Abstract
Ochratoxin A (OTA) is a well-known mycotoxin with wide distribution in food and feed. Fungal genome sequencing has great utility for identifying secondary metabolites gene clusters for known and novel compounds. A comparative analysis of the OTA-biosynthetic cluster in A. steynii, A. westerdijkiae, A. niger, A. carbonarius, and P. nordicum has revealed a high synteny in OTA cluster organization in five structural genes (otaA, otaB, ota, otaR1, and otaD). Moreover, a recent detailed comparative genome analysis of Aspergilli OTA producers led to the identification of a cyclase gene, otaY, located in the OTA cluster between the otaA and otaB genes, encoding for a predicted protein with high similarity to SnoaLs domain. These proteins have been shown to catalyze ring closure steps in the biosynthesis of polyketide antibiotics produced in Streptomyces. In the present study, we demonstrated an upregulation of the cyclase gene in A. carbonarius under OTA permissive conditions, consistent with the expression trends of the other OTA cluster genes and their role in OTA biosynthesis by complete gene deletion. Our results pointed out the involvement of a cyclase gene in OTA biosynthetic pathway for the first time. They represent a step forward in the understanding of the molecular basis of OTA biosynthesis in A. carbonarius.
Keywords: OTA, biosynthetic cluster, SnoaL domain, polyketide cyclases, CRISPR/Cas9
1. Introduction
Ochratoxin A (OTA) is a well-known mycotoxin with wide distribution in food and feed, including cereal products, grapes and by-products, coffee, beverages, cocoa, nuts, dried fruits, and cured meat [1,2]. Ochratoxin A is a secondary fungal metabolite with several toxicological effects (such as hepatotoxic, nephrotoxic, teratogenic, embryotoxic, etc.) [3,4,5]. Ochratoxin A is produced by many species of the genus Aspergillus and Penicillium [6]. One of the main OTA producing species is Aspergillus carbonarius, known not only for its high capacity for producing OTA and its high percentage of toxigenic strains but also because it is considered the main species for OTA contamination of grapes in the vineyard worldwide [7].
Fungal genome sequencing has great utility for the identification of secondary metabolite gene clusters for known and novel compounds [8,9]. This kind of genomic approach has led to the identification of the clustered OTA biosynthetic genes, primarily in A. niger, and subsequently in A. carbonarius [10,11] and recently in several other OTA producing species [12,13]. In particular, the genomic analysis of A. carbonarius revealed the crucial role of three genes (otaA-AcOTApks, otaB-AcOTAnrps, and otaD-AcOTAhal) in the OTA biosynthesis [14,15,16]. Further, the involvement of a P450 cytochrome oxidase (otaC) and a bZIP transcription factor (otaR1) has been evidenced in six different OTA-producing species [12,17]. The essential role of these five core genes in OTA biosynthesis has also been consolidated by the gene knockout approach. Moreover, a comparative analysis of OTA-biosynthetic clusters in A. steynii, A. westerdijkiae, A. niger, A. carbonarius, and P. nordicum has revealed a high synteny in cluster organization [13]. A detailed comparative analysis of Aspergilli genomes has recently led us to identify a gene whose sequence encoded a predicted protein similar to bacterial polyketide cyclases. This gene is located in the OTA cluster between the otaA and otaB genes; it is present in the genome sequences of all currently sequenced OTA-producing fungi, and it was hypothesized to be involved in OTA production [12]. In particular, this gene encodes a protein with high similarity to SnoaLs domain. Largely characterized in bacteria, SnoaL domain-containing proteins have been characterized as small polyketide cyclases composed of about 140 amino acids. These proteins have been shown in Streptomyces to catalyze cyclization steps in the biosynthesis of polyketide antibiotics [18]. Concerning the OTA biosynthetic pathway, the cyclization process that leads to the formation of the heterocyclic structure of OTβ, in the initial biosynthetic steps, has remained unclear. Our identification of this SnoaL like protein—named otaY—along with the gene expression analysis suggested its putative involvement in the ring closure step of the OTA polyketide backbone [12]. Based on these results, we confirmed the role of otaY gene in OTA biosynthesis by gene disruption. In order to do this, we used a CRISPR/Cas9 mutagenesis approach to disrupt the cyclase gene in the strain of A. carbonarius ITEM 5010.
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)–Cas9 (CRISPR-associated protein 9) is used by bacteria and archaea as a defense against viral infection and has grown into a revolutionary genome editing tool, which induces targeted DNA double strand breaks (DSBs). This gene editing system consists of two components: the Cas9 nuclease and a single guide RNA (sgRNA) that drives the Cas9/RNA complex to a specific target. Once the DNA double-strand is cleaved at the target site by Cas9 nuclease activity, both ends can subsequently be repaired by the non-homologous end joining (NHEJ) or homology directed repair (HDR) pathway. Due to the simplicity, efficiency, and the possibility of introducing mutations in multiple genes in one step, CRISPR/Cas9 has become a fast-growing genome editing tool in fungi [19]. It has been developed into a powerful technology that has been applied to various filamentous fungi, including A. nidulans and A. aculeatus [20], A. oryzae [21], A. fumigatus [22,23], A. niger [24], A. carbonarius [25], Neurospora crassa [26], Alternaria alternata [27], Fusarium oxysporum [28], and F. graminearum [29]. One of the recent advances in this technique is the direct delivery of Cas9/RNA complex into the cell [30,31,32,33,34]. Therefore, the need to express the Cas9 protein in the fungal host cell and assemble specific expression cassettes for each sgRNA is avoided by the direct transformation of fungal protoplasts with in vitro-assembled ribonucleoproteins (RNPs). The CRISPR/Cas9 system is particularly effective for complete gene deletions when coupled with dual in vitro-assembled Cas9–RNPs that target the upstream and downstream regions of a gene.
This technique offers the advantage of minimizing the risk of modifying adjacent gene sequences and targeting clustered genes located very close to each other, as in the genomic region hosting secondary metabolite clusters.
In the present study, we demonstrate the role of the otaY gene by complete gene deletion using the CRISPR/Cas9 approach. Moreover, the expression level of the cyclase gene is investigated to demonstrate its correlation to the kinetics of OTA accumulation and the expression profile of the other OTA biosynthetic genes. Our results point out for the first time the involvement of a new gene in the biosynthetic pathway of OTA in A. carbonarius and represent a knowledge advancement in the molecular basis of OTA biosynthesis.
2. Results
2.1. Gene Expression Analysis
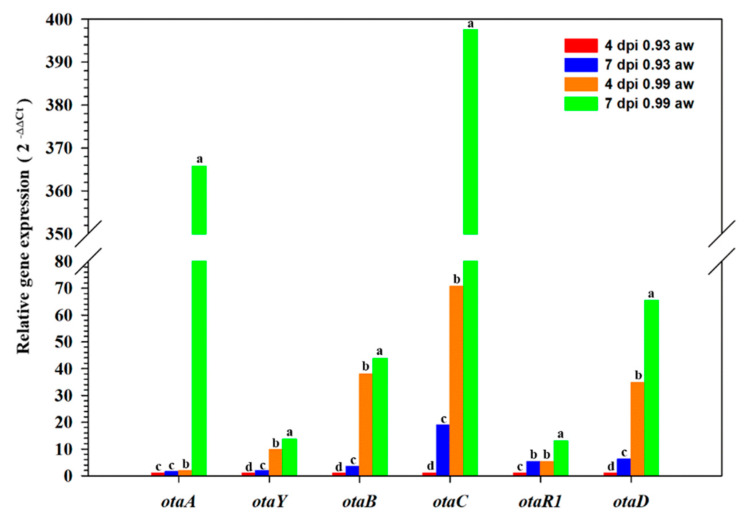
While expression of otaY was confirmed previously from various OTA-producing fungi [12], we explored the expression of this cyclase-encoding gene relative to the other OTA biosynthetic cluster genes at multiple time points in A. carbonarius ITEM 5010. The qRT-PCR analysis of otaY gene and other OTA cluster genes (otaA, otaB, otaC, otaR1, and otaD) demonstrated an upregulation of the cyclase gene in A. carbonarius ITEM 5010 under OTA permissive conditions, in terms of growth conditions (medium and temperature). Furthermore, the modulation of the cyclase-encoding gene expression levels was consistent with the expression trends of the other OTA cluster genes at 4 and 7 dpi (Figure 1).
Figure 1.
qRT-PCR analysis of OTA related genes in A. carbonarius ITEM 5010 at 18–31 °C. The reported gene expression under OTA permissive (0.99 aw) and not permissive (0.93 aw) was related to OTA not permissive (0.93 aw) conditions at 4-days post-inoculation (dpi). Relative gene expression analysis was performed in triplicates. Letters indicate significantly different means according to Tukey’s test (p ≤ 0.05).
2.2. Selection and Analysis of ∆otaY Mutants
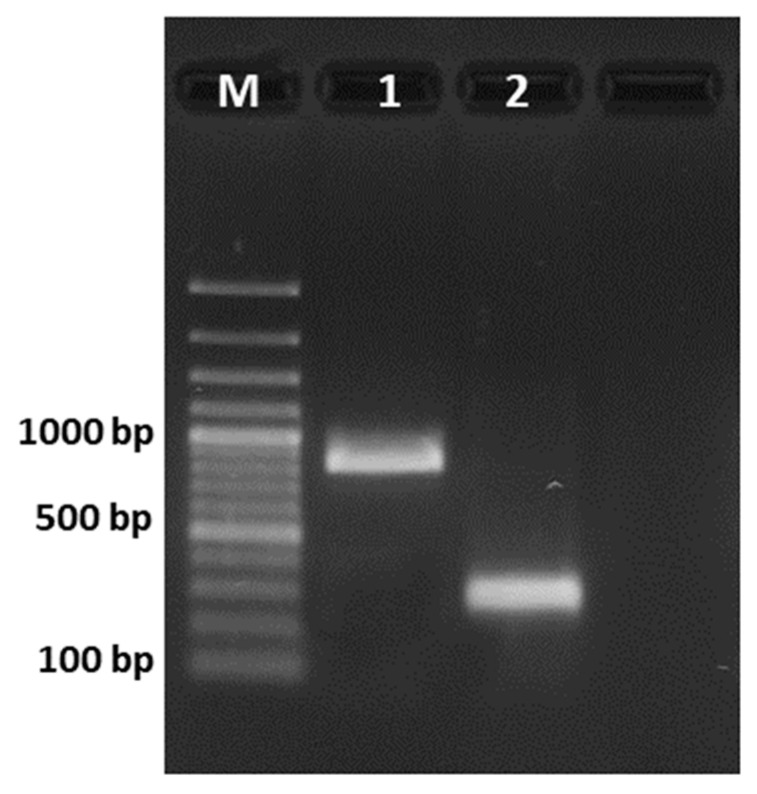
To delete the coding sequence of the cyclase gene otaY, we designed two sgRNAs (Table 1) that directed Cas9 RNPs complex to the 5’ and 3’-UTR regions of the otaY coding sequence. Based on predicted PAM sites and sgRNA scores, two sgRNAs were selected: the sgRNAotaY_up, located 172 bp upstream of the otaY start codon, and the sgRNAotaY_dw, located 32 bp downstream of the stop codon. After protoplast transformation, eight putative ∆otaY transformants were isolated on hygB medium. The putative ∆otaY mutants were PCR screened by amplification of the genomic sequence surrounding the excision sites. The sequencing of amplified regions obtained from the wild-type strain and ∆otaY mutant strains, represented by the selected strain AC2021 in Figure 2, resulted in amplicons of 916 bp and 292 bp, respectively, revealing the complete deletion of otaY gene and the lack of integration of the hygB resistance cassette in the otaY deletion site. Nevertheless, all the analyzed mutants were able to grow on hygB selective media and did not produce OTA (data not shown). In addition, sequence analysis of the amplified regions did not evidence any sequence alteration of UTRs regulatory regions of the genes adjacent to gene otaY (otaA and otaB). No significant phenotypic differences in terms of sporulation rate, pigmentation, and growth rate were observed among the wild-type and mutant strains grown on Czapek yeast extract agar (CYA), malt extract agar (MEA), and yeast extract sucrose (YES) agar at 25 °C in the dark after three and five days of incubation (Figure S1).
Table 1.
sgRNAs used in this study.
| ID | Protospacer Sequence 5′→3′ | PAM Site |
|---|---|---|
| sgRNAotaY_up | ATTAGCCCTACACGTCAACC | GGG |
| sgRNAotaY_dw | GTTAGTTGGGATTCGCTGCT | GGG |
Figure 2.
PCR amplification from genomic DNA of deletion site from wild-type ITEM 5010 (1) and AC2021 ∆otaY mutant strain (2). M = 100 bp ladder.
2.3. Genome Sequencing
The genome of the selected AC2021 ΔotaY mutant strain was sequenced and the de-novo assembly of quality filtered reads obtained from the sequencing generated 1461 contigs. The full sequencing dataset is available at Bioproject PRJNA738506. The analysis enabled the exact mapping of the excision site of the two RNP complexes. In detail, the comparative analysis with the sequence genome of A. carbonarius ITEM 5010 (Aspca3- https://mycocosm.jgi.doe.gov/Aspca3/Asp-ca3.home.html, accessed on 17 November 2021) revealed the complete deletion of otaY gene occurring between the position 951,737 and 952,362 of the original scaffold_12 of the wild-type strain, resulting in a deletion of 624 bp in the mutant strain. The deleted chromosomal segment included the full length otaY gene and was congruent with PCR analysis and Sanger sequencing of the region spanning the predicted deletion sites as reported in Figure 2. Analysis of UTRs regions of otaA and otaB gene surrounding the cyclase gene as well as sequence analysis of the entire genomic region of OTA cluster did not reveal any other nucleotide modification in the ΔotaY mutant strain. With regard to the hygB marker gene, its integration occurred between the positions 1,805,697 and 1,805,702 of the original scaffold_7 of the wild-type strain 5010. Upstream of the integration site is a gene encoding a peroxisomal NUDIX hydrolase (protein ID 396812) and downstream is a gene encoding a DNA binding protein (protein ID 207537). The integration of the hygB gene did not alter the coding sequences of genes surrounding the hygB integration site.
2.4. Expression Analysis of OTA Cluster Genes in ∆otaY Mutant Strain
The RT-PCR analysis of transcription of OTA cluster genes in strains ITEM 5010 and AC2021 after 4 dpi on MM agar medium (see oligonucleotides used in Table 2) revealed the expression of the complete set of genes in wild-type strain ITEM 5010 and the absence of the otaY transcript only in the mutant strain AC2021 (data not shown).
Table 2.
Primers for qRT-PCR and RT-PCR used in this study.
| Primer | Sequence (5′–3′) | Concentration | Target Gene | Reference |
|---|---|---|---|---|
| RT_OTApks_Ac_FOR | CGTGTCCGATACTGTCTGTGA | 200 nM | otaA | [35] |
| RT_OTApks_Ac_REV | GCATGGAGTCCTCAAGAACC | 200 nM | ||
| RT_Ac_otaY_for | ACCATCCTCACCACCCTTGT | 200 nM | otaY | [14] |
| RT_Ac_otaY_rev | GGGACTCTGGGCTAACACCT | 200 nM | ||
| RT_nrps_Ac_FOR | ACGGGTCGCTGCTCTATATC | 200 nM | otaB | [14] |
| RT_nrps_Ac_REV | ACTCACCACATCAACCACGA | 200 nM | ||
| RT_AcOTAp450_F | GTGGTTATCCCGCCCAATAC | 200 nM | otaC | [14] |
| RT_AcOTAp450_R | TGCCAGATTCATCCCGATAC | 200 nM | ||
| RT_Ac_OTAbZIP_for | AATGGAACCAGCATTGATCTC | 250 nM | oraR1 | [14] |
| RT_Ac_OTAbZIP_rev | GACCCAAGCATTCGCTCTA | 250 nM | ||
| RT_hal_Ac_FOR | GAACGCCAGTAGAGGGACAG | 200 nM | otaD | [14] |
| RT_hal_Ac_REV | ATGGAGGTGGTGTTGTTGTG | 200 nM | ||
| RT3 BT Ac_F | CAAACCGGCCAGTGTGGTA | 200 nM | BenA | [14] |
| RT3 BT Ac_R | CGGAGGTGCCATTGTAAACA | 200 nM |
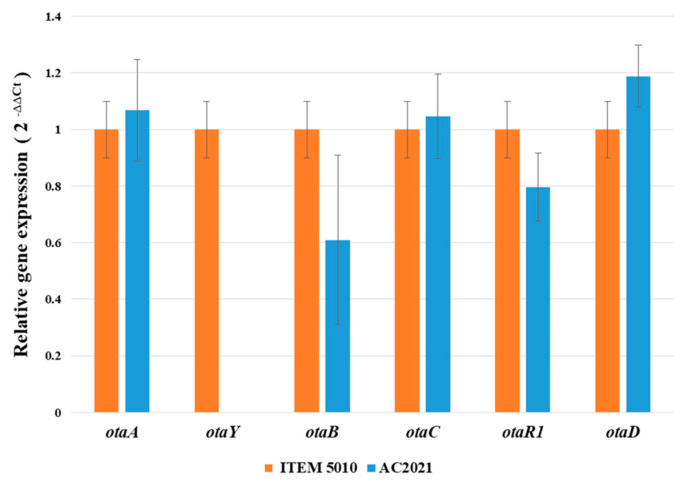
Moreover, qRT-PCR analysis of otaA, otaB, otaC, otaR1, and otaD genes confirmed that the deletion of otaY gene did not prevent or significantly alter the expression of the other OTA cluster biosynthetic genes (Figure 3).
Figure 3.
qRT-PCR analysis of OTA related genes in the wild-type ITEM 5010 and AC2021 ∆otaY mutant strain of A. carbonarius. Gene expressions were evaluated on MM agar plates after 4 days of incubation at 25 °C in the dark. The reported relative gene expression (2−∆∆Ct) was related to expression levels of each gene in wild-type ITEM 5010. Relative gene expression analysis was performed in triplicates. Bars indicate standard error.
2.5. OTA Analysis
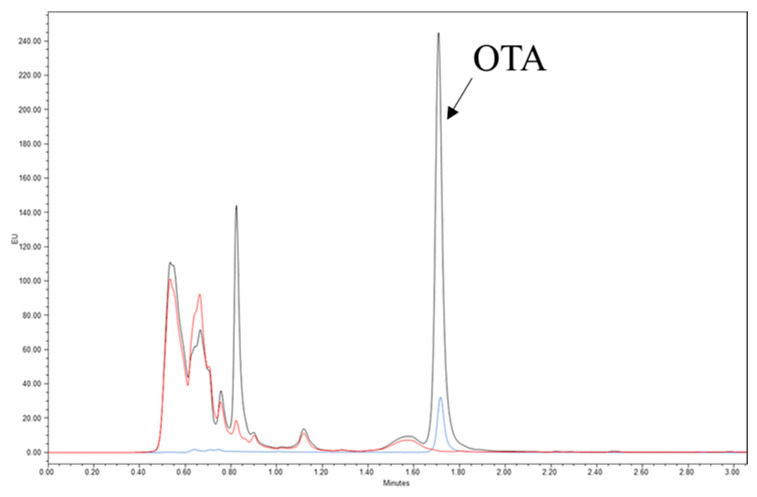
The production of OTA by UPLC-FLD in mycelium collected after growth on 0.99 and 0.93 aw Grape Juice Medium (GJM) at 18/31 °C under 10 h/14 h dark/light photoperiod accounted for the OTA permissive condition (0.99 aw GJM) in 472.23 and 452.82 ng/g at 4 and 7 dpi, respectively. OTA was not detected (<LOD) in mycelium from the non-permissive condition (0.93 aw GJM) at both collection points. In addition, analysis of OTA in mutant strains, lacking the otaY gene, after seven days of incubation in the dark on MM agar at 25 °C revealed the absence of the toxin in culture extracts. In comparison, the production of OTA was confirmed in A. carbonarius wild-type strain ITEM 5010 cultured under the same conditions (6467 ng/g) (Figure 4). The chromatograms reported in Figure 4 clearly show the presence of OTA in the diluted extract of the wild-type strain and the absence of OTA in the diluted extract of AC2021 mutant strain, selected as representative ∆otaY mutant strain. Another prominent peak, eluting at 0.85 min, was present in the chromatogram of the wild type, which is probably one of the precursors of OTA, as previously reported by Gallo et al., 2012 [15]. This peak was also present in the chromatogram of the diluted extract of the mutant strain but at a much lower concentration. Further studies are necessary to identify this compound, its possible involvement in the biosynthetic pathway of OTA, and its relationship with the otaY gene. In order to confirm the lack of OTA production by the mutant strain shown in Figure 4, even at very low concentrations, the fungal extract was purified and concentrated through an immunoaffinity column specific for OTA and analyzed by UPLC-FLD. No OTA was detected in the purified/concentrated extract of the mutant strain.
Figure 4.
UPLC-FLD chromatograms of OTA standard solution (1018 ng/g) (blue trace), extract of A. carbonarius wild-type strain ITEM 5010 (6467 ng/g) (black trace), and extract of A. carbonarius ∆otaY deletion mutant strain AC2021 (red trace). Retention times = OTA 1.72 min.
3. Discussion
Ochratoxin A is a fungal toxin known to be produced by more than twenty Aspergillus species of Sect. Nigri and Circumdati, and by Penicillium nordicum, P. thymicola, P. verrucosum. The recent availability of genome sequences from 21 OTA-producing species (19 Aspergillus and 2 Penicillium), revealed some differences in gene attributes reported by different authors. However, it established the clear involvement of five core genes (otaA, otaB, otaC, otaD, and otaR1) in the biosynthetic pathway of OTA [12,14,15,35]. Recently, the extensive analysis of the genomic region related to the biosynthetic cluster of OTA indicated the presence of a new gene, encoding a “SnoaL-like cyclase” domain located between the polyketide synthase (otaA) and non-ribosomal peptide synthase (otaB) genes [12]. This finding was interesting because the cyclization process of the heterocyclic structure of OTβ, in the first steps of OTA biosynthesis pathway, has not yet been clarified. Indeed, the evidence of the presence of a gene encoding a polyketide cyclase in the OTA clusters points out its possible role in the cyclization of OTA polyketide backbone. SnoaLs proteins have been characterized as belonging to a family of small polyketide cyclases, consisting of about 140 amino acids, which catalyze in Streptomyces cyclization steps in the biosynthesis of polyketide antibiotics [18]. The role of cyclization in polyketide biosynthesis is complex and important. However, there is limited data regarding the role of specific cyclase genes in secondary metabolite biosynthesis from filamentous fungi. More often, cyclization is thought to happen either spontaneously via intramolecular interactions within the growing polyketide or mediated by a cyclization domain that could be part of the domain structure of PKSs [36]. A second hypothesis could be the involvement of a separate gene from the pks gene, namely cyclase, which could be the case of the OTA biosynthesis. As example, the biosynthetic pathway for aurovertin in Calcarisporium arbuscula seems to be increased by a cyclase protein (aurE), with homology to a SnoaL-like bacterial polyketide cyclase [37,38]. While the involvement of SnoaL-like polyketide cyclases has been clearly demonstrated in the biosynthesis of polyketide antibiotics, only a few proteins with this functional domain have been reported in filamentous fungi [12,39].
The analysis of the otaY gene expression under permissive and non-permissive OTA conditions showed regulation of otaY gene congruent with the expression trend of the other OTA cluster genes, confirming an upregulation of its transcription level under OTA permissive conditions compared to non-permissive conditions. These findings supported the hypothesis that a different gene is involved in the cyclization of OTA molecule instead of otaA gene, responsible for polyketide biosynthesis. In this study, the role of cyclase gene otaY in the biosynthesis of OTA was demonstrated by using the CRISPR/Cas9 approach. The use of CRISPR/Cas9 system for the deletion of otaY gene was successful and revealed the usefulness of this genome editing tool for the deletion of genes located at genomic level very close to each other. Considering the genomic region where the otaY gene is placed, the otaA gene is located at about 350 bp upstream of the cyclase gene and the otaB gene is located at about 200 bp downstream. This short distances between important structural genes for OTA biosynthesis and the presence in this intergenic spacer of the respective UTRs regions would have made the deletion of the cyclase gene difficult with a traditional approach. The classical homologous recombination with a deletion cassette requires the fusion of 600–1000 bp of sequence flanking the target gene at both ends of a marker gene. Indeed, the CRISPR/Cas9 tool application guaranteed an almost “surgical” deletion of otaY gene without altering any adjacent gene coding sequences and the related gene regulation regions. Since the hygB resistance was not integrated as expected in the otaY deletion site, in order to dispel any doubts about any possible alteration of OTA cluster genes, we: (i) sequenced the entire otaY deletion site; (ii) verified the transcription of OTA cluster genes by RT-PCR; (iii) verified any significant change in expression levels of OTA cluster genes by qRT-PCR. Moreover, the integration of hygB resistance in a different site, probably due to the high microhomology of the 50 bp sequence fused to the HygB cassette with a different genomic region, led us to sequence the full genome of AC2021 strain, selected as representative mutant strain. Genome sequence analysis confirmed the occurrence of the complete deletion of otaY gene and the absence of any mutation in OTA cluster genes and related regulatory regions. The sequencing also confirmed that the integration of hygB resistance in a different genome region did not modify the coding sequence of the genes surrounding the integration site. To our knowledge, this is the first evidence of the involvement of a cyclase gene in the biosynthesis of OTA. The complete deletion of otaY gene demonstrated the inability of ΔotaY mutant to synthesize the OTA molecule.
It is well known that fungal polyketide synthases are multifunctional enzymes involved in the biosynthesis of fungal polyketides, harboring various functional domains with acyl transferase, ketosynthase, thioesterase, methyltransferase, and other activities.
They often use acetyl CoA as the starting substrate, malonyl CoA as the extension unit, and repeated Claisen condensation to extend the polyketide chain [40]. In general, the cyclization process of fungal polyketide metabolite is due to a Thioesterase (TE) domain in PKS multifunctional proteins [41]. In this regard, the otaA gene coding for a polyketide synthase, crucial for the OTA biosynthesis, lacks the TE domain, and this supports our hypothesis of the SnoaL-like cyclase involvement in the early cyclization step of OTA formation [12]. SnoaL is a polyketide cyclase that adopts a distorted alpha-beta barrel fold, and the polyketide cyclases thus form a family of enzymes with a unique catalytic strategy for aldol condensation. As detailed above, SnoaL-like cyclases are well described in the antibiotic synthesis in Streptomyces and other bacteria, but not in fungal secondary metabolism. Recently, a putative ester cyclase (named UvEC1) presenting a SnoaL domain has been shown to be important for the virulence of Ustilaginoidea virens but remarkably, its expression is inversely related to toxin production in this pathogen [39]. In conclusion, our results indicate for the first time the involvement of a cyclase gene in the biosynthesis of a polyketide mycotoxin, previously the cyclization step was always associated with the presence of a specific domain in the fungal PKS or NRPS enzymes [41]. Our results are supported by the prediction of the presence of this gene within the cluster genes in all the ochratoxigenic fungi [12] and support the original idea of an essential role of this gene in the OTA biosynthetic pathway. Our findings allowed the definition of a core cluster of six genes (otaA, otaY, otaB, otaC, otaD, and otaR1) involved in the biosynthesis of OTA. However, some questions remain that concern essentially precursor and intermediate molecules in the early stages of the biosynthetic pathway during the formation of the polyketide structure which the analyses conducted in this current work could not determine. Further investigation would permit the identification of other key pathway intermediates upstream of otβ, which is linked to phenylalanine in the reaction catalyzed by the NRPS encoded by otaB gene, and OTB, which is formed in the binding reaction and then converted into OTA by the action of the halogenase protein encoded by the otaD gene [14,15].
A logical next step will be to perform similar experiments to demonstrate the role of otaY gene identified in other ochratoxigenic fungi in our previous work [12]. A more in-depth knowledge can be obtained from the study of the OTA pathway in the ochratoxigenic species belonging to different sections of filamentous fungi to verify the evolutionary process responsible for the structure of OTA clusters.
4. Materials and Methods
4.1. Fungal Strains and Growth Conditions
The wild-type A. carbonarius ITEM 5010 from the Agro-Food Microbial Culture Collection of the Institute of Sciences of Food Production, CNR, Bari, Italy (www.ispa.cnr.it/Collection, accessed on 17 November 2021) and the mutant strain AC2021 (ΔotaY), generated from ITEM 5010, were used in this study. Fungal strains were grown on minimal medium (MM) agar plates [14]. The knockout ΔotaY strain was grown on MM supplemented with 100 mg/L hygromycin B (hygB).
4.2. Cyclase Gene Expression Analysis
The analysis of otaY gene expression in relation to the other OTA biosynthetic genes (otaA, otaB, otaC, otaR1, and otaD) was conducted under water activity conditions permissive (aw 0.99) and not permissive (aw 0.93) for OTA production. In detail, 100 μL of a conidial suspension (106 conidia/mL) from wild-type strain ITEM 5010 of A. carbonarius were inoculated on Grape Juice Medium (GJM) prepared according to [42]. The water activity (aw) of GJM was adjusted to 0.93 by adding an appropriate amount of glucose. The aw of GJM was measured using an AquaLab 4TE aw meter at 25 °C. Inoculated plates were incubated at 18/31 °C under 10 h/14 h dark/light photoperiod. Triplicate cultures were prepared and analyzed for each experiment. Fungal mycelium was harvested after 4- and 7-day post-inoculation (dpi) and stored at −80 °C for RNA extraction and OTA analysis. Total RNA was extracted from frozen mycelium ground in liquid nitrogen using the RNeasy kit (Qiagen, Hilden, Germany) and treated with RNase-free DNase I (Qiagen) according to the manufacturer’s protocol. First-strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen, San Diego, CA, USA) according to the manufacturer’s protocol. The expression levels of OTA biosynthetic genes in A. carbonarius strains 5010 were analyzed using real-time quantitative reverse transcription-PCR (qRT-PCR); β-tubulin was used as an internal reference gene. The list of primers used in the qPCR assays is shown in Table 2. Real-time PCR was performed using the Viia 7 Real Time PCR system according to [42]. Relative gene expression analysis was performed in triplicates. The relative quantification (2−ΔΔCT) of gene expression was assessed by Quant-Studio™ RT-PCR Software (Thermo-Fisher Scientific, Waltham, MA, USA), considering as reference condition 4 dpi aw 0.93 at 18/31 °C under 10 h/14 h dark/light photoperiod. Analysis of variance (ANOVA) was carried out to determinate significant differences in gene expression among different growth conditions for each analyzed gene. Tukey’s test was applied to compare the mean values. Statistical significance was set at p ≤ 0.05. Statistical analysis was performed using the software Statistica 7.0 (StatSoft Inc., Tulsa, OK, USA).
4.3. Cyclase Gene Deletion: sgRNAs Selection and Fungal Transformation
The sgRNAs (Table 1) for the complete deletion of otaY gene (NCBI accession number MT706047 Figure S2) were designed with the Eukaryotic Pathogen CRISPR guide RNA/DNA Design Tool—EuPaGDT (grna.ctegd.uga.edu). The annotated sequence of reference strain ITEM 5010 was used to predict PAM sites and off-targets. The intergenic region between PKS and NRPS genes was selected as target region for sgRNAs prediction. In order to select optimal guide RNAs for upstream and downstream regions of otaY gene, any sgRNAs targeting the UTRs regions of PKS and NRPS genes and the coding sequence of otaY gene were excluded. The sgRNA DNA templates were PCR-assembled, transcribed, and pre-loaded on TrueCut Cas9 Protein v2 (Thermo-Fisher Scientific) as previously described by [34]. The nucleotide sequences of primer pairs IVT-cycUP-fwd/IVT-cycUP-rev and IVT-cycDW-fwd/IVT-cycDW-rev used to assemble the sgRNAs DNA templates are reported in Table 3. The two ribonucleoprotein (RNP) complexes assembled in vitro were used to transform protoplasts of A. carbonarius ITEM 5010 obtained according to [14]. In detail, protoplasts aliquots (200 µL, 2 × 106 protoplasts) were co-transformed with 1 µg of the two RNPs (1:1) and 3μg of purified HDR-HygB repair template prepared by fusion PCR with the primer pair 5harm_cyc_Hyg_F/3harm_cyc_HygB_R (Table 3) as reported by [33]. Then, 50 µL of polyethylene glycol (PEG)-CaCl2 buffer (40% [w/v] PEG 4000, 50 mM CaCl2·H2O, 50 mM Tris-HCl, pH 7.5) were added, and the mixture was incubated on ice for 30 min. Afterward, 1.5 mL PEG-CaCl2 buffer supplemented with 1 M sorbitol was added, and the mixture was incubated at room temperature for 20 min. Subsequently, the mixture was diluted to a total volume of 3 mL with MM supplemented with 1 M sorbitol and incubated at 30 °C with gentle shaking for 1 h. After incubation, protoplasts were plated by inclusion in non-selective 1 M sorbitol MM soft agar (0.8% w/v agar) to allow regeneration of the fungal cell wall. After 16–18 h at 25 °C, a top layer of MM agar 1.2% w/v supplemented with 100 mg/L hygB was overlaid on transformant plates. Resistant colonies were selected after 3–6 days of incubation at 25 °C. Putative transformants were transferred to PDA containing 100 mg/L of hygromycin B. Selected mutants were subcultured as a single spore on selective MM agar supplemented with 100 mg/L of hygromycin. Monoconidial A. carbonarius ∆otaY mutant strains were cultured on selective PDA for further analysis and stored in sterile 15% glycerol at −80 °C as conidial suspensions.
Table 3.
Primers used in this study. Primer sequences specific for amplification of the hygB cassette are in bold.
| Primer ID | Nucleotide Sequence 5′→3′ |
|---|---|
| IVT-cycUP-fwd | TAATACGACTCACTATAGATTAGCCCTACACGTC |
| IVT-cycUP-rev | TTCTAGCTCTAAAACGGTTGACGTGTAGGGCTAA |
| IVT-cycDW-fwd | TAATACGACTCACTATAGGTTAGTTGGGATTCGC |
| IVT-cycDW-rev | TTCTAGCTCTAAAACAGCAGCGAATCCCAACTAA |
| 5harm_cyc_Hyg_F | CTCACCAGGCTGTGGCAAGCAGTTGGCGTGTATATTAGCCCTACACGTCAACAGTTTAGCTTGCCTCGTC |
| 3harm_cyc_HygB_R | TACTTCATATATCCAACCAAACAAAACACATACCCTAGTTTCTCACCCAGTCCAGTATAGCGACCAGCATT |
| cyc_out_F | CATTGTGCTGGACTTTGGGC |
| cyc_out_R | CCGGCTTTACTTTCATGGCG |
4.4. PCR Analysis of Putative A. Carbonarius ∆otaY Mutants
Genomic DNA of wild-type ITEM 5010 and A. carbonarius ∆otaY mutants was extracted by using the Gene JET Plant Genomic DNA Purification Kit (Invitrogen, San Diego, CA, USA). The genomic region spanning the deletion site was PCR amplified with primers cyc_out_F/cyc_out_R (Table 3). PCR amplifications were carried out with Platinum SuperFi II PCR Master Mix (Invitrogen) and 500 nM of each primer. PCR amplification conditions were: 98 °C for 30 s, then 30 cycles of 98 °C for 10 s, 60 °C for 15 s and 72°C for 1 min, and a final extension at 72 °C for 5 min. Amplification products were evaluated by gel electrophoresis. Amplicons were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystem, Waltham, MA, USA). The obtained nucleotide sequences were analyzed using CLC Genomics Workbench 7.5 (CLC, Inc., Aarhus, Denmark).
4.5. Gene Expression Analysis of OTA Cluster Genes in ∆otaY Mutant
An end-point Reverse Transcription (RT)-PCR and qRT-PCR were used to confirm the expression of OTA biosynthetic genes (otaA, otaB, otaC, otaR1 and otaD) and the absence of otaY transcript in deletion strain.The wild-type ITEM 5010 and the selected ∆otaY strain AC2021 of A. carbonarius were grown on MM agar plates for 4 days in the dark at 25 °C. Total RNA and cDNA were obtained as described above (Chapter 4.2). PCR amplifications were performed with Platinum SuperFi II PCR Master Mix (Invitrogen) on 100 ng of cDNA and 500 nM of each primer. PCR amplification conditions were: 98 °C for 30 s, then 30 cycles of 98 °C for 10 s, 58 °C for 15 s and 72 °C for 30 s, and a final extension at 72 °C for 5 min. The primers Bt2a/Bt2b [43] were used to monitor β-tubulin gene expression as endogenous control. These primers span three introns, which also allowed checking for DNA contamination. Real-time PCR analysis was performed as reported in chapter 4.2. The relative quantification (2−ΔΔCT) of gene expression was assessed by Quant-Studio™ RT-PCR Software (Thermo-Fisher Scientific, Waltham, MA, USA), considering as reference condition the expression level of each gene in wild-type strain ITEM 5010.
4.6. Genome Sequencing and Assembly
The genome of AC2021 A. carbonarius ∆otaY mutant strain was sequenced by Illumina HiSeq 2 × 150 paired end reads. After quality filtering, the reads were de-novo assembled by using the de Bruijn graph-based algorithm implemented in MaSuRCA Assembler—v3.2.9 [44].
Obtained scaffolds were used to map the excision site of otaY gene and the integration site of hygromycin B gene marker. Genes surrounding otaY excision and hygB integration sites were manually annotated to check if the editing procedure and induced DNA repairing events modified any other locus of OTA cluster and/or known genes.
4.7. OTA Analysis
The production of OTA by strain ITEM 5010 was monitored on 0.99 and 0.93 aw GJM after 4- and 7-day incubation at 18/31 °C under 10 h/14 h dark/light photoperiod.
The method of Gallo et al., 2014 [35], for the determination of OTA in mycelium was slightly modified and used herein for OTA determination. The samples were analyzed directly by UPLC-FLD system without immunoaffinity cleanup. In particular, 500 mg of frozen mycelium grounded in liquid nitrogen and stored at −80 °C (the same used for gene expression analysis) were transferred to a 12 mL test tube. About 0.5 g of mycelium were extracted with 10 mL of mixture MeOH:ACN:H2O (30:30:40, v/v/v) by shaking for 120 min at room temperature and centrifuged for 15 min at 3000× g. An aliquot of 600 µL of supernatant was diluted with 400 µL of mixture ACN:H2O:Acetic Acid (35:62.5:2.5, v/v/v) and 10 µL, corresponding about 0.00023 g of solid sample, were injected into Waters Acquity UPLC system (Milford, MA, USA) full loop injection system. The chromatographic column was an Acquity UPLC BEH RP-18 (100 mm × 2.1 mm i.d., 1.7 μm column, Acquity) with an Acquity UPLC column in-line filter (0.2 μm).
Moreover, OTA production was verified for wild-type strain ITEM 5010 and four independent ΔotaY strains (AC2021-AC2024) grown on MM agar plates in the dark at 25 °C, performing the cleanup step reported in Official Method AOAC 2001.01 [45]. After 7 days of growth, 5 agar plugs with mycelium (about 500 mg) were collected from each replicate and extracted as reported above. An aliquot of 6 mL of sample extracts in MeOH:ACN:H2O (30:30:30, v/v/v) were diluted with 44 mL of a water solution containing PEG (1%) and NaHCO3 (5%), mixed, filtered through a glass microfiber filters GF/A and 10 mL of diluted sample cleaned-up by OchraTest immunoaffinity columns (Vicam, Watertown, MA, USA). The column was washed with 10 mL of an aqueous solution containing NaCl (2.5%) and NaHCO3 (0.5%), followed by 10 mL of ultrapure water at a flow rate of 1−2 drops/s. The eluates were discarded, and the OTA was recovered in a vial by passing 2 mL of methanol through the column. The eluate was collected in a glass vial, evaporated to dryness at 50 °C under a gentle stream of nitrogen, and redissolved in 500 µL of a UPLC mobile phase (H2O:ACN:Acetic Acid, 99:99:2, v/v/v). The purified extracts were filtered through a 0.22 µm pore filter. Ten microliters, corresponding to about 0.001 g of mycelium, were injected into the UPLC-FLD.
Acknowledgments
The authors thank Domenico Genchi for his IT support.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/toxins13120892/s1, Figure S1: Colonies (clockwise from the top) on Czapek yeast extract agar (CYA), malt extract agar (MEA), and yeast extract sucrose (YES) after three and five days of incubation at 25°C in the dark of A. carbonarius wild type strain ITEM 5010 (A and C) and A. carbonarius ∆otaY deletion mutant strain AC2021 (B and D), Figure S2: Nucleotide sequence of Aspergillus carbonarius otaY polyketide cyclase (NCBI accession number MT706047).
Author Contributions
Conceptualization, M.F., A.G., S.E.B. and G.P.; Data curation, M.F., A.G., C.C. and L.G.; Formal analysis, M.F., C.C. and L.G.; Investigation, M.F., C.C. and L.G.; Methodology, M.F., A.G., C.C. and L.G.; Software, M.F.; Validation, M.F. and C.C.; Writing—original draft, M.F., A.G., C.C., L.G. and G.P.; Writing—review & editing, M.F., M.S., S.E.B. and G.P. All authors have read and agreed to the published version of the manuscript.
Funding
Funding support for SEB is acknowledged from EMSL, a National Scientific user facility sponsored by US Department of Energy’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory operated by Battelle for the US DOE under contract AC06-76RLO 1830 and the DOE Joint BioEnergy Institute supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. Regione Puglia: PSR Puglia 2014–2020 M16.2 project name “Sistemi di supporto alle decisioni (DSS) per il miglioramento della sostenibilità in viticoltura (DIGITAL-GRAPE)” DdS n. 94250033571 CUP B29J20000070009.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequence data are available at NCBI SRA under BioProject ID: PRJNA738506.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
The role in OTA biosynthesis of a new gene, otaY, coding for a polyketide cyclase, was demonstrated for the first time by CRISPR/Cas9 mediated deletion in A. carbonarius. Our findings allowed the definition of a core cluster of six genes (otaA, otaY, otaB, otaC, otaD, and otaR1) involved in the biosynthesis of OTA.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jørgensen K. Occurrence of ochratoxin A in commodities and processed food—A review of EU occurrence data. Food Addit. Contam. 2005;22:26–30. doi: 10.1080/02652030500344811. [DOI] [PubMed] [Google Scholar]
- 2.el Khoury A.E., Atoui A. Ochratoxin a: General overview and actual molecular status. Toxins. 2010;2:461–493. doi: 10.3390/toxins2040461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castegnaro M., Canadas D., Vrabcheva T., Petkova-Bocharova T., Chernozemsky I.N., Pfohl-Leszkowicz A. Balkan endemic nephropathy: Role of ochratoxins A through biomarkers. Mol. Nutr. Food Res. 2006;50:519–529. doi: 10.1002/mnfr.200500182. [DOI] [PubMed] [Google Scholar]
- 4.IARC . Monographs on the Evaluation of Carcinogenic Risks to Humans: Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. International Agency for Research on Cancer; Lion, France: 1993. [Google Scholar]
- 5.Malir F., Ostry V., Pfohl-Leszkowicz A., Malir J., Toman J. Ochratoxin A: 50 years of research. Toxins. 2016;8:191. doi: 10.3390/toxins8070191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houbraken J., Kocsubé S., Visagie C.M., Yilmaz N., Wang X.C., Meijer M., Kraak B., Hubka V., Bensch K., Samson R.A., et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020;95:5–169. doi: 10.1016/j.simyco.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perrone G., Gallo A., Susca A., Varga J. Aspergillus in grapes: Ecology, biodiversity and genomics. In: Varga J., Samson R.A., editors. Aspergillus in the Genomic Era. Wageningen Academic Publishers; Wageningen, The Netherlands: 2008. pp. 179–212. [DOI] [Google Scholar]
- 8.Vesth T.C., Brandl J., Andersen M.R. FunGeneClusterS: Predicting fungal gene clusters from genome and transcriptome data. Synth. Syst. Biotechnol. 2016;1:122–129. doi: 10.1016/j.synbio.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kjaerbølling I., Vesth T.C., Frisvad J.C., Nybo J.L., Theobald S., Kuo A., Bowyer P., Matsuda Y., Mondo S., Lyhne E.K., et al. Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proc. Natl. Acad. Sci. USA. 2018;115:E753–E761. doi: 10.1073/pnas.1715954115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pel H.J., de Winde J.H., Archer D.B., Dyer P.S., Hofmann G., Schaap P.J., Turner G., de Vries R.P., Albang R., Albermann K., et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 11.Baker S.E., Perrone G., Gallo A., Mulè G., Susca A. The Aspergillus carbonarius genome: Analysis of potential secondary metabolite biosynthetic gene clusters; Proceedings of the Book of Abstract of 25th Fungal Genetics Conference; Asilomar, CA, USA. 17–22 March 2009; p. 106. [Google Scholar]
- 12.Ferrara M., Gallo A., Perrone G., Magistà D., Baker S.E. Comparative Genomic Analysis of Ochratoxin A Biosynthetic Cluster in Producing Fungi: New Evidence of a Cyclase Gene Involvement. Front. Microbiol. 2020;11:581309. doi: 10.3389/fmicb.2020.581309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gil-Serna J., Vázquez C., Patiño B. The Genomic Regions That Contain Ochratoxin A Biosynthetic Genes Widely Differ in Aspergillus Section Circumdati Species. Toxins. 2020;12:754. doi: 10.3390/toxins12120754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrara M., Perrone G., Gambacorta L., Epifani F., Solfrizzo M., Gallo A. Identification of a halogenase involved in the biosynthesis of ochratoxin A in Aspergillus carbonarius. Appl. Environ. Microbiol. 2016;82:5631–5641. doi: 10.1128/AEM.01209-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallo A., Bruno K., Solfrizzo M., Perrone G., Mulè G., Visconti A., Baker S. New insight in the ochratoxin A biosynthetic pathway by deletion of an nrps gene in Aspergillus carbonarius. Appl. Environ. Microbiol. 2012;78:8208–8218. doi: 10.1128/AEM.02508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallo A., Ferrara M., Perrone G. Recent advances on the molecular aspects of ochratoxin A biosynthesis. Curr. Opin. Food Sci. 2017;17:49–56. doi: 10.1016/j.cofs.2017.09.011. [DOI] [Google Scholar]
- 17.Gerin D., Garrapa F., Ballester A.-R., González-Candelas L., De Miccolis Angelini R.M., Faretra F., Pollastro S. Functional Role of Aspergillus carbonarius AcOTAbZIP Gene, a bZIP Transcription Factor within the OTA Gene Cluster. Toxins. 2021;13:111. doi: 10.3390/toxins13020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sultana A., Kallio P., Jansson A., Wang J.S., Niemi J., Mäntsälä P., Schneider G. Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation. EMBO J. 2004;23:1911–1921. doi: 10.1038/sj.emboj.7600201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ullah M., Xia L., Xie S., Sun S. CRISPR/Cas9-based genome engineering: A new breakthrough in the genetic manipulation of filamentous fungi. Biotechnol. Appl. Biochem. 2020;67:835–851. doi: 10.1002/bab.2077. [DOI] [PubMed] [Google Scholar]
- 20.Nødvig C.S., Nielsen J.B., Kogle M.E., Mortensen U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE. 2015;10:e0133085. doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katayama T., Tanaka Y., Okabe T., Nakamura H., Fujii W., Kitamoto K., Maruyama J. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 2016;38:637–642. doi: 10.1007/s10529-015-2015-x. [DOI] [PubMed] [Google Scholar]
- 22.Fuller K.K., Chen S., Loros J.J., Dunlap J.C. Development of the CRISPR/Cas9 System for Targeted Gene Disruption in Aspergillus fumigatus. Eukaryot. Cell. 2015;14:1073–1080. doi: 10.1128/EC.00107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber J., Valiante V., Nødvig C.S., Mattern D.J., Slotkowski R.A., Mortensen U.H., Brakhage A.A. Functional reconstitution of a fungal natural product gene cluster by advanced genome editing. ACS Synth. Biol. 2017;6:62–68. doi: 10.1021/acssynbio.6b00203. [DOI] [PubMed] [Google Scholar]
- 24.Leynaud-Kieffer L., Curran S.C., Kim I., Magnuson J.K., Gladden J.M., Baker S.E., Simmons B.A. A new approach to Cas9-based genome editing in Aspergillus niger that is precise, efficient and selectable. PLoS ONE. 2019;14:e0210243. doi: 10.1371/journal.pone.0210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weyda I., Yang L., Vang J., Ahring B.K., Lübeck M., Lübeck P.S. A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius. J. Microbiol. Methods. 2017;135:26–34. doi: 10.1016/j.mimet.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Matsu-Ura T., Baek M., Kwon J., Hong C. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol. Biotechnol. 2015;2:4. doi: 10.1186/s40694-015-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wenderoth M., Pinecker C., Voß B., Fischer R. Establishment of CRISPR/Cas9 in Alternaria alternata. Fungal Genet. Biol. 2017;101:55–60. doi: 10.1016/j.fgb.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Wang Q., Cobine P.A., Coleman J.J. Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes. Fungal Genet Biol. 2018;117:21–29. doi: 10.1016/j.fgb.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardiner D.M., Kazan K. Selection is required for efficient Cas9-mediated genome editing in Fusarium graminearum. Fungal Biol. 2017;122:131–137. doi: 10.1016/j.funbio.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C., Meng X., Wei X., Lu L. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet. Biol. 2016;86:47–57. doi: 10.1016/j.fgb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Kuivanen J., Wang Y.J., Richard P. Engineering Aspergillus niger for galactaric acid production: Elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microb. Cell Fact. 2016;15:210. doi: 10.1186/s12934-016-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu R., Chen L., Jiang Y., Zhou Z., Zou G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015;1:15007. doi: 10.1038/celldisc.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pohl C., Kiel J.A., Driessen A.J., Bovenberg R.A., Nygård Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth. Biol. 2016;5:754–764. doi: 10.1021/acssynbio.6b00082. [DOI] [PubMed] [Google Scholar]
- 34.Ferrara M., Haidukowski M., Logrieco A.F., Leslie J.F., Mulè G. A CRISPR-Cas9 System for Genome Editing of Fusarium proliferatum. Sci. Rep. 2019;9:19836. doi: 10.1038/s41598-019-56270-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallo A., Knox B.P., Bruno K.S., Solfrizzo M., Baker S.E., Perrone G. Identification and characterization of the polyketide synthase involved in ochratoxin A biosynthesis in Aspergillus carbonarius. Int. J. Food Microbiol. 2014;79:10–17. doi: 10.1016/j.ijfoodmicro.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Herbst D.A., Townsend C.A., Maier T. The architectures of iterative type I PKS and FAS. Nat. Prod. Rep. 2018;35:1046–1069. doi: 10.1039/C8NP00039E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hang L., Liu N., Tang Y. Coordinated and Iterative Enzyme Catalysis in Fungal Polyketide Biosynthesis. ACS Catal. 2016;6:5935–5945. doi: 10.1021/acscatal.6b01559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao X.-M., Zhan Z.-J., Grayson M.N., Tang M.-C., Xu W., Li Y.-Q., Yin W.-B., Lin H.-C., Chooi Y.-H., Houk K.N., et al. Efficient Biosynthesis of Fungal Polyketides Containing the Dioxabicyclo-octane Ring System. J. Am. Chem. Soc. 2015;137:11904–11907. doi: 10.1021/jacs.5b07816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X., Pei Z., Li P., Li X., Duan Y., Liu H., Chen X., Zheng L., Luo C., Huang J., et al. Quantitative Proteomics Analysis Reveals the Function of the Putative Ester Cyclase UvEC1 in the Pathogenicity of the Rice False Smut Fungus Ustilaginoidea virens. Int. J. Mol. Sci. Artic. 2021;22:4069. doi: 10.3390/ijms22084069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schümann J., Hertweck C. Advances in cloning, functional analysis and heterologous expression of fungal polyketide synthase genes. J. Biotechnol. 2006;124:690–703. doi: 10.1016/j.jbiotec.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 41.Adrover-Castellano M.L., Schmidt J.J., Sherman D.H. Biosynthetic Cyclization Catalysts for the Assembly of Peptide and Polyketide Natural Products. ChemCatChem. 2021;13:2095–2116. doi: 10.1002/cctc.202001886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cervini C., Gallo A., Piemontese L., Magistà D., Logrieco A.F., Ferrara M., Solfrizzo M., Perrone G. Effects of temperature and water activity change on ecophysiology of ochratoxigenic Aspergillus carbonarius in field-simulating conditions. Int. J. Food Microbiol. 2020;315:108420. doi: 10.1016/j.ijfoodmicro.2019.108420. [DOI] [PubMed] [Google Scholar]
- 43.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zimin A.V., Març Ais G., Puiu D., Roberts M., Salzberg S.L., Yorke J.A. Genome analysis The MaSuRCA genome assembler. Bioinformatics. 2013;29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visconti A., Pascale M., Centonze G. Determination of Ochratoxin A in Wine and Beer by Immunoaffinity Column Cleanup and Liquid Chromatographic Analysis with Fluorometric Detection: Collaborative Study. J. AOAC Int. 2001;84:1818–1827. doi: 10.1093/jaoac/84.6.1818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data are available at NCBI SRA under BioProject ID: PRJNA738506.