To the editor - with this correspondence we aim to introduce Global Natural Product Social Molecular Networking (GNPS) to your readership and to highlight the quick-start interface we have recently developed. GNPS is a chemistry-focused, community-curated, small molecule mass spectrometry analysis ecosystem for untargeted metabolomics data.1 GNPS was designed for the analysis and capture of mass spectrometry knowledge and data by the community. These capabilities are possible due to the deep integration of GNPS with the metabolomics data repository MassIVE. Co-localizing metabolomics data, knowledgebases, and computational infrastructure allows for maximal reuse by the community in the spirit of open data. As of February 2021, GNPS/MassIVE has grown to 1,800 public data sets (>490,000 mass spectrometry files, >1.2 billion tandem mass spectra) with on average over 300,000 accesses to the GNPS analysis ecosystem per month by users from more than 160 countries.
The GNPS ecosystem has been used in different fields, including natural products, exposomics, nutrition science, forensics, marine science, environmental science and clinical metabolomics (28.67% of all public files with metadata are associated with a disease condition and 21.36% of the human files are associated with a disease). Approximately 50 analytical tools are connected to the GNPS ecosystem/platform. These tools are accessible without having to convert or move raw data out of the platform. Uniquely, the tools at GNPS enable users to directly match their data to all public MS/MS reference libraries for annotation1, and perform molecular networking, which enables the discovery of related metabolites (including transformations due to metabolism) without needing an isotopic label (Fig. 1a–f).1–3 The processed data tables and results from GNPS jobs can be exported into other downstream analysis infrastructures such as Cytoscape4, Metaboanalyst5 or QIIME26, which provide interactive network, statistical, machine learning or multivariate analysis and visualization capabilities (Fig. 1g). In addition, all public data deposited at GNPS/MassIVE can be searched using MS/MS spectra (MASST)7 or using controlled vocabularies or annotations (ReDU).8 MASST and ReDU are allowing researchers to leverage the information in the entire GNPS/MassIVE repository to provide a global context to their data. Finally, all tools within GNPS aim to improve data analysis reproducibility by facilitating easy replication with identical parameters and the sharing of data analysis job results as URL links (Fig. 1).
Fig. 1 |. The quick-start interface of GNPS can be used for Molecular Networking and multivariate analysis.
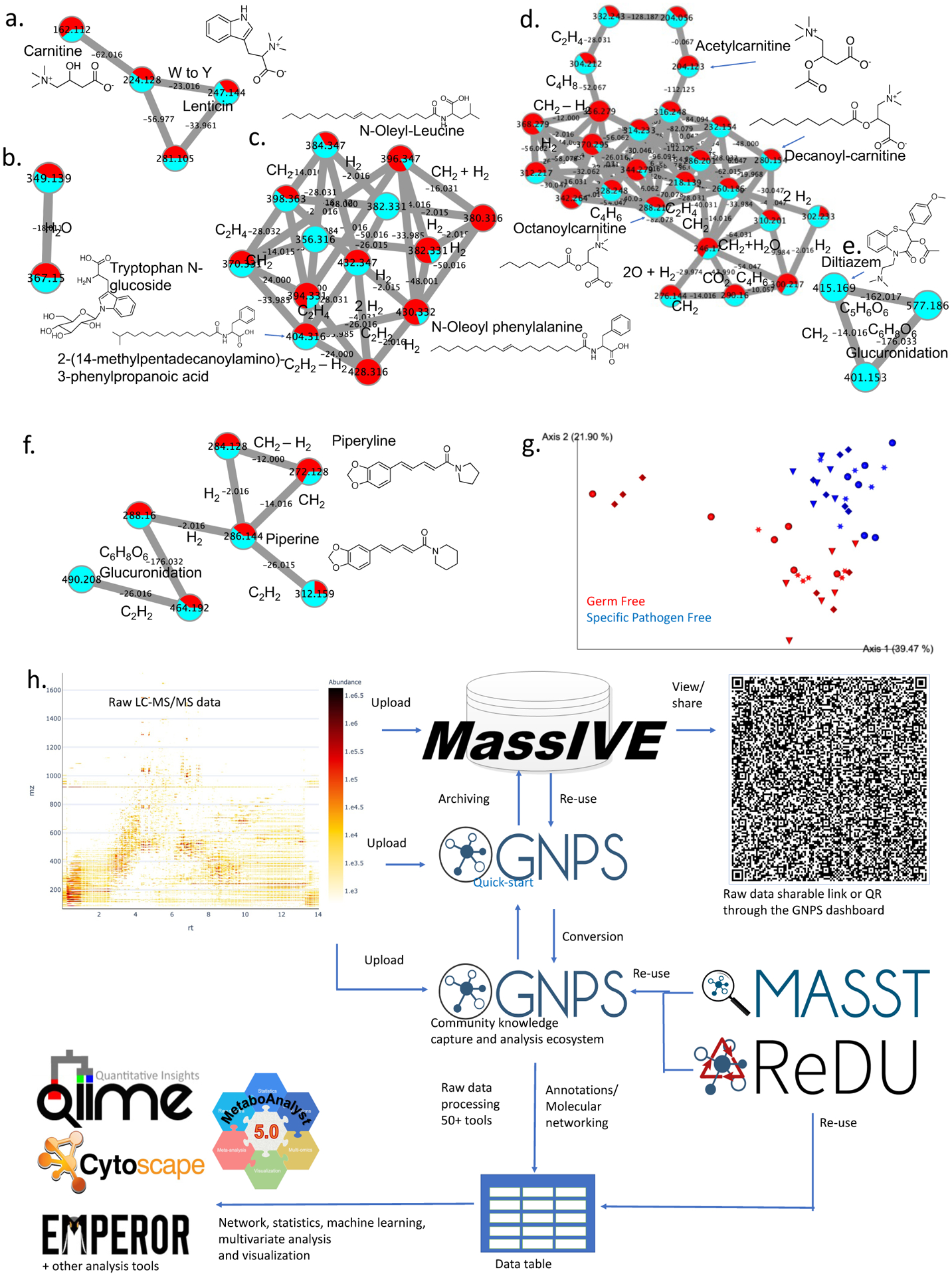
Representative molecular families obtained via Classical MS/MS Molecular Networking of plasma from an Alzheimer’s disease (AD) (public MassIVE data set MSV000085256, DOI 10.25345/C5D410 and complete analysis job can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=ab00b95c6f2149bf92973c1a6bf19be6). The molecular network is visualized with Cytoscape.4 Representative molecular families (sub-networks) reveal metabolites (a,b), lipids (c,d), drugs (e) or food-derived molecules (f) and their natural analogs (e.g. CH=CH, CH2) and metabolites from phase I (e.g. reduction, demethylation or oxidation) and/or Phase II metabolism (e.g. glucuronidation). The families are annotated based on spectral matches against MS/MS reference libraries.1 Blue are the relative levels in normal controls while red are the levels observed in the AD patients (relative levels approximated via spectral count). g, PCoA visualization of the binary Jaccard similarity of the data from multiple duodenum sections associated with germ free and specific pathogen free mice (public MassIVE data set MSV000083437, DOI 10.25345/C51G8P) using Qiime 26 and Emperor10 for visualization (An interactive link to the PCoA can be found here https://view.qiime2.org/visualization/?type=html&src=https%3A%2F%2Fcors.bridged.cc%2Fhttps%3A%2F%2Fgnps.ucsd.edu%2FProteoSAFe%2FDownloadResultFile%3Ftask%3D65a02c388b224f5dbd8311343731a8f2%26file%3Dqiime2_output%2Fqiime2_emperor.qzv%26block%3Dmain, n=4 mice for each condition, 6 sections each, each mouse is indicated by color and symbol). h, overview of the GNPS analysis ecosystem, including the new GNPS quick-start, analysis ecosystem and how it interfaces with the MassIVE repository, the GNPS search engines ReDU and MASST and the processing of data tables and external data table analysis and visualization tools. The QR code leads to the GNPS dashboard to visualize raw data.
However, GNPS is admittedly not very easy to use. Users must learn to convert data to open formats, how to transfer files via ftp clients into GNPS, and how to submit jobs before they can visualize the data as a molecular network (Fig. 1a–f) or as an interactive PCoA plot (Fig. 1g). We thus developed an easier-to-use quick start infrastructure (https://gnps-quickstart.ucsd.edu) for data conversion, validation of library batch files, public data set creation, classic and Feature-Based Molecular Networking (FBMN) for up to 50 files (87% of GNPS’s usage, Fig. 1h). These improvements aim to make the infrastructure more accessible to a wider scientific community, including users interested in the study of cellular metabolic pathways or lipids. Complementing the quick-start, we have created documentation for all GNPS’ tools (https://ccms-ucsd.github.io/GNPSDocumentation/).
Data conversion and upload is the first step to use GNPS, a pain point for many beginner users of mass spectrometry. Our online data conversion quick-start infrastructure, leveraging ProteoWizard9, enables the conversion of LC-MS/MS files from most vendor formats to the open data format .mzML, the preferred format for GNPS analysis. Once converted, the data can be used in two other quick-start modules: Classical1 and FBMN3. Using Classical Molecular Networking, users can drag-and-drop files into up to three possible cohorts and select the type of instrument used (high vs low resolution). That is all that is necessary for users to run their first Classical GNPS Molecular Networking workflow. For FBMN, users can drag-and-drop a quantification table and a MS/MS spectral file(e.g. .mgf) that is generated according to the FBMN instructions (pre-processing instructions at https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking/). Another GNPS quick-start module enables data deposition into MassIVE (https://massive.ucsd.edu/). The last quick-start is to help build the knowledgebase in GNPS by allowing the community to deposit public MS/MS libraries. This quick-start allows validation of the library batch files to ensure compatibility with the GNPS ecosystem before uploading MS/MS reference spectra to the GNPS spectral database. None of the quick-start pages require login to run jobs. However, if the user creates an account, it can store jobs and results in the user space; otherwise, the jobs without a user account will be cleared periodically.
The inherent richness of untargeted metabolomics data makes analysis difficult. GNPS, a resource for metabolomics, lipidomics and natural products communities, has been developed for and by these communities to put forth a set of tools that help to tackle the complexities of untargeted LC-MS/MS data analysis. As a community resource, GNPS offers many innovative solutions/tools for data analysis that are continually growing due to community contributions. We hope the ever-expanding capabilities of GNPS can be made more accessible and reach a broader audience with the introduction here of the GNPS quick-start set of tools and interfaces.
Acknowledgements
This research was supported, in part, by the National Institutes of Health (NIH) awards U19AG063744, GM107550, 1RF1AG051550, R01AG061066, 1R01LM013115, P41GM103484 and R24GM127667, and National Science Foundation (NSF) award no. ABI 1759980. C.C. was funded by the NIH fellowship F31AT010419.
Footnotes
Competing Interests
P.C.D is a scientific advisor to Sirenas, Galileo and Cybele and co-founder and scientific advisor to Ometa and Enveda with approval by the University of California San Diego. M.W. is a co-founder of Ometa Labs LLC.
Code availability
The GNPS quick-start page can be accessed at https://gnps-quickstart.ucsd.edu and the github repository for the code is available at https://github.com/mwang87/GNPS_quickstart. We used the public MassIVE datasets MSV000085256 (DOI 10.25345/C5D410) and MSV000083437 (DOI 10.25345/C51G8P).
References
- 1.Wang M et al. Nat. Biotechnol 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aksenov AA et al. Nat. Biotechnol 39, 169–173 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nothias LF et al. Nat. Methods 17, 905–908 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shannon P et al. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chong J et al. Nucleic Acids Res. 46, W486–W494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolyen E et al. Nat. Biotechnol 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M et al. Nature Biotechnology 38, 23–26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarmusch AK et al. Nat. Methods 17, 901–904 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kessner D, et al. Bioinformatics 21, 2534–2536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vázquez-Baeza Y, et al. Gigascience 2:16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The GNPS quick-start page can be accessed at https://gnps-quickstart.ucsd.edu and the github repository for the code is available at https://github.com/mwang87/GNPS_quickstart. We used the public MassIVE datasets MSV000085256 (DOI 10.25345/C5D410) and MSV000083437 (DOI 10.25345/C51G8P).


