Abstract
Spatiotemporal regulation of viral capsid assembly ensures selection of the viral genome for encapsidation. The porcine circovirus 2 is the smallest autonomously replicating pathogenic virus, yet it remains unknown how PCV2 capsid assembly is regulated to occur within the nucleus. We report that pure PCV2 capsid proteins, in the absence of nucleic acids, require acidic conditions to assemble into empty capsids in vitro. By employing Constant pH Replica Exchange Molecular Dynamics, we unveil the atomistic mechanism of pH-dependency for capsid assembly. The results show that the appropriate protonation configuration for a cluster of acidic amino acids is necessary to properly position the GH-loop for driving capsid assembly. We demonstrate that assembly is prohibited at neutral pH because deprotonation of these residues results in their electrostatic repulsion, shifting the GH-loop to a position incompetent with capsid assembly. We propose that encapsulation of nucleic acid overcomes this repulsion to properly position the GH-loop. Our findings provide the first atomic resolution mechanism of capsid assembly regulation. The findings are useful for development of therapeutics that inhibit PCV2 self-assembly.
Graphical Abstract
INTRODUCTION
Porcine circovirus (PCV) types 1-3 are 20nm T=1 icosahedral virion that possess circular single stranded DNA genomes and belong to the Circovirus genus of the Circoviridae family 1, 2. These viruses are morphologically similar, but genetically, antigenically and pathogenically distinct. PCV1 is apathogenic and was first detected in porcine kidney cells (PK-15). PCV2 is highly pathogenic, infects nearly every organ of the host, and responsible for porcine circovirus-associated disease (PCVAD) or porcine circovirus disease (PCVD), which culminates in the immunosuppression of the host and death from secondary infections 3-7. Naturally occurring PCV2 infections of animals in the wild include rodents, calves, foxes, minks, goats, racoon dogs, boars, and swine 8-13. Controlled PCV2 infections under laboratory conditions include BALB/c mice, Kumming mice, and multiple human cell lines 14-18. Infections in calves and rodents results in immunosuppression of the host 8, 9, 12, 13, 19. PCV3 was recently shown to associate with multiple diseases in swine that include reproductive failure, dermatitis, and nephropathy syndrome 20-22. Swine farms are billion-dollar industries in swine rearing countries, and PCV2 breakouts can be economically devastating 16. The ability of PCV2 to overcome the species barrier, induce its pathogenic phenotype, and infect human cells in culture is suggestive of a pathogen that is on the precipitous of becoming zoonotic and potentially a threat to human health. Additionally, PCV2 is a deterrent for xenotransplantation, as swine are considered a viable source of organs for humans 23-25. While a vaccination program for PCV2 has been established, PCV2’s high mutation (1.2x10−6 substitutions site−1 year−1) and recombination rates are responsible for recent genotypes demonstrating resistance to vaccination 26, 27. Consequently, alternative therapeutics that disrupt one or multiple stages of PCV2 life-cycle are needed. A promising strategy is to disrupt the self-assembly of viruses within the cell; however, this requires knowledge of the critical factors needed for the successful assembly of infectious virion. Studies describing PCV2 capsid assembly are limited 19, 28, 29. Recent work demonstrated that capsid assembly occurs within the nucleus of cells expressing the capsid protein (CP) 30. Knowledge of the PCV2 assembly mechanism will increase our understanding of virus capsid assembly and provide an avenue for developing therapeutics to inhibit PCV2 self-assembly.
The PCV2 CP (27.8 kDa) possesses a forty-two amino acid N-terminus where nearly half of the amino acids are arginines. Hereafter, we refer to this N-terminus as the arginine rich motif (ARM). The ARMs possess multiple nuclear localization signals (NLS) that are responsible for trafficking the virus to the nucleus during the entry process and/or genome packaging 31. Other functions associated with the ARM include disruption of liposomal membrane for cellular entry, and interaction with the negative charged phosphates of the genome 32. Expression of the CP in Trichoplusia ni (insect) or HEK293 (human) cells results in the formation of nucleic acid containing capsids within the cells 30, 33, 34. The capsid structures from these expression systems are nearly indistinguishable and demonstrate the CP to adopt the canonical viral jelly roll fold prevalent in many viral capsid proteins. However, the ARMs cannot be modeled in any of these studies 30, 33, 35-37. This is most likely because the ARMs either do not adopt the icosahedral symmetry imposed during structure determination, or that the ARMs are unstructured.
PCV2 capsids assemble within the cell to enclose nucleic acid; however, PCV2 variants designed to eliminate this interaction are unable to assemble into capsids within the cell 30. Others have shown that capsids can assemble in vitro to enclose nucleic acid -suggesting that the interaction between CP and nucleic acid is a requisite for capsid assembly 38. We have been able to establish an in vitro protocol for assembling PCV2 capsids in the absence of nucleic acid. This allows us the unique opportunity to study capsid assembly in the absence of nucleic acid. By combining our biochemical assays with in silico studies, we demonstrate that ARM is not necessary for in vitro capsid assembly, and report that a protonation event of the CP is responsible for regulating capsid assembly. In particular, the CPs assemble into capsids at pH 5, and not at pH 3 or 7. This suggests that pH 5 somehow mimics cellular conditions necessary for capsid self-assembly, and the CP at this pH is likely to resemble the assembly competent structure prior to cellular capsid assembly. Concurrently, the in silico approach demonstrates the structural changes at pH 3 and 7 responsible for inhibiting assembly. We emphasize that it is not trivial to determine the atomic structure of monomeric CP at multiple pH values using experimental approaches. We have shown that full-atom Molecular Dynamics (MD) simulation of entire PCV2 capsid faithfully represents its structure and dynamics and, thus, can be used as a reliable source of information complementing and explaining experimental data 39-42. Here, we use MD to study the conformational landscape of the CP at different pH values. The use of conventional MD for studying the effect of pH on structure is problematic as this approach maintains a single protonation state for each residue throughout simulation. Thus, the dependence between changes in protonation states and changes in the structure are lost, and potentially inaccurate conformations and dynamics are sampled. In the computational part of this work, we interpret the biochemical results to determine what happens to the monomeric CP structure at these pH values by performing recently developed Constant pH Molecular Dynamics simulation (CpHMD) with Replica Exchange (RE) techniques in pH-dimension (CpH REMD) 43. Unlike the classical MD approach, CpHMD simulation allows for the change of amino acid protonation states during the simulation. The RE technique provides enhanced sampling. Capsid assembly originates from monomeric CPs assembling into larger oligomers; thus, atomistic knowledge of the CP monomer is a requisite for understanding capsid assembly. We study the pH-dependent structural features of CP using CpH REMD.
We demonstrate that CpH REMD provides interpretation for the observed phenomena of pH driven in vitro capsid assembly. The CP conformation suited for capsid assembly is reliant on the position of the GH-loop, located at the 3-fold icosahedral axes of symmetry (iaos), and this position is regulated by a protonation/deprotonation event involving two pairs of acidic amino acids that we refer to as the dyads. Our results can also explain regulation of PCV2 self-assembly within the cell. We recently demonstrated that the GH-loop interacts with nucleic acid at the 3-fold iaos of nucleic acid containing VLP 30. We hypothesize that cytoplasmic capsid assembly is prohibited by electrostatic repulsion of these dyads, and that interaction of the GH-loop with nucleic acid inside the nucleus at the 3-fold iaos may be the driving factor for overcoming this repulsion. To the best of our knowledge, this is first description of how PCV2 regulates its capsid assembly.
RESULTS
In vitro capsid assembly is driven by pH
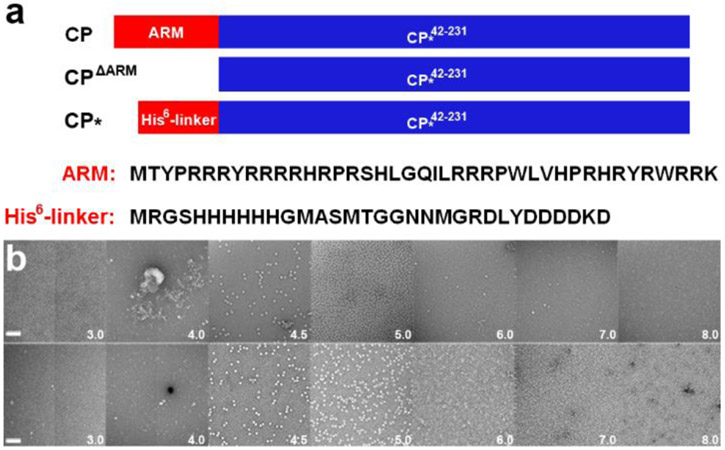
The PCV2 CP (GenBank: AAO39759.1) can be expressed and purified from E. coli to homogeneity as monomeric subunits 32, 33. These samples are devoid of nucleic acid as determined by a 260 nm / 280 nm ratio. Incubation of CP in low pH conditions for several weeks produces capsids; however, deletion or modification of the ARM results in robust and rapid capsid assembly 32. This may be because nucleic acid is needed to neutralize the positive charge associated with the ARM. Therefore, we chose to substitute and exclude the ARM from our assembly studies. Here we discuss two variants: 1) CP*, which replaces the ARM with a hexa-histidine affinity tag followed by a twenty-six amino acid linker (His6-linker); and 2) CPΔARM, which excludes the first forty-two amino acids (Fig. 1). The capsid assembly buffer was determined by optimizing the buffer composition used to produce crystals of PCV2 capsids for X-ray crystallography 33. Reducing the pH of the assembly buffer from 8.0 to 3.0 produces the greatest number of capsids at pH 5 (Fig. 1b). Assembled capsids are not observed at pH values greater than 6.0 despite increasing the CP or PEG concentration. This suggests that protonation of one or more titratable residues (Asp, Glu or His) may be responsible for regulation of capsid assembly. To further explore this possibility, we used molecular dynamics to study the conformations of CP* at different pH values. We note that the ability of CPΔARM to assemble with a similar pH profile as the CP* suggests that the N-terminus sequence in CP* does not affect capsid assembly (Fig. 1).
Fig. 1.
In vitro PCV2 capsid assembly. (a) Schematic representations of full-length CP, CPΔARM, and CP∗ describing differences between proteins. The amino acid sequences of the N-termini are written below. (b) Transmission electron micrographs of negative stained in vitro assembled PCV2 capsid. Top, effect of pH (white text) in assembly of CP∗ capsid. Bottom, effect of pH (white text) in assembly of CPΔARM capsid.
Conformational sampling of CP∗ with CpH-REMD simulation
To understand the relationship between capsid assembly and pH, we investigated the representative CP* conformations at pH 3, 5, and 7 using CpHMD simulations in combination with pH-REMD to enhance conformational sampling. We set 10 replicas, 1 μs each, spanning the pH range from 3.0 to 7.5 at 0.5 pH-unit increments. There are 32 amino acids (Asp, Glu, His) in the CP sequence that are titratable in this pH range. These residues were the target of titration during the MD simulation.
Coordinates for the N-terminus are not present in any of the experimental structures of PCV2 30, 32-34, 38; therefore, we modeled it using MODELLER 44. The convergence of simulation was monitored by observing the evolution of free energy surfaces (FES) as a function of variables Rg vs. SASA and Rg vs. RMSD (Supplementary Fig. 1-11). The comparison of the Rg vs. SASA FES suggests the CP to adopt a more compact shape at pH 7 -having smaller Rg and SASA. RMSD was calculated from the alignment of each conformation to the capsid CP structure (PDB entry 3R0R). Initial alignments produced large (> 20 Å) RMSD and Rg values (< 29 Å). We hypothesized that a flexible N-terminus may be responsible for the large Rg, SASA and RMSD values. Exclusion of the His6-linker reduced the RMSD (< 7.5 Å) and Rg (< 22 Å) values significantly (Supplementary Fig. 4 - 6). Analysis of FES indicated that our CpH-REMD simulation converged within 1 μs. We note that because our biochemical experiments demonstrate CP∗ and CPΔARM to exhibit comparable assembly profiles, we conclude that the pH-dependent conformational changes regulating self-assembly must involve the segment CP∗42-231. We focus on the section without the His6-linker (CP∗42-231).
Free energy surfaces at different pH values demonstrate different CP conformations
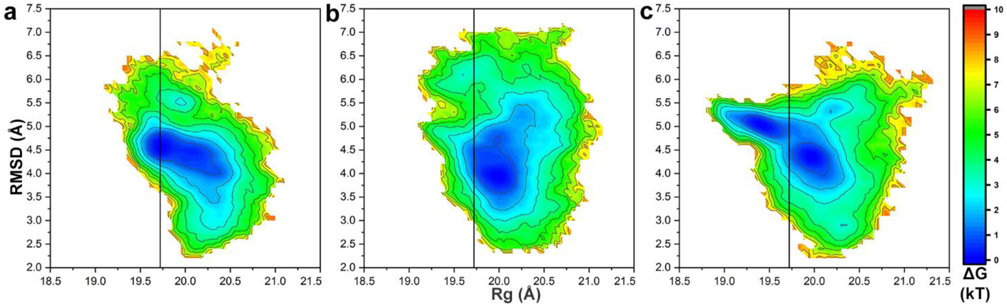
Figure 2 shows the conformational landscapes for each pH by the free energy surfaces in Rg and RMSD values. Highly populated regions (blue) corresponding to minimum energy of CP conformations were identified during the 1 μs simulation. The distribution of these regions is unique for each pH value. A minimum is observed at Rg value of 19.7 Å and RMSD value of 4.6 Å for pH 3. A minimum for pH 5 is observed at Rg 20.0 Å and RMSD 4.0 Å. Two minima are observed at pH 7 with Rg 20.0 Å and RMSD of 4.4 Å (min 1), and Rg 19.5 Å RMSD 5.0 Å (min 2).
Fig. 2.
Conformational landscape sampled by the capsid protein CP∗ 42-231. Free energy surface as function of parameters RMSD (CP∗ 42-231 Cα atoms) vs. Rg (CP∗ 42-231 Cα atoms) from the simulation at (a) pH 3, (b) pH 5, (c) pH 7. RMSD was calculated by comparing conformations generated from 1 μsec of simulation to the reference CP in the assembled capsid (PDB entry 3R0R). Vertical black lines signify Rg value of PDB entry 3R0R.
The existence of two minima suggests the presence of at least two predominant conformations at pH 7. Remarkably, the RMSD value of the minimum at pH 5 is lower than at pH 3 or 7, meaning that the conformation of CP∗42-231 at pH 5 may be more similar to the conformation observed in the assembled capsid.
pH changes the conformation of CD and GH-loops
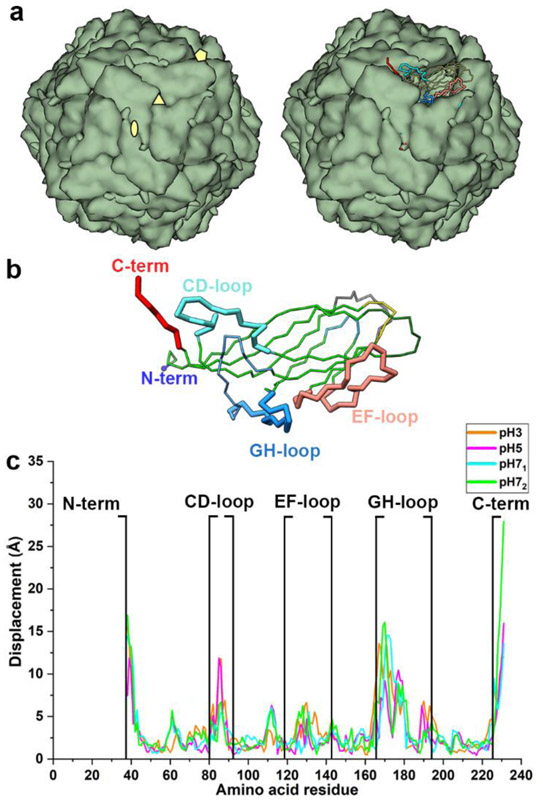
Representative CP conformations for each pH were identified using structural clustering of the extracted structures from the minima at each pH. To determine the effect of pH on the CP structure, we compared the representative conformation of each cluster (centroid) to the capsid CP structure (Fig. 3a) 33, 37. Four regions deviate significantly from the capsid CP structure: the N-terminus (His6-linker), the CD-loop (residues 80-92), the GH-loop (residues 166-194), and the C-terminus (residues 225-231) (Figs. 3a, b and 4). There are thirteen titratable residues in the N-terminus (7 His and 6 Asp). Our pKa calculations demonstrated that the N-terminus His and Asp residues are fully and partially protonated at pH 3, respectively (Table 1). Increasing of pH from 3 results in partial deprotonation of these His residues and complete deprotonation of these Asp residues at pH 5, and complete deprotonation of both at pH 7. In the assembled capsid, the N-terminus helps define the CP:CP interface at the 2-fold iaos (Fig. 3a, 4a). At pH 3 and 5, the N-terminus is positioned near the 2-fold iaos (Figs. 4b, c). On the other hand, at pH 7 it is positioned near the face that defines the inner surface of the capsid, due to the ionic interactions between the negatively charged Asp in the N-terminus and the positively charged Arg and Lys at the capsid inner surface (Fig. 4d, e).
Fig. 3.
Conformational variability of CP at pH 3, 5 and 7. (a) left: PCV2 capsid with CP shown as green blobs. The icosahedral axes of symmetry (iaos) are shown as pentagon (5-fold), triangle (3-fold), and ellipse (2-fold). Right: one of the blobs (subunits) is shown as a Cα tube representation with the important loops (N-term, CD, and GH as thick tubes). (b) PCV2 CP Cα tube representation with important loops (thick tubes) labelled. (c) distances between equivalent Cα atoms of the representative conformations at pH 3, 5, 7 after alignment to capsid structure (PDB entry 3R0R). Panels a and b were made using the structure of the capsid. In the context of the assembled capsid, the N-terminus is in the interior of the capsid near the 2-fold iaos, CD-loop is located on the surface of the capsid near the 2-fold iaos, GH-loop is buried near the 3-fold iaos, and the C-terminus is located on the capsid surface near the 2-fold iaos.
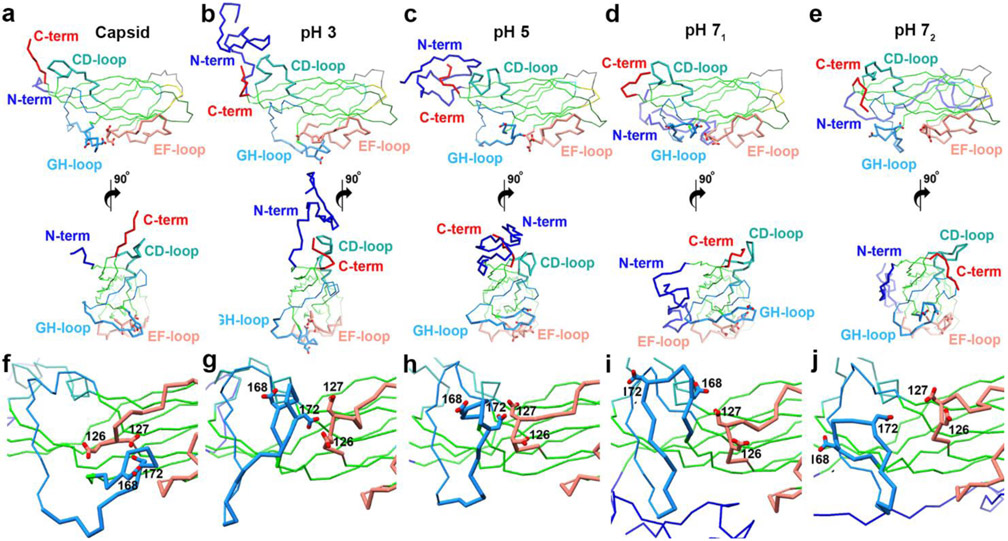
Fig. 4.
Representative conformations from the first cluster at each pH value. Regions with significant deviation from the capsid structure are highlighted according to Fig. 3b. a-e top versus bottom) Cα trace of two orientations related by an anticlockwise 90° rotation about the y-axis. a) CP extracted from assembled capsids, b) representative of most populated cluster at pH 3, c) representative of most populated cluster at pH 5, d) representative of most populated cluster from first peak at pH 7, e) representative of most populated cluster from second peak at pH 7, f-j) close-up of EF-(pink) and GH-loops (blue) showing their acidic dyads f) CP extracted from assembled capsids, g) representative of most populated cluster at pH 3, h) representative of most populated cluster at pH 5, i) representative of most populated cluster from first peak at pH 7, j) representative of most populated cluster from second peak at pH 7. RMSD values for the representative structures: b – 4,6 Å; c – 3,9 Å; d – 4.3 Å; e – 5.1 Å.
Table 1.
Titratable amino acid calculated pKa values.—pKa values were calculated using the conformations extracted from the minima of pH 3, 5 and 7. Blue and red indicate protonated and deprotonated, respectively. Amino acids Asp126, Asp127, Asp168, and Asp172 are in bold.
| pH 3 | pH 5 | pH 71 | pH 72 | pH 3 | pH 5 | pH 71 | pH 72 | ||
|---|---|---|---|---|---|---|---|---|---|
| HIS 10 | 5.1 | 5.6 | 6.0 | 5.5 | GLU 104 | 3.3 | 3.0 | 3.3 | 3.4 |
| HIS 11 | 5.3 | 5.5 | 6.1 | 5.5 | ASP 115 | 3.9 | 4.3 | 5.3 | 4.8 |
| HIS 12 | 4.7 | 5.4 | 5.8 | 5.5 | ASP 126 | 3.5 | 4.0 | 4.6 | 4.2 |
| HIS 13 | 5.1 | 5.5 | 5.9 | 5.7 | ASP 127 | 3.6 | 4.4 | 5.0 | 5.1 |
| HIS 14 | 4.9 | 5.5 | 6.3 | 6.4 | ASP 139 | 3.6 | 4.4 | 4.9 | 4.1 |
| HIS 15 | 5.0 | 5.7 | 6.0 | 5.8 | HIS 148 | 4.9 | 5.2 | 5.4 | 5.2 |
| ASP 29 | 3.0 | 3.9 | 3.9 | 3.8 | HIS 157 | 6.6 | 6.3 | 6.9 | 7.4 |
| ASP 32 | 2.8 | 3.9 | 4.5 | 5.2 | ASP 168 | 3.7 | 3.9 | 4.6 | 4.5 |
| ASP 33 | 2.9 | 3.6 | 3.9 | 4.3 | ASP 172 | 6.1 | 5.8 | 5.6 | 4.2 |
| ASP 34 | 3.0 | 4.1 | 5.0 | 3.4 | ASP 194 | 2.0 | 2.8 | 3.1 | 3.3 |
| ASP 35 | 3.5 | 4.1 | 4.6 | 2.6 | HIS 195 | 6.3 | 6.5 | 6.8 | 7.3 |
| ASP 37 | 3.3 | 4.0 | 4.3 | 3.6 | GLU 203 | 4.3 | 5.7 | 5.9 | 6.1 |
| HIS 38 | 5.6 | 6.1 | 6.3 | 6.2 | ASP 208 | 2.8 | 3.4 | 3.5 | 3.9 |
| ASP 70 | 4.9 | 5.1 | 5.2 | 5.6 | GLU 210 | 4.6 | 5.1 | 5.1 | 5.0 |
| ASP 78 | 2.3 | 3.2 | 3.8 | 3.8 | GLU 223 | 4.3 | 4.9 | 5.3 | 5.3 |
| GLU 94 | 2.9 | 3.7 | 4.4 | 4.0 | ASP 228 | 4.4 | 5.0 | 5.3 | 4.9 |
In the assembled capsid, the CD-loop contacts an equivalent loop in the neighboring subunit (Fig. 3a). Our MD results demonstrate the CD-loop to adopt distinct conformations at each pH (Fig. 4); however, the lack of titratable residues in the loop suggests that the observed variability may be intrinsic to the loop. Biochemical studies have demonstrated that substitution and truncation of the CD-loop do not abrogate capsid assembly, demonstrating that CD-loop is likely not a deciding factor in capsid assembly 28. The GH-loop positions are more interesting: the loop adopts the same position as in assembled capsid at pH 5, however, the loop is closer to the EF-loop at pH 3, or twists and moves closer to the CD-loop at pH 7 (Figs. 4f-j). From the analysis of the experimental capsid structure, the GH-loop contributes a third (1000 Å2) of the CP surface that is buried upon capsid assembly 45. We explore the significance of the GH-loop further below. In the assembled capsid, the C-terminus also helps define the CP:CP interface at the 2-fold iaos. Our MD results demonstrate the C-terminus to adopt distinct positions at each pH value (Fig. 4a-e, and supplementary Figs. 16, 18-21). There is a single titratable residue (Asp228) in the C-terminus, and pKa calculations suggest that it becomes deprotonated at pH values greater than 5 (see below). Zhan et al. recently published that only the main chain atoms of amino acids 225-231 are necessary for capsid assembly, as deletion but not substitution of this region abolished capsid assembly 29. It is likely that the C-terminus adopts the necessary position and conformation when two subunits interact at the 2-fold iaos.
Two aspartic dyads located in the EF- and GH-loops regulate capsid assembly
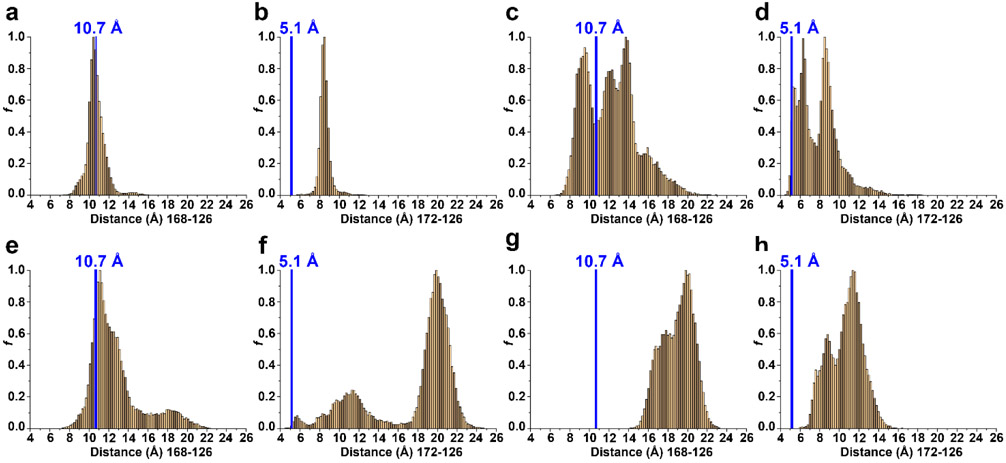
In the assembled capsid, two acidic residues in the EF-loop (Asp126 and Asp127) and two in the GH-loop (Asp168 and Asp172) are located in close proximity (Fig. 4f-j). Their protonation state may be responsible for the observed conformational variability of the GH-loop, so they were titrated in the simulation. To quantify the position of the GH-loop relative to the EF-loop at the three pH values, we calculated distances between the amino acids of these two loops for the extracted conformations (Fig. 2). Figure 5 represents the distribution of distances between the centers of mass for residues 168 and 126 (hereafter denoted as 168:126) (Figs. 5a, c, e, g) and residues 172:126 (Figs. 5b, d, f, h).
Fig. 5.
Distribution of distances between centers of mass for residues 126 and 168 (a, c, e, g) and residues 126 and 172 (b, d, f, h); at (a, b) pH 3; (c, d) pH5; (e, f) pH 7 (minimum 1); and (g, h) pH7 (minimum 2). The blue vertical line on each graph represents the distance between these residues in the capsid structure (10.7 Å between 168 and 126; 5.1 Å between 172 and 126). The distribution was calculated for all structures extracted from free energy surface minima at each pH
We compared the observed distance distribution at each pH with the distances in the experimentally determined capsid structure. At pH 3, the monomodal distribution between residues 168:126 (10-11.2 Å) was similar to the distance observed in the capsid structure (10.7 Å); however, distances between residues 172:126 were distributed monomodally (8.1-8.7 Å) and were considerably greater than that of the capsid structure (5.1 Å) (Figs. 5a, b). At pH 5, the multimodal distribution between residues 168:126 (8.5-10.5 Å and 10.9-14.5 Å) and 172:126 (5.1-6.7 Å and 8.1-9.3 Å) was similar to the distances observed in the capsid structure (Figs. 5c, d). At pH 7 minima 1, the multimodal distribution between residues 168:126 (10.3-13.1 Å and 16.0-20.0 Å) was similar to the distance observed in the capsid structure; however, the multimodal distribution between residues 172:126 (8.2-14.4 Å and 18.7-21.3 Å) is significantly greater than that observed in the capsid structure (Figs. 5e, f). These values reflect the observed twisting of the GH-loop in Fig. 5d, where Asp168 adopts the same position as in the assembled capsid, while Asp172 is pushed away from the EF-loop. At pH 7 minima 2, the bimodal distributions between amino acids 168:126 and 172:126 are considerably greater than that of the capsid structure (Figs. 5g, h). The increased distances between these dyads at pH 7 versus 5 demonstrates that the GH-loop moves away from the EF-loop at neutral pH.
The most important factor responsible for this movement could be repulsive forces between the two deprotonated dyads. To address this possibility, we calculated the pKa values of titratable amino acids from the conformations extracted from each minimum (Fig. 2, Table 1). Based on the calculated pKa values shown in Table 1, Asp126 and Asp127 (EF-loop) are predominantly protonated at pH 3, partially deprotonated at pH 5, and predominantly deprotonated at pH 7 for both minima. Asp168 (GH-loop) is predominantly protonated at pH 3 and deprotonated at pH 5 and pH 7 (for both minima). Asp172 is protonated at pH 3, protonated at pH 5, and deprotonated at pH 7 (for both minima). Protonation of the four aspartic acids (two dyads) at pH 3 allows the EF- and GH-loops to be kept in close proximity (Fig. 4g). At pH 5, Asp126, Asp127 and Asp168 are deprotonated while Asp172 is protonated. The deprotonation event of these three aspartic acids results in electrostatic repulsion pushing the GH-loop away from the EF-loop (Fig. 4h) to the position observed in the capsid structure (Fig. 4f). At pH 7, all four aspartic acids are deprotonated, and additional electrostatic repulsion pushes GH-loop further from EF-loop (Figs. 4i, j). For nearly all representative conformations in two minima at pH 7 (Fig. 2c), we observed twisting of the GH-loop (Figs. 4i, j).
DISCUSSION
The ability of virion to regulate the assembly of their CP to generate infectious virion may provide them greater evolutionary fitness over virion whose CP assemble uncontrollably into aggregates, empty, or cellular nucleic acid filled capsids within the infected cell. Some of these regulatory mechanisms may be inherent to the viral CP or capsid. Understanding the mechanisms responsible for this regulation are requisite to understanding viral life cycles, and the development of therapeutics to disturb formation of infectious virion. It has been shown that expression of PCV2 CP in mammalian cells results in capsid assembly solely within the nucleus 30. This suggests that PCV2 is able to regulate capsid formation by inhibiting assembly within the cytoplasm, and triggering it within the nucleus where PCV2 genome replication occurs. In this work we begin to study how PCV2 performs this regulation by using biochemical and computational approaches.
Within the cell, CP assembles near neutral pH to enclose nucleic acid. In vitro, CP assembles near neutral pH only in the presence of nucleic acid; thus, the interaction between CP and nucleic acid is a requisite for capsid assembly 38. The CP:nucleic acid is likely driven by the high Arg content of the ARM. To diminish the CP:nucleic interaction, and to focus on CP:CP interaction during capsid assembly, we chose to substitute and exclude the ARM from our assembly studies. Here we demonstrate that PCV2 CP devoid of nucleic acid assemble in vitro only under acidic conditions (pH 5, Fig. 1). Earlier structural studies demonstrated these capsids to be indistinguishable from capsids assembled within insect and mammalian cells 30, 32, 33, 37. Consequently, in the absence of nucleic acid, pH 5 can substitute as the driving factor for CP assembly. To understand how pH 5 influences capsid assembly, we simulated CP at different pH values using Constant pH MD simulation with Replica Exchange technique in pH-dimension. The CpH-REMD simulations demonstrated that the GH-loop adopts the position observed in the capsid only at pH 5 (Fig. 4f, h). Calculation of pKa values using the CP conformations generated by CpH-REMD demonstrated that the GH-loop adopts the proper position at pH 5 because Asp126/Asp127 (EF-loop) and Asp168 (GH-loop) are deprotonated while Asp172 (GH-loop) is protonated (Fig. 4h). However, these aspartic acids are deprotonated at pH 7, and electrostatic repulsion between them pushes the GH-loop away from the EF-loop to suppresses capsid assembly (Fig. 4i, j). Alignment of all GenBank PCV2 CP sequences demonstrated that at least three acidic amino acids (Asp/Glu) are present in the four positions, and that Asp172 is absolutely conserved 30. This is in support of our hypothesis that PCV2 utilizes Asp172 for regulating capsid assembly. In the context of infection, the electrostatic repulsion described by the CpH-REMD study remains to be addressed because the pH of the cytoplasm and nucleus are anticipated to be close to neutral.
An electrostatic potential map of PCV2 capsid demonstrated its inner surface to be positively charged 40. Amino acids responsible for these charges are likely to interact with the negative charged phosphates of nucleic acid packaged within the capsid 41. Indeed, it has been shown that amino acid substitutions that interfere with CP-nucleic acid phosphate interaction abolish capsid assembly 30. Given that Khayat et al. have demonstrated the GH-loop (Lys179) to interact with the backbone phosphates of nucleic acid and the importance of this loop for capsid assembly (as demonstrated by these studies), we propose that the interaction between the GH-loop and nucleic acid may be responsible for properly positioning the GH-loop for capsid assembly 30. While others have demonstrated capsid assembly of E. coli expressed CP at pH 7, it remains to be determined if their material contained nucleic acid that may have facilitated the assembly process 29, 46. Our results also demonstrate that the presence of CP ARM is not necessary for in vitro capsid assembly at pH 5.
Reports of PCV2 vaccine failure outbreaks and genotype shifts suggest that the development of antiviral therapeutics is necessary 19, 33, 47. Our results show that specific positioning of the GH-loop is important for PCV2 capsid assembly, and distraction from this position may inhibit assembly. Therefore, compounds capable of controlling the position of the GH-loop may serve as potential candidates for inhibiting PCV2 capsid assembly. We propose that two families of compounds may achieve this task. One family would position the GH-loop such that assembly is not possible. The alternative family would promote the CP conformation that is capable of assembling into capsids within the cytoplasm, where the PCV2 genome is absent. Such assemblies are noninfectious particles. These compounds may be effective inhibitors by promoting capsid assembly in the absence of nucleic acid. Thus, our observed CP conformations have physiological importance.
In conclusion, we believe that our finding is of fundamental and practical importance, as it provides an understanding of how a virus regulates its self-assembly and provides new opportunity for development of antiviral therapeutics. To the best of our knowledge, this is first explanation for the PCV2 assembly mechanism.
MATERIALS AND METHODS
Purification of PCV2 capsid protein
The preparation of the CP∗ protein (Fig. 1) is as previously described 33. The CP lacking the ARM (CPΔARM) expression construct was generated by genetically fusing a His6 small-ubiquitin related modifier (SUMO) at the N-terminus of the CP (amino acids 42-233) (pET28a plasmid, Novagen) using the services of GenScript. Both constructs were expressed in the Escherichia coli Rosetta DE3 strain (Novagen). Cells were grown in Terrific Broth (TB) media at 37 °C until mid-log phase in the presence of appropriate antibiotic. Protein expression was induced with 0.25 mM Isopropyl β-D- 1-thiogalactopyranoside (IPTG) for 16 h at 20 °C. The cells were harvested by centrifugation (30 min, 4,000 rpm, 4 °C). The pellet was lysed in 50 mM N- cyclohexyl-3-aminopropanesulfonic acid (CAPS) pH 10.5, 50 mM sodium chloride (NaCl), 0.2 mM phenylmethanesulfonyl fluoride (PMSF), 0.5 mM tris (2-carboxyethyl) phosphine (TCEP) and 25 units of benzonase (MilliporeSigma). The lysate was centrifuged (30 min, 16.000 rpm, 4 °C) and the supernatant was subjected to Ni-NTA gravity flow chromatography. The protein was eluted with the lysis buffer supplemented with 1 M imidazole pH 10.5. The elution was treated with 25 units of benzonase while dialyzing in lysis buffer overnight. The protein was further purified using tandem cation-anion ion-exchange chromatography (HiTrap SP HP and HiTrap Q FF column, GE healthcare). Protein was bound to the HiTrapQ FF column and eluted around 200 mM NaCl. 250 mM L-Arg was added to the eluent and the protein was concentrated to 20 mg ml−1. Protein concentration was determined using the method of Gill and von Hippel 48, aliquoted into 50 μl, flash frozen in N2(l), and stored at −80 °C.
SUMO-specific protease expression and purification
The laboratory of Dr. Patrick J. Loll provided the expression construct for the SUMO-specific protease (dtuD1). The protein was expressed and purified following the publications protocol 49.
In vitro capsid assembly
The CPΔARM protein was incubated with dtuD1 to release the free CPΔARM. The composition of the digested material was confirmed using SDS-PAGE analysis. To determine the capsid assembly condition, 3 mg ml−1 capsid protein was mixed with different assembly buffers in a 1:1 ratio and incubated at 4 °C for 48 hours. The first group of assembly buffer was 12% polyethylene glycol (PEG) 3350, 5% iso-propanol, 0.6 M Ammonia Citrate (pH 3.0, pH 4.0, pH 4.5, pH 5.0, pH 6.0, pH 7.0 and pH 8.0). The second group of assembly buffer was 0.6 M Ammonia Citrate (pH 5.0), 5% iso-propanol, 3%, 6%, 9%, 12% and 20% PEG 3350.
Negative stained grids were prepared by applying 5 μl of the sample at 0.1 mg ml−1 to a 400-mesh carbon coated grid for 3 seconds. The specimen was wicked away and stained with 2% uranyl formate (UF). Three equivalent grids were prepared and imaged for each condition. A JEOL 1230 electron microscope at the New York Structural Biology Center (NYSBC) was used for imaging the specimen at a magnification of 60,000x. Imaging was performed using the Leginon package 50.
Molecular dynamics simulation
All simulations were performed using AMBER 18 package 51. The starting structure for simulations was derived from the crystal structure of a CP∗ subunit of the PCV2 capsid (PDB entry 3R0R) 33. The unresolved first 32 amino acids of the structure were reconstructed with topology modelling in MODELLER 44 and added to the CP structure. The initial solvated system of the simulation was prepared with the leap module of AmberTools. The protein was solvated with TIP3P water model in a cubic box of the size providing 10 Å between protein and edges. To neutralize the system and mimic 0.15 M ionic strength, 128 Cl− and 115 Na+ ions were added. The total number of atoms in the system was 129762. The force field parameters of AMBER FF10 51 was assigned for the protein. The SHAKE algorithm was applied to bonds involving hydrogens bonds 52. The long-range Coulomb interactions were treated with the particle mesh Ewald (PME) method 53 and the direct space cutoff distance was set to 8 Å. The van der Waals interactions were calculated using 8 Å cutoff.
Energy minimization of the solvated system was performed with steepest descent followed by conjugate gradient algorithm for 5000 steps. The energy-minimized system was then heated for 400 ps from 10 K to 300 K using Langevin dynamics (collision frequency of 5 ps−1). Then, 4 ns simulation of the system was performed in NPT ensemble. The temperature and pressure were kept constant at 300 K and 1 atm by Langevin thermostat and Berendsen barostat respectively. Furthermore, an additional 4 ns simulation was performed in NVT ensemble with the Langevin thermostat maintaining temperature 300 K.
For constant pH MD simulation, we used discrete protonation state model proposed by Mongan and developed by Swails for MD propagated in explicit solvent 43. After each 200 fs of MD simulation performed in explicit solvent, the protonation change event was attempted for all titratable residues in GB implicit solvent 54, solvent relaxation time was set to 200 fs. In CpHMD simulation, all Asp, Glu and His residues (a total of 32 ionizable residues) were titrated. We performed 10 replicas in pH-dimension spanning the pH range 3.0 – 7.5 at 0.5 pH-unit intervals, each replica was simulated for 1 μs in NVT ensemble, resulting in a total of 10 μs of simulation. Attempts to exchange between replicas were set to every 200 fs of the simulation. The CpHMD simulations were performed using PMEMD with GPU acceleration 55-59. Exchange plots for each replica presented in SI(Supplementary Fig 13).
Data Analysis of the simulation
Trajectory reconstructions for data analysis were performed for pH 3, 5 and 7 using Cpptraj program in AMBERTools. For each free energy surface, the free energy of the global minimum was set to 0 kT. Structural analysis at each pH was performed on structures extracted from free energy minima at each pH (Fig. 2). Hierarchical agglomerative approach was applied for clustering the extracted conformations, all-vs-all Cα atoms, with 2 Å cutoff 60. Processing of protonation states was performed with cphstats in AMBER 55-59 and pKa values were calculated using Henderson–Hasselbalch equation based on the ratio of time points with a protonated vs. deprotonated amino acid with inhouse written program 54. Cpptraj in AMBERTools was used for distance calculations between the specified residues center of mass, solvent accessible surface area (SASA), radius of gyration (Rg), and Cα root mean square deviation (RMSD) 60. Buried surface area (BSA) values were calculated using PDBePISA server 61. Contour plots for population maps, distances histograms, and ribbon diagrams were made in Origin 2018b (OriginLab), MOE (Chemical Computing Group, Inc) and UCSF Chimera 62.
Supplementary Material
ACKNOWLEDGEMENTS
The calculations were performed on the HOKUSAI Big Waterfall system at RIKEN; E.T. acknowledges a JSPS International Research Fellowship, grant number PE17023. We would like to thank Dr. Yoshinori Hirano, Dr. Gentaro Morimoto for their technical support, and Yumie Ohyama for administrative support. Funds responsible for supporting these studies were provided by NIH National Institute of General medical Sciences and National institute of Allergy and Infectious Diseases (5SC1AI114843) to R.K., by Grant Number 5G12MD007603-30 from the National Institute on Minority Health and Health Disparities to R.K. Electron microscopy was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310).
Footnotes
CONFLICT OF INTEREST
The authors declare that no conflict of interest exists.
Reference
- 1.Todd D, Avian Pathol, 2000, 29, 373–394. [DOI] [PubMed] [Google Scholar]
- 2.Rosario K, Breitbart M, Harrach B, Segales J, Delwart E, Biagini P and Varsani A, Arch Virol, 2017, 162, 1447–1463. [DOI] [PubMed] [Google Scholar]
- 3.Mankertz A, Mankertz J, Wolf K and Buhk HJ, J Gen Virol, 1998, 79 (Pt 2), 381–384. [DOI] [PubMed] [Google Scholar]
- 4.Opriessnig T, Meng X-J and Halbur PG, Journal of Veterinary Diagnostic Investigation, 2007, 19, 591–615. [DOI] [PubMed] [Google Scholar]
- 5.Ramamoorthy S and Meng XJ, Anim Health Res Rev, 2009, 10, 1–20. [DOI] [PubMed] [Google Scholar]
- 6.Segalés J, Virus Res, 2012, 164, 10–19. [DOI] [PubMed] [Google Scholar]
- 7.Deng ZB, Yuan AW, Luo W, Wang ND, Gong QL, Yu XL and Xue LQ, Acta Vet Hung, 2013, 61, 234–243. [DOI] [PubMed] [Google Scholar]
- 8.Li L, Shan T, Soji OB, Alam MM, Kunz TH, Zaidi SZ and Delwart E, J Gen Virol, 2011, 92, 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halami MY, Freick M, Shehata AA, Muller H and Vahlenkamp TW, Vet Microbiol, 2014, 173, 125–131. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Ma Y, Khalil H, Wang R, Lu T, Zhao W, Zhang Y, Chen J and Chen T, Int J Nanomedicine, 2016, 11, 4025–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Li W, Xu X, Wang W, He K and Fan H, Gene, 2018, 651, 57–61. [DOI] [PubMed] [Google Scholar]
- 12.Song T, Hao J, Zhang R, Tang M, Li W, Hui W, Fu Q, Wang C, Xin S, Zhang S, Rui P, Ren H and Ma Z, BMC Vet Res, 2019, 15, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song T, Zhang S, Hao J, Xin S, Hui W, Tang M, Li W, Tian R, Liu X, Rui P, Ren H, Wang C, Fu Q and Ma Z, Transbound Emerg Dis, 2019, 66, 1–6. [DOI] [PubMed] [Google Scholar]
- 14.Kiupel M, Stevenson GW, Choi J, Latimer KS, Kanitz CL and Mittal SK, Vet Pathol, 2001, 38, 74–82. [DOI] [PubMed] [Google Scholar]
- 15.Hattermann K, Roedner C, Schmitt C, Finsterbusch T, Steinfeldt T and Mankertz A, Xenotransplantation, 2004, 11, 284–294. [DOI] [PubMed] [Google Scholar]
- 16.Pinheiro AL, Bulos LH, Onofre TS, de Paula Gabardo M, de Carvalho OV, Fausto MC, Guedes RM, de Almeida MR and Silva Junior A, Res Vet Sci, 2013, 94, 764–768. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Ouyang T, Ma T, Ouyang H, Pang D and Ren L, BMC Veterinary Research, 2018, 14, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Ouyang T, Ouyang H, Liu X, Niu G, Huo W, Yin W, Pang D and Ren L, Sci Rep, 2019, 9, 5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang N, Zhan Y, Wang A, Zhang L, Khayat R and Yang Y, J Gen Virol, 2016, 97, 3331–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tischer I, Rasch R and Tochtermann G, Zentralbl Bakteriol Orig A, 1974, 226, 153–167. [PubMed] [Google Scholar]
- 21.Tischer I, Gelderblom H, Vettermann W and Koch MA, Nature, 1982, 295, 64–66. [DOI] [PubMed] [Google Scholar]
- 22.Palinski R, Piñeyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E and Hause BM, Journal of Virology, 2017, 91, e01879–01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekser B, Rigotti P, Gridelli B and Cooper DK, Transpl Immunol, 2009, 21, 87–92. [DOI] [PubMed] [Google Scholar]
- 24.Pierson RN 3rd, Dorling A, Ayares D, Rees MA, Seebach JD, Fishman JA, Hering BJ and Cooper DK, Xenotransplantation, 2009, 16, 263–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denner J and Mankertz A, Viruses, 2017, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duffy S, Shackelton LA and Holmes EC, Nat Rev Genet, 2008, 9, 267–276. [DOI] [PubMed] [Google Scholar]
- 27.Xiao CT, Halbur PG and Opriessnig T, J Gen Virol, 2015, 96, 1830–1841. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Zhang S, Zou Y, Yu W, Jiang Y, Zhan Y, Wang N, Dong Y and Yang Y, Front Cell Infect Microbiol, 2018, 8, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhan Y, Yu W, Cai X, Lei X, Lei H, Wang A, Sun Y, Wang N, Deng Z and Yang Y, J Virol, 2020, DOI: 10.1128/JVI.00042-20. [DOI] [Google Scholar]
- 30.Khayat R, Wen K, Alimova A, Gavrilov B, Katz A, Galarza JM and Gottlieb P, Virology, 2019, 537, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shuai J, Wei W, Jiang L, Li X. l., Chen N and Fang W, Acta Biochimica et Biophysica Sinica, 2008, 40, 71–77. [DOI] [PubMed] [Google Scholar]
- 32.Dhindwal S, Feng S and Khayat R, J Mol Biol, 2019, 431, 3261–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khayat R, Brunn N, Speir JA, Hardham JM, Ankenbauer RG, Schneemann A and Johnson JE, J Virol, 2011, 85, 7856–7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Guo F, Wang F, Li TC and Jiang W, Structure, 2016, 24, 319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrison SC, Olson AJ, Schutt CE, Winkler FK and Bricgone G, Nature, 1978, 279, 368–373. [DOI] [PubMed] [Google Scholar]
- 36.Crowther RA, Berriman JA, Curran WL, Allan GM and Todd D, J Virol, 2003, 77, 13036–13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhindwal S, Avila B, Feng S and Khayat R, J Virol, 2019, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mo X, Li X, Yin B, Deng J, Tian K and Yuan A, PLoS Pathog, 2019, 15, e1007562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarasova E, Farafonov V, Khayat R, Okimoto N, Komatsu TS, Taiji M and Nerukh D, J Phys Chem Lett, 2017, 8, 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarasova E, Korotkin I, Farafonov V, Karabasov S and Nerukh D, Journal of Molecular Liquids, 2017, 245, 109–114. [Google Scholar]
- 41.Tarasova E, Farafonov V, Taiji M and Nerukh D, Journal of Molecular Liquids, 2018, 265, 585–591. [Google Scholar]
- 42.Tarasova E and Nerukh D, The Journal of Physical Chemistry Letters, 2018, 9, 5805–5809. [DOI] [PubMed] [Google Scholar]
- 43.Swails JM, York DM and Roitberg AE, J Chem Theory Comput, 2014, 10, 1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sali A and Blundell TL, J Mol Biol, 1993, 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 45.Krissinel E and Henrick K, J Mol Biol, 2007, 372, 774–797. [DOI] [PubMed] [Google Scholar]
- 46.Wu P-C, Lin W-L, Wu C-M, Chi J-N, Chien M-S and Huang C, Applied Microbiology and Biotechnology, 2012, 95, 1501–1507. [DOI] [PubMed] [Google Scholar]
- 47.Franzo G, Cortey M, Segales J, Hughes J and Drigo M, Mol Phylogenet Evol, 2016, 100, 269–280. [DOI] [PubMed] [Google Scholar]
- 48.Gill SC and von Hippel PH, Anal Biochem, 1989, 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 49.Weeks SD, Drinker M and Loll PJ, Protein Expr Purif, 2007, 53, 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, Pulokas J and Reilein A, Journal of Structural Biology, 2000, 132, 33–45. [DOI] [PubMed] [Google Scholar]
- 51.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A and Simmerling C, Proteins, 2006, 65, 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryckaert J-P, Ciccotti G and Berendsen HJC, Journal of Computational Physics, 1977, 23, 327–341. [Google Scholar]
- 53.Darden T, York D and Pedersen L, The Journal of Chemical Physics, 1993, 98, 10089–10092. [Google Scholar]
- 54.Mongan J, Case DA and McCammon JA, Journal of Computational Chemistry, 2004, 25, 2038–2048. [DOI] [PubMed] [Google Scholar]
- 55.Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S and Walker RC, Journal of Chemical Theory and Computation, 2012, 8, 1542–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Grand S, Götz AW and Walker RC, Computer Physics Communications, 2013, 184, 374–380. [Google Scholar]
- 57.Salomon-Ferrer R, Götz AW, Poole D, Le Grand S and Walker RC, J Chem Theory Comput, 2013, 9, 3878–3888. [DOI] [PubMed] [Google Scholar]
- 58.Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham I, E. T, Cruzeiro VWD, Darden TA, Duke RE, Ghoreishi D, Gilson MK, Gohlke H, Goetz AW, Greene D, Harris R, Homeyer N, Huang Y, Izadi S, Kovalenko A, Kurtzman T, Lee TS, LeGrand S, Li P, Lin C, Liu J, Luchko T, Luo R, Mermelstein DJ, Merz KM, Miao Y, Monard G, Nguyen C, Nguyen H, Omelyan I, Onufriev A, Pan F, Qi R, Roe DR, Roitberg A, Sagui C, Schott-Verdugo S, Shen J, Simmerling CL, Smith JG, SalomonFerrer R, Swails JM, Walker RC, Wang J, Wei H, Wolf RM, Wu X, Xiao L, York DM and Kollman PA, Journal, 2018. [Google Scholar]
- 59.Mermelstein DJ, McCammon JA and Walker RC, J Mol Recognit, 2019, 32, e2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roe DR and Cheatham TE, Journal of Chemical Theory and Computation, 2013, 9, 3084–3095. [DOI] [PubMed] [Google Scholar]
- 61.Krissinel E and Henrick K, Acta Crystallographica Section D, 2004, 60, 2256–2268. [DOI] [PubMed] [Google Scholar]
- 62.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC and Ferrin TE, J Comput Chem, 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.