Abstract
Fall armyworm, Spodoptera frugiperda, entered Thailand in late 2018 and has now spread in several regions, with devastating effects in maize and rice production, which are some of the most important cereals in the world. Since then, farmers have utilized the available chemical insecticides to try to control it, but their efforts have been futile. Instead, they have ended up using extraordinary dosages, hence threatening non-target species and other fauna and flora, as well as being costly. In this regard, research has been ongoing, aiming to come up with eco-friendly solutions for this insect. We surveyed and collected various isolates of native entomopathogenic fungi intending to test their efficacy against fall armyworm. Six isolates of entomopathogenic fungi were obtained and identified to Beauveria bassiana based on morphological characteristics and multi-gene phylogenetic analyses. Thereafter, the six isolates of B. bassiana were used to perform efficacy experiments against fall armyworm. Additionally, the glycosyl transferase-like protein 1 (GAS1) gene was analyzed. Consequently, all the isolates showed efficacy against S. frugiperda, with isolate BCMU6 causing up to 91.67% mortality. Further, molecular analysis revealed that all the isolates possess the GAS1 gene, which contributed to their virulence against the insect. This is the first report of utilizing native entomopathogenic B. bassiana to manage S. frugiperda in Thailand, with the revelation of GAS1 as a factor in inducing virulence and cuticle penetration. This study has provided valuable information on the potential development of Beauveria bassiana as an eco-friendly bioinsecticide for the management of fall armyworm in Thailand.
Keywords: Spodoptera frugiperda, Beauveria bassiana, multi-gene, GAS1 gene, efficacy
1. Introduction
Maize ranks among the topmost commercially produced and utilized cereals globally. Firstly, it is used as human and animal feed. Additionally, maize is used in the food and beverage, paper, bioplastics, pharmaceutical, and textile industries, among others [1,2]. The agricultural sector in Thailand contributes about 9.9% to the Gross Domestic Product (GDP) in Thailand, and maize is among the top five crops on commercial production that contributes economically to the GDP (https://www.intracen.org, accessed on 29 November 2021). Lately, maize production has not met the needs of Thailand, causing more to be imported [3]. On the other hand, rice is a staple food in many countries in the world, including Thailand. Rice contributes immensely to the economy of Thailand. Due to pests and diseases, global warming, and increased drought, maize and rice production are threatened [1]. Until 2016 when it gained entry into West Africa and spread like bush fire, probably due to the favorable climatic conditions, fall armyworm (Spodoptera frugiperda) was a native of the Americas [4]. By the year 2018, it had traversed several territories to be reported in India, and once in India, somehow Asia was conquered [4,5]. In late 2018, some of the fields of maize and rice in several provinces of Thailand and Myanmar were already invaded by the fall armyworm, and since then, losses have been incurred [6,7]. This prompted farmers to struggle to control this insect using chemical insecticides, and their efforts have been futile. Eventually, they ended up using a lot of chemicals, which leads to the buildup of the same in the environment and is also likely to enhance the development of resistance by the insect (http://exchange.growasia.org/, accessed on 12 November 2021) [7,8].
Regarding the above, the search for an effective biological control agent to complement the existing chemical control began in the early 1980s after discovering that most of the insects being managed were developing resistance, and also the chemicals had detrimental residues in plants. Additionally, the environment had been impacted negatively [8]. Today, researchers have been able to identify over 750 species of entomopathogenic microorganisms, including fungi, nematodes, bacteria, and viruses, drawn from about 85 genera [8,9,10]. Interestingly, of all the entomopathogens, only fungi do not require ingestion or the openings like nematodes to gain entry into their host, but instead use enzymes to penetrate through the cuticle [11]. Among the identified species of fungi, the most utilized are in the genera Beauveria, Cordyceps, Isaria, Lecanicillium, Metarhizium, and Nomuraea, among others, of which several bio-insecticides are attributed [12,13,14]. Due to their ubiquitous nature and ability to survive in different kinds of environments, these entomopathogenic fungi have been tried against insects in several parts of the world and have emerged victorious in many aspects [12,15,16]. Admittedly, most of them can overwinter in the soil, waiting to infect the next generation of insect hosts, which makes it a perfect biological management choice that is eventually less costly [17]. Several entomopathogenic fungi have been used to successfully control economically important insects in many countries in the world. Reportedly, Thailand has been at the forefront in the isolation and utilization of entomopathogens [11]. When fall armyworm was first reported in Thailand and the neighboring countries, a regional action plan known as ASEAN FAW was set up to find a solution to this destructive pest (https://www.aseanfawaction.org, accessed on 29 November 2021). However, since the entry of fall armyworm in the territory, there is no report of entomopathogenic fungi suggested to manage it. These fungi gain entry into the insect through the cuticle by the use of secreted enzymes or glycosyl transferase-like protein to penetrate through the cuticle. In addition, this protein aids in causing virulence to the insect, leading to mycosis. Research has determined that the GAS1 gene is responsible for the synthesis of this protein [18,19,20]. Therefore, this study aimed to isolate native entomopathogenic fungi in northern Thailand and investigate their pathogenic ability against the invasive fall armyworm. The obtained fungi were identified through morphological characteristics and multi-gene phylogenetic analyses. Moreover, the GAS1 gene involved in fungal virulence and aiding the penetration of the insect cuticle was investigated.
2. Materials and Methods
2.1. Collection and Isolation of Entomopathogenic Fungi
A survey was conducted in Chiang Mai and Lampang provinces of northern Thailand. During the field collection and identification of entomopathogens, the mycosis that is displayed due to the colonization of the insect after the emergence of the hyphae through the cuticle to cause white muscardine disease was employed. The infected cadavers were then taken to the laboratory and isolation of the fungus was performed. The isolation of the native entomopathogens from the insect cadavers and soil samples was conducted using the bait method according to the protocol proposed by Meyling [21]. Pure cultures were kept in the Insect Pathology Laboratory, Department of Entomology and Plant Pathology, Faculty of Agriculture, Chiang Mai University, for further studies.
2.2. Identification of Entomopathogenic Fungi
2.2.1. Morphological Study
Morphological observations were performed using pure fungal cultures on potato dextrose agar (PDA) for two weeks at 25 ± 1 °C with a photoperiod of 12:12 h (Dark:Light). Macroscopic observations were determined through colony characteristics obtained by the shape, elevation, growth pattern, color, and texture [9,22,23]. Using a sterile needle, fungal structures were picked up and mounted in lactic acid on glass slides and observed using a compound microscope (ZEISS AX10) by the program ZEN 3.3 (blue edition) to observe hyphal conidiophore and measure conidial sizes. For each isolate, the sizes of 20 conidia were measured following the methods described in previous studies [22].
2.2.2. Molecular Study
Genomic DNA was extracted from fresh fungal mycelium grown on PDA at 28 °C for one week using the DNA Extraction Mini Kit (FAVOGEN, Ping-Tung, Taiwan). Four gene loci: the internal transcribed spacer (ITS), the translation elongation factor 1-alpha (TEF-1), the RNA polymerase II largest subunit (RPB1) and the partial RNA polymerase second largest subunit (RPB2) were amplified using polymerase chain reaction (PCR), with ITS4/ITS5, 983F/2218R, RPB1-Af/RPB1-Cr, and fRPB2-5F2/fRPB2-7cR primers, respectively (Table 1). The PCR amplification conditions for ITS and TEF-1 are: initial denaturation at 94 °C for 3 min, with 30 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s and extension at 72 °C for 45 s, and final extension at 72 °C for 5 min. In addition, PCR conditions for RPB1 and RPB2 amplifications are: initial denaturation at 94 °C for 2 min, with 35 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 60 s and extension at 72 °C for 1 min, and final extension at 72 °C for 5 min. PCR products were purified using Gel Extraction NucleoSpin® Gel and the PCR Clean-up Kit (Macerey-Nagel, Dueren, Germany). PCR products were sent to commercial sequencing at 1st BASE Company (Kembangan, Malaysia).
Table 1.
Primers used in this study.
| Gene | Primer Name | Primer Sequence | Reference |
|---|---|---|---|
| ITS | ITS1F | CTTGGTCATTTAGAGGAAGTAA | [24] |
| ITS4 | TCCTCCGCTTATTGATATGC | [25] | |
| TEF-1 | 983F | GCTCCYGGHCAYCGTGAYTTYAT | [26] |
| 2218R | ATGACACCRACRGCRACRGTYTG | [26] | |
| RPB1 | RPB1-Af | GAR TGYCCDGGDCAYTTYGG | [27] |
| RPB1-Cr | CCNGCDATNTCRTTRTCCATRTA | [27] | |
| RPB2 | fRPB2-5f2 | GAYGAYMGWGATCAYTTYGG | [28] |
| fRPB2-7cR | CCCATRGCTTGYTTRCCCAT | [28] |
The related taxa were identified using the BLAST search (http://blast.ddbj.nig.ac.jp/top-e.html, accessed on 2 November 2021) results and previous publications. Details of the sequences used for the phylogenetic analyses are provided in Table 2. Preliminarily, individual DNA sequence matrixes were aligned by MUSCLE [29] and improved where necessary using BioEdit v.6.0.7 [30]. A phylogenetic tree was constructed under maximum likelihood (ML) and Bayesian inference (BI) methods. The ML analysis was carried out using RAxML-HPC2 on XSEDE (8.2.10) in CIPRES Science Gateway V.3.3 [31] using the GTRCAT model with 25 categories and 1000 bootstrap (BS) replications. The optimum nucleotide substitution model was obtained using the jModel test v.2.3 [32] under the Akaike information criterion (AIC) method. The BI analysis was performed using MrBayes 3.2.6 software for Windows [33]. The selected optimal model of each gene is similar to the GTR+I+G model. Six simultaneous Markov chains were run with one million generations, starting from random trees and keeping one tree every 100th generation until the average standard deviation of split frequencies was below 0.01. The value of burn-in was set to discard 25% of trees when calculating the posterior probabilities. Bayesian posterior probabilities (PP) were obtained from the 50% majority-rule consensus of the trees kept. The tree topologies were visualized in FigTree v1.4.0 [34].
Table 2.
GenBank sequences data of fungal isolates used in this study.
| Strain and Voucher No. | Taxon | Country | Host/substratum | GenBank Accession Number | Reference | |||
|---|---|---|---|---|---|---|---|---|
| ITS | TEF-1 | RPB1 | RPB2 | |||||
| BCMU1 | Beauveria bassiana | Thailand | Bactocera dorsalis | OL375165 | OL410297 | OL410303 | OL410309 | This study |
| BCMU2 | Beauveria bassiana | Thailand | Coffeeberry borer | OL375167 | OL410298 | OL410304 | OL410310 | This study |
| BCMU3 | Beauveria bassiana | Thailand | Ant | OL375168 | OL410299 | OL410305 | OL410311 | This study |
| BCMU4 | Beauveria bassiana | Thailand | Coffeeberry borer | OL375169 | OL410300 | OL410306 | OL410312 | This study |
| BCMU5 | Beauveria bassiana | Thailand | Coffee stem borer | OL375170 | OL410301 | OL410307 | OL410313 | This study |
| BCMU6 | Beauveria bassiana | Thailand |
Bactocera
dorsalis |
OL375173 | OL410302 | OL410308 | OL410314 | This study |
| ARSEF 1564 T | Beauveria bassiana | Italy |
Hyphantria
cunea |
GU734762 | EF222318 | HQ880833 | HQ880905 | [35] |
| CHE-CNRCB 168 | Beauveria bassiana | Mexico | Diaphorina citri | KU725691 | KU725693 | KU725699 | KU725703 | [36] |
| Isolate 4511 | Beauveria bassiana | China | Soil | KX901310 | KX901322 | KX901328 | KY464981 | [37] |
| Isolate 4508 | Beauveria bassiana | China | Soil | KX901307 | KX901319 | KX901325 | KY464978 | [37] |
| 2898 T | Beauveria medogensis | China | Soil | KU994837 | KU994833 | KU994835 | KU994834 | [37] |
| BUB426 | Beauveria medogensis | China | Soil | MG642832 | MG642904 | MG642859 | MG642874 | [37] |
| RCEF5500 T | Beauveria lii | China | Henosepilachna vigintioctopunctata | JN689372 | JN689371 | JN689374 | JN689370 | [38] |
| ARSEF 8257 T | Beauveria verroae | France | Varroa destructor | NR111599 | HQ881002 | HQ880872 | HQ880944 | [39] |
| ARSEF 2694 | Beauveria verroae | Switzerland | Larinus sp. | HQ880802 | HQ881004 | HQ880874 | HQ880946 | [39] |
| ARSEF 4598 T | Beauveria australis | Australia | Soil | NR111597 | HQ880995 | HQ880861 | HQ880933 | [39] |
| ARSEF 4580 | Beauveria australis | Australia | Orthoptera: Acrididae |
HQ880788 | HQ880994 | HQ880860 | HQ880932 | [39] |
| ARSEF 7032 T | Beauveria kipukae | USA | Not provided | NR111600 | HQ881005 | HQ880875 | HQ880947 | [39] |
| ARSEF 7760 T | Beauveria malawiensis | Malawi |
Phoracantha
semipunctata |
DQ376247 | DQ376246 | HQ880897 | HQ880969 | [40] |
| Bwetak89 | Beauveria malawiensis | New Zealand | Not provided | MW027837 | MW030946 | MW027830 | MW027829 | [35] |
| ARSEF 4850 T | Beauveria asiastica | Korea | Coleoptera: Cerambycidae |
NR111596 | KJ523141 | HQ880859 | HQ880931 | [39] |
| BCC13243 | Beauveria asiastica | Thailand | NR | MN401629 | MN401455 | MN401553 | NR | [41] |
| ARSEF 2567 T | Beauveria caledonica | Scotland | Soil | HQ880817 | EF469057 | HQ880889 | HQ880961 | [39] |
| BUB421 | Beauveria caledonica | China | Soil | MG642831 | MG642903 | MG642858 | MG642873 | [39] |
| GZU12141 T | Beauveria majiangensis | China | Coleoptera | MG052642 | MG052640 | MG052644 | NR | [42] |
| GZU12142 | Beauveria majiangensis | China | Coleoptera | MG052643 | MG052641 | MG052645 | NR | [42] |
| ARSEF 617 T | Beauveria brongniartii | France | Coleoptera: Scarabaeidae |
NR111595 | HQ880991 | HQ880854 | HQ880926 | [39] |
| ARSEF 7516 | Beauveria brongniartii | Japan | Coleoptera: Scarabaeidae |
HQ880766 | HQ880976 | HQ880838 | HQ880910 | [39] |
| Bt99 | Beauveria hoplocheli | Reunion Island | Coleoptera: Melolonthidae |
KC339698 | KC339710 | KM453949 | KM453958 | [43] |
| ARSEF 3405 T | Beauveria pseudobassiana | Kentucky, USA | Lepidoptera: Tortricidae |
NR111598 | NR | HQ880864 | HQ880936 | [39] |
| ARSEF 1855 | Beauveria pseudobassiana | Canada | Coleoptera: Scolytidae |
HQ880796 | HQ880999 | HQ880868 | HQ880940 | [39] |
| ARSEF 2922 T | Beauveria vermiconia | Chile | Soil | NR151832 | NR | HQ880894 | HQ880966 | [44] |
| ARSEF 2641 T | Beauveria amorpha | Brazil | Hymenoptera: Formicidae |
NR111601 | NR | HQ880880 | HQ880952 | [39] |
| ARSEF 7542 | Beauveria amorpha | Colorado, USA | Hymenoptera: Formicidae |
HQ880805 | HQ881007 | HQ880877 | HQ880949 | [39] |
| CBS 350.85 | Lecanicillium antillanum | Cuba | Hymenomycete: Agaric |
MH861888 | DQ522350 | DQ522396 | DQ522450 | [45] |
| ARSEF 4029 | Isaria farinosa | Denmark | Coleoptera: Carabidae |
HQ880828 | HQ881019 | HQ880900 | HQ880972 | [39] |
“NR” = Not reported. Superscript “T” indicates type strain
2.3. Efficacy Test
2.3.1. Insect Source and Rearing
Spodoptera frugiperda used in this study were collected from the demonstration field of the Faculty of Agriculture and Mae-Hia Agricultural Training and Research Center, Chiang Mai University, during the maize growing season. The collected adults and larvae were maintained at the Insect Pathology Laboratory, Department of Entomology and Plant Pathology, Faculty of Agriculture, Chiang Mai University, Thailand. Disease-free neonate larvae were placed in plastic containers (19 cm in width by 27 cm in length by 8 cm in height) and fed on baby corn under laboratory room conditions at 26 ± 1 °C, with a photoperiod of 12:12 h (Dark:Light) and 65% ± 5% humidity [17]. The egg batches oviposited were maintained under controlled laboratory conditions, and upon hatching, the neonate larvae were individually placed in plastic containers measuring 6 × 3 cm, and 3 oz.
2.3.2. Fungal Inoculum Preparation
For each isolate, stock suspension was prepared in 250 mL reagent bottles with the addition of the mass sporulating culture in 100 mL of distilled water, with 0.01% Tween 80. Thereafter, the surface was softly scraped to dislodge the conidia using a sterile loop [9]. The suspensions were pipetted from the plates. The mixture was then vigorously shaken for 3 min and then filtered. The hyphae were isolated through a sterilized millipore cloth after blending. An improved Neubauer hemocytometer was used to determine conidial concentration under a light microscope at 400× magnification [22]. After this, two different concentrations, 1 × 106 and 1 × 108 mL−1 conidia, were obtained by dissolving the original solution in sterilized distilled water with 0.01% Tween 80 [46]. These concentrations were used for the efficacy bioassay [13].
2.3.3. Insect Bioassay
Two days after hatching, the second instar larvae were dipped into the two different fungal concentrations (1 × 106 and 1 × 108 mL−1 conidia) obtained in the process described above and returned into the plastic container supplied with about 5 g of baby corn. For the control, sterilized distilled water with 0.01% Tween 80 was used. This process was repeated three times after every three days, during which the food was also changed [47]. Mortality data were recorded every three days. The experiment followed a completely randomized block design with three replications for each concentration. Each replicate had 30 larvae. The experiments were independently repeated twice. Mortality data were analyzed using one-way ANOVA and presented as a percentage, as indicated in Table 4. The treatment means were compared using Tukey’s test for their significance at the 0.05% probability level. The mortalities were compared by the F-test. Differences were considered significant at p < 0.05. The IBM SPSS Statistical Software package version 23.0 (IBM Corp., Armonk, NY, USA, 2015) was used to conduct the statistical analyses.
2.4. Molecular Characterization of GAS1 Gene
Fungal DNA of each isolate was extracted from mycelia covered on fall armyworm by the DNA Extraction Mini Kit (FAVOGEN, Ping-Tung, Taiwan) following the manufacturer’s protocol. The GAS1 gene was amplified using the specific primers GTF2 (5′-CCCGTCA TCTCCTTGCTCATCAG-3′) and GTR2 (5′-GTCATCAACGAAAAGGGCAACGAG-3′), following the study of Zhang et al. [19]. The PCR amplification conditions for GAS1 followed the initial denaturation at 95 °C for 3 min, with 35 cycles of denaturation at 95 °C for 30 s, annealing at 56 °C for 30 s and extension at 72 °C for 1 min, and final extension at 72 °C for 10 min. PCR products were purified using Gel Extraction NucleoSpin® Gel and the PCR Clean-up Kit (Macerey-Nagel, Dueren, Germany). PCR products were sent to commercial sequencing at 1st BASE Company (Kembangan, Malaysia). Purified PCR products were sequenced using a commercial provider obtained from the 1st BASE Company (Kembangan, Malaysia). The sequences were assembled and then subjected to BLAST search in the GenBank database (http://blast.ddbj.nig.ac.jp/top-e.html, accessed on 10 November 2021).
3. Results
3.1. Collection and Morphological Characterization of Entomopathogenic Fungi
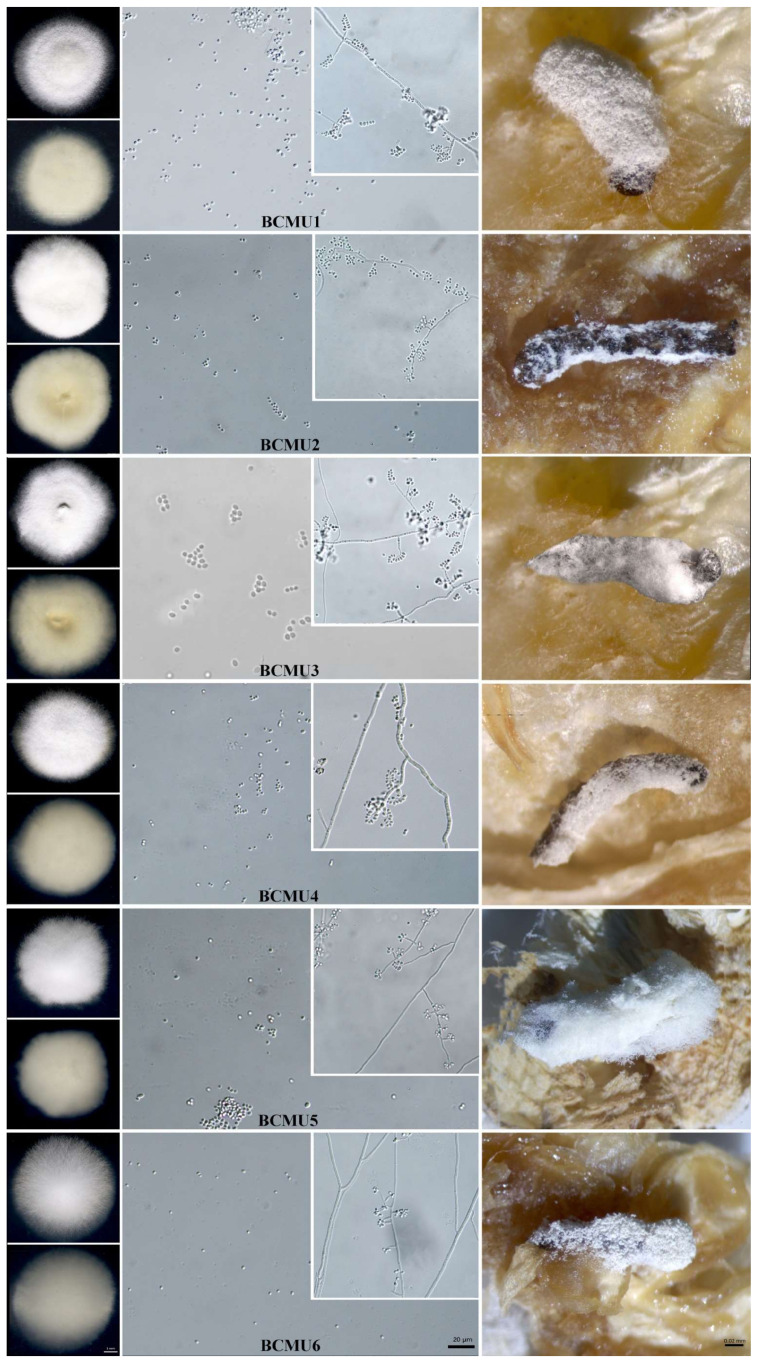
After carrying out preliminary observations on the isolates collected during the field survey, six fungal isolates were obtained, namely BCMU1, BCMU2, BCMU3, BCMU4, BCMU5, and BCMU6, as shown in Table 2. Morphologically, all six isolates examined were typical of the Beauveria genus due to the features displayed. Microscopic and macroscopic observations of the morphology confirmed the typical characteristics of B. bassiana, displaying white to the yellowish coloration on the mycelium on the PDA media (Figure 1 and Supplementary Table S1).
Figure 1.
Pictorial presentation of BCMU1–BCMU6 colony on the obverse and reverse sides on PDA media, conidia, and the hyphae and the mycosis caused on Spodoptera frugiperda. Scale Bar = 1 µm, 20 µm, and 0.02 mm respectively. The isolates were cultured on potato dextrose agar for 14 days at 25 ± 1 °C with a photoperiod of 12:12 h (Dark: Light). Once the larvae had died, they were placed on moist conditions to allow mycosis.
We observed conidiogenous cells that are short and globose. Additionally, the conidiophores had whorls and clustered compactly. The mycelium was observed to be cottony and closely appressed to the media, with all of the isolates white on the top side except for BCMU4, which was off-white, while on the underside, most of them displayed a yellowish coloration save for BCMU5, which was brownish in appearance. In terms of shape, four of the isolates were round in shape while two were almost oval. They were all raised in terms of elevation and smooth in texture. Three of them had a dispersed growth pattern while three were dense and disperse. However, fungal identification was confirmed by multi-gene phylogenetic analyses.
3.2. Phylogenetic Results of Obtained Entomopathogenic Fungi
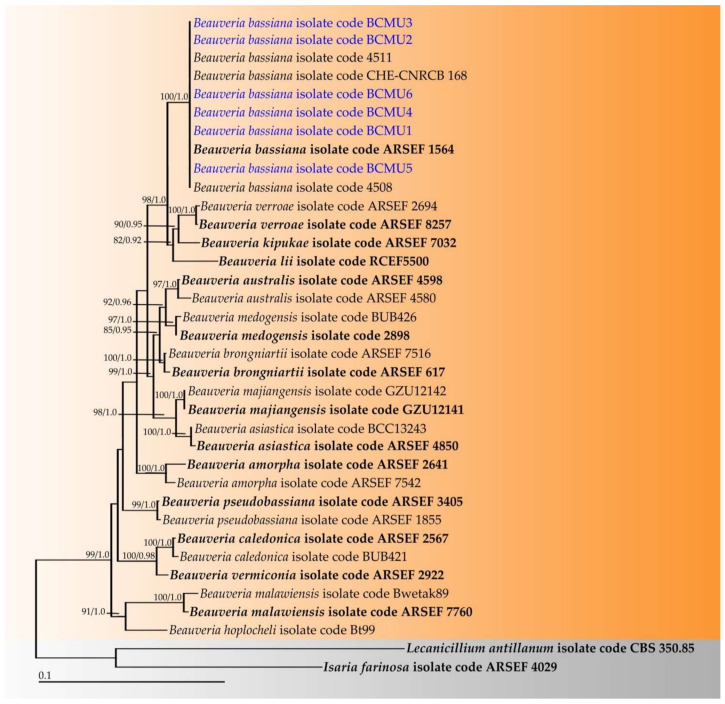
The sequences of six fungal strains in this study were deposited in the GenBank database (Table 2). The alignment of a combination of ITS, TEF-1, RPB1, and RPB2 genes contained 3185 characters, including gaps (ITS: 1–563, TEF-1: 564–1501, RPB1: 1502–2257, and RPB2: 2258–3185). RAxML analysis of the combined dataset yielded a best-scoring tree with a final ML optimization likelihood value of −10,287.872. The matrix contained 622 distinct alignment patterns with 8.74% undetermined characters or gaps. Estimated base frequencies were recorded as follows: A = 0.2431, C = 0.2867, G = 0.521, T = 0.2179, and substitution rates of: AC = 1.3261, AG = 4.2754, AT = 1.2198, CG = 0.8715, CT = 10.6195, and GT = 1.0000. The gamma distribution shape parameter alpha was equal to 0.2350 and the Tree-Length was equal to 0.4903. In addition, the final average standard deviation of the split frequencies at the end of the total MCMC generations was calculated as 0.00837 through BI analysis. Phylograms of the ML and BI analyses were similar in terms of topology (data not shown). Therefore, the phylogram obtained from the ML analysis was selected and presented for this study. The phylogram was comprised of 34 sequences of Beauveria strains and two sequences (Isaria farinosa ARSEF 4029 and Lecanicillium antillanum CBS 350.85) of the outgroup (Figure 2). Our analysis confirmed that all fungal strains in this study (BCMU1, BCMU2, BCMU3, BCMU4, BCMU5, and BCMU6) belonged to the B. bassiana, with high support values (BS = 100% and PP = 1.0). Beauveria bassiana formed the sister clade to B. kipukae, B. lii, and B. varroae, with high support (BS = 98% and PP = 1.0).
Figure 2.
Phylogram derived from maximum likelihood analysis of 36 sequences of the combined ITS, TEF-1, RPB1, and RPB2 sequences. Isaria farinosa ARSEF 4029 and Lecanicillium antillanum CBS 350.85 were used as the outgroup. The numbers above branches represent bootstrap percentages (left) and Bayesian posterior probabilities (right). Bootstrap values > 75% and Bayesian posterior probabilities > 0.90 are shown. The scale bar represents the expected number of nucleotide substitutions per site. Sequences obtained in this study are in blue. Type strains are indicated in bold.
3.3. Efficacy Test
Generally, all six isolates caused mortality to fall armyworm to a greater extent, as displayed in Table 3. Apparently, there were significant differences in their efficacy, with the most efficacious isolate being BCMU6 at a concentration of 1 × 108 mL−1. Interestingly, by the third day, this isolate had caused about 43% mortality to the larvae of S. frugiperda, which was an impressive outcome. After twelve days of observation, the same isolate had caused mortality of 91.67%, while the least mortality observed was on the isolate BCMU1 in both concentrations of 1 × 106 and 1 × 108 mL−1, respectively.
Table 3.
Percentage of cumulative mortalities caused of S. frugiperda by the six isolates of B. bassiana.
| Isolates | Time (Days) | |||
|---|---|---|---|---|
| 3 | 6 | 9 | 12 | |
| BCMU1 108 | 3.33 ± 1.67 ab* | 5.00 ± 0.00 a | 6.67 ± 1.67 a | 41.67 ± 3.33 c |
| BCMU1 106 | 3.33 ± 3.33 ab | 3.33 ± 3.33 a | 3.33 ± 3.33 a | 10.00 ± 2.89 a |
| BCMU2 108 | 15.00 ± 0.00 bc | 20.00 ± 2.87 b | 31.67 ± 3.33 cd | 41.67 ± 3.33 c |
| BCMU2 106 | 0.00 ± 0.00 a | 3.33 ± 3.33 a | 5.00 ± 2.88 a | 35.00 ± 2.89 bc |
| BCMU3 108 | 36.67 ± 3.33 ef | 40.00 ± 2.87 c | 43.33 ± 3.33 e | 73.33 ± 3.33 ef |
| BCMU3 106 | 3.33 ± 1.67 ab | 11.67 ± 1.67 ab | 11.67 ± 1.67 ab | 55.00 ± 2.89 d |
| BCMU4 108 | 30.00 ± 2.89 de | 53.33 ± 3.33 d | 55.00 ± 2.87 f | 60.00 ± 2.89 d |
| BCMU4 106 | 21.67 ± 1.67 cd | 21.67 ± 1.67 b | 21.67 ± 1.67 bc | 23.33 ± 1.67 b |
| BCMU5 108 | 28.33 ± 4.41 de | 56.67 ± 7.26 d | 66.67 ± 4.41 g | 83.33 ± 6.01 fg |
| BCMU5 106 | 3.33 ± 1.67 ab | 8.33 ± 1.67 ab | 8.33 ± 1.67 a | 35.00 ± 0.00 bc |
| BCMU6 108 | 43.33 ± 6.00 f | 71.67 ± 4.41 e | 76.67 ± 3.33 g | 91.67 ± 1.67 g |
| BCMU6 106 | 15.00 ± 2.87 bc | 23.33 ± 3.33 b | 33.33 ± 1.67 de | 61.67 ± 3.33 de |
| Control | 0.00 a | 0.00 a | 0.00 a | 0.00 a |
| df1 | 12 | 12 | 12 | 12 |
| df2 | 26 | 26 | 26 | 26 |
| F-test 0.05 | 27.798 | 50.594 | 91.074 | 81.081 |
* The lower case letters a, b and c show significant differences in mortalities caused by the different concentration of the isolates.
3.4. Molecular Characterization of GAS1 Gene
Molecular characterization of the GAS1 gene from fungal genomic DNA was performed by PCR amplification. The result showed that the GAS1 gene obtained from each fungal isolate showed 100% similarity to B. bassiana ARSEF2860 (Table 4). Additionally, the obtained GAS1 gene showed 89.31% and 88.12% similarity to Cordyceps militaris ATCC 34164 and Isaria fumosorosea ARSEF2679, respectively. Therefore, this study confirms the presence of the GAS1 gene from obtained fungi.
Table 4.
Details of the GAS1 sequences obtained from entomopathogenic fungi in this study.
| Fungal Isolate | Length (bp) | GenBank Accession Number |
Closeted Species/Accession Number | Similarity (%) |
|---|---|---|---|---|
| BCMU1 | 346 | OL469003 | Beauveria bassiana ARSEF 2860/XM008599737 | 100 |
| BCMU2 | 358 | OL469004 | Beauveria bassiana ARSEF 2860/XM008599737 | 100 |
| BCMU3 | 356 | OL469005 | Beauveria bassiana ARSEF 2860/XM008599737 | 100 |
| BCMU4 | 340 | OL469006 | Beauveria bassiana ARSEF 2860/XM008599737 | 100 |
| BCMU5 | 346 | OL469007 | Beauveria bassiana ARSEF 2860/XM008599737 | 100 |
| BCMU6 | 350 | OL469008 | Beauveria bassiana ARSEF 2860/XM008599737 | 100 |
4. Discussion
This study sought to molecularly and morphologically characterize six isolates of Beauveria bassiana that were previously isolated from insect cadavers, and also to determine their efficacy against the invasive Spodoptera frugiperda. The search for biological control against fall armyworm intensifies by the day in order to avoid the development of resistance to the chemical control measures that have been observed in its native region [48]. The morphology of the six isolates studied was similar to B. bassiana, as described in previous studies [46,49]. For a long time, morphological analysis had been the traditional tool used in the characterization of various entomopathogenic fungi, until it was discovered that the conidial features can easily change in the process of culturing. For example, B. bassiana and B. asiastica have been observed to be morphologically similar. Therefore, the use of multi-gene molecular analysis comes in handy as a confirmatory tool [50]. Since the year 1990, molecular work has been put into action for the identification of entomopathogenic fungi, especially of the genus Beauveria [51].
Beauveria bassiana has been in the limelight for a long time as far as efficacy is concerned. In the present study, all the isolates used in the study caused mortality, but at differing rates, with the highest instigating up to 91.67% after twelve days since the initial inoculation. The second highest isolate caused 83.33% mortality after twelve days. Previous studies have recorded mortalities of 100%, which is incredible, but mortalities of 60% and above are good enough in controlling insect populations [52]. Beauveria bassiana has been an entomopathogen in many insect orders, including Lepidoptera, Hemiptera, Coleoptera, and Diptera, among others. Particularly, Lepidopteran insects have been successfully controlled by this fungus, as can be ascertained by the study performed on Plutella xylostella, where it caused over 74% mortality [53]. Additionally, when inoculated against Galleria mellonella, B. bassiana caused mortalities within a short time, which was a promising gesture toward the management of this particular insect [54]. Further, it has been previously confirmed that the lethal action of B. bassiana is proportionally dependent on the concentration. The higher the concentration, the more efficacious it is to the insect [55]. Looking at our results, this fact is corroborated. In all the isolates studied, the highest mortalities were achieved with concentration of 1 × 108 mL−1. However, this is not a cause for alarm as these regimens are safer both for the environment and for animals, including humans [55]. Although our study was laboratory-based, we investigated some records on the endophytic influence that may be caused by B. bassiana on plants, and it is clear that it is friendly and does not cause any significant changes in the plant fresh and dry weight as well as the nutrient elements [56]. In fact, the endophytic strains have been reported to promote plant growth and at the same time help in controlling insect pests [57,58].
Finally, the GAS1 gene was found to be present in the six isolates of B. bassiana upon analysis. This points to the fact that the virulence caused against the fall armyworm is aided by the presence of this gene [19]. Xiao et al. observed that B. bassiana has many species-specific virulence genes, with GAS1 being among them [59]. Lai et al. [60] also observed the upregulation of the BBGAS1 gene prior to B. bassiana gaining entry into the hemocoel of the Anopheles mosquito. These records further corroborate the role the GAS1 gene plays in the infection and virulence process. Additionally, the GAS1 gene is known to code for conidial thermotolerance [20]. To further prove that the GAS1 gene is key to the infection process, a study carried out by Cao et al. in 2012 resulted in reduced cuticle penetration when they deleted the Magas1 gene in Metarhizium acridum [20]. Therefore, our findings are proof that these isolates of B. bassiana possess an important gene that is a key contributor to the insect infection process and virulence. Furthermore, the conidial thermotolerance role that the gene plays is valuable when B. bassiana-based commercial biopesticides are used in an extreme environment. In addition, B. bassiana has been reported to produce secondary metabolites, such as beauvericin, tenellin, oosperein, bassianin, and bassianolide, which are capable of enervating the immune systems of insect hosts [61,62].
5. Conclusions
We confirmed that our indigenous fungal isolates are B. bassiana through morphological observations as well as multi-gene molecular analyses. This is the first report of B. bassiana instigating efficacy against S. frugiperda in Thailand, which contributes valuable knowledge towards the search for an eco-friendly solution to the invasive insect. Our data will be helpful in the identification and characterization of B. bassiana isolates that are ubiquitous and effective in the control of several invasive insects around the world. Additionally, the finding that one of the isolates could cause mortality of up to 91.67% to the fall armyworm is a great inspiration towards the continuous search for a biological solution to fall armyworm. Notwithstanding, the GAS1 gene, which is known to code for cuticle penetration, conidial thermotolerance, and virulence against insect hosts, was found to be present in all the isolates investigated. Moreover, our research provides a pedestal for similar studies in the future. However, we recommend further research to be performed, including field experiments to determine whether there are effects attributed to ultraviolet rays and any other biotic and abiotic factors. We hope that this will lead to the development of a strategy for the commercialization of an indigenous B. bassiana-based bio-insecticide against S. frugiperda.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof7121073/s1. Table S1: The macroscopic and microscopic colony characteristics of the six isolates of Beauveria bassiana.
Author Contributions
Conceptualization, J.R., S.P., N.S., J.K., M.T., S.M., A.A.P. and P.K.; methodology, J.R., S.P., N.S., J.K., M.T. and P.K.; software, J.R., N.S. and J.K.; validation J.R., S.P., N.S., J.K., M.T., S.M., A.A.P. and P.K.; formal analysis, J.R., S.P., N.S., J.K. and P.K.; investigation, J.R., S.P., N.S., J.K., M.T. and P.K.; resources, N.S., J.K., M.T., S.M., A.A.P. and P.K.; data curation, J.R., S.P., N.S., J.K. and P.K.; writing—original draft preparation, J.R., S.P., N.S., J.K. and P.K.; writing—review and editing, J.R., N.S., J.K. and P.K.; visualization, J.R., S.P., N.S., J.K., M.T., S.M. and P.K.; supervision, M.T., S.M., A.A.P. and P.K.; project administration, M.T., S.M., A.A.P. and P.K.; funding acquisition, P.K. and M.T. All authors have read and agreed to the published version of the manuscript.
Funding
We would like to acknowledge the Presidential Scholarship from the Graduate School, Chiang Mai University. This research work was partially supported by Chiang Mai University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The DNA sequences data obtained in this study have been deposited in GenBank under accession numbers ITS (OL375165, OL375167, OL375168, OL375169, OL375170, OL375173, GU734762), TEF-1 (OL410297, OL410298, OL410299, OL410300, OL410301, OL410302), RPB1 (OL410303, OL410304, OL410305, OL410306 OL410307 OL410308), RPB2 (OL410309, OL410310, OL410311, OL410312, OL410313, OL410314), and GAS1 (OL469003, OL469004, OL469005, OL469006, OL469007, OL469008).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rosegrant M.R., Ringler C., Sulser T.B., Ewing M., Palazzo A., Zhu T., Nelson G.C., Koo J., Robertson R., Msangi S., et al. Agriculture and Food Security under Global Change: Prospects for 2025/2050. International Food Policy Research Institute (IFPR); Washington, DC, USA: 2009. pp. 145–178. [Google Scholar]
- 2.Womack E.D., Williams W.P., Smith J.S., Warburton M.L., Bhattramakki D., Hesler L. Mapping Quantitative Trait Loci for Resistance to Fall Armyworm (Lepidoptera: Noctuidae) Leaf-Feeding Damage in Maize Inbred Mp705. J. Econ. Entomol. 2020;113:956–963. doi: 10.1093/jee/toz357. [DOI] [PubMed] [Google Scholar]
- 3.Ekasingh B., Gypmantasiri P., Thong-Ngam K., Grudloyma P. Maize in Thailand: Production Systems, Constraints, and Research Priorities. CIMMYT; Bangkok, Thailand: 2014. [Google Scholar]
- 4.Hernandez-Mendoza J.L., López-Barbosa E.C., Garza-González E., Mayek-Perez N. Spatial distribution of Spodoptera frugiperda (Lepidoptera: Noctuidae) in maize landraces grown in Colima, Mexico. J. Trop. Insect Sci. 2008;28:126–129. doi: 10.1017/S1742758408096112. [DOI] [Google Scholar]
- 5.Sartiami D., Harahap I.S., Kusumah Y.M., Anwar R. First record of fall armyworm (Spodoptera frugiperda) in Indonesia and its occurrence in three provinces. IOP Conf. Ser. Earth Environ. Sci. 2020;468:012021. doi: 10.1088/1755-1315/468/1/012021. [DOI] [Google Scholar]
- 6.Abel C.A., Scott M.P. Evaluation of 21 Thailand Maize Germplasms for Resistance to Leaf Feeding Spodoptera frugiperda (Lepidoptera: Noctuidae) J. Kansas Entomol. Soc. 2020;93:97–102. doi: 10.2317/0022-8567-93.1.97. [DOI] [Google Scholar]
- 7.Early R., González-Moreno P., Murphy S.T., Day R. Forecasting the global extent of invasion of the cereal pest Spodoptera frugiperda, the fall armyworm. NeoBiota. 2018;40:25–50. doi: 10.3897/neobiota.40.28165. [DOI] [Google Scholar]
- 8.Aktar W., Sengupta D., Chowdhury A. Impact of pesticides use in agriculture: Their benefits and hazards. Interdiscip. Toxicol. 2009;2:1–12. doi: 10.2478/v10102-009-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Norjmaa U., Nasamdulam D., Enkhjargal B., Banzragch D. Morphological and molecular identification of Beauveria bassiana from agricultural soils. Mong. J. Agric. Sci. 2019;27:20–24. doi: 10.5564/mjas.v27i02.1280. [DOI] [Google Scholar]
- 10.Sandhu S.S., Sharma A.K., Beniwal V., Goel G., Batra P., Kumar A., Jaglan S., Sharma A.K., Malhotra S. Myco-Biocontrol of Insect Pests: Factors Involved, Mechanism, and Regulation. J. Pathog. 2012;2012 doi: 10.1155/2012/126819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajula J., Rahman A., Krutmuang P. Entomopathogenic fungi in Southeast Asia and Africa and their possible adoption in biological control. Biol. Control. 2020;151:104399. doi: 10.1016/j.biocontrol.2020.104399. [DOI] [Google Scholar]
- 12.Draganova S., Donkova R., Georgieva D. Impact of Strains of Entomopathogenic Fungi on Some Main Groups of Soil Microorganisms. J. Plant Prot. Res. 2008;48:169–179. doi: 10.2478/v10045-008-0020-y. [DOI] [Google Scholar]
- 13.Kulu I.P., Abadi A.L., Afandhi A. Morphological and molecular identification of Beauveria bassiana as entomopathogen agent from central kalimantan Peatland, Indonesia. Int. J. ChemTech Res. 2015;8:2079–2084. [Google Scholar]
- 14.Ruiz-Nájera R.E., Ruiz-Estudillo R.A., Sánchez-Yáñez J.M., Molina-Ochoa J., Skoda S.R., Coutiño-Ruiz R., Pinto-Ruiz R., Guevara-Hernández F., Foster J.E. Occurrence of Entomopathogenic Fungi and Parasitic Nematodes on Spodoptera frugiperda (Lepidoptera: Noctuidae) Larvae Collected in Central Chiapas, México. Fla. Entomol. 2013;96:498–503. doi: 10.1653/024.096.0215. [DOI] [Google Scholar]
- 15.Acosta R.I.T., Humber R.A., Sánchez-Peña S.R. Zoophthora radicans (Entomophthorales), a fungal pathogen of Bagrada hilaris and Bactericera cockerelli (Hemiptera: Pentatomidae and Triozidae): Prevalence, pathogenicity, and interplay of environmental influence, morphology, and sequence data on fungal identification. J. Invertebr. Pathol. 2016;139:82–91. doi: 10.1016/j.jip.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Thaochan N., Sausa-Ard W. Occurrence and effectiveness of indigenous Metarhizium anisopliae against adults Zeugodacus cucurbitae (Coquillett)(Diptera: Tephritidae) in Southern Thailand. Songklanakarin J. Sci. Technol. 2017;39:325–334. doi: 10.14456/sjst-psu.2017.35. [DOI] [Google Scholar]
- 17.Wanjiru J., Sunday K. Ovicidal effects of entomopathogenic fungal isolates on the invasive Fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae) J. Appl. Entomol. 2019;143:626–634. doi: 10.1111/jen.12634. [DOI] [Google Scholar]
- 18.Keppanan R., Sivaperumal S., Hussain M., Bamisile B.S., Aguila L.C.R., Qasim M., Mekchaya S., Wangade L., Krutmuang P. Molecular characterization of pathogenesis involving the GAS 1 gene from Entomopathogenic fungus Lecanicillium lecanii and its virulence against the insect host Diaphorina citri. Pestic. Biochem. Physiol. 2019;157:99–107. doi: 10.1016/j.pestbp.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S., Xia Y., Keyhani N.O. Contribution of the gas1 gene of the entomopathogenic fungus Beauveria bassiana, encoding a putative glycosylphosphatidylinositol-anchored β-1,3-glucanosyltransferase, to conidial thermotolerance and virulence. Appl. Environ. Microbiol. 2011;77:2676–2684. doi: 10.1128/AEM.02747-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y., Zhu X., Jiao R., Xia Y. The Magas1 gene is involved in pathogenesis by affecting penetration in Metarhizium acridum. J. Microbiol. Biotechnol. 2012;22:889–893. doi: 10.4014/jmb.1111.11055. [DOI] [PubMed] [Google Scholar]
- 21.Meyling N.V. Methods for Isolation of Entomopathogenic Fungi from the Soil Environment. 1–18 January 2007. [(accessed on 4 November 2021)]. Available online: http://orgprints.org/11200.
- 22.Wang Y., Tang D.X., Duan D.E., Wang Y.B., Yu H. Morphology, molecular characterization, and virulence of Beauveria pseudobassiana isolated from different hosts. J. Invertebr. Pathol. 2020;172:107333. doi: 10.1016/j.jip.2020.107333. [DOI] [PubMed] [Google Scholar]
- 23.Affandi A., Chailani S.R., Mimbar S.M., Wiroatmodjo B. Isolation and Phenotypic Characterization of Morphology in Fungus Beauveria bassiana (Balsamo) Vuillemin Colony Naturally From Leaf Surface, Soil, and Insect as Host in Tomato Plantation. AGRIVITA J. Agric. Sci. 2012;34:303–310. doi: 10.17503/agrivita-2012-34-3-p303-310. [DOI] [Google Scholar]
- 24.Gardes M., Bruns T.D. ITS primers with enhanced specificity for basidiomycetes application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 25.White T.J., Bruns T., Lee S.J.W.T., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990;18:315–322. [Google Scholar]
- 26.Rehner S.A. Primers for Elongation Factor 1-Alpha (EF1-Alpha) 2001. [(accessed on 11 December 2021)]. Available online: http://ocid.NACSE.ORG/research/deephyphae/EF1primer.pdf.
- 27.Castlebury L.A., Rossman A.Y., Sung G.H., Hyten A.S., Spatafora J.W. Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 2004;108:864–872. doi: 10.1017/S0953756204000607. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 29.Edgar R.C. Muscle: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall T. BioEdit. [(accessed on 24 August 2021)]. Available online: http://www.mbio.ncsu.edu/bioedit/bioedit.html.
- 31.Miller M.A., Pfeiffer W., Schwartz T. Gateway Computing Environments Workshop, GCE 2010. [(accessed on 4 November 2021)]. Available online: http://www.proceedings.com/10226.html.
- 32.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and high-performance computing Europe PMC Funders Group. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 34.Rambaut A. FigTree Tree Figure Drawing Tool Version 131. Institute of Evolutionary 623 Biology, University of Edinburgh. [(accessed on 4 November 2021)]. Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 35.Rehner S.A., Minnis A.M., Sung G.H., Luangsa-ard J.J., Devotto L., Humber R.A. Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia. 2011;103:1055–1073. doi: 10.3852/10-302. [DOI] [PubMed] [Google Scholar]
- 36.Serna-Domínguez M.G., Andrade-Michel G.Y., Arredondo-Bernal H.C., Gallou A. Two efficient methods for isolation of high-quality genomic DNA from entomopathogenic fungi. J. Microbiol. Methods. 2018;148:55–63. doi: 10.1016/j.mimet.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 37.Imoulan A., Wu H.J., Lu W.L., Li Y., Li B.B., Yang R.H., Wang W.J., Wang X.L., Kirk P.M., Yao Y.J. Beauveriamedogensis sp. nov., a new fungus of the entomopathogenic genus from China. J. Invertebr. Pathol. 2016;139:74–81. doi: 10.1016/j.jip.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Zhang S.L., He L.M., Chen X., Huang B. Beauveria lii sp. nov. isolated from Henosepilachna vigintioctopunctata. Mycotaxon. 2013;121:199–206. doi: 10.5248/121.199. [DOI] [Google Scholar]
- 39.Rehner J.F., de Muro S.A., Bischoff M.A. Description and phylogenetic placement of Beauveria malawiensis sp. nov. (Clavicipitaceae, Hypocreales) Mycotaxon. 2006;98:137–145. [Google Scholar]
- 40.Glare T., Campbell M., Biggs P., Winter D., Durrant A., McKinnon A., Cox M. Mitochondrial evolution in the entomopathogenic fungal genus Beauveria. Arch. Insect Biochem. Physiol. 2020;105:e21754. doi: 10.1002/arch.21754. [DOI] [PubMed] [Google Scholar]
- 41.Khonsanit A., Luangsa-ard J.J., Thanakitpipattana D., Noisripoom W., Chaitika T., Kobmoo N. Cryptic diversity of the genus Beauveria with a new species from Thailand. Mycol. Prog. 2020;19:291–315. doi: 10.1007/s11557-020-01557-9. [DOI] [Google Scholar]
- 42.Chen W.H., Liu M., Huang Z.X., Yang G.M., Han Y.F., Liang J.D., Liang Z.Q. Beauveria majiangensis, a new entomopathogenic fungus from Guizhou, China. Phytotaxa. 2018;333:243–250. doi: 10.11646/phytotaxa.333.2.8. [DOI] [Google Scholar]
- 43.Robène-Soustrade I., Jouen E., Pastou D., Payet-Hoarau M., Goble T., Linderme D., Lefeuvre P., Calmès C., Reynaud B., Nibouche S., et al. Description and phylogenetic placement of Beauveria hoplocheli sp. nov. used in the biological control of the sugarcane white grub, Hoplochelus marginalis, on Reunion Island. Mycologia. 2015;107:1221–1232. doi: 10.3852/14-344. [DOI] [PubMed] [Google Scholar]
- 44.Rehner S.A., Buckley E.A. Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 45.Spatafora J.W., Sung G.H., Sung J.M., Hywel-Jones N.L., White J.F. Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol. Ecol. 2007;16:1701–1711. doi: 10.1111/j.1365-294X.2007.03225.x. [DOI] [PubMed] [Google Scholar]
- 46.Imoulan A., Elmeziane A. Pathogenicity of Beauveria bassiana isolated from Moroccan Argan forests soil against larvae of Ceratitis capitata (Diptera: Tephritidae) in laboratory conditions. World J. Microbiol. Biotechnol. 2014;30:959–965. doi: 10.1007/s11274-013-1514-y. [DOI] [PubMed] [Google Scholar]
- 47.Sade C., Da B., Roberto J., Parra P. New method for rearing Spodoptera frugiperda in laboratory shows that larval cannibalism is not obligatory. Rev. Bras. Entomol. 2013;57:347–349. [Google Scholar]
- 48.Bolzan A., Padovez F.E., Nascimento A.R., Kaiser I.S., Lira E.C., Amaral F.S., Kanno R.H., Malaquias J.B., Omoto C. Selection and characterization of the inheritance of resistance of Spodoptera frugiperda (Lepidoptera: Noctuidae) to chlorantraniliprole and cross-resistance to other diamide insecticides. Pest Manag. Sci. 2019;75:2682–2689. doi: 10.1002/ps.5376. [DOI] [PubMed] [Google Scholar]
- 49.Safavi S. Isolation, identification and pathogenicity assessment of a new isolate of entomopathogenic fungus, Beauveria bassiana in Iran. J. Plant Prot. Res. 2010;50:158–163. doi: 10.2478/v10045-010-0027-z. [DOI] [Google Scholar]
- 50.Imoulan A., Hussain M., Kirk P.M., El Meziane A., Yao Y.J. Entomopathogenic fungus Beauveria: Host specificity, ecology and significance of morpho-molecular characterization in accurate taxonomic classification. J. Asia Pac. Entomol. 2017;20:1204–1212. doi: 10.1016/j.aspen.2017.08.015. [DOI] [Google Scholar]
- 51.Hegedus D.D., Khachatourians G.G. Construction of cloned DNA probes for the specific detection of the entomopathogenic fungus Beauveria bassiana in grasshoppers. J. Invertebr. Pathol. 1993;62:233–240. doi: 10.1006/jipa.1993.1105. [DOI] [Google Scholar]
- 52.Hassan F.R., Abdullah S.K., Assaf L.H. Pathogenicity of the entomopathogenic fungus, Beauveria bassiana (Bals.) Vuill. endophytic and a soil isolate against the squash beetle, Epilachna chrysomelina (F.) (Coleoptera: Coccinellidae) Egypt. J. Biol. Pest Control. 2019;29:1–7. doi: 10.1186/s41938-019-0169-x. [DOI] [Google Scholar]
- 53.Talaei-Hassanloui R., Kharazi-Pakdel A., Goettel M., Mozaffari J. Variation in virulence of Beauveria bassiana isolates and its relatedness to some morphological characteristics. Biocontrol Sci. Technol. 2006;16:525–534. doi: 10.1080/09583150500532758. [DOI] [Google Scholar]
- 54.Oreste M., Bubici G., Poliseno M., Triggiani O., Tarasco E. Pathogenicity of Beauveria bassiana (Bals.-Criv.) Vuill. and Metarhizium anisopliae (Metschn.) sorokin against Galleria mellonella L. and Tenebrio molitor L. in laboratory assays. Redia. 2012;95:43–48. [Google Scholar]
- 55.Yasin M., Wakil W., Ghazanfar M.U., Qayyum M.A., Tahir M., Bedford G.O. Virulence of entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae against red palm weevil, Rhynchophorus ferrugineus (Olivier) Entomol. Res. 2019;49:3–12. doi: 10.1111/1748-5967.12260. [DOI] [Google Scholar]
- 56.Ekesi S., Akpa A.D., Onu I., Ogunlana M.O. Entomopathogenicity of Beauveria bassiana and Metarhizium anisopliae to the cowpea aphid, Aphis craccivora koch (Homoptera: Aphididae) Arch. Phytopathol. Plant Prot. 2000;33:171–180. doi: 10.1080/03235400009383341. [DOI] [Google Scholar]
- 57.Macuphe N., Oguntibeju O.O., Nchu F. Evaluating the endophytic activities of Beauveria bassiana on the physiology, growth, and antioxidant activities of extracts of lettuce (Lactuca sativa L.) Plants. 2021;10:1178. doi: 10.3390/plants10061178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barra-Bucarei L., González M.G., Iglesias A.F., Aguayo G.S., Peñalosa M.G., Vera P.V. Beauveria bassiana multifunction as an endophyte: Growth promotion and biologic control of Trialeurodes vaporariorum, (Westwood) (Hemiptera: Aleyrodidae) in tomato. Insects. 2020;11:591. doi: 10.3390/insects11090591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russo M.L., Scorsetti A.C., Vianna M.F., Allegrucci N., Ferreri N.A., Cabello M.N., Pelizza S.A. Effects of endophytic Beauveria bassiana (Ascomycota: Hypocreales) on biological, reproductive parameters and food preference of the soybean pest Helicoverpa gelotopoeon. J. King Saud Univ.-Sci. 2019;31:1077–1082. doi: 10.1016/j.jksus.2018.11.009. [DOI] [Google Scholar]
- 60.Xiao G., Ying S.H., Zheng P., Wang Z.L., Zhang S., Xie X.Q., Shang Y., Leger R.J., Zhao G.P., Feng M.G. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2012;2:483. doi: 10.1038/srep00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lai Y., Chen H., Wei G., Wang G., Li F., Wang S. In vivo gene expression profiling of the entomopathogenic fungus Beauveria bassiana elucidates its infection stratagems in Anopheles mosquito. Sci. China Life Sci. 2017;60:839–851. doi: 10.1007/s11427-017-9101-3. [DOI] [PubMed] [Google Scholar]
- 62.Ávila-Hernández J.G., Carrillo-Inungaray M.L., Cruz-Quiroz R.D., Wong-Paz J.E., Muñiz-Márquez D.B., Parra R., Aguilar C.N., Aguilar-Zárate P. Beauveria bassiana secondary metabolites: A review inside their production systems, biosynthesis, and bioactivities. Mex. J. Biotechnol. 2020;5:1–33. doi: 10.29267/mxjb.2020.5.4.1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA sequences data obtained in this study have been deposited in GenBank under accession numbers ITS (OL375165, OL375167, OL375168, OL375169, OL375170, OL375173, GU734762), TEF-1 (OL410297, OL410298, OL410299, OL410300, OL410301, OL410302), RPB1 (OL410303, OL410304, OL410305, OL410306 OL410307 OL410308), RPB2 (OL410309, OL410310, OL410311, OL410312, OL410313, OL410314), and GAS1 (OL469003, OL469004, OL469005, OL469006, OL469007, OL469008).