Smad2 via its last three BRCT domain-containing regions in an activin signal-dependent fashion.
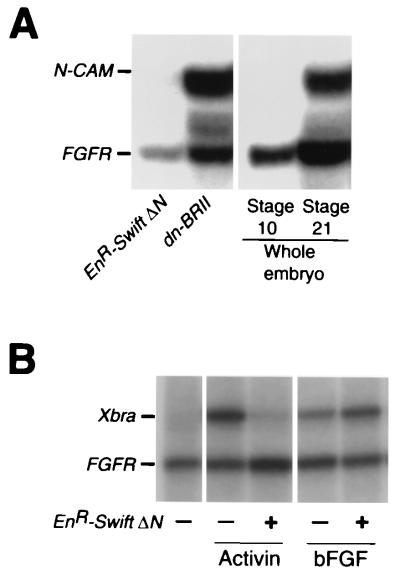
FIG. 7.
(A) Effects of EnR-Swift ΔN on the BMP pathway. EnR-Swift ΔN (2 ng) or dn-BRII (0.5 ng) mRNA was injected into the ventral part of the animal region at the two-cell stage. Animal caps were dissected at stage 8.5; at stage 21, N-CAM gene induction was analyzed by RNase protection assays. (B) Effects of EnR-Swift ΔN on the FGF pathway. EnR-Swift ΔN mRNA (0.6 ng) was injected into the animal pole of the two-cell-stage embryo. At stage 8.5, animal caps were explanted, treated with a low dose of activin (0.2 ng/ml) or bFGF (0.2 μg/ml), and analyzed for expression of Xbra at stage 10.25 by RNase protection assays. bFGF was purchased from R&D Systems Inc.
Swift enhances activin-Smad2-dependent transcription.
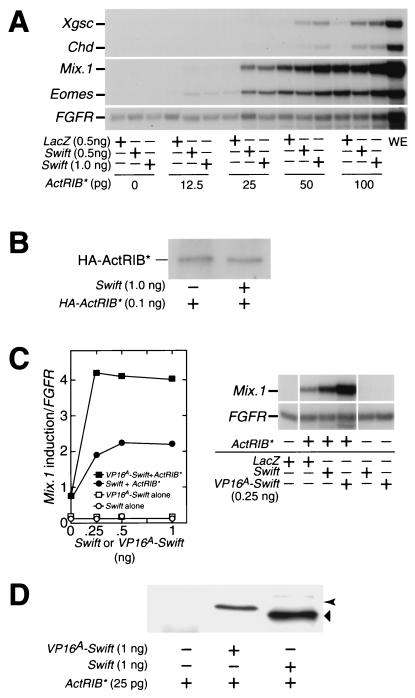
If Swift functionally interacts with Smad2, Swift should have some effect on activin-induced gene transcription in Xenopus blastula cells. Overexpressed Swift alone in animal caps does not activate gene expression (Fig. 4A). We therefore examined the effect of overexpressed Swift on gene expression induced by a constitutively active activin receptor IB (ActRIB*). ActRIB* transduces activin dose-dependent gene responses in the same way as activin (2). We injected the indicated amount of ActRIB* mRNA with or without Swift mRNA in the animal pole of the one-cell-stage embryo, dissected animal caps at stage 8.5, and measured gene expression at stage 10.25 using RNase protection assays. ActRIB* activates Xgsc, Chd, Mix.1, and Eomes gene transcription in a dose-dependent manner (Fig. 4A). The coinjection of Swift mRNA with ActRIB* mRNA results in a synergistic activation of the ActRIB*-induced transcription of all the tested genes (Fig. 4A), including Xbra (data not shown).
FIG. 4.
Swift is an activin-dependent transcriptional cofactor of Smad2. (A) Effects of Swift on activin-induced gene transcription. Each embryo was coinjected with the indicated amount of ActRIB* mRNA with Swift or lacZ mRNA at stage 1, animal caps were dissected at stage 8.5, and then at stage 10.25 gene induction was analyzed by RNase protection assays. lacZ was used as a negative control. (B) Effects of coinjected Swift mRNA on expression levels of ActRIB* protein. HA-ActRIB* mRNA (0.1 ng) was injected with or without Swift mRNA (1 ng) in the animal pole of the one-cell-stage embryo, and animal caps were dissected at stage 8.5. At stage 10.5, an animal cap was homogenized and proteins were resolved by SDS-PAGE and then analyzed by Western blotting using an anti-HA antibody (control from noninjected embryos in panel D). (C) Effects of wild-type Swift and VP16A-Swift on activin-induced gene transcription. Each embryo was injected with the indicated amount of Swift or VP16A-Swift mRNA with or without ActRIB* mRNA at stage 1, animal caps were dissected at stage 8.5, and then at stage 10.25 Mix.1 gene induction was analyzed by RNase protection assays. Left, quantitation of gel analysis; right, gel analyses. (D) Expression levels of Swift and VP16A-Swift proteins. Each embryo was injected with Swift or VP16A-Swift mRNA (1 ng) with ActRIB* mRNA (25 pg) at stage 1. At stage 10.25, an embryo was homogenized; proteins were resolved by SDS-PAGE and then analyzed by Western blotting using an anti-HA antibody. The arrow and arrowhead indicate HA-tagged VP16A-Swift and HA-tagged Swift proteins, respectively.
It is possible that the coinjected Swift mRNA increases the expression level of ActRIB* protein and indirectly enhances transcription. To test this possibility, we injected HA-ActRIB* mRNA with or without Swift mRNA at stage 1 and dissected animal caps at stage 8.5. At stage 10.25, animal cap proteins were resolved by SDS-PAGE and analyzed by Western blotting using an anti-HA antibody. The coinjected Swift mRNA does not have any affect on the expression level of HA- ActRIB* protein (Fig. 4B). HA-ActRIB* had the same effect on mesendoderm gene expression as ActRIB* (data not shown). If the coinjected Swift mRNA affects the endogenous level of ActRIB protein and if the increased levels of endogenous ActRIB protein activate mesendoderm gene expression, overexpressed Swift alone should activate mesendoderm gene transcription. However, overexpressed Swift alone does not activate mesendoderm gene expression (Fig. 4A). These results indicate that the coinjected Swift mRNA exerts its effect directly on transcription but not on the expression levels of ActRIB* or endogenous ActRIB protein.
We also confirmed that this synergy occurred at the level of Smad2. When we coinjected Swift mRNA with Smad2 mRNA instead of ActRIB* mRNA, Swift had a similar, but somewhat weaker, activating effect on Smad2-induced gene transcription (data not shown). It is not surprising that Swift more effectively enhances gene transcription induced by ActRIB* than by Smad2, because overexpressed Smad2 may not be properly phosphorylated, while ActRIB* efficiently phosphorylates and activates endogenous Smad2 (9). Therefore, we used ActRIB* instead of Smad2 in our subsequent experiments. We conclude that Swift is involved in directly stimulating activin-Smad2-induced gene transcription. Furthermore, the fact that Swift has similar effects on all of the response genes which we tested suggests that Swift may be a general component of TGFβ/activin signaling in Xenopus embryos.
Swift is a transcriptional cofactor of Smad2.
We show that Swift interacts with Smad2 in an activin signal-dependent manner (Fig. 3D). Therefore, if Swift does indeed behave as a cofactor of Smad2 at the promoter level, Swift fused to a VP16 transactivation domain from herpes simplex virus (VP16A-Swift) (34) should potentiate an activin response more strongly than wild-type Swift. In contrast, if Swift acts at a level upstream of the promoter, then VP16A-Swift should not be more effective than wild-type Swift. We injected the indicated amount of wild-type Swift or VP16A-Swift mRNA with or without a small amount of ActRIB* mRNA (25 pg) that only weakly induced transcription of Mix.1 at stage 1 and assayed Mix.1 gene expression in animal caps at stage 10.25. Indeed, VP16A-Swift synergistically activates Mix.1 gene transcription with ActRIB* more strongly than wild-type Swift (Fig. 4C). In addition, like wild-type Swift, VP16A-Swift alone does not activate Mix.1 gene transcription (Fig. 4A and C). VP16A-Swift had a similar activating effect on ActRIB*-induced gene transcription of Xgsc, Chd, Eomes, and Xbra (data not shown).
To confirm that the effect of VP16A-Swift on ActRIB*-induced gene transcription is not due to the higher expression levels of VP16A-Swift protein than wild-type Swift protein, we coinjected VP16A-Swift or Swift mRNA with ActRIB* mRNA at stage 1. At stage 10.25, embryos were homogenized and proteins were resolved by SDS-PAGE and then analyzed by Western blotting using an anti-HA antibody. The expression level of VP16A-Swift protein is somewhat less than that of Swift protein (Fig. 4D). This result indicates that the synergistic activation of mesendoderm genes by VP16A-Swift with ActRIB* more strongly than by wild-type Swift is not due to the higher expression levels of VP16A-Swift protein than wild-type Swift. We conclude that Swift is an activin signal-dependent transcriptional cofactor of Smad2.
Swift has intrinsic transactivation activity.
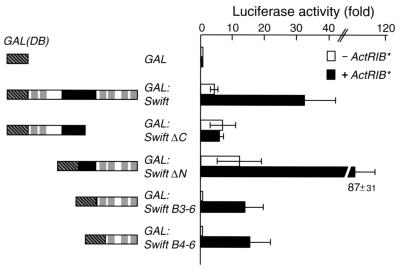
How does Swift synergize with Smad2 at the promoter level? We found that Swift has a glutamine-rich region (Fig. 1); in some transcription factors such as Sp1 and Oct1, glutamine-rich regions mediate transcriptional activation (8). We examined whether Swift has a transactivation activity by fusing full-length Swift to a GAL4 DNA-binding domain. The ability of the fusion, termed GAL:Swift, to activate a GAL4 reporter construct was tested in embryos. We coinjected GAL:Swift mRNA and GAL4-luciferase reporter plasmid DNA with or without ActRIB* mRNA at stage 1 and measured the luciferase activity at stage 10. GAL:Swift has intrinsic transactivation activity that is enhanced by ActRIB* (Fig. 5). We next defined the transactivation domain by testing various regions of Swift in this GAL4 fusion assay. GAL:Swift ΔC, which contains a part of the glutamine-rich region, has intrinsic transactivation activity that is not enhanced by ActRIB*. GAL:Swift ΔN, which contains a part of the glutamine-rich region and four BRCT domains, has intrinsic transactivation activity that is enhanced by ActRIB*. GAL:Swift B3−6, which contains only the last four BRCT domains, does not have any intrinsic transactivation activity, but some transcriptional activity is observed in the presence of ActRIB*. GAL:Swift B4−6 has an effect similar to that of GAL:Swift B3−6. These results indicate that the Swift intrinsic transactivation activity is located between amino acids 567 and 782 in the glutamine-rich region and that its activity is enhanced by activin signaling. In addition, the last three BRCT domains are required for activin signaling-dependent stimulation, suggesting that in the presence of activin signaling, other coactivators may be recruited via Smad2 and the BRCT domains interaction. Therefore, both the glutamine-rich region and the last three BRCT domains of Swift are necessary for the Swift transactivation activity in response to an activin signal.
FIG. 5.
Determination of the Swift intrinsic transactivation domain. Each embryo was coinjected with mRNA encoding various regions of Swift fused to a GAL4 DNA-binding domain and with 5xGAL4-luciferase promoter plasmid DNA with or without ActRIB* mRNA at stage 1, and luciferase activity was measured at stage 10. The values are means ± standard errors of three independent experiments. Closed and open bars show luciferase activities with and without ActRIB* mRNA, respectively.
Swift is involved in TGFβ signaling in embryos.
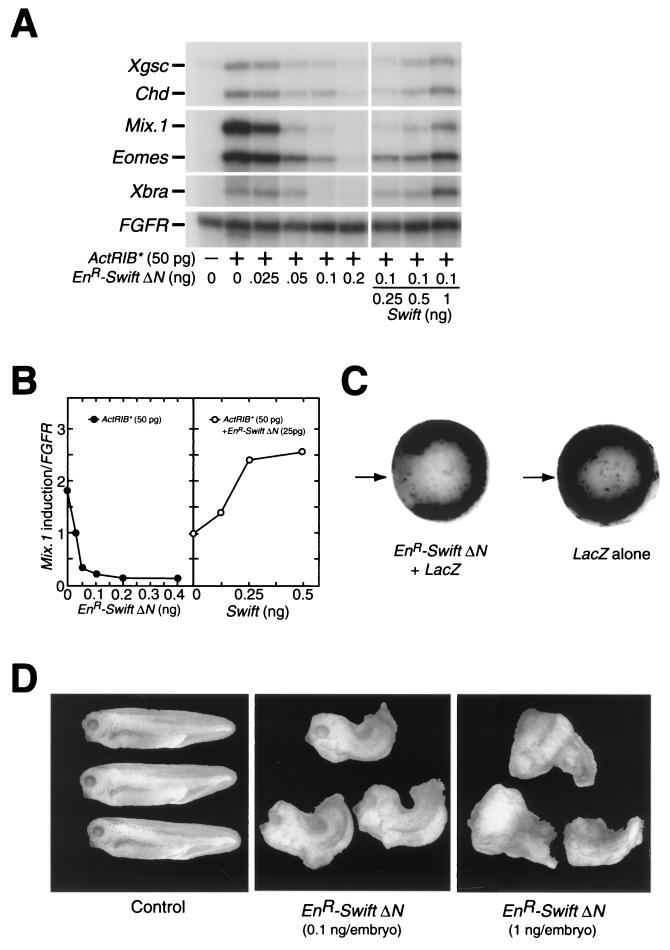
To test if Swift is involved in TGFβ signaling in embryos, we used the Drosophila Engrailed repressor (22) attached to Swift. If Swift is involved in TGFβ signaling in embryos, Swift fused to an Engrailed repressor should suppress TGFβ-induced gene expression. Expression of full-length Swift protein fused to the Engrailed repressor was not detectable in mRNA-injected embryos by Western blotting (data not shown). The transactivation activity of Swift ΔN is similar to that of full-length Swift (Fig. 5), and Swift ΔN also binds to Smad2 (Fig. 3B and C). Therefore, we used Swift ΔN fused to the Engrailed repressor (EnR-Swift ΔN), and expression of this protein was detectable (data not shown). We coinjected EnR-Swift ΔN mRNA with ActRIB* mRNA in the one-cell-stage embryo and assayed gene expression in animal caps at stage 10.25. The coinjection of EnR-Swift ΔN mRNA with ActRIB* mRNA results in the suppression of ActRIB*-induced transcription of Xgsc, Chd, Mix.1, Eomes, and Xbra in a dose-dependent manner (Fig. 6A). To establish whether the suppression of ActRIB*-induced gene transcription is specific for the EnR-Swift ΔN activity, we coinjected wild-type Swift mRNA with EnR-Swift ΔN and ActRIB* mRNAs. Wild-type Swift rescues the suppression of ActRIB*-induced gene transcription by EnR-Swift ΔN in a dose-dependent manner (Fig. 6A). We confirmed that wild-type Swift completely rescues the suppression of ActRIB*-induced transcription of Mix.1 by EnR-Swift ΔN (Fig. 6B). To test if endogenous Mix.1 expression is suppressed by EnR-Swift ΔN, we injected EnR-Swift ΔN mRNA into the equatorial region of a blastomere of the four-cell-stage embryo and assayed expression of endogenous Mix.1 by in situ hybridization at stage 10.25. The injection of EnR-Swift ΔN mRNA results in the complete suppression of endogenous Mix.1 gene expression (Fig. 6C).
FIG. 6.
Swift is involved in TGFβ signaling in embryos. (A) Suppression of activin-induced gene expression by EnR-Swift ΔN. Each embryo was coinjected with various combinations of EnR-Swift ΔN, ActRIB*, and wild-type Swift mRNAs at stage 1; and at stage 10.25, gene induction in animal caps was analyzed by RNase protection assays. (B) Quantitation of RNase protection assays of Mix.1 gene induction. (C) In situ hybridization of the Mix.1 gene. EnR-Swift ΔN (0.4 ng) mRNA was injected into the equatorial region of a blastomere of the four-cell-stage embryo, and expression of Mix.1 was assayed by in situ hybridization. Left, coinjection of EnR-Swift ΔN mRNA (0.4 ng) and lacZ mRNA (0.5 ng); right, injection of lacZ mRNA (0.5 ng) alone. EnR-Swift ΔN (n = 47), 100% Mix.1 suppression; LacZ alone (n = 22), 0%. Arrows indicate sites of injection. (D) Phenotypic effects of EnR-Swift ΔN. A low (0.1 ng/embryo) or high (1 ng/embryo) dose of EnR-Swift ΔN mRNA was injected radially in all blastomeres of the four-cell-stage embryo, and the phenotypes were observed at stage 36. Left, control (no injection); center, EnR-Swift ΔN (0.1 ng) (n = 25, 72% trunk defect); right, EnR-Swift ΔN (1 ng) (n = 40, 100% severe head defect).
Since EnR-Swift ΔN blocks expression of activin-induced mesendoderm genes in animal cap explants, we expect that overexpressed EnR-Swift ΔN would cause loss of mesoderm-derived tissues in embryos and that its phenotype would be same as those observed by blocking activin, Smad2, and FAST functions. To test this possibility, we injected EnR-Swift ΔN mRNA radially in all blastomeres of the four-cell-stage embryo and observed the phenotypes at stage 36. The injection of a low concentration of mRNA (0.1 ng/embryo) results in defective trunk development (Fig. 6D, center). At a high dose of injected mRNA (1 ng/embryo), embryos show a complete loss of axial structure (Fig. 6D, right). These phenotypes are reminiscent of those observed using a dominant negative activin receptor, a dominant negative Smad2, or EnR-FAST in embryos (18, 20, 41), but not those observed using a dominant negative fibroblast growth factor (FGF) receptor (1) or by blocking bone morphogenic protein (BMP) signaling (11, 38). Moreover, this dose-responsive severity of developmental defects is observed in another TGFβ signaling component, FAST. The injection of a low dose of EnR-FAST results in a trunk defect, while high doses lead to both head and trunk defects (41). Taken together, these observations lead us to conclude that Swift is involved in embryonic TGFβ signaling.
Swift functions specifically in TGFβ signaling during early development.
Using in vitro binding assays, we have shown that Swift binds to Smad1 as well as Smad2 (Fig. 3E). Although the phenotypes induced by EnR-Swift ΔN suggest that Swift is interacting with TGFβ signaling but not with BMP or FGF signaling (Fig. 6D), it is important to directly verify that Swift does not function with Smad1 in early embryos. Smad1 mediates signaling of BMP2/4 (other TGFβ family members) in early Xenopus development and regulates epidermal/neural as well as ventral mesoderm gene transcription (14, 43). Inhibition of BMP signals in Xenopus causes neuralization of an epidermal cell (reviewed in reference 42). If Swift interacts functionally with Smad1 in the BMP pathway in the gastrula, overexpressed EnR-Swift ΔN should suppress BMP signaling and induce expression of the N-CAM gene, a pan-neural marker. We injected the ventral side of two-cell-stage embryos with mRNA encoding EnR-Swift ΔN or a dominant negative BMP receptor II (dn-BRII) and dissected animal caps at stage 8.5. Animal caps were cultured until stage 21 and analyzed for expression of N-CAM by RNase protection assays. EnR-Swift ΔN in ventral animal caps does not induce N-CAM gene transcription, while dn-BRII does (Fig. 7A). We conclude that Swift does not function in the BMP pathway in early embryos.
Next we asked if Swift functions in FGF signaling. It is reported that mesoderm induction by activin requires FGF-mediated intracellular signals and that FGF induces some mesoderm genes, including Xbra (26). If Swift functions in FGF signaling as well as TGFβ signaling, overexpression of EnR-Swift ΔN should result in the suppression of FGF-induced Xbra gene expression. To examine this possibility, we injected EnR-Swift ΔN mRNA (0.6 ng) into the animal pole of the two-cell-stage stage embryo and explanted animal caps at stage 8.5. Animal caps were treated with a low concentration of activin (0.2 ng/ml) or basic FGF (bFGF)(0.2 μg/ml) and analyzed for expression of Xbra by RNase protection assays. Activin-induced Xbra gene expression is suppressed by EnR-Swift ΔN, while bFGF-induced Xbra gene transcription is not (Fig. 7B). Taken together, these findings lead us to conclude that Swift functions specifically in the TGFβ pathway but not in the BMP/Smad1 or FGF pathways in early embryos.
Swift is required for early development.
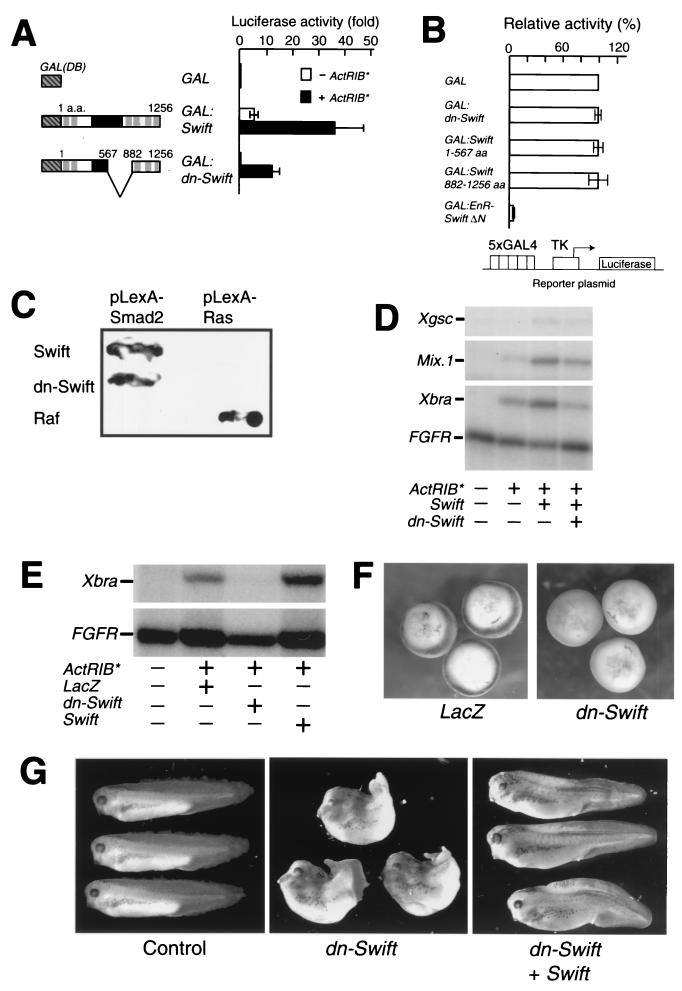
Our finding that Swift is involved in TGFβ signaling in embryos does not necessarily mean that Swift is required for early Xenopus development. To test if Swift is required for early development, we designed a very subtle dominant negative form of Swift (dn-Swift) to interfere with endogenous Swift function. We prepared dn-Swift lacking the transactivation domain located between amino acids 567 and 782. We confirmed using luciferase assays that dn-Swift has neither intrinsic transactivation activity nor transcriptional repression activity (Fig. 8A and B), suggesting that dn-Swift is a weaker dominant negative than EnR-Swift ΔN. To confirm that dn-Swift interacts with Smad2, we transformed pLexA-Smad2 with pACTIIHK-dn-Swift or pACTIIHK-Swift in yeast, where dn-Swift and Swift bind to Smad2 but not to the negative control (Fig. 8C). If dn-Swift functions as a dominant negative, overexpressed dn-Swift should suppress Swift-enhanced mesendoderm gene expression. To test this possibility, we coinjected dn-Swift mRNA with Swift and/or ActRIB* mRNAs at stage 1 and measured mesendoderm gene expression in animal caps at stage 10.25 by RNase protection assays. Swift enhances ActRIB*-induced gene expression of Xgsc, Mix.1, and Xbra (Fig. 8D). dn-Swift suppresses the effects of wild-type Swift (Fig. 8D). dn-Swift had the same effects on Chd and Eomes gene expression (data not shown). We conclude that dn-Swift functions as a dominant negative of wild-type Swift.
FIG. 8.
Swift is required for early development. (A) dn-Swift lacks intrinsic transactivation activity. dn-Swift consists of nucleotides 1 to 1700 and 2649 to 3765. Each embryo was coinjected with mRNA encoding dn-Swift or wild-type Swift fused to a GAL4 DNA-binding domain and 5xGAL4-luciferase reporter plasmid DNA with or without ActRIB* mRNA at stage 1, and luciferase activity was measured at stage 10. The values are means ± standard errors of three independent experiments. Closed and open bars show luciferase activities with and without ActRIB* mRNA, respectively. (B) dn-Swift does not have transcriptional repression activity. Each embryo was coinjected with mRNA encoding a GAL fusion construct and 5xGAL4-TK-luciferase reporter plasmid DNA at stage 1, and luciferase activity was measured at stage 10. The activity of the reporter in the absence of GAL fusion was normalized to a value of 100. The values are means ± standard errors of three independent experiments. (C) Interaction of dn-Swift with Smad2 in yeast. Interaction of dn-Swift with Smad2 using yeast two-hybrid assays was examined by qualitative assays for β-galactosidase activity. (D) dn-Swift functions as a dominant negative. Each embryo was coinjected with various combinations of ActRIB* (25 pg), Swift (0.5 ng), and dn-Swift (8 ng) mRNAs at stage 1. Animal caps were explanted at stage 8.5 and analyzed for expression of mesendoderm genes by RNase protection assays at stage 10.25. (E) dn-Swift suppresses activin-induced Xbra gene transcription. Each embryo was coinjected with lacZ, dn-Swift, or wild-type Swift mRNA (4 ng) with ActRIB* mRNA (50 pg) at stage 1; at stage 10.25, Xbra gene induction in animal caps was analyzed by RNase protection assays. (F) In situ hybridization of Xbra. Embryos were injected radially in all blastomeres of the four-cell-stage embryo with 2 ng of dn-Swift or lacZ mRNA per blastomere. Expression of Xbra gene was assayed by in situ hybridization. dn-Swift (n = 35), 89% Xbra suppression; LacZ (n = 24), 0%. (G) dn-Swift inhibits trunk development. dn-Swift (4 ng/embryo) and/or wild-type Swift (0.2 ng/embryo) mRNAs with lacZ mRNA (0.5 ng/embryo) were injected into dorsal blastomeres of the four-cell-stage embryo, and phenotypes were observed at stage 36. dn-Swift (n = 36), 67% defective trunk development; dn-Swift+Swift (n = 25), 0%. The lineage tracing with β-galactosidase was detected by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining (grey).
If endogenous Swift transactivation activity is required for TGFβ-induced gene expression in embryos, overexpressed dn-Swift should suppress expression of the target genes. To test this possibility, we coinjected dn-Swift mRNA (4 ng) with ActRIB* mRNA (50 pg) at stage 1 and measured mesendoderm gene expression in animal caps. dn-Swift suppresses ActRIB*-induced Xbra gene expression, while wild-type Swift enhances ActRIB*-induced Xbra gene expression (Fig. 8E). dn-Swift did not suppress ActRIB*-induced gene transcription of Xgsc, Chd, Mix.1, or Eomes (data not shown). Because dn-Swift is a more subtle dominant negative than EnR-Swift ΔN and Xbra expression may be very sensitive to Swift function, the effects of dn-Swift might be obvious on Xbra. To test if endogenous Swift is required for endogenous Xbra transcription, we injected dn-Swift mRNA radially at the four-cell stage and characterized Xbra expression by in situ hybridization at stage 10.25. The injection of dn-Swift mRNA into embryos results in down-regulation of Xbra (Fig. 8F). The embryos injected with dn-Swift mRNA into dorsal blastomeres of the four-cell-stage embryo were allowed to develop until tailbud stages and show defective trunk development (Fig. 8G). The same phenotypes were observed when dn-Swift mRNA was injected radially (data not shown). These phenotypes are similar to those observed using a low dose of EnR-Swift ΔN mRNA (0.1 ng/embryo) (Fig. 6D, center), a result consistent with dn-Swift being a weaker dominant negative than EnR-Swift ΔN. The phenotype looks similar to that observed when Xbra function is inhibited (6). The dn-Swift-induced defect of trunk development is rescued by coinjection of wild-type Swift mRNA, indicating that the effects are due to specific inhibition of endogenous Swift. These results indicate that Swift is required at least for embryonic TGFβ-induced Xbra gene expression and normal mesoderm development.
DISCUSSION
In this study we show that Swift is a novel coactivator of Smad2 in Xenopus. Smad2 binds to the Swift C-terminal region via the last three BRCT domains in an activin signaling-dependent manner. The glutamine-rich region of Swift has latent transactivation capacity that is potentiated by activin signaling. We show that Swift enhances activin-induced gene expression at the promoter level by interacting with Smad2. Our data suggest that Swift itself is unlikely to directly bind to DNA in the promoters of responding genes in the absence of an activin signal, because VP16A-Swift alone does not induce gene expression. We favor a model in which, upon TGFβ signaling, phosphorylated Smad2 enters the nucleus and binds to Swift along with DNA-binding proteins to form a TGFβ-responsive transcriptional complex assembled on mesendoderm gene promoters. Thus, Swift exerts its function as a ligand-dependent transcriptional coactivator of the complex.
The specificity of Swift.
We have shown that Swift functions in the TGFβ pathway, but not in the BMP or FGF pathway, during early development (Fig. 7). These results indicate that Swift interacts functionally with Smad2 but not with Smad1, another receptor-regulated Smad, in early embryos. Does Swift functionally interact with other Smads during early Xenopus development? Smad4 is a common Smad and forms a complex with Smad1 as well as with Smad2 (30). Swift is not likely to interact with Smad4, because Swift does not function in the BMP pathway. Xenopus Smad3 has been neither identified nor characterized. Smad3 null mice are viable and survive to adulthood, suggesting that Smad3 is not required during gastrulation (45). Thus, Swift is not likely to function with the putative Xenopus Smad3 during gastrulation. In summary, Swift may interact functionally with Smad2 but not Smad1/3/4 in early embryos. Since we show that Smad1 binds to Swift in vitro (Fig. 3E), it is possible that Swift and Smad1 interact in some other contexts but not in early embryos.
EnR-Swift ΔN and dn-Swift.
In general, EnR-Swift ΔN and dn-Swift have the same effects on development (Fig. 6 and 8), but the effects of EnR-Swift ΔN are stronger, because EnR-Swift ΔN has a strong transcriptional repressor activity whereas dn-Swift does not (Fig. 8B). We show that low levels of EnR-Swift ΔN block trunk development in a way that is indistinguishable from the effects of high levels of dn-Swift (Fig. 6D and 8G), while high levels of EnR-Swift ΔN block both head and trunk development (Fig. 6D). This dose-responsive severity of developmental defects has been observed in another TGFβ signaling component, FAST (41). Because dn-Swift is less potent than EnR-Swift ΔN, we would have to inject very large amounts of dn-Swift mRNA to see a severe head deficiency. However, the injection of such large levels of mRNA results in nonspecific and toxic effects.
Smad coactivators.
p300 and CBP are reported as coactivators of Smad2 and Smad3 in mammalian cells (10, 21, 32). p300 and CBP function to regulate transcription and chromatin structure as general transcriptional activators (35). In Xenopus, p300 and CBP regulate neurogenesis, and inhibition of their activity blocks mesendoderm induction (23), suggesting that like Swift, p300 and CBP function as coactivators of Smads during early Xenopus development. How do both Smad2 coactivators, Swift and p300/CBP, cooperate in TGFβ signaling? We consistently find that Xbra expression and trunk development are very sensitive to Swift function (Fig. 6A and D and 8D to G) and this may be why the effects of EnR-Swift ΔN and dn-Swift are very obvious on Xbra and/or trunk development. This may reflect some aspect of specificity of Swift function between different TGFβ responses. It is possible that Swift functions together or in parallel with p300 and CBP in embryonic TGFβ signaling to efficiently activate gene expression.
The generality of Swift.
Swift may define a new class of BRCT domain proteins that function as transcriptional cofactors in TGFβ signaling. GenBank searches during this study revealed two mammalian Swift homologues, mouse PTIP and human CAGF28 (Fig. 1B). Their C-terminal regions share about 80% amino acid sequence identity with that of Swift and contain BRCT domains. The C-terminal BRCT domains of Swift are required for Smad2 binding (Fig. 3B and C), suggesting that PTIP and CAGF28 may also bind to Smad2 and may regulate TGFβ signaling. Interestingly, mouse PTIP binds to the Pax2 DNA-binding protein (27), suggesting that Swift may interact with other DNA-binding proteins as well as with Smad2. FASTs, the other Smad2-binding proteins, identified first in Xenopus (4) and then in mice and humans (25, 44), were found to have a general role in TGFβ signaling in vertebrate development. Thus, like FAST, Swift may be a general component of TGFβ signaling during vertebrate development.
ACKNOWLEDGMENTS
We thank Yoshinori Takei, Shinya Kuroda, Jon Pines, and Henrietta J. Standley for advice and comments on the manuscript.
This work was supported by the Cancer Research Campaign. K.S. was also supported by the Japan Society for the Promotion of Science. A.M.Z. is a Wellcome Trust fellow.
REFERENCES
- 1.Amaya E, Musci T J, Kirshner M W. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- 2.Armes N A, Smith J C. The ALK-2 and ALK-4 activin receptors transduce distinct mesoderm-inducing signals during early Xenopus development but do not co-operate to establish thresholds. Development. 1997;124:3797–3804. doi: 10.1242/dev.124.19.3797. [DOI] [PubMed] [Google Scholar]
- 3.Callebaut I, Mornon J. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Rubock M J, Whitman M. A transcriptional partner for MAD proteins in TGF-β signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 6.Conlon F L, Sedgwick S G, Weston K M, Smith J C. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development. 1996;122:2427–2435. doi: 10.1242/dev.122.8.2427. [DOI] [PubMed] [Google Scholar]
- 7.Ecochard V, Cayrol S, Rey S, Foulquier F, Caillol D, Lemaire P, Duprat A M. A novel Xenopus mix-like gene milk involved in the control of the endomesodermal fates. Development. 1998;125:2577–2585. doi: 10.1242/dev.125.14.2577. [DOI] [PubMed] [Google Scholar]
- 8.Escher D, Bodmer-Glavas M, Barberis A, Schaffner W. Conservation of glutamine-rich transactivation function between yeast and humans. Mol Cell Biol. 2000;20:2774–2782. doi: 10.1128/mcb.20.8.2774-2782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faure S, Lee M A, Keller T, ten Dijke P, Whitman M. Endogenous patterns of TGFβ superfamily signaling during early Xenopus development. Development. 2000;127:2917–2931. doi: 10.1242/dev.127.13.2917. [DOI] [PubMed] [Google Scholar]
- 10.Feng X, Zhang Y, Wu R, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frisch A, Wright C V E. XBMPRII, a novel Xenopus type II receptor mediating BMP signaling in embryonic tissues. Development. 1998;125:431–442. doi: 10.1242/dev.125.3.431. [DOI] [PubMed] [Google Scholar]
- 12.Fuks F, Burgers W A, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 13.Germain S, Howell M, Esslemont G M, Hill C S. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 2000;14:435–451. [PMC free article] [PubMed] [Google Scholar]
- 14.Graff J M, Bansal A, Melton D A. Xenopus mad proteins transduce distinct subsets of signals for the TGFβ superfamily. Cell. 1996;85:479–487. doi: 10.1016/s0092-8674(00)81249-0. [DOI] [PubMed] [Google Scholar]
- 15.Green J B, New H V, Smith J C. Responses of embryonic Xenopus cells to activin and FGF are separated by multiple dose thresholds and correspond to distinct axes of the mesoderm. Cell. 1992;71:731–739. doi: 10.1016/0092-8674(92)90550-v. [DOI] [PubMed] [Google Scholar]
- 16.Green J B A, Smith J C. Graded changes in dose of a Xenopus activin A homologue elicit stepwise transitions in embryonic cell fate. Nature. 1990;347:391–394. doi: 10.1038/347391a0. [DOI] [PubMed] [Google Scholar]
- 17.Gurdon J B, Harger P, Mitchell A, Lemaire P. Activin signalling and response to a morphogen gradient. Nature. 1994;371:487–492. doi: 10.1038/371487a0. [DOI] [PubMed] [Google Scholar]
- 18.Hemmati-Brivanlou A, Melton D A. A truncated activin receptor inhibits mesoderm induction and formation of axial structures in Xenopus embryos. Nature. 1992;359:609–614. doi: 10.1038/359609a0. [DOI] [PubMed] [Google Scholar]
- 19.Henry G L, Melton D A. Mixer, a homeobox gene required for endoderm development. Science. 1998;281:91–96. doi: 10.1126/science.281.5373.91. [DOI] [PubMed] [Google Scholar]
- 20.Hoodless P A, Tsukazaki T, Nishimatsu S, Attisano L, Wrana J L, Thomsen G H. Dominant-negative Smad2 mutants inhibit activin/Vg1 signaling and disrupt axis formation in Xenopus. Dev Biol. 1999;207:364–379. doi: 10.1006/dbio.1998.9168. [DOI] [PubMed] [Google Scholar]
- 21.Janknecht R, Wells N J, Hunter T. TGF-β-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaynes J, O'Farrell P. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991;10:1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato Y, Shi Y, He X. Neuralization of the Xenopus embryo by inhibition of p300/CREB-binding protein function. J Neurosci. 1999;19:9364–9373. doi: 10.1523/JNEUROSCI.19-21-09364.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koonin E V, Altschul S F, Bork P. BRCA1 protein products…functional motifs… Nat. Genet. 1996;13:266–267. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 25.Labbé E, Silvestri C, Hoodless P A, Wrana J L, Attisano L. Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 26.LaBonne C, Whitman M. Mesoderm induction by activin requires FGF-mediated intracellular signals. Development. 1994;120:463–472. doi: 10.1242/dev.120.2.463. [DOI] [PubMed] [Google Scholar]
- 27.Lechner M S, Levitan I, Dressler G R. PTIP, a novel BRCT domain-containing protein interacts with Pax2 and is associated with active chromatin. Nucleic Acids Res. 2000;28:2741–2751. doi: 10.1093/nar/28.14.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margolis R L, Abraham M R, Gatchell S B, Li S, Kidwai A S, Breschel T S, Stine O C, Callahan C, McInnis M G, Ross C A. cDNAs with long CAG trinucleotide repeats from human brain. Hum Genet. 1997;100:114–122. doi: 10.1007/s004390050476. [DOI] [PubMed] [Google Scholar]
- 29.Massagué J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 30.Massagué J, Wotton D. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDowell N, Zorn A M, Crease D J, Gurdon J B. Activin has a direct long range signalling activity and can form a concentration gradient by diffusion. Curr Biol. 1997;7:671–681. doi: 10.1016/s0960-9822(06)00294-6. [DOI] [PubMed] [Google Scholar]
- 32.Pouponnot C, Jayaraman L, Massagué J. Physical and functional interactions of Smads and p300/CBP. J Biol Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- 33.Ryan K, Garrett N, Mitchell A, Gurdon J B. Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell. 1996;87:989–1000. doi: 10.1016/s0092-8674(00)81794-8. [DOI] [PubMed] [Google Scholar]
- 34.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 35.Shikama N, Lyon J, La Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu K, Gurdon J B. A quantitative analysis of signal transduction from activin receptor to nucleus and its relevance to morphogen gradient interpretation. Proc Natl Acad Sci USA. 1999;96:6791–6796. doi: 10.1073/pnas.96.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu K, Ohtsuka T, Takai Y. Cell-free assay system for Ras-and Rap1-dependent activation of MAP-kinase cascade. Methods Mol Biol. 1998;84:173–183. doi: 10.1385/0-89603-488-7:173. [DOI] [PubMed] [Google Scholar]
- 38.Smith W C, Harland R M. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- 39.Takaishi K, Sasaki T, Kotani H, Nishioka H, Takai Y. Regulation of cell-cell adhesion by rac and rho small G proteins in MDCK cells. J Cell Biol. 1997;139:1047–1059. doi: 10.1083/jcb.139.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe M, Whitman M. FAST-1 is a key maternal effector of mesoderm inducers in the early Xenopus embryo. Development. 1999;126:5621–5634. doi: 10.1242/dev.126.24.5621. [DOI] [PubMed] [Google Scholar]
- 42.Weinstein D C, Hemmati-Brivanlou A. Neural induction in Xenopus laevis; evidence for the default model. Curr Opin Neural. 1997;7:7–12. doi: 10.1016/s0959-4388(97)80114-6. [DOI] [PubMed] [Google Scholar]
- 43.Wilson P A, Lagna G, Suzuki A, Hemmati-Brivanlou A. Concentration-dependent patterning of the Xenopus ectoderm by BMP4 and its signal transducer Smad1. Development. 1997;124:3177–3184. doi: 10.1242/dev.124.16.3177. [DOI] [PubMed] [Google Scholar]
- 44.Zhou S, Zawel L, Lengauer C, Kinzler K W, Vogelstein B. Characterization of human FAST-1, a TGF-β and activin signal transducer. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Y, Richardson J A, Parada L F, Graff J M. Smad3 mutant mice develop metastatic colorectal cancer. Cell. 1998;94:703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]
- 46.Zorn A M, Barish G D, Williams B O, Lavender P, Klymkowsky M W, Varmus H E. Regulation of Wnt signaling by Sox proteins: XSox17α/β and XSox3 physically interact with β-catenin. Mol Cell. 1999;4:487–498. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]