Abstract
Some patients with B-cell-negative severe combined immune deficiency (SCID) carry mutations in RAG-1 or RAG-2 that impair V(D)J recombination. Two recessive RAG-1 mutations responsible for B-cell-negative SCID, R621H and E719K, impair V(D)J recombination without affecting formation of single-site recombination signal sequence complexes, specific DNA contacts, or perturbation of DNA structure at the heptamer-coding junction. The E719K mutation impairs DNA cleavage by the RAG complex, with a greater effect on nicking than on transesterification; a conservative glutamine substitution exhibits a similar effect. When cysteine is substituted for E719, RAG-1 activity is enhanced in Mn2+ but remains impaired in Mg2+, suggesting an interaction between this residue and an essential metal ion. The R621H mutation partially impairs nicking, with little effect on transesterification. The residual nicking activity of the R621H mutant is reduced at least 10-fold upon a change from pH 7.0 to pH 8.4. Site-specific nicking is severely impaired by an alanine substitution at R621 but is spared by substitution with lysine. These observations are consistent with involvement of a positively charged residue at position 621 in the nicking step of the RAG-mediated cleavage reaction. Our data provide a mechanistic explanation for one form of hereditary SCID. Moreover, while RAG-1 is directly involved in catalysis of both nicking and transesterification, our observations indicate that these two steps have distinct catalytic requirements.
The central event in the generation of immunological diversity is the assembly of T-cell receptor and immunoglobulin genes from discrete DNA segments, V, D, and J, during lymphocyte development. This process, termed V(D)J recombination, is the only known form of site-specific DNA rearrangement in vertebrates. V(D)J recombination is mediated by heptamer and nonamer signal sequences, which are separated by spacer regions of 12 or 23 bp (28), and is initiated by the recombination-activating proteins RAG-1 and RAG-2 (33, 39), which act in concert to cleave DNA at the junctions between antigen receptor coding segments and conserved recombination signals that specify sites of recombination (31, 48).
RAG-mediated DNA cleavage occurs in two steps (31). First, one DNA strand is nicked between the recombination signal sequence (RSS) heptamer and the coding sequence. This is followed by a transesterification reaction in which the free hydroxyl group at the 3′ end of the coding sequence attacks a phosphodiester on the opposite strand (49, 50). As a result, two DNA ends are produced: a signal end, terminating in a blunt, 5′-phosphorylated, double-strand break, and a coding end, terminating in a hairpin.
RAG-1 and RAG-2 are both essential for initiation of V(D)J recombination. RAG-2 alone has no detectable DNA binding activity in vitro but collaborates with RAG-1 in RSS recognition (2, 11, 20, 29, 44, 45). In the absence of RAG-2, RAG-1 interacts weakly with the RSS through the nonamer element (11, 20, 42, 45). In contrast, assembly of a complex containing the RSS, RAG-1, and RAG-2 is strongly heptamer dependent (2, 11, 20, 45) and is accompanied by kinking or unwinding of substrate DNA near the scissile bond at the heptamer-coding boundary (45). Chemical interference and footprinting have shown that in the presence of RAG-1 alone, DNA contacts are restricted to the vicinity of the nonamer (45). Occupancy of the heptamer requires RAG-2 (45), which induces RAG-1 to approach the scissile bond (44). The proximity of RAG-1 to the site of DNA cleavage in the presence of RAG-2 has suggested a direct role for RAG-1 in DNA cleavage. The relative roles of RAG-1 and RAG-2 in catalysis of nicking and transesterification, however, remain incompletely understood.
The chemistry of DNA cleavage by the RAG proteins is formally equivalent to that employed by classical transposases such as Mu A (37, 49, 50). Several additional lines of evidence indicate that V(D)J recombination represents a specialized form of DNA transposition, including the formal equivalence of hybrid joint formation to the retroviral disintegration reaction and the ability of the RAG proteins to integrate signal ends into nonhomologous DNA (1, 21). The similarity between V(D)J rearrangement and other types of transposition suggests that the RAG catalytic core may resemble that of other transposases. The transposases of retroviruses, retrotransposons, IS3 elements, and Mu, as well as the TnsB transposase component of Tn7, all contain an array of acidic amino acids, the DDE motif (7, 35), which binds an essential divalent cation such as Mg2+ at or near the active sites for DNA cleavage and strand transfer (3, 5, 12, 24, 36, 38). Direct participation of the DDE motif residues in metal binding has been demonstrated for the human immunodeficiency virus and avian sarcoma virus integrases by crystallographic analysis (5, 12) and for TnsB of Tn7 by the observation that cysteine substitution results in altered metal specificity (38). Although the RAG proteins lack significant homology to the retroviral integrase superfamily, precluding identification of a putative active center by sequence alignment, recent studies employing iron-induced hydroxyl radical cleavage and site-directed mutagenesis have identified three acidic residues within RAG-1, D600, D708, and E962, that are essential for nicking and transesterification activity (14, 23, 27). Substitution of cysteine at two of these residues, D600 and D708, was associated with metal ion-specific recovery of DNA cleavage activity, implicating these amino acids in the binding of an essential divalent cation.
About one-third of all patients with severe combined immunodeficiency (SCID) lack detectable B cells. A number of B-cell-negative SCID patients have been shown to carry debilitating mutations in RAG-1, RAG-2, or both, resulting in impairment of V(D)J recombination (40). In this communication we have examined the biochemical properties of SCID-associated RAG-1 alleles, with the goal of defining the precise molecular defects arising from these mutations.
Two RAG-1 mutations associated with B-cell-negative SCID, R621H and E719K, impair V(D)J recombination in vivo without affecting formation of single-site RSS complexes in vitro. The E719K mutation impaired both the nicking and transesterification steps of DNA cleavage. In contrast, the R621H mutation partially impaired nicking, with little or no effect on transesterification. Further analysis suggested that RAG-mediated strand scission requires a positively charged residue at position 621 of RAG-1. These data provide a mechanistic explanation for one form of hereditary SCID and indicate that RAG-dependent nicking and transesterification are likely to have distinct catalytic requirements.
MATERIALS AND METHODS
DNA constructs and site-directed mutagenesis.
Maltose-binding protein (MBP) fusions containing RAG-1 (residues 384 to 1008) or RAG-2 (residues 1 to 387) cores, tagged at the carboxyl terminus with a myc epitope and polyhistidine, have been described previously (31, 48). DNA fragments encoding these proteins (gifts of Dik van Gent and Martin Gellert) were cloned between the BamHI and NotI sites of pcDNA-1 (Invitrogen) to create pcDNAR1 and pcDNAR2 (45).
RAG-1 cDNA, cloned into pBluescript SK(II), was the starting material for mutagenesis. Mutations R621H, E719K, and Y935Stop were introduced by standard PCR; E719C was introduced using divergent PCR (30). Products were confirmed by nucleotide sequencing. For expression in mammalian cells, DNA cassettes spanning the mutations were exchanged for the corresponding wild-type fragment of pcDNAR1.
Cell culture, transfection, and protein purification.
The 293 cell line was maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. RAG expression constructs were cotransfected with pRSV-T into 293 cells by the calcium phosphate method.
For purification of RAG fusion proteins, pcDNAR1 or a corresponding mutant construct was cotransfected with pcDNAR2 into 293 cells (10 μg of each plasmid per 10-cm-diameter culture plate). At 48 h after transfection, RAG-1 and RAG-2 fusion proteins were copurified by amylose affinity chromatography as described elsewhere (29, 45).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were carried out as described previously (45). Binding reaction mixtures (10 μl) contained 0.02 pmol of 32P-labeled substrate DNA, 100 nM nonspecific duplex oligonucleotide DAR81/82 (20), 20 ng of RAG-1, and 15 ng of RAG-2 in 60 mM potassium acetate, 60 mM KCl, 25 mM morpholinepropanesulfonic acid-KOH (pH 7.0), 10 mM Tris Cl (pH 7.6), 1 mM CaCl2, 0.8 mM dithiothreitol, 100 μg of bovine serum albumin/ml, 20% dimethyl sulfoxide, and 4% glycerol. Except where indicated, reaction mixtures were incubated at 37°C for 20 min and chilled on ice for 5 min, after which time 4 μl of ice-cold 25% glycerol and 0.01% bromophenol blue was added. Samples were fractionated by 4% polyacrylamide gel electrophoresis in 0.5× Tris-borate-EDTA (TBE). Gels and running buffer were equilibrated to 4°C before loading, and electrophoresis was carried out at 4°C. 32P was detected using a phosphorimager.
DNA cleavage assays.
Except where indicated, DNA cleavage reactions were performed in Mg2+ as described previously (19), using substrates DAR39/DAR40 (containing a 12-spacer RSS), DAR61/DAR62 (containing a 23-spacer RSS), or both, as indicated in Results. Reaction mixtures (10 μl) contained 0.02 pmol of radiolabeled substrate, 20 ng of RAG-1, and 15 ng of RAG-2. In assays involving the sulfur-substituted RAG-1 mutant E719C, reactions were conducted in 6 mM Mg2+ or 1 mM Mn2+. The prenicked substrate was constructed by annealing oligonucleotides SD2049 (5′-GATCTGGCCTGTCTTA-3′), SD2050 (5′-CACAGTGCTACAGACTGGAACAAAAACCCTGCAG-3′), and SD2082 (5′-CTGCAGGGTTTTTGTTCCAGTCTGTAGCACTGTGTAAGACAGGC CAGATC-3′). SD2050 was phosphorylated by T4 polynucleotide kinase at its 5′ end before annealing to simulate a physiologic nicking product. HMG-1 protein (8 μg/ml, final concentration) was added to some cleavage reaction mixtures as indicated in Results. Reaction products were detected with a phosphorimager and quantitated using ImageQuant software.
Assays for V(D)J recombination and DNA cleavage in vivo.
The wild-type RAG-1 expression construct pcDNAR1 (5 μg) or corresponding mutant plasmids were contransfected into 293 cells with 10 μg of the recombination substrate pJH200 (17) and 5 μg of pcRAG-2. Rearrangement of the recombination substrate was assayed as described elsewhere (17). Signal ends at the 12-spacer RSS were assayed by ligation-mediated PCR using the oligonucleotide linker FM25/11 (48); amplification was performed with primers 1233 and FM25 as described previously (48). To control for recovery of the recombination substrate, primers 1233 and DR1 (48) were used to amplify a portion of the pJH200 backbone.
Modification interference assays.
Modification interference assays were performed as described previously (45). In brief, a 12-spacer substrate was formed by annealing oligonucleotides SD2609 (5′-CGTGATCTGGCCTGTCT TACACAGTGCTACAGACTGGAACAAAAACCCTGCAGTGTAACGTAG) and SD2610 (5′-CTACGTTACACTGCAGGGTTTTTGTTCCAGTCTGTAGCACTGTGTAAGACAGGCCAGATCACG); heptamer and nonamer sequences are underlined. 5′-32P-labeled oligonucleotides were modified with dimethyl sulfate (DMS) or KMnO4 (41, 46), annealed to the unlabeled complementary strand, and purified (29). Preparative EMSA was carried out as described above. DNA was electrophoretically transferred to DEAE cellulose paper (DE81; Whatman) in 0.5× TBE at 20 V and 4°C for 18 h. The paper was washed in 50 mM NaCl, 10 mM Tris (pH 7.6), and 1 mM EDTA for 5 min at 25°C, and DNA was eluted in 1 M NaCl, 10 mM Tris (pH 7.6), and 1 mM EDTA at 65°C for 30 min. The eluate was filtered through 0.22-μm-pore-size cellulose acetate (Spin-X; Costar), and DNA was recovered by ethanol precipitation. DNA was cleaved at modified positions in 10% piperidine. Cleavage products were fractionated by denaturing gel electrophoresis and detected with a phosphorimager.
RESULTS
SCID-associated RAG-1 mutations R621H and E719K impair accumulation of free signal ends in vivo.
It has previously been shown by photo-cross-linking that RAG-1 is positioned near the scissile bond in a precleavage complex containing RAG-2 and an RSS (13, 32, 44), suggesting that RAG-1 might participate directly in DNA cleavage. This was borne out by evidence that residues D600, D708, and E962 of RAG-1 play an intimate role in catalysis of DNA cleavage (14, 23, 27). It has been estimated that about 10% of patients with SCID carry mutations in RAG-1 or RAG-2 that impair V(D)J recombination (40). The SCID-associated RAG-1 point mutations R624H and E722K profoundly impair both signal joint formation and coding joint formation in an extrachromosomal V(D)J recombination assay (40). Both mutations occur at residues that are phylogenetically invariant. Defective recombination does not result from mislocalization of mutant proteins or defects in protein accumulation (40).
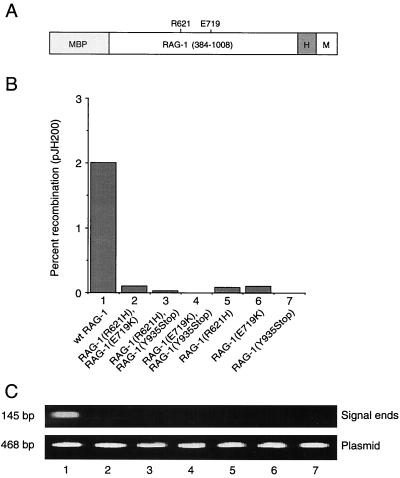
To determine whether the R624H and E722K mutations exert their effects before or after DNA cleavage, the corresponding amino acid substitutions, R621H and E719K, were introduced into a mouse RAG-1 core (amino acids 384 to 1008), fused at the amino terminus to MBP and at the carboxyl terminus to polyhistidine and a c-myc epitope (Fig. 1A) (31, 45, 48). A similar fusion protein representing a SCID-associated RAG-1 truncation at residue Y935 (corresponding to human RAG-1 residue Y938 [40]) was also constructed. The extrachromosomal V(D)J recombination substrate pJH200 was cotransfected into 293 cells with expression plasmids encoding wild-type or mutant RAG-1 and wild-type RAG-2 core (residues 1 to 387) fusion proteins. Recombination frequency was scored at 48 h (17). Plasmid DNA was examined in parallel by ligation-mediated PCR for the presence of signal breaks at the 12-spacer RSS (48). As expected, each of the RAG-1 mutations impaired recombination in vivo (Fig. 1B, lanes 5 to 7). Moreover, recombination was not rescued by coexpression of any two mutant alleles of RAG-1 (Fig. 1B, lanes 2 to 4), indicating a lack of interallelic complementation. Although similar amounts of pJH200 plasmid DNA were recovered from all transfections, only wild-type RAG-1 supported detectable production of free signal ends (Fig. 1C, compare lane 1 to lanes 2 through 7). While RAG-1(Y935Stop) was poorly soluble (data not shown), RAG-1(R621H) and RAG-1(E719K) were expressed in soluble form at levels equivalent to that of wild-type RAG-1 (Fig. 2 and data not shown). From these results we infer that the R621H and E719K mutations act at or prior to DNA cleavage.
FIG. 1.
Impairment of V(D)J recombination and DNA cleavage in vivo by RAG-1 mutations associated with B-cell-negative SCID. (A) Diagram of the RAG-1 fusion protein. The RAG-1 core (amino acids 384 to 1008), MBP, polyhistidine tag (H), and c-Myc epitope (M) are indicated. Positions of SCID-associated point mutations are indicated. (B) Wild-type or mutant RAG-1 fusion protein was coexpressed with the RAG-2 core in 293 cells and signal joint formation was quantitated by using the extrachromosomal V(D)J recombination substrate pJH200. Percent recombination was calculated as described by Hesse et al. (17); values represent the means of at least three independent experiments. Lanes: 1, wild-type RAG-1; 2, RAG-1(R621H) and RAG-1(E719K); 3, RAG-1(R621H) and RAG-1(Y935Stop); 4, RAG-1(E719K) and RAG-1(Y935Stop); 5, RAG-1(R621H); 6, RAG-1(E719K); 7, RAG-1(Y935Stop). All assays included wild-type RAG-2 core. (C) Signal ends (upper panel) were assayed by ligation-mediated PCR. Products were detected by staining with ethidium bromide. Total pJH200 was assayed by PCR amplification of a backbone sequence (lower panel), as described in Materials and Methods. Lanes are numbered as for panel B.
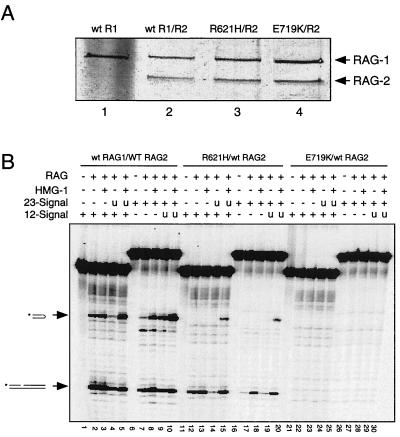
FIG. 2.
Impairment of RSS cleavage in vitro by SCID-associated RAG-1 mutations. (A) Wild-type or mutant RAG-1 core fusion proteins, diagrammed in Fig. 1A, were coexpressed with an MBP-tagged RAG-2 core in 293 cells and purified by affinity chromatography as described elsewhere (29, 45). Equal volumes of purified protein (25 μl) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by silver staining. Lane 1, wild-type RAG-1; lane 2, wild-type RAG-1 and RAG-2; lane 3, RAG-1(R621H) and RAG-2; lane 4, RAG-1(E719K) and RAG-2. Positions of RAG-1 and RAG-2 fusion proteins are indicated by arrows. (B) In vitro cleavage assay. Cleavage of 32P-labeled 12-spacer (lanes 1 to 5, 11 to 15, and 21 to 25) or 32P-labeled 23-spacer (lanes 6 to 10, 16 to 20, and 26 to 30) substrate (0.02 pmol in a 10-μl reaction volume) was assayed in the presence of Mg2+ as described elsewhere (19). Lanes 2 to 5 and 7 to 10, wild type RAG-1 and RAG-2; lanes 12 to 15 and 17 to 20, RAG-1(R621H) and RAG-2; lanes 22 to 25 and 27 to 30, RAG-1(E719K) and RAG-2; lanes 1, 6, 11, 16, 21, and 26, and RAG added. Lanes 4, 5, 14, 15, 24, and 25, addition of equimolar unlabeled 23-spacer substrate (u). Lanes 3, 5, 8, 10, 13, 15, 18, 20, 23, 25, 28, and 30, addition of HMG-1 to 8μg/ml. Positions of nicked and hairpin products are indicated by arrows at left.
R621H and E719K mutations impair substrate DNA cleavage in vitro.
To examine the effects of the R621H and E719K mutations on RAG-1 activity in vitro, the chimeric wild-type and mutant RAG-1 proteins were coexpressed with a chimeric RAG-2 core (amino acids 1 to 387 [31, 45, 48]) in 293 cells and purified by amylose affinity chromatography as described previously (45). All three RAG-1 proteins were obtained in similar yield and in similar proportion to the RAG-2 fusion protein (Fig. 2A).
Purified RAG proteins were assayed in the presence of Mg2+, which favors pairwise cleavage of 12-spacer and 23-spacer substrates in vitro. As expected, wild-type RAG-1, in combination with RAG-2, supported nicking and transesterification of single 12-spacer and 23-spacer substrates (Fig. 2B, lanes 2, 3, 7, and 8). Paired cleavage of 12-spacer and 23-spacer substrates was also observed, as evidenced by a significant increase in the yield of hairpin product in the presence of both substrates and HMG-1 protein (Fig. 2B, lanes 5 and 10). Consistent with published results, the efficiency of hairpin formation from a radiolabeled 12-spacer substrate was reduced when unlabeled 23-spacer substrate was present in the absence of HMG-1 (Fig. 2B, lane 4), possibly by unproductive, competitive binding of RAG proteins. As expected, HMG-1 stimulated cleavage of a radiolabeled 23-spacer substrate, even in the absence of unlabeled 12-spacer oligonucleotide (Fig. 2B, lane 8). DNA cleavage in Mg2+ was most efficient, however, when 12- and 23-spacer substrates were combined in the presence of HMG-1 (Fig. 2B, lanes 5 and 10), consistent with previous observations (47).
While DNA cleavage was impaired by both SCID-associated point mutations, a more severe defect was seen with the E719K mutant (Fig. 2B, lanes 11 to 30; Fig. 3). RAG-1(R621H) supported some nicking of 12- and 23-spacer substrates in the absence or presence of HMG-1 (Fig. 2B, lanes 12, 13, 17, and 18). Hairpin product was detected in paired cleavage reactions with RAG-1(R621H) in the presence of HMG-1 (Fig. 2B, lanes 15 and 20), reflecting the stimulatory effect of paired RSS signals in reaction mixtures containing Mg2+. In contrast to RAG-1(R621H), RAG-1(E719K) exhibited impairment of DNA cleavage under all conditions tested (Fig. 2B, lanes 21 to 30).
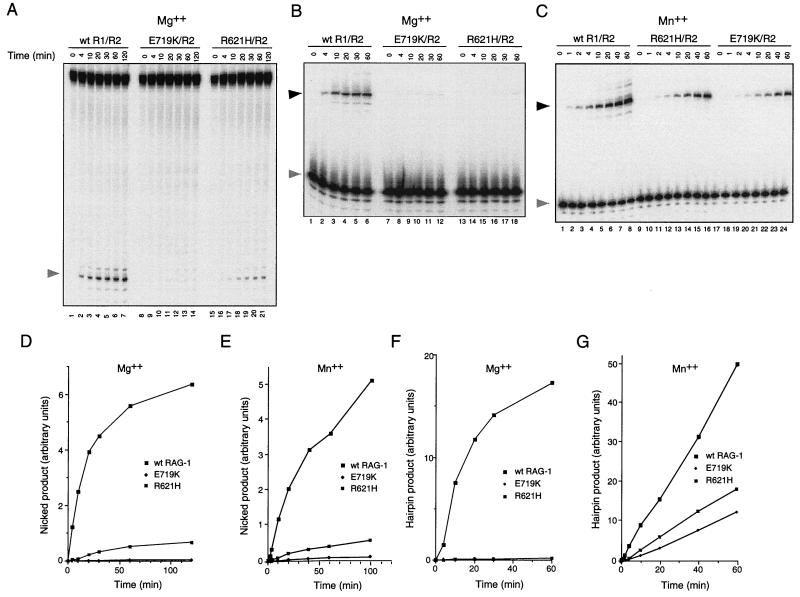
FIG. 3.
Effects of the R621H and E719K mutations on nicking and transesterification. (A) Kinetic analysis of RSS nicking by RAG-1(R621H) and RAG-1(E719K) in Mg2+. Lanes 1 to 7, wild-type RAG-1 and RAG-2; lanes 8 to 14, RAG-1(E719K) and RAG-2; lanes 15 to 21, RAG-1(R621H) and RAG-2. Assays were carried out in Mg2+ as described in Materials and Methods by using the mutant 12-spacer substrate C17A, which undergoes nicking in the absence of transesterification (29). Samples were withdrawn at times indicated. The position of nicked product is indicated by the shaded arrow at left. (B) Kinetic analysis of hairpin formation by RAG-1(R621H) and RAG-1(E719K) in Mg2+. Lanes 1 to 6, wild-type RAG-1 and RAG-2; lanes 7 to 12, RAG-2(E719K) and RAG-1; lanes 13 to 18, RAG-1(R621H) and RAG-2. Assays were carried out in Mg2+ using a prenicked substrate as described in Materials and Methods. Samples were withdrawn at times indicated. The positions of prenicked substrate (shaded arrow) and hairpin product (filled arrow) are indicated at left. (C) Kinetic analysis of hairpin formation by RAG-1(R621H) and RAG-1(E719K) in Mn2+. Lanes 1 to 8, wild-type RAG-1 and RAG-2; lanes 9 to 16, RAG-1(R621H) and RAG-2; lanes 17 to 24, RAG-1(E719K) and RAG-2. Assays were carried out in Mn2+ using a prenicked substrate as described in Materials and Methods. Samples were withdrawn at times indicated. The positions of prenicked substrate (shaded arrow) and hairpin product (filled arrow) are indicated at left. (D and F) The yield of nicked products in panel A and hairpin products in panel B was quantitated by phosphorimager and plotted as a function of time. Filled squares, wild-type RAG-1; filled diamonds, RAG-1(E719K); dotted squares, RAG-1(R621H). (E and G) The kinetics of nicking (E) or hairpin formation (G) by RAG-1(R621H) and RAG-1(E719K) were assayed for panels D and F except that reactions were performed in the presence of Mn2+. Products were quantitated by phosphorimager and plotted as a function of time. Symbols are as defined for panel D.
Tests of nicking and transesterification by RAG-1 R621H and E719K mutants.
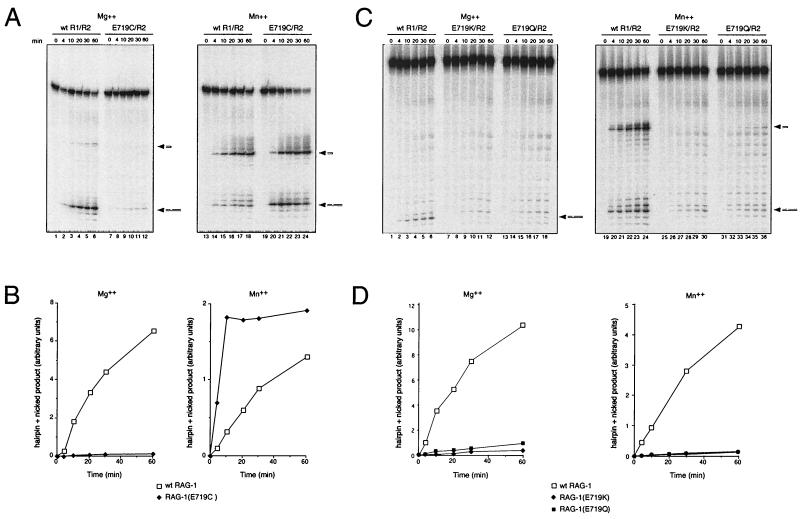
We next quantified the effects of the R621H and E719K mutations on the kinetics of nicking in Mg2+ by using a mutant 12-spacer substrate (C17A) in which the cytosine residue in the first heptamer position had been mutated to adenosine. Because the C17A substrate undergoes nicking without subsequent transesterification (29), accumulation of nicked products is a direct indicator of the kinetics of nicking. Substrate was incubated with RAG-2 and wild-type RAG-1 (Fig. 3A, lanes 1 to 7), RAG-1(E719K) (Fig. 3A, lanes 8 to 14), or RAG-1(R621H) (Fig. 3A, lanes 15 to 21) in the presence of Mg2+ under standard reaction conditions (see Materials and Methods), and nicked products were assayed at various times up to 2 h. The initial reaction rate, relative to that of the wild type, was decreased more than 20-fold by the R621H mutation and more than 450-fold by the E719K mutation (Fig. 3D). Purified, chimeric mutant proteins were also assayed in the presence of RAG-2 and Mg2+ for the ability to convert a prenicked, 12-spacer substrate to a hairpin product. Transesterification under these conditions was severely impaired by both mutations (Fig. 3B and F).
The same mutants were assayed for the ability to support nicking and hairpin formation in Mn2+, because under these conditions transesterification of a single-site RSS substrate in vitro is more efficient than in the presence of Mg2+. As assayed with the C17A substrate, the initial rate of nicking in Mn2+ was reduced about 10-fold by the R621H mutation and more than 40-fold by the E719K mutation (Fig. 3E). Transesterification was less severely impaired in Mn2+ than in Mg2+, with decreases in initial reaction rate of about four- to five fold for RAG-1(R621H) and seven- to nine fold for RAG-1(E719K) (Fig. 3C and G). Taken together, these results are consistent with participation of residues E719 and R621 in DNA cleavage by the V(D)J recombinase, preferentially at the nicking step.
Wild-type RAG-1, RAG-1(R621H), and RAG-1(E719K) show similar patterns of binding to the RSS.
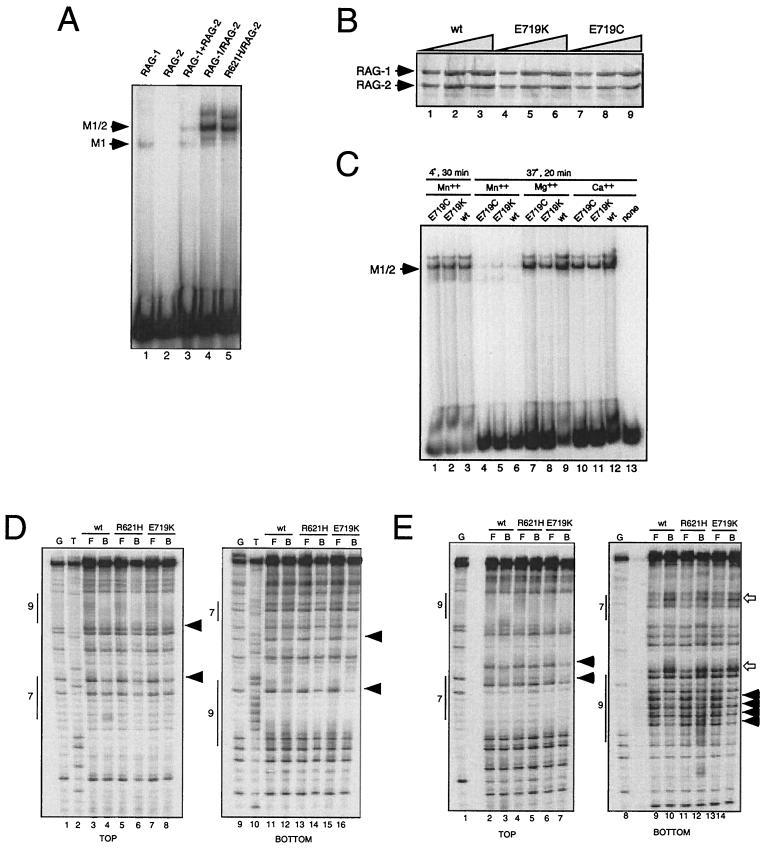
The deleterious effects of the R621H and E719K mutations on formation of free signal ends in vivo and DNA cleavage in vitro could result from defects in catalysis or at some prior step, such as RAG-2 association or DNA binding. Wild-type or mutant RAG-1 fusion proteins were tested for binding to radiolabeled 12-spacer substrates with or without wild-type RAG-2 in calcium, as described previously (45). Under these conditions RAG-1, RAG-2, and substrate DNA form a stable preinitiation complex, termed M1/2, composed of a RAG-1 dimer, monomeric RAG-2, and DNA (45). M1/2 represents a functional complex as assessed by its ability to support nicking and transesterification (20), as well as by the coordinate impairment of M1/2 formation, in vitro DNA cleavage, and recombination activity by a subset of RAG-1 mutations (45). Binding reaction mixtures containing separately coexpressed RAG proteins were analyzed in parallel with reaction mixtures containing coexpressed RAG proteins (Fig. 4A). In the absence of RAG-2, wild-type RAG-1 forms a mobility shift complex termed M1 (45) (Fig. 4A, lane 1). RAG-2 alone exhibits undetectable binding activity (Fig. 4A, lane 2). When RAG-2 is combined with wild-type RAG-1, the slower migrating M1/2 complex is observed in addition to the M1 species (45) (Fig. 4A, lane 3).
FIG. 4.
(A to C) RAG-1(R621H), RAG-1(E719K), and RAG-1(E719C) retain the ability to form RAG-2-dependent RSS complexes. Binding to a 32P-labeled 12-spacer substrate and EMSAs were carried out as described in Materials and Methods. (A) EMSAs of wild-type RAG-1 and RAG-1(R621H). Lane 1, wild-type RAG-1 alone; lane 2, RAG-2 alone; lane 3, wild-type RAG-1 and RAG-2, expressed separately and combined; lane 4; wild-type RAG-1 and RAG-2, coexpressed and copurified; lane 5, RAG-1(R621H) and RAG-2, coexpressed and copurified. Positions of the M1 and M1/2 complexes are indicated by arrows at left. (B) Wild-type or mutant MBP–RAG-1 fusion proteins, coexpressed and copurified with MBP–RAG-2, were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by silver staining. Increasing amounts of purified complexes containing wild-type RAG-1 (lanes 1 to 3), RAG-1(E719K) (lanes 4 to 6), and RAG-1(E719C) (lanes 7 to 9) were loaded. Positions of RAG-1 and RAG-2 are indicated at left. (C) EMSAs of wild-type RAG-1 (lanes 3, 6, 9, and 12), RAG-1(E719C) (lanes 1, 4, 7, and 10), and RAG-1(E719K) (lanes 2, 5, 8, and 11) or in the absence of added protein (lane 13). Equal amounts of protein, copurified with RAG-2 and quantitated for panel B, were added. Reactions were carried out in the presence of Mn2+ (lanes 1 to 6), Mg2+ (lanes 7 to 9), or Ca2+ (lanes 10 to 13) at 4°C for 30 min (lanes 1 to 3) or at 37°C for 20 min (lanes 4 to 13). (D and E) RAG-1(R621H) and RAG-1(E719K) yield modification interference patterns identical to that of wild-type RAG-1. (D) DMS modification interference. Cleavage products from free or bound DNA, radiolabeled on the top strand (left panel) or bottom strand (right panel), were fractionated by gel electrophoresis and detected with a phosphorimager. The RSS heptamer (7) and nonamer (9) are indicated by vertical bars. The arrowheads mark positions of strongest interference. Lanes 1 and 9, G-specific sequencing tracts; lanes 2 and 10, T-specific sequencing tracts; lanes 3, 5, 7, 11, 13, and 15, products from free DNA; lanes 4, 6, 8, 12, 14, and 16, products from bound fractions. Lanes 3, 4, 11, and 12, wild-type RAG-1 and RAG-2; lanes 5, 6, 13, and 14, RAG-1(R621H) and RAG-2; lanes 7, 8, 15, and 16, RAG-1(E719K) and RAG-2. (E) KMnO4 modification interference. Cleavage products from free or bound DNA, radiolabeled on the top strand (left panel) or bottom strand (right panel), were fractionated by gel electrophoresis and detected by phosphorimager analysis. The heptamer (7) and nonamer (9) are indicated by vertical bars. Closed and open arrowheads mark positions at which modification is underrepresented or overrepresented, respectively, in the bound fraction. Lanes 1 and 8, G-specific sequencing tracts; lanes 2, 4, 6, 9, 11, and 13, products from free DNA; lanes 3, 5, 7, 10, 12, and 14, products from bound fractions. Lanes 2, 3, 9, and 10, wild-type RAG-1 and RAG-2; lanes 4, 5, 11, and 12, RAG-1(R621H) and RAG-2; lanes 6, 7, 13, and 14, RAG-1(E719K) and RAG-2.
As expected from previous observations (20, 45), the yield of the M1/2 complex was enhanced when wild-type RAG-1 and RAG-2 were coexpressed and copurified (Fig. 4A, lane 4). RAG-1(R621H) (Fig. 4A, lane 5) and RAG-1(E719K) (Fig. 4C, lane 11) also supported formation of M1/2 complexes, suggesting that these mutants retain the ability to dimerize, associate with RAG-2, and bind substrate. Consistent with previous observations (19), a complex of slower mobility than M1/2 was observed in reaction mixtures containing wild-type RAG-1 (Fig. 4A, lane 4, and Fig. 4C, lane 12), the R621H mutant (Fig. 4A, lane 5), or the E719K mutant (Fig. 4C, lane 11).
To compare RSS binding by wild-type and mutant RAG-1 proteins, we assessed the effects of guanine- and thymine-specific modification on formation of the M1/2 complex by binding interference (41, 46), reasoning that differences in the interactions between protein and DNA would be revealed by this approach. Top- or bottom-strand oligonucleotides were treated with DMS (Fig. 4D) or potassium permanganate (Fig. 4E), which react with G or T residues to produce N7 methylguanine or a C5, C6 glycol of thymine, respectively. N7-methylation of guanine affects major groove contacts (41). Glycolization displaces the modified thymine (26), potentially disrupting major and minor groove contacts. After chemical modification and annealing, M1/2 complexes were formed in the presence of Ca2+. Bound and free DNA fragments were separated by gel electrophoresis and cleaved by piperidine. Underrepresented cleavages in the bound fraction indicated positions at which modification interfered with formation of the M1/2 complex. In combination with RAG-2, wild-type RAG-1, RAG-1(R621H), and RAG-1(E719K) yielded identical patterns, with prominent interferences occurring at residues 6 (hT6) and 7 (hG7) of the heptamer, at residues 2 and 4 through 7 of the nonamer (nG2 and nT4 to nT7), and in the heptamer-proximal half of the spacer region (sT2, sG4), as numbered in the sense orientation (Fig. 4D and E). (The faint band at the heptamer-coding junction seen in Fig. 4D, lane 4, probably represents a small amount of nicked product formed in the presence of Ca2+ and wild-type protein; migration of this species at a position midway between piperidine cleavage products is consistent with this interpretation.)
These observations are consistent with our previous mapping of DNA-protein contacts in a wild-type RAG-DNA complex (45) and strongly suggest that such interactions are not significantly disrupted by the R621H or E719K mutations. Moreover, the characteristic overrepresentation of thymine modifications at the heptamer-coding junction and the nonamer-spacer junction also remains unaltered in the two mutant complexes (Fig. 4E), indicating that RAG-1(R621H) and RAG-1(E719K), like their wild-type counterpart, retain the ability to perturb the DNA structure at these sites. Taken together, these results indicate that the defects in single-site DNA cleavage observed for RAG-1(R621H) and RAG-1(E719K) do not result from disruption of RSS contacts in the prenicking complex.
Evidence that E719 of RAG-1 interacts with a divalent cation essential for catalysis of DNA cleavage.
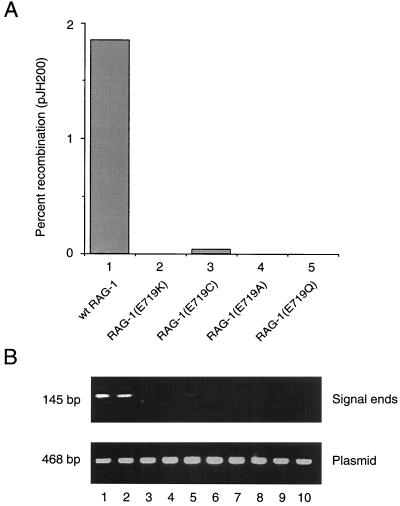
RAG-1(E719K) exhibits impairment of nicking and transesterification activity in Mg2+, although its contacts with the DNA substrate are similar or identical to those of wild-type RAG-1. We considered the possibility that the deleterious effect of the E719K mutation results specifically from the nonconservative substitution of lysine for glutamate. To address this, we tested the more conservative mutants RAG-1(E719Q) and RAG-1(E719A) for V(D)J recombination and DNA cleavage in vivo by using the extrachromosomal assay and ligation-mediated PCR as described above. Like E719K, the E719Q and E719A mutations profoundly impaired V(D)J recombination (Fig. 5A, compare lanes 2, 4, and 5 to lane 1). Although similar amounts of the pJH200 substrate were recovered from all transfections (Fig. 5B, lower panel), signal ends were undetectable with RAG-1(E719Q), RAG-1(E719A), or RAG-1(E719K) (Fig. 5B, upper panel, lanes 3, 4, and 7 to 10). Thus, we excluded the possibility that the E719K defect in vivo results specifically from the introduction of a positive charge.
FIG. 5.
Impairment of V(D)J recombination and DNA cleavage in vivo by diverse substitutions at E719 of RAG-1. (A) Wild-type or mutant RAG-1 fusion protein was coexpressed with RAG-2 core in 293 cells, and signal joint formation was quantitated using the extrachromosomal V(D)J recombination substrate pJH200. Percent recombination was calculated; values represent the means of two independent experiments. Lanes: 1, wild-type RAG-1; 2, RAG-1(E719K); 3, RAG-1(E719C); 4, RAG-1(E719A); 5, RAG-1(E719Q). (B) Signal ends were assayed by ligation-mediated PCR (upper panel); total pJH200 was assayed by PCR amplification of a backbone sequence (lower panel), as described in Materials and Methods. Lanes 1 and 2, wild-type RAG-1; lanes 3 and 4, RAG-1(E719K); lanes 5 and 6, RAG-1(E719C); lanes 7 and 8, RAG-1(E719A); lanes 9 and 10, RAG-1(E719Q). All transfections included wild-type RAG-2 core.
In classical transposases, the acidic residues aspartate and glutamate form a catalytic triad, the DDE motif, which provides a binding site for an essential divalent cation at or near the active site for DNA hydrolysis and strand transfer (3, 5, 7, 12, 24, 35, 36, 38). Residues D600 and D708 of RAG-1 have been implicated in the binding of a divalent cation essential for DNA cleavage in vivo (23, 27).
To address whether E719 of RAG-1 might also participate in metal binding, we pursued a strategy previously used to demonstrate direct interaction of an essential metal ion with the substrate in RNA self-splicing (8, 34); with the DDE motif of TnsB, a subunit of the Tn7 transposase (38); or with other acidic residues of RAG-1 (14, 22, 26). This approach exploits the differential chemistry of metal-sulfur and metal-oxygen binding. In TnsB, for example, replacement of D114 with cysteine altered the metal specificity of the 5′ processing reaction from Mg2+ to Mn2+, implying that in the wild-type protein, D114 participates in metal binding (38). An analogous mutation, by which E719 was replaced with cysteine (E719C), was introduced into the chimeric RAG-1 core protein. Unlike E719K, E719Q, and E719A, the E719C mutant supported detectable, albeit greatly reduced, V(D)J recombination and DNA cleavage in vivo (Fig. 5A, column 3, and 5B, faint bands in lanes 5 and 6).
RAG-1(E719C) and RAG-2 were coexpressed, copurified, and assayed for cleavage of the 12-signal substrate in vitro in the presence of 6 mM Mg2+ or 1 mM Mn2+. Wild-type RAG-1 and RAG-2 were assayed in parallel. The wild-type RAG proteins readily supported nicking in Mg2+ (Fig. 6A, lanes 1 to 6); transesterification was inefficient under these conditions, consistent with published results (49, 50). In contrast, RAG-1(E719C) showed very little nicking activity (about 50-fold reduction in initial rate, relative to that of the wild type) in the presence of RAG-2 and Mg2+ (Fig. 6A, lanes 7 to 12; Fig. 6B, left panel). In the presence of Mn2+, as expected, the combination of wild-type RAG-1 and RAG-2 core proteins supported nicking and hairpin formation (Fig. 6A, lanes 13 to 18). Strikingly, in the presence of Mn2+, RAG-1(E719C) exhibited a marked (approximately fivefold) enhancement of cleavage activity relative to that of wild-type protein (Fig. 6A, lanes 18 to 24; Fig. 6B, right panel).
FIG. 6.
Substitution of RAG-1 E719 by cysteine, but not by glutamine or lysine, confers preferential cleavage activity in Mn2+. (A) Kinetic analysis of RSS cleavage by purified wild-type RAG-1 or RAG-1(E719C) and wild-type RAG-2. Assays were carried out against a 12-spacer substrate in 6 mM Mg2+ (left panel) or 1 mM Mn2+ (right panel) as described in Materials and Methods. Lanes 1 to 6 and 13 to 18, wild-type RAG-1 and RAG-2; lanes 7 to 12 and 19 to 24, RAG-1(E719C) and RAG-2. Samples were withdrawn at times indicated. Positions of nicked and hairpin products are indicated at right. (B) The total yield of nicked and hairpin products shown in panel A, as quantitated by phosphorimager, is plotted as a function of time. Open squares, wild-type RAG-1; filled diamonds, RAG-1(E719C); left panel, cleavage activity in Mg2+; right panel, cleavage activity in Mn2+. (C) RAG-1(E719K) and RAG-1(E719Q) do not exhibit altered metal ion specificity. Kinetic analysis of RSS cleavage was carried out by purified proteins against a 12-spacer substrate in 6 mM Mg2+ or 1 mM Mn2+ as in panel A. Lanes 1 to 6 and 19 to 24, wild-type RAG-1 and RAG-2; lanes 7 to 12 and 25 to 30, RAG-1(E719K) and RAG-2; lanes 13 to 18 and 31 to 36, RAG-1(E719Q) and RAG-2. Positions of nicked and hairpin products are indicated at right. (D) The total yield of nicked and hairpin products in panel C, as quantitated by phosphorimager, is plotted as a function of time. Open squares, wild-type RAG-1; filled diamonds, RAG-1(E719K); filled squares, RAG-1(E719Q); left panel, cleavage activity in Mg2+; right panel, cleavage in Mn2+.
We considered the possibility that the differential activities of RAG-1(E719C), RAG-1(E719K), and wild-type RAG-1 in Mg2+ and Mn2+ might reflect differences in RSS binding. Wild-type RAG-1, RAG-1(E719K), or RAG-1(E719C) was copurified with RAG-2. Following quantitation by gel electrophoresis and silver staining (Fig. 4B), equivalent amounts of protein were assayed for binding to a radiolabeled 12-spacer substrate in the presence of Ca2+ (Fig. 4C, lanes 10 to 12), Mg2+ (Fig. 4C, lanes 7 to 9), or Mn2+ (Fig. 4C, lanes 1 to 6). For comparison, probe was incubated in the presence of Ca2+ but in the absence of protein (Fig. 4C, lane 13). Binding reactions were carried out under standard conditions of time and temperature (20 min at 37°C; Fig. 4C, lanes 4 to 13) or for 30 min at 4°C (Fig. 4C, lanes 1 to 3). In the presence of each divalent cation, RAG-1(E719C), RAG-1(E719K), and wild-type RAG-1 supported formation of the M1/2 complex. The yields of M1/2 complexes in reaction mixtures containing Mn2+ were substantially increased when binding was carried out at 4°C for 30 min rather than at 37°C for 20 min (Fig. 4C, compare lanes 1 to 3 and lanes 4 to 6). Importantly, under conditions in which RAG-1(E719C) is significantly more active for DNA cleavage than either RAG-1(E719K) or wild-type RAG-1, all three proteins supported formation of the M1/2 complex to a similar extent. Moreover, the ability of the three proteins to form M1/2 complexes, relative to each other, was not substantially altered by changes in the divalent cation included in the binding reaction mixture.
To confirm that the enhanced activity of RAG-1(E719C) in the presence of Mn2+ is a specific property of the cysteine mutant, RAG-1(E719K) and RAG-1(E719Q) were assayed in parallel in Mg2+ or Mn2+ (Fig. 6C and D). In Mg2+, the conservative E719Q mutant exibited a reduction in activity similar to that seen for E719K (Fig. 6C; Fig. 6D, left) or E719C (Fig. 6B, left). In contrast to its effect on RAG-1(E719C), Mn2+ failed to restore activity of either RAG-1(E719K) (Fig. 6C, lanes 25 to 30) or RAG-1(E719Q) (Fig. 6C, lanes 31 to 36). We conclude that the rescue of RAG-1(E719C) activity by Mn2+ is a specific property of the thiol substitution at that position. Taken together, these results indicate that residue E719 is intimately involved in binding of an essential divalent metal ion by RAG-1 and suggest that this residue lies at or near the catalytic site for DNA nicking.
Differential dependence of nicking and transesterification on presence of positive charge at residue 621 of RAG-1.
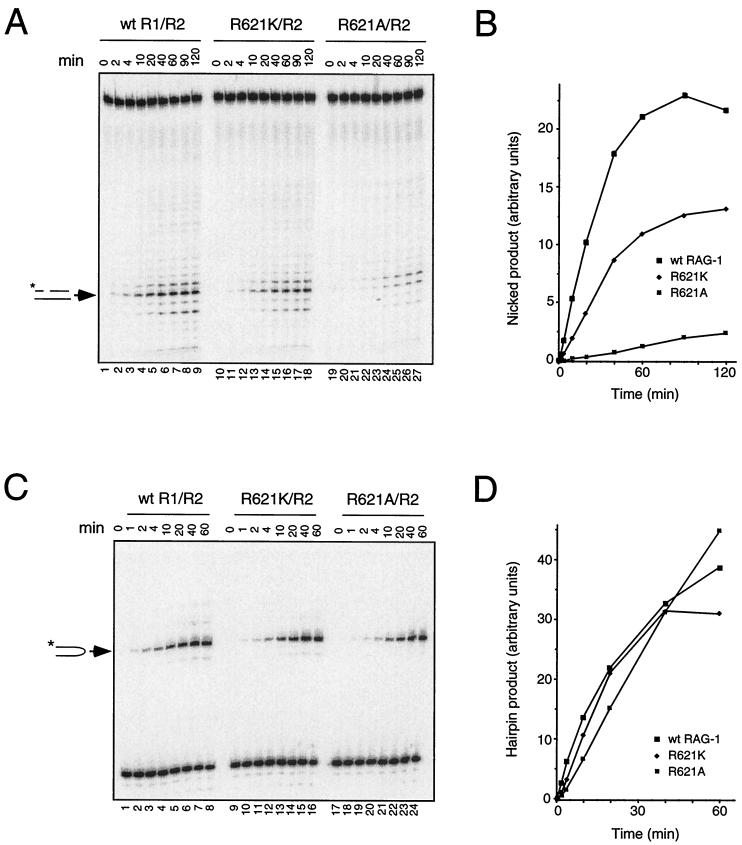
The retention of partial nicking activity by the RAG-1 R621H mutant prompted us to ask whether catalysis of this step was correlated with the presence of a positive charge at this position. Lysine or alanine was substituted for R621 and the resulting MBP fusion proteins were expressed and purified as described above. RAG-1(R621K) and RAG-1(R621A), like RAG-1(R621H), were similarly capable of associating with RAG-2 and binding to a single RSS substrate, as assessed by EMSA (data not shown). The ability of these mutants to support nicking of the C17A substrate was assessed in the presence of RAG-2 and Mn2+ at standard pH (7.0). RAG-1(R621K) exhibited a two- to three fold diminution in the initial rate of nicking, compared to that of the wild-type protein, while in a reaction containing RAG-1(R621A) the initial rate was reduced 20- to 100-fold (Fig. 7A and B and Table 1). When assayed under the same conditions for transesterification, however, the R621K and R621A mutants retained substantial activity, with decreases in initial reaction rates of only about 1.5- to 4-fold (Fig. 7C and D and Table 1).
FIG. 7.
Impairment of nicking but not transesterification by a nonconservative substitution at R621 of RAG-1. (A) Kinetic analysis of RSS nicking by RAG-1(R621K) and RAG-1(R621A). Lanes 1 to 9, wild-type RAG-1 and RAG-2; lanes 10 to 18, RAG-1(R621K) and RAG-2; lanes 19 to 27, RAG-1(R621A) and RAG-2. Assays were carried out in Mn2+ as described in Materials and Methods by using the mutant 12-spacer substrate C17A, which undergoes nicking in the absence of transesterification (29). Samples were withdrawn at times indicated above. The position of nicked product is indicated by the arrow at left. (B) The yield of nicked products in panel A was quantitated by phosphorimager and plotted as a function of time. Filled squares, wild-type RAG-1; filled diamonds, RAG-1(R621K); dotted squares, RAG-1(R621A). (C) Kinetic analysis of hairpin formation by RAG-1(R621K) and RAG-1(R621A). Lanes 1 to 8, wild-type RAG-1 and RAG-2; lanes 9 to 16, RAG-1(R621K) and RAG-2; lanes 17 to 24, RAG-1(R621A) and RAG-2. Assays were carried out in Mn2+ using a prenicked substrate as described in Materials and Methods. Samples were withdrawn at times indicated above. The positions of hairpin product are indicated at left. (D) The yield of hairpin products in panel C was quantitated by phosphorimager and plotted as a function of time. Symbols are as defined for panel B.
TABLE 1.
Relative initial rates of nicking and hairpin formation by RAG-1 proteins bearing amino acid substitutions at arginine 621
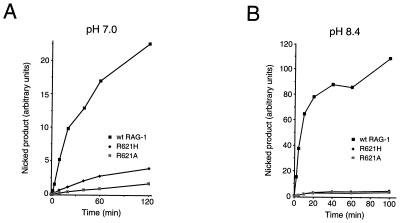
To obtain further support for the participation of a positively charged residue at position 621 in the nicking reaction, the activities of RAG-1(R621H) and RAG-1(R621A) were compared at standard pH conditions (pH 7.0), relatively near the pKa of free histidine (≈ pH 6.5), and at pH 8.4, well above the pKa of free histidine but below that of arginine. At pH 7.0 (Fig. 8A and Table 1), the initial reaction rate for RAG-1(R621H) was reduced to between 13 and 18% of wild type, while the rate for RAG-1(R621A) was about 1 to 5% that of wild type. At pH 8.4, however, the initial reaction rates for RAG-1(R621H) and RAG-1(R621A) were apparently identical, both being about 2% that of wild-type RAG-1 (Fig. 8B and Table 1). While other interpretations are possible, these results suggest that a positively charged side chain at residue 621 participates preferentially in the nicking step of DNA cleavage.
FIG. 8.
Comparison of nicking by RAG-1(R621H) and RAG-1(R621A) at pH 7.0 and at pH 8.4. (A) Assay at pH 7.0. Assays were carried out in Mn2+ under standard conditions (pH 7.0) using the mutant 12-spacer substrate C17A. Samples were withdrawn at various times and products were fractionated by gel electrophoresis. The yield of nicked products was quantitated by phosphorimager and plotted as a function of time. Filled squares, wild-type RAG-1; filled diamonds, RAG-1(R621H); dotted squares, RAG-1(R621A). (B) As in panel A, except that reactions were carried out at pH 8.4.
DISCUSSION
Participation of RAG-1 in catalysis of nicking and transesterification.
DNA-protein photo-cross-linking had demonstrated that in the presence of RAG-2, RAG-1 approaches the scissile bond at the heptamer-coding junction (13, 32, 44). This suggested that RAG-1 might play a direct catalytic role in one or both steps of DNA cleavage, an idea that was borne out by mutational analysis (14, 23, 27). The B-cell-negative SCID mutations analyzed here, R621H and E719K, also confer biochemical defects consistent with direct involvement of RAG-1 in catalysis of DNA cleavage. Recombination, accumulation of signal ends in vivo, and DNA cleavage in vitro are impaired despite the ability of the mutant proteins to form precleavage complexes that preserve the specific DNA-protein contacts observed with wild-type protein. Mutations in RAG-1 are also associated with Omenn syndrome, an immunodeficiency disorder with less severe impairment of lymphocyte development than classical B-cell-negative SCID (51). These mutations, in distinction to those occurring at R621 and E719, exert debilitating effects on V(D)J recombination by impairing DNA binding or RAG-1–RAG-2 association (51).
Evidence presented here suggests that residue E719 of RAG-1 may interact with an essential metal ion. The E719K mutation is associated with a decrease of more than 450-fold in the initial rate of substrate nicking in vitro relative to that of the wild type. This effect is not a particular property of the nonconservative lysine substitution that occurs in SCID patients, as the conservative E719Q mutation impaired RAG-1 activity to a similar extent. When cysteine is substituted for E719, the resulting RAG-1 mutant exhibits enhanced activity in Mn2+ but 50-fold reduced activity in Mg2+, relative to that of the wild type. The enhanced activity of RAG-1(E719C) in Mn2+ likely reflects an alteration in metal ion specificity conferred by the substitution of thiol for carboxylate, rather than a more general effect, because the activities of neither RAG-1(E719K) nor RAG-1(E719Q) were rescued by Mn2+. A similar, Mn2+-specific increase in enzymatic activity is associated with cysteine replacement mutations in the catalytic triad of TnsB (38). Taken together, these results provide evidence that residue E719 of RAG-1 interacts with a metal ion essential for DNA cleavage by the V(D)J recombinase.
It is likely that two catalytic regions are present in a RAG-1 dimer. These regions could be distributed between the individual subunits in one of two ways. In the first model, each site could be constructed from residues contributed by both subunits, so that an intact catalytic region would straddle the dimer interface, as in the site-specific recombinase Flp (6). In the second model, each of the two catalytic regions would reside in a single subunit. Recent observations are consistent with the latter model and indicate, moreover, that DNA cleavage is catalyzed in trans by the RAG-1 subunit residing opposite the bound subunit (43). The present data do not eliminate the possibility that RAG-2 also contributes a portion of the catalytic site. Previous studies have indicated essential roles for RAG-2 in formation of stable RAG-RSS complexes and in promoting contact between RAG-1 and the heptamer-coding junction. Whether RAG-2 plays a more direct role in catalysis of DNA cleavage remains an open question.
Comparison to transposases.
The initial steps of V(D)J recombination and transposition are chemically identical, with the exception that in V(D)J recombination transesterification occurs between neighboring DNA strands, while in transposition it occurs between donor and target duplexes (see reference 35 for review). This formal chemical identity is reinforced by the ability of the RAG proteins to promote the transposition of a donor DNA segment flanked by RSSs into a nonspecific target DNA molecule in vitro (1, 21). Thus, RAG-1 and RAG-2 can be considered to represent the subunits of a specialized, multicomponent transposase.
On this basis, the catalytic DDE motif characteristic of classical transposases, or its functional equivalent, might be expected to occur within one or both RAG proteins. The DDE motif cannot be reliably assigned on the basis of sequence homology, however, because it contains only three highly conserved amino acid residues whose spacing is variable. Several acidic residues in RAG-1, including D600, D708, and E962, have been identified previously as important for catalysis of DNA cleavage and may serve a metal-binding function similar to that of the DDE motif (14, 23, 27). DNA cleavage is abolished by nonconservative mutations at these three positions (14, 23, 27), in contrast to mutations at E719, which allow partial catalytic activity.
Recent evidence indicates that transposase proteins are more structurally diverse than previously appreciated. The Tn7 transposase contains two components, TnsA and TnsB. TnsA catalyzes DNA cleavage at the 5′ ends of the transposon, while TnsB is responsible for cleavage of the 3′ ends and strand transfer (38). Unlike TnsB, which resembles other members of the retroviral integrase superfamily, the catalytic fold of TnsA is structurally related to that of type II restriction endonucleases. Moreover, the catalytic center of TnsA contains a cluster of amino acid residues—E63, D114, K132 and E149—characteristic of restriction endonuclease active sites; with respect to their spatial arrangement, these four residues correspond closely to E71, K190, D134, and E204 of the restriction enzyme Cfr10I (18). These observations define a close relationship between TnsA and type II restriction enzymes and suggest that identification of essential catalytic residues may be insufficient to allow classification of a transposase in the absence of sequence homology or direct structural analysis.
Effects of mutations at R621.
Loss of a positive charge at residue 621 of RAG-1 is positively correlated with impairment of RSS-mediated DNA nicking. This effect is not likely to reflect a general structural perturbation, as hairpin formation is relatively impervious to nonconservative mutations at R621. While the present article was under review, RAG-1 mutations that selectively impair the transesterification step of DNA cleavage were described elsewhere (22). These complementary observations clearly indicate that the nicking and transesterification steps of RAG-mediated DNA cleavage have distinct catalytic requirements.
In addition to R621, R713 has been found to be important for RAG-1 catalytic activity (23). While the functions of these basic residues in RAG-mediated DNA cleavage are unknown, several possibilities are suggested by consideration of classical transposases and restriction endonucleases. In the Tn5 transposase and related enzymes, a conserved arginine residue appears to stabilize the hairpin conformation essential for cut-and-paste transposition (15). In the Tn5 synaptic complex this residue, R322, interacts with the 5′ phosphate group of a thymine residue in the nontransferred DNA strand, stabilizing a bend in the phosphodiester backbone (9, 10); in the related IS10 transposase, an alanine substitution at the corresponding position blocks nicking and strand transfer activities (4). In the human immunodeficiency virus type-1 integrase and in Tn5, two conserved basic residues near the active site make specific contacts with DNA (10, 16).
Other possibilities are suggested by type II restriction endonuclease. In these enzymes a conserved, essential lysine (e.g., K92 in EcoRV, corresponding to K190 of Cfr10I) orients and stabilizes a hydroxide ion for attack of the scissile phosphate and is postulated to stabilize the negative charge of pentavalent phosphorus in the transition state (25). In TnsA, K132 occupies an analogous position in the three-dimensional structure, suggesting that it may play a similar role in Tn7 transposition. Whether RAG-1 incorporates features of TnsA, in addition to those of the retroviral integrase family, remains to be determined.
ACKNOWLEDGMENTS
We thank Joanne Hesse, Dik van Gent, and Martin Gellert for reagents and the Howard Hughes Medical Institute Biopolymers Facility at Johns Hopkins for oligonucleotides. We are grateful to Patrick Swanson, Jinhak Lee, Amit Golding, Ashley Ross, and our colleagues in the Department of Molecular Biology and Genetics for stimulating discussions. Nick Dordai provided expert technical assistance.
This work was supported by grant CA16519 from the National Cancer Institute and by the Howard Hughes Medical Institute.
REFERENCES
- 1.Agrawal A, Eastman Q M, Schatz D G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 2.Akamatsu Y, Oettinger M A. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequence. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker T A, Luo L. Identification of residues in the Mu transposase essential for catalysis. Proc Natl Acad Sci USA. 1994;91:6654–6658. doi: 10.1073/pnas.91.14.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolland S, Kleckner N. The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell. 1996;84:223–233. doi: 10.1016/s0092-8674(00)80977-0. [DOI] [PubMed] [Google Scholar]
- 5.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure. 1996;4:89–96. doi: 10.1016/s0969-2126(96)00012-3. [DOI] [PubMed] [Google Scholar]
- 6.Chen J W, Lee J, Jayaram M. DNA cleavage in trans by the active site tyrosine during Flp recombination: switching protein partners before exchanging strands. Cell. 1992;69:647–658. doi: 10.1016/0092-8674(92)90228-5. [DOI] [PubMed] [Google Scholar]
- 7.Craig N L. Transposon Tn7. Curr Top Microbiol Immunol. 1996;204:27–48. doi: 10.1007/978-3-642-79795-8_2. [DOI] [PubMed] [Google Scholar]
- 8.Dahm S C, Uhlenbeck O C. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- 9.Davies D R, Braam L M, Reznikoff W S, Rayment I. The three-dimensional structure of a Tn5 transposase-related protein determined to 2.9-A resolution. J Biol Chem. 1999;274:11904–11913. doi: 10.1074/jbc.274.17.11904. [DOI] [PubMed] [Google Scholar]
- 10.Davies D R, Goryshin I Y, Reznikoff W S, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 11.Difilippantonio M J, McMahan C J, Eastman Q M, Spanopoulou E, Schatz D G. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 12.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 13.Eastman Q M, Villey I J, Schatz D G. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fugmann S D, Villey I J, Ptaszek L M, Schatz D G. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 15.Haren L, Ton-Hoang B, Chandler M. Integrating DNA: transposases and retroviral integrases. Annu Rev Microbiol. 1999;53:245–281. doi: 10.1146/annurev.micro.53.1.245. [DOI] [PubMed] [Google Scholar]
- 16.Hazuda D J, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler J A, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller M D. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 17.Hesse J, Lieber M, Gellert M, Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V(D)J joining signals. Cell. 1987;49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- 18.Hickman A B, Li Y, Mathew S V, May E W, Craig N L, Dyda F. Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol Cell. 2000;5:1025–1034. doi: 10.1016/s1097-2765(00)80267-1. [DOI] [PubMed] [Google Scholar]
- 19.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 20.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 21.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 22.Kale S B, Landree M A, Roth D B. Conditional RAG-1 mutants block the hairpin step of V(D)J recombination. Mol Cell Biol. 2001;21:459–466. doi: 10.1128/MCB.21.2.459-466.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D R, Dai Y, Mundy C L, Yang W, Oettinger M A. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim K, Namgoong S Y, Jayaram M, Harshey R M. Step-arrest mutants of phage Mu transposase. Implications in DNA-protein assembly, Mu end cleavage, and strand transfer. J Biol Chem. 1995;270:1472–1479. doi: 10.1074/jbc.270.3.1472. [DOI] [PubMed] [Google Scholar]
- 25.Kovall R A, Matthews B W. Type II restriction endonucleases: structural, functional and evolutionary relationships. Curr Opin Chem Biol. 1999;3:578–583. doi: 10.1016/s1367-5931(99)00012-5. [DOI] [PubMed] [Google Scholar]
- 26.Kung H C, Bolton P H. Structure of a duplex DNA containing a thymine glycol residue in solution. J Biol Chem. 1997;272:9227–9236. doi: 10.1074/jbc.272.14.9227. [DOI] [PubMed] [Google Scholar]
- 27.Landree M A, Wibbenmeyer J A, Roth D B. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis S. The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 29.Li W, Swanson P, Desiderio S. RAG-1- and RAG-2-dependent assembly of functional complexes with V(D)J recombination substrates in solution. Mol Cell Biol. 1997;17:6932–6939. doi: 10.1128/mcb.17.12.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Dordai D I, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 31.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 32.Mo X, Bailin T, Sadofsky M. RAG-1 and RAG-2 cooperate in specific binding to the recombination signal sequence in vitro. J Biol Chem. 1999;274:7025–7031. doi: 10.1074/jbc.274.11.7025. [DOI] [PubMed] [Google Scholar]
- 33.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 34.Piccirilli J A, Vyle J S, Caruthers M H, Cech T R. Metal ion catalysis in the Tetrahymena ribozyme reaction. Nature. 1993;361:85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- 35.Polard P, Chandler M. Bacterial transposases and retroviral integrases. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 36.Rice P, Mizuuchi K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell. 1995;82:209–220. doi: 10.1016/0092-8674(95)90308-9. [DOI] [PubMed] [Google Scholar]
- 37.Roth D B, Craig N L. VDJ recombination: a transposase goes to work. Cell. 1998;94:411–414. doi: 10.1016/s0092-8674(00)81580-9. [DOI] [PubMed] [Google Scholar]
- 38.Sarnovsky R J, May E W, Craig N L. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- 39.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz K, Gauss G H, Ludwig L, Pannicke U, Li Z, Lindner D, Friedrich W, Seger R A, Hansen-Hagge T E, Desiderio S, Lieber M R, Bartram C R. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 41.Siebenlist U, Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci USA. 1980;77:122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spanopoulou E, Zaitseva F, Wang F-H, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 43.Swanson P C. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol Cell Biol. 2001;21:449–458. doi: 10.1128/MCB.21.2.449-458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swanson P C, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swanson P C, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 46.Truss M, Chalepakis G, Beato M. Contacts between steroid hormone receptors and thymines in DNA: an interference method. Proc Natl Acad Sci USA. 1990;87:7180–7184. doi: 10.1073/pnas.87.18.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Gent D C, Hiom K, Paull T T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 49.van Gent D C, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 50.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 51.Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, Gatta L B, Ochs H D, Schwarz K, Notarangelo L D, Vezzoni P, Spanopoulou E. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93:885–896. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]