Abstract
While the transactivation function of the tumor suppressor p53 is well understood, less is known about the transrepression functions of this protein. We have previously shown that p53 interacts with the corepressor protein mSin3a (hereafter designated Sin3) in vivo and that this interaction is critical for the ability of p53 to repress gene expression. In the present study, we demonstrate that expression of Sin3 results in posttranslational stabilization of both exogenous and endogenous p53, due to an inhibition of proteasome-mediated degradation of this protein. Stabilization of p53 by Sin3 requires the Sin3-binding domain, determined here to map to the proline-rich region of p53, from amino acids 61 to 75. The correlation between Sin3 binding and stabilization supports the hypothesis that this domain of p53 may normally be subject to a destabilizing influence. The finding that a synthetic mutant of p53 lacking the Sin3-binding domain has an increased half-life in cells, compared to wild-type p53, supports this premise. Interestingly, unlike retinoblastoma tumor suppressor protein, MDMX, and p14ARF, Sin3 stabilizes p53 in an MDM2-independent manner. The ability of Sin3 to stabilize p53 is consistent with the model whereby these two proteins must exist on a promoter for extended periods, in order for repression to be an effective mechanism of gene regulation. This model is consistent with our data indicating that, unlike the p300-p53 complex, the p53-Sin3 complex is immunologically detectable for prolonged periods following exposure of cells to agents of DNA damage.
The p53 tumor suppressor protein is widely believed to monitor the cellular stress response to genotoxic damage, as well as unfavorable environmental conditions such as hypoxia, inadequate growth factor levels, and unscheduled cellular division (31). In response to these stimuli, p53 becomes posttranslationally stabilized and activated as a transcription factor (for review, see references 15, 27, and 31). The cellular outcome of this response is p53-mediated growth arrest at the G1 and G2/M checkpoints or induction of programmed cell death (apoptosis). In part, cell type and environmental signals mediate the decision between growth arrest and apoptosis, but the level of p53 induced in the cell also plays a role (9). Given the significance of the outcome of unregulated amounts of p53 protein in a cell (growth arrest or cell death), elucidation of the parameters that control p53 levels in vivo continues to be an important area of study.
In normal cells p53 protein has a very short half-life (5 to 20 min), and there is good evidence that it is subject to ubiquitin-mediated degradation via the 26S proteasome (32, 44). Cells with a temperature-sensitive E1 enzyme show high levels of p53 at the restrictive temperature (10), and ubiquitin conjugates of p53 are evident in many cell types (32). There exist several proteins that control the level of p53 in the cell; not surprisingly, the functions of many of these are altered in human cancer. Perhaps chief in importance among these proteins is MDM2. MDM2 binds to p53 and enhances its ubiquitin-mediated degradation by acting directly as an E3 ubiquitin ligase; this activity requires MDM2's nuclear export function (18, 21, 22, 29, 39, 48). This appears to occur only for the p53 oligomer, as the p53 monomer is unable to interact with MDM2 (33). Nontetrameric p53 also requires its own nuclear export sequence, which is deeply imbedded in the oligomerization domain and consequently masked on the monomer (14, 45). These data implicate the existence of multiple mechanisms controlling p53 stability.
Inhibition of normal MDM2 function, by antibody binding (which breaks the p53-MDM2 complex), by expression of antisense RNA to MDM2, or by the drug leptomycin B (which inhibits nucleocytoplasmic shuttling), leads to accumulated p53 in the cell and subsequent apoptosis (6, 8, 12). Additionally, much of the posttranslational stabilization of p53 following exposure to DNA-damaging agents results from inhibition of the MDM2-p53 interaction. This interaction is weakened by phosphorylation of p53 at serine 20 following genotoxic stress (7, 38, 43), as well as by phosphorylation of MDM2 by kinases of the ATM family (26, 37). Similarly, there are at least three cellular proteins that stabilize p53 by antagonizing MDM2 function. The p14ARF tumor suppressor protein directly antagonizes the ability of MDM2 to degrade p53 by relocalizing the p14ARF-MDM2-p53 complex in a manner that interferes with degradation of p53 (25, 52) and by inhibiting MDM2's ubiquitin ligase activity (22). The retinoblastoma tumor suppressor protein (pRB) and the MDM2 homologue MDMX have also been shown to stabilize p53; like p14ARF, these proteins accomplish this by inhibiting the function of MDM2 (23, 42). Additionally, however, control of p53 stability by mechanisms that are independent of MDM2, by calpain 1, Jun N-terminal kinase (JNK), and β-catenin, has also been observed (11, 13, 28).
We previously described the interaction of p53 with the corepressor protein mSin3a (hereafter designated Sin3); this interaction is necessary for the ability of p53 to repress transcription (36). Sin3 is a ubiquitous nuclear corepressor protein that is utilized by many other transcriptional repressors (for a review, see reference 3). When analyzing the ability of p53 to cooperate with Sin3 to repress gene expression, we noted a consistent increase in both endogenous and exogenous p53 in cells in which Sin3 is introduced. We report here that interaction with Sin3 stabilizes p53 protein by inhibiting proteasome-mediated degradation of this protein. This stabilization requires the Sin3-binding domain of p53 (amino acids 61 to 75) and specifically requires the proline residue at amino acid 71 of p53. Interestingly, we show that unlike pRB, MDMX, and p14ARF, Sin3 can stabilize p53 in MDM2-null cells. The combined data suggest that the Sin3-binding domain of p53, which maps within the proline-rich region of p53, may normally be subject to interaction with a destabilizing protein. Interaction with Sin3 would be predicted to inhibit this destabilization, thereby facilitating the existence of the p53-Sin3 repression complex on the promoters of p53-repressed genes.
MATERIALS AND METHODS
Cell culture.
The H1299 human lung adenocarcinoma cell line was maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS) and 100 U of penicillin-streptomycin/ml. MCF-7 human breast carcinoma cells were maintained in RPMI 1640 medium supplemented with 10% FBS and 100 U of penicillin-streptomycin/ml. 174-1 cells are murine embryo fibroblasts that are null for both p53 and MDM2 and were provided courtesy of Gigi Lozano, M. D. Anderson Cancer Center. These cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% FBS and 100 U of penicillin-streptomycin/ml. All cells were grown at 37°C in a 5% CO2 humidified atmosphere.
Plasmid constructs and transfections.
Kevin Ryan and Karen Vousden (National Cancer Institute) kindly provided the Tyr175 mutant of p53. The deletion mutants of p53 (Δ1–40TZ, Δ44–61TZ, Δ61–75TZ, and Δ1–100TZ) were generated by PCR of the parent plasmid p53TZ (kindly provided by Thanos Halazonetis, The Wistar Institute) and subcloned into the vector pCR3.1 (Invitrogen), and the correct sequence was confirmed by sequence analysis. The p53 point mutants P71R and P80L were generated in this vector with the QuickChange site-directed mutagenesis protocol (Stratagene). All p53 constructs utilized in this study were of human origin and were driven by the same cytomegalovirus immediate-early promoter. The human wild-type (wt) p53 construct was cloned in pRc/CMV and was courtesy of Arnold Levine (Rockefeller University). The human p14ARF construct was generated by reverse transcription-PCR of mRNA from human thymus, sequenced, and cloned into CMV-neo-Bam3. The human Mdm2 cDNA in CMV-neo-Bam3, pCHDM1A, was provided by Jiandong Chen (H. Lee Moffitt Cancer Center, University of South Florida). The p53-inducible plasmid construct SpVII contains a p53 consensus site (5′ GGGCGTGCGCCGACATGCCC 3′) linked to the minimal promoter construct E1B-TATA, courtesy of James Manfredi, Mount Sinai School of Medicine.
For transfections, H1299 cells were seeded at 2 × 106 cells per 10-cm dish, allowed to recover overnight, and transfected for 24 h with calcium phosphate or FuGene, according to protocols provided by the manufacturer (Gibco/BRL and Roche Molecular Systems, respectively). MCF-7 and 174-1 cells were transfected for 24 h with Lipofectin or FuGene, with protocols provided by the manufacturer (Gibco/BRL and Roche Molecular Systems, respectively). A total of 0.5 to 1 μg of p53 plasmid was transfected with 7.5 to 8 μg of Sin3 (pCMX-mSin3a, kindly provided by Ron Evans, The Salk Institute) or p14ARF, 4 μg of MDM2, and 1 μg of CMV-β-galactosidase as a control for transfection efficiency. For treatment with proteasome inhibitors, transfected cells were treated with 200 μM MG132 (Calbiochem), 45 μM ALLN (N-acetyl-Leu-Leu-norleucinal; Sigma), 25 μm lactacystin (Calbiochem), or dilution vehicle alone (dimethyl sulfoxide [DMSO]) for 2 h.
Immunoprecipitation (IP) and Western analysis.
Western analysis was performed essentially as previously described (36). Briefly, subconfluent cells were harvested and lysed in radioimmunoprecipitation assay buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], and 1% sodium deoxycholate) supplemented with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 10 μg of pepstatin/ml, 10 μg of aprotinin/ml, and 5 μg of leupeptin/ml). Protein concentrations were determined with the Bio-Rad DC Protein assay. Equal amounts of protein (between 50 and 120 μg) were run on SDS–10 or 12% polyacrylamide gel electrophoresis (PAGE) gels and transferred overnight onto polyvinylidene difluoride membranes (Bio-Rad). Western blots were incubated in antibody in 5% nonfat dry milk in phosphate-buffered saline (PBS) supplemented with 0.2% Tween 20. Blots were incubated with primary antibody (in parentheses) at the following dilutions: p53 (Ab-6; Calbiochem), 1:1,000; actin (Santa Cruz), 1:400; actin (AC-15; Sigma), 1:5,000; and MDM2 (Ab-1; Calbiochem), 1:1,000. Blots were washed with 5% nonfat dry milk in PBS supplemented with 0.2% Tween 20, incubated in horseradish peroxidase-linked secondary antibody (Jackson ImmunoResearch Laboratories), and developed via the chemiluminescence protocol provided by the manufacturer (NEN). Autoradiographs were quantitated with NIH Image software.
For IP-Western analyses, cells were lysed in NP-40 lysis buffer and equal amounts of protein (1,000 to 2,000 μg) were immunoprecipitated with antisera to Sin3 (AK-11; Santa Cruz Biotechnology). Each IP mixture was washed twice in NP-40 buffer, followed by four to six washes in radioimmunoprecipitation assay buffer. IPs were run on SDS–7.5 or 10% PAGE gels and transferred overnight onto Immuno-Blot polyvinylidene difluoride membranes (Bio-Rad). Blots were incubated with 1 μg of antibody (Ab-6; Calbiochem)/ml for 1 h at room temperature, followed by washing with PBS–0.2% Tween 20, incubation in peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories), and chemiluminescence detection (NEN). Autoradiographs were quantitated with NIH Image software.
Pulse-chase analysis and half-life experiments.
For half-life experiments, H1299 cells were transfected with 0.25 μg of p53 or the Δ61–75 mutant for 24 h with FuGene, as per protocols derived from the manufacturer (Roche Molecular Systems). Cells were then radiolabeled for 14 h with [35S]methionine (35S-EXPRESS; NEN) and chased with cold 2 mM l-methionine for the time points indicated. Equal counts per minute of the lysates were immunoprecipitated with anti-p53 polyclonal antisera (Santa Cruz Biotechnology), and SDS-PAGE gels were fluorographed (Enhance; NEN) and exposed to film overnight. For cycloheximide experiments, cells were incubated with 40 μg of cycloheximide/ml for the indicated time points, and equal amounts of lysate, in micrograms, were subjected to Western analyses. Autoradiographs were quantitated with NIH Image software.
Immunofluorescence.
MCF-7 cells were seeded at 25% confluence on coverslips in six-well tissue culture plates and allowed to recover overnight. Cells were transfected with 0.5 μg of pCMX-mSin3a or 0.5 μg of p14ARF and 2 μg of pEGFP with Lipofectin, via the protocol derived from the manufacturer (Gibco/BRL). Twenty-four hours later, cells were fixed in 4% paraformaldehyde solution for 10 min at room temperature, followed by a 10-min incubation with 0.2% Triton X-100 diluted in PBS. Immunofluorescence was performed with p53 rabbit polyclonal antisera (Santa Cruz Biotechnology) at 1 μg/ml and rhodamine-X-conjugated secondary antibody (Jackson ImmunoResearch Laboratories). Green fluorescent protein (GFP) was visualized at an excitation wavelength of 490 nm with an emission wavelength of 505 nm, while the p53-rhodamine X was observed at an excitation wavelength of 555 nm with an emission wavelength of 605 nm. Images were acquired and analyzed with Isee software (Innovision) and a Quantix 12-bit cooled charged-coupled device camera (Photometrics).
Northern analysis and luciferase assays.
Total RNA was isolated from cells by CsCl purification (35) or with TRIzol, as per the manufacturer (Gibco/BRL). Northern analyses were performed as described (35). Probes for Northern analyses were radiolabeled with random primers (Prime-It-II; Stratagene) and [α-32P]dCTP (NEN). Autoradiographs were quantitated with NIH Image software. The data depicted are representative of at least three independent experiments. For luciferase assays, H1299 cells were seeded in six-well plates at 2 × 105 cells/well and allowed to settle overnight. Cells were transfected with 1.25 μg of firefly luciferase reporter construct SpVII (containing a consensus p53 binding site in the minimal promoter vector pE1B-TATA, courtesy of James Manfredi, Mount Sinai School of Medicine), along with the indicated amounts of p53 expression plasmid (in pRc/CMV) and 100 ng of the transfection control pEGFP (Clontech). Transfections were performed using FuGene, according to protocols derived from the manufacturer (Roche). After 24 to 36 h, the cells were harvested and lysed, and luciferase assays were performed as per the protocol derived from the manufacturer (Promega) with a Monolight 2010 luminometer (Analytical Luminescence Laboratory).
RESULTS
Sin3 stabilizes p53 and protects p53 from MDM2-mediated degradation.
We previously reported that p53 protein interacts directly with the corepressor protein Sin3. The p53-Sin3 interaction results in the recruitment of histone deacetylases to the promoters of p53-repressed genes like Map4. This recruitment leads to subsequent deacetylation of the histones associated with these promoters (36). In an effort to examine the ability of Sin3 to cooperate with p53 in the repression of transcription, we noted that transfection with Sin3, but not vector alone, led to consistent increases in p53 protein levels. These data led us to test the possibility that Sin3 expression and/or binding influences p53 stability.
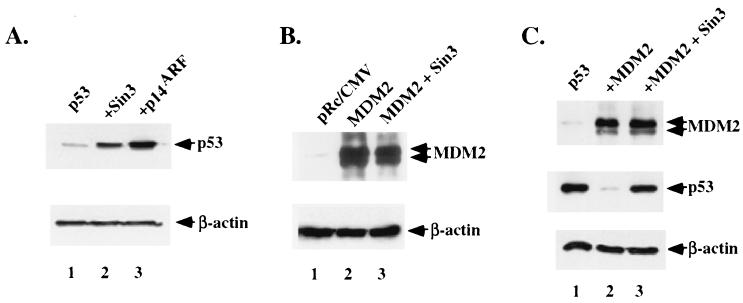
To investigate the ability of Sin3 to alter p53 protein levels, H1299 cells (p53-null human lung adenocarcinoma cells) were transfected with wt p53 in the presence of parental vector alone (pRc/CMV), Sin3, or the positive regulator of p53 stability, p14ARF. p14ARF stabilizes p53 by inhibiting MDM2 function (25, 46, 52). Western analysis of transfected cells revealed that wt p53 was expressed at four- to fivefold-higher levels when cells were transfected with Sin3 (Fig. 1A, lane 2). This was comparable to the level of p53 induced by p14ARF transfection (lane 3). In contrast, transfection with Sin3 did not lead to increased levels of a cotransfected β-galactosidase gene, glutathione S-transferase (data not shown), or MDM2 (Fig. 1B, lane 3); the latter is, like p53, a short-lived protein that is degraded by the proteasome. Increased p53 levels following cotransfection with Sin3 occurred in several different p53-null cell lines, with independent plasmid preparations of p53 and Sin3 (data not shown). Northern analysis of RNA isolated from transfected cells revealed no increase in p53 transcript levels, indicating that the effect of Sin3 on p53 was posttranscriptional (data not shown).
FIG. 1.
Sin3 stabilizes p53 and protects it from MDM2-mediated degradation. (A) Western analysis of p53 (Ab-6; Calbiochem) in lysates made from H1299 cells transiently transfected with wt p53 in the presence of parental vector alone (pRc/CMV) (lane 1), pCMX-Sin3 (lane 2), or p14ARF (lane 3). Equivalent protein loading between lanes was confirmed by Western analysis of β-actin levels. (B) Western analysis of MDM2 (Ab-1; Calbiochem) in H1299 cells transfected with parental vector alone (pRc/CMV) (lane 1) or a cytomegalovirus (CMV)-driven MDM2 expression construct, in the presence (lane 3) and absence (lane 2) of an equal amount, in micrograms, of cotransfected Sin3. Equivalent protein loading was confirmed by Western analysis of β-actin. (C) Western analysis of MDM2 and p53 in H1299 cells transfected with wt p53 in the presence of MDM2 (lane 2) or MDM2 and Sin3 (lane 3). While transfection with MDM2 leads to significant decreases in p53 steady-state levels (lane 2), this effect is reversed by transfection with Sin3 (lane 3). Equal levels of protein loading were confirmed by Western analysis of β-actin levels; the occasional differences in p53 levels relative to β-actin represent the use of different p53 antisera (Ab-6 or p53 fl1-393 [Santa Cruz Biotechnology]) as well as different actin antisera (AC-15 [Sigma] versus antiactin polyclonal Santa Cruz [Biotechnology]). While these antisera occasionally revealed different levels of p53 and/or actin, the trends in p53 stabilization in the presence of Sin3 were consistent.
To address the possibility that Sin3 transfection counteracts MDM2-mediated degradation of p53, H1299 cells were transfected with wt p53 in the presence or absence of MDM2 or of MDM2 and Sin3 combined. As depicted in Fig. 1C, cotransfection of wt p53 with its negative regulator MDM2 markedly decreased the amount of detectable p53 protein, consistent with published results (18, 29). Significantly, cotransfection of Sin3 was able to abrogate this effect, and p53 returned to its original levels (Fig. 1C, lane 3). In general, the level of cotransfected MDM2 remained largely unchanged, though in some cases it appeared to increase with increased p53 levels (Fig. 1C and data not shown). These data indicate that Sin3 expression can increase p53 levels and that this activity is sufficient to modulate the effect of MDM2 on p53 stability. It should be noted, however, that these data do not indicate that Sin3 and MDM2 are directly antagonistic, as these proteins may be affecting separate pools of p53. This possibility is addressed further below.
Sin3 inhibits proteasome-mediated degradation of p53.
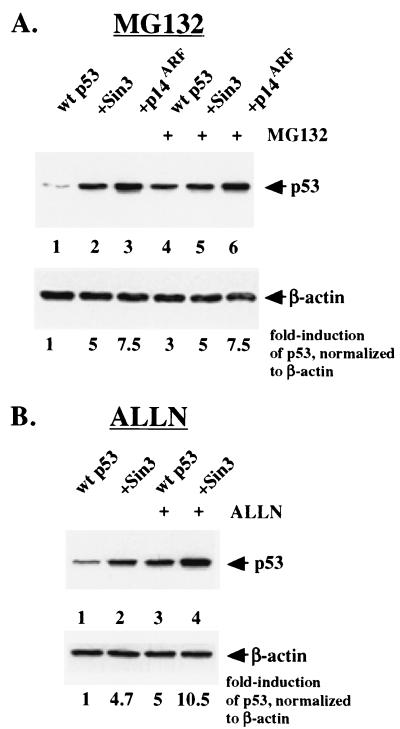
p53 protein is rapidly turned over in unstressed cells, primarily due to proteasome-mediated degradation, although the protease calpain 1 also plays a role in p53 degradation (28). To address the possibility that Sin3 inhibits proteasome-mediated degradation of p53, H1299 cells were transiently transfected with combinations of expression constructs for p53, Sin3, and p14ARF. Twenty-four hours after transfection, cells were treated with the proteasome inhibitor MG132 or dilution vehicle alone (DMSO) for 2 h. The finding that MG132 could not further increase the level of Sin3-stabilized p53 would support the conclusion that these two agents act on the same pathway by inhibiting proteasome-mediated degradation of p53.
As shown in Fig. 2, MG132 treatment led to a threefold increase in transfected p53 (Fig. 2A, compare lanes 1 and 4). Consistent with previous findings (46), p53 was stabilized by p14ARF, and this stabilization was not enhanced by MG132 (Fig. 2A, lanes 3 and 6). Significantly, MG132 was likewise unable to function additively with Sin3 to increase the level of p53 in Sin3-transfected cells, indicating that the stabilization of p53 by both Sin3 and MG132 occurs via inhibition of proteasome-mediated degradation (Fig. 2A, lanes 2 and 5). Results identical to these were obtained using the proteasome inhibitor lactacystin (data not shown). In contrast, the calpain 1 inhibitor ALLN was able to further increase the level of Sin3-stabilized p53 (Fig. 2B). Treatment of transfected cells with the calpain 1 inhibitor ALLN at concentrations previously reported to inhibit calpain-mediated degradation of p53 (28) led to a fivefold increase in p53 protein (Fig. 2B, lane 3). Notably, when combined with Sin3 transfection, ALLN and Sin3 functioned in an additive manner, leading to a 10-fold increase in p53 levels (Fig. 2B, lane 4). The combined data support the hypothesis that Sin3 expression leads to stabilization of p53 protein and that this stabilization is due to inhibition of proteasome-mediated degradation of p53.
FIG. 2.
Sin3 inhibits proteasome-mediated degradation of p53. (A) Western analysis of p53 levels in H1299 cells transfected with wt p53 in the presence of cotransfected Sin3 (lanes 2 and 5) or p14ARF (lanes 3 and 6). Lanes 4 to 6 include a 2-h posttransfection incubation with a 200 μM concentration of the proteasome inhibitor MG132, while lanes 1 to 3 are treated with dilution vehicle (DMSO). While MG132 is able to stabilize transfected p53 approximately threefold (compare lanes 1 and 4), it has an insignificant effect on the level of p53 stabilized by Sin3 (compare lanes 2 and 5), indicating that Sin3 and MG132 likely function through redundant pathways (inhibition of proteasome-mediated degradation). (B) The calpain 1 inhibitor ALLN is able to further stabilize Sin3-stabilized p53 (compare lanes 2 and 4). Both Sin3 transfection and ALLN treatment (45 μM) result in fivefold increases in p53 levels; the two agents together function additively (lane 4). Equal protein loading was confirmed by Western analysis for β-actin.
Stabilization of p53 by Sin3 requires the Sin3-binding domain of p53 (amino acids 61 to 75).
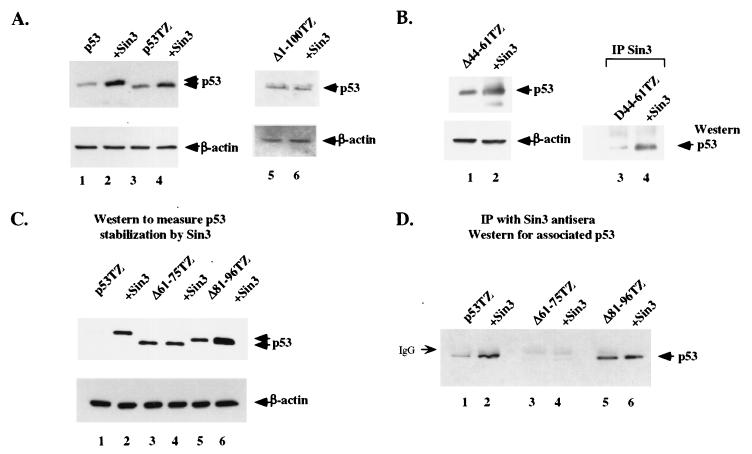
The accumulated data support the notion that Sin3 expression can influence the degradation of p53. However, it remained formally possible that Sin3 expression influenced p53 levels indirectly, by inducing a stress response. In order to distinguish between direct and indirect effects of Sin3 on p53 levels, the requirement for an interaction between Sin3 and p53 for stabilization was determined. Specifically, deletion constructs of p53 lacking the Sin3-binding domain were generated and tested for their ability to be stabilized by Sin3. We previously mapped the Sin3-binding domain of p53 to amino acids 40 to 100 of p53 (36). In the present study, we generated a series of synthetic p53 mutants with internal deletions encompassing this domain; these include internal deletions of amino acids 44 to 61 (Δ44–61), 61 to 75 (Δ61–75), and 81 to 96 (Δ81–96), as well as the extensive deletion of amino acids 1 to 100 (Δ1–100). Because Sin3 also interacts weakly but consistently with the oligomerization domain of p53 (36), these deletion mutants were created with an artificial tetramerization domain (TZ) from Saccharomyces cerevisiae GCN4 (51). This artificial TZ still allows p53 to oligomerize, transactivate, suppress cell growth, and be ubiquitinated normally (33, 51).
Transfection of H1299 cells with either wt p53 or the full-length p53TZ construct (containing an artificial oligomerization domain) resulted in a three- to fourfold increase in both proteins in the presence of Sin3, as assessed by Western analysis (Fig. 3A, lanes 1 to 4). In contrast, the Δ1–100TZ mutant, which we have previously shown fails to interact with Sin3 (36), was unaffected by Sin3 expression (Fig. 3A, lanes 5 and 6). Deletion of amino acids 44 to 61 of p53 (Δ44–61TZ) failed to affect Sin3 stabilization (Fig. 3B, lanes 1 and 2) or binding (lanes 3 and 4). Two other deletion mutants of p53, Δ61–75TZ and Δ81–96TZ, were also analyzed for their ability to be stabilized (Fig. 3C) and interact (Fig. 3D) with Sin3. As depicted in Fig. 3, the Δ81–96TZ mutant can bind (Fig. 3D, lanes 5 and 6) and be stabilized by (Fig. 3C, lanes 5 and 6) Sin3, to a level comparable to that of the full-length p53TZ control (Fig. 3A). In contrast, the Δ61–75TZ mutant was consistently unable to bind (Fig. 3D, lanes 3 and 4) or be stabilized by (Fig. 3C, lanes 3 and 4) Sin3. These data narrow down the region of interaction between p53 and Sin3 to amino acids 61 to 75 of p53. Further, they indicate that this minimal interaction domain is required for stabilization of p53 by Sin3.
FIG. 3.
Stabilization of p53 by Sin3 requires the Sin3-binding domain of p53 (amino acids 61 to 75). (A) Western analysis of H1299 cells transiently transfected with wt p53 (lanes 1 and 2), the p53TZ construct encoding wt p53 but containing an artificial TZ (lanes 3 and 4), and a deletion mutant of this protein that lacks amino acids 1 to 100 and fails to interact with Sin3 (Δ1–100TZ) (lanes 5 and 6). Odd-numbered lanes are cotransfected with parental vector, and even-numbered lanes are cotransfected with an equal amount of Sin3 expression construct. (B) Western analysis of a p53 deletion mutant with amino acids 44 to 61 deleted (Δ44–61TZ). This mutant is stabilized by exogenous Sin3 (lanes 1 and 2) and binds to Sin3 in transfected H1299 cells that are immunoprecipitated with antiserum to Sin3 (AK-11) and immunoblotted with p53 antiserum (Ab-6; Calbiochem) (lanes 3 and 4). (C) Western analysis of H1299 cells transfected with the p53TZ cDNA or with internal deletions created in the p53TZ backbone (Δ61–75TZ) (lanes 3 and 4) and Δ81–96TZ (lanes 5 and 6). Odd-numbered lanes are cotransfected with parental vector, and even-numbered lanes are cotransfected with Sin3 expression construct. Only the Δ61–75 mutant fails to be stabilized by exogenous Sin3 (lane 4). (D) IP-Western analysis of the interaction of Sin3 in vivo with deletion mutants of p53, including the p53 mutant that fails to be stabilized by Sin3 (Δ61–75TZ) (lanes 3 and 4). Odd-numbered lanes are cotransfected with parental vector, and even-numbered lanes are cotransfected with Sin3 expression construct.
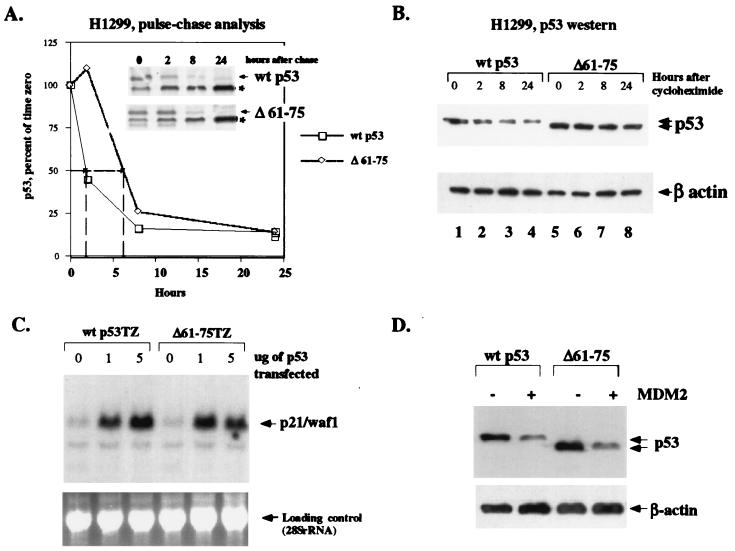
A deletion mutant of p53 lacking the Sin3-binding domain (p53Δ61–75) has an increased half-life in vivo.
The above data support the hypothesis that interaction with Sin3 may protect p53 from degradation. A corollary to this hypothesis would be that the Sin3-binding domain of p53, from amino acids 61 to 75, is normally subject to a destabilizing influence and that Sin3 abrogates this influence by interacting with this domain. In order to test this hypothesis, we measured the half-life of wt p53 and the Δ61–75 deletion mutant in transfected cells. H1299 cells were transfected with wt p53 or the Δ61–75 mutant, and transfected cells were radiolabeled with [35S]methionine and chased with cold methionine for 24 h. Cells were harvested and subjected to IP with polyclonal antisera to p53. As indicated in Fig. 4A, both wt p53 and the Δ61–75 mutant show similar first-order decay kinetics, but the Δ61–75 mutant demonstrates a longer half-life. In this study, the half-life of wt p53 was estimated to be approximately 2 h, consistent with published results of this kind (17), while the Δ61–75 mutant had an estimated half-life of approximately 6 h. Similar half-life experiments, using cycloheximide to block de novo protein synthesis in transfected cells, likewise revealed a consistent increase in the half-life of the Δ61–75 mutant, relative to that of wt p53 (Fig. 4B). These data support the hypothesis that amino acids 61 to 75 of p53 may normally be subject to a destabilizing influence.
FIG. 4.
A p53 deletion mutant lacking the Sin3-binding domain (Δ61–75) shows enhanced stability in vivo. (A) Pulse-chase analysis of 35S-methionine radiolabeled H1299 cells transfected with wt p53 or the p53 mutant lacking the Sin3-binding domain (Δ61–75). Cells were radiolabeled with [35S]methionine and chased for the indicated time points with excess unlabeled methionine. Equal counts per minute of lysate were immunoprecipitated with polyclonal antisera to p53 (Santa Cruz Biotechnology), as indicated in Materials and Methods. The asterisk (*) indicates β-actin, which is frequently nonspecifically immunoprecipitated by polyclonal antisera to p53. The graph depicts densitometric analysis of this representative experiment, which indicates that wt p53 has a 2-h half-life, while the Δ61–75 mutant has an approximately 6-h half-life. (B) Western analysis of the half-life of wt p53 (lanes 1 to 4) and the Δ61–75 mutant (lanes 5 to 8) following transfection of H1299 cells for 24 h and treatment with 40 μg of cycloheximide/ml for the times indicated to halt new protein synthesis. The data shown are representative of at least three independent experiments; β-actin is shown to control for protein loading. (C) Northern analysis of H1299 cells transiently transfected with 0, 1, and 5 μg of the wt p53TZ cDNA or the Δ61–75 deletion mutant (Δ61–75TZ). Thirty-six hours following transfection, cells were harvested and analyzed by Northern analysis for the level of endogenous p21, which was up-regulated equally well by both forms of p53. A picture of the ethidium bromide-stained gel used as a control for RNA loading and integrity is included. (D) Western analysis of p53 levels in cells transfected with parental vector alone (odd-numbered lanes) or vector expressing MDM2 (even-numbered lanes). Both wt p53 (lanes 1 and 2) and the Δ61–75 mutant (lanes 3 and 4) are effectively degraded by MDM2.
We next sought to test if the p53 mutant lacking the Sin3-binding domain (Δ61–75) retained some of the functions of wt p53, specifically transactivation and degradation by MDM2. As depicted in Fig. 4C, the Δ61–75 mutant of p53 retained the ability to transactivate the endogenous p21 (waf1) gene when transfected into p53-null H1299 cells, to a level comparable to that of wt p53 (Fig. 4C). Additionally, this mutant likewise was degraded by MDM2 to a level comparable to that of wt p53 (Fig. 4D). Therefore, deletion of the Sin3-binding domain from amino acids 61 to 75 does not denature or otherwise inactivate p53.
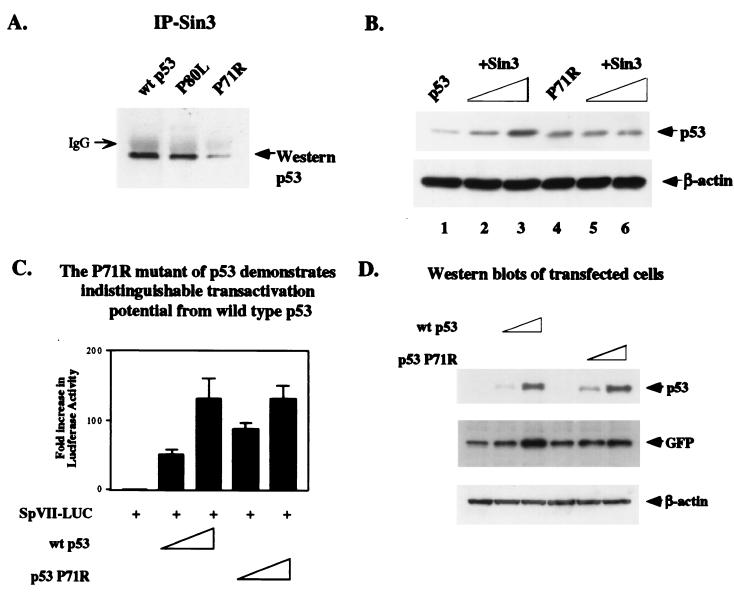
The P71R mutant of p53 fails to bind to Sin3 or be stabilized by this protein.
In order to extend the mapping of the Sin3-p53 interaction domain, we created point mutations in several of the conserved amino acids in the Sin3-binding region of p53. Three of these point mutants, encoding leucine at amino acid 80 (P80L) and glycine at amino acid 62 or 63 (E62G or A63G, respectively), were able to be stabilized and bind to Sin3 (Fig. 5A and data not shown). One point mutant, however, encoding arginine instead of proline at amino acid 71 (P71R), consistently demonstrated impaired binding to Sin3 in vivo (Fig. 5A) and failed to be stabilized by exogenous Sin3 (Fig. 5B, lanes 4 to 6). Identical results were obtained when leucine was substituted for proline at amino acid 71 (P71L) (data not shown). The proline to arginine substitution at amino acid 71 did not affect the ability of p53 to function as a transactivator, as the P71R mutant transactivated the p53-responsive luciferase construct SpVII to levels equivalent to those of wt p53 (Fig. 5C). As a control for these studies, Western analysis indicated that both forms of p53 were expressed at equivalent levels in these transfected cells (Fig. 5D).
FIG. 5.
A point mutant of p53 containing arginine instead of proline at amino acid 71 (P71R) fails to interact with Sin3 in vivo or to be stabilized by exogenous Sin3. This mutant shows transactivation potential, however, that is indistinguishable from wt p53. (A) Transfection of H1299 cells with wt p53 or point mutants containing leucine at amino acid 80 (P80L) or arginine at amino acid 71 (P71R) followed by IP with Sin3 antiserum (AK-11; Santa Cruz Biotechnology) and immunoblotting with p53 antiserum (Ab-6; Calbiochem). The data indicate that the P71R mutant shows impaired interaction with Sin3 in transfected cells. (B) Transfection of H1299 cells with 50 ng of p53 or the P71R mutant, along with increasing concentrations of Sin3 expression plasmid (0.5 and 2.5 μg, lanes 2, 3, 5, and 6). Immunoblot analysis of p53 levels indicates that p53 is stabilized by Sin3 in a dose-dependent manner but that neither dose of Sin3 is capable of stabilizing the P71R mutant, which shows a defective interaction with Sin3. A β-actin control is included for protein loading. (C) Luciferase activity of the p53-inducible luciferase vector SpVII (a minimal promoter containing the E1B TATA box and a single p53 consensus element) transiently transfected into H1299 cells in the presence of parental vector (pRc/CMV) or increasing concentrations of wt p53 (25 and 100 ng) or the P71R point mutant of p53. The data depicted represent the fold increase in luciferase activity obtained after transfection, averaged from five independent experiments. Error bars depict the standard error of the mean for the five experiments. (D) Western analysis depicting comparable p53 levels expressed in the transfected cells analyzed in the results shown in panel C. A total of 1.25 μg of the SpVII luciferase reporter construct was cotransfected with either vector alone or increasing concentrations of wt p53 (25 and 100 ng), P71R (25 and 100 ng), and 100 ng of pEGFP (transfection control) in six-well plates and analyzed for p53 levels by Western analysis. Western analysis of the GFP levels between samples is used to show relatively equal transfection efficiencies, and equivalent protein loading is depicted by Western analysis for β-actin.
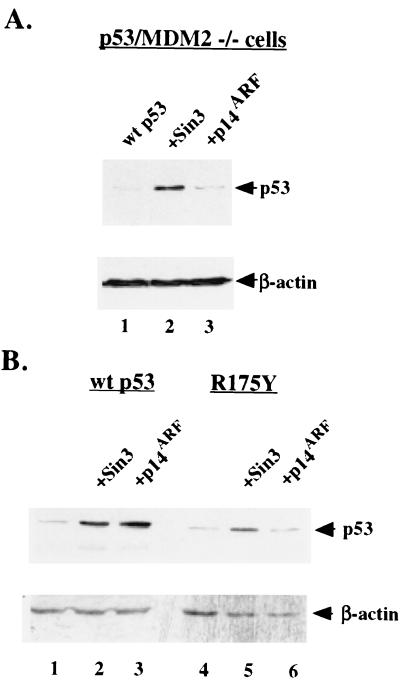
Stabilization of p53 by Sin3 does not require MDM2.
The MDM2 binding domain of p53 has been mapped to amino acids 17 to 23; this domain is believed to form an induced-fit amphipathic helix that fits into a hydrophobic pocket at the amino terminus of MDM2 (30). The juxtaposition of this region to the Sin3-binding domain at residues 61 to 75 raised the possibility that Sin3 stabilizes p53 by sterically hindering MDM2 from binding and degrading p53. A testable prediction from this hypothesis would be that Sin3 is unable to stabilize p53 in MDM2-null cells. To address this issue, murine embryo fibroblasts generated from mice nullizygous for both p53 and MDM2 (174-1 cells) were transfected with p53 along with parental vector alone, Sin3, or p14ARF. Western analysis revealed that Sin3 transfection led to increased p53 levels in these cells, while p14ARF had no effect (Fig. 6A, lanes 2 and 3). Therefore, unlike p14ARF, MDMX, and pRB, Sin3 does not appear to stabilize p53 by binding or inhibiting MDM2. Consistent with these data, we have found by IP-Western analysis that a complex between p53, MDM2, and Sin3 is immunologically detectable in MCF-7 cells (data not shown).
FIG. 6.
Stabilization of p53 by Sin3 occurs in an MDM2-independent manner. (A) Western analysis of 174-1 cells (p53 −/− MDM2−/− transfected with wt p53 in the presence of parental vector alone (lane 1), Sin3 expression construct (lane 2), or p14ARF (lane 3). (B) Western analysis of H1299 cells transfected with wt p53 (lanes 1 to 3) or the tumor-derived mutant R175Y (lanes 4 to 6) in the presence of vector alone (lanes 1 and 4), Sin3 expression construct (lanes 2 and 5), and p14ARF (lanes 3 and 6). Equal protein loading among the lanes was confirmed by Western analysis for β-actin.
It became of interest to test the ability of Sin3 to stabilize human tumor-derived mutant forms of p53, which we have previously shown are capable of interacting with Sin3 (36). Using transient transfection and Western blot analyses, we found that cotransfection of Sin3 with the DNA-binding domain mutant R175Y of p53 led to slight but consistent increases in the steady-state level of this protein (Fig. 6B, lanes 4 and 5). In contrast, p14ARF was incapable of stabilizing this p53 mutant, consistent with the inability of the R175Y mutant to transactivate the endogenous mdm2 gene, which is necessary for stabilization by p14ARF (Fig. 6B, lane 6). These data (Fig. 6) support the premise that stabilization of p53 by Sin3 occurs independently of MDM2 function.
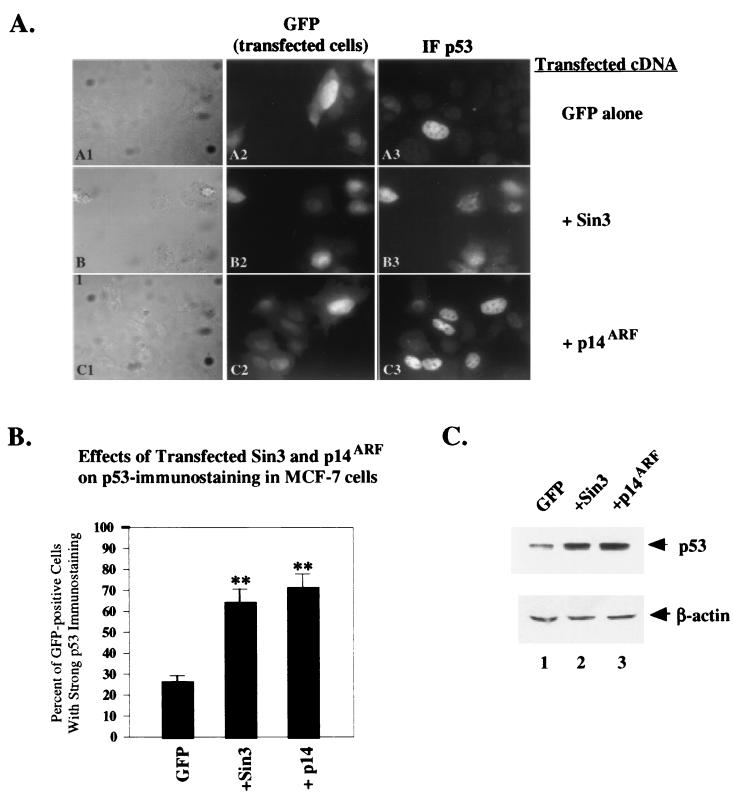
Sin3 stabilizes endogenous p53.
To assess the ability of Sin3 to affect the localization and stabilization of endogenous p53, the human breast carcinoma cell line MCF-7 was utilized. This cell line contains wt p53, but much of this protein is poorly detectable by immunofluorescence, possibly because this protein is sequestered in the cytoplasm (4, 8). MCF-7 cells were transfected with Sin3 or p14ARF; as a marker for transfected cells, cells were cotransfected with an expression construct encoding GFP. Following transfection, cells were assayed by confocal microscopy for the presence of GFP (a marker for transfected cells), as well as for p53 immunostaining. In each of three experiments, over 100 GFP-positive cells were scored for the presence of strong p53 immunostaining, which would be indicative of p53 stabilization.
Control MCF-7 cells transfected with GFP alone showed strong immunostaining for p53 in 27% of cells (Fig. 7A, panel A3); this is comparable to the percentage with untransfected cells and is consistent with previously published reports about this cell line (4). Cotransfection with Sin3 resulted in a significant increase in the number of GFP-positive cells showing strong p53 immunostaining (from 27% for GFP alone to 65% in Sin3-transfected cells) (Fig. 7A, panels B2 and B3), consistent with the endogenous p53 protein being stabilized. Transfection with the positive control p14ARF resulted in a similar increase in cells with strong immunostaining for p53 (from 27% for GFP alone to 74% in p14ARF-transfected cells) (Fig. 7A, panels C2 and C3). These results, which were obtained in three independent experiments scored blindly, were statistically significant (P < 0.004 and P < 0.003 for Sin3 and p14ARF, respectively) (Fig. 7B). Notably, Western analysis of duplicate plates of transfected cells confirmed these results, indicating that the endogenous p53 protein level in this cell line was increased 2.5- to 3-fold following transfection with Sin3 (Fig. 7C, lane 2) but not GFP alone (lane 1). Similar results were obtained with the human melanoma cell line CaCl (wt p53) (data not shown).
FIG. 7.
Sin3 stabilizes endogenous p53 in MCF-7 cells. (A) Immunofluorescence of MCF-7 cells transfected with a GFP expression construct (pEGFP), analyzed by differential interference contrast (A1 and C1), for GFP fluorescence (A2 and C2) and for p53 staining (A3 and C3) with polyclonal antisera to p53. GFP-positive cells were scored as transfected cells, and cells with high levels of p53 immunostaining were scored as positive for p53. In three independent blinded studies, over 100 GFP-positive cells were scored for strong p53 immunostaining; the combined results from these experiments are presented in panel B. (B) The averaged data from three independent experiments indicate that transfection with Sin3 and p14ARF leads to significant increases in the number of transfected cells with strong immunopositivity for p53. While 27% of cells transfected with GFP alone show strong immunostaining for p53, 65 and 74% of cells transfected with Sin3 and p14ARF, respectively, showed increased p53 immunostaining. The asterisks indicate a statistically significant difference between the indicated treatment and control (P < 0.005, Student's t test). The error bars indicate the standard error of the mean for the combined three experiments. (C) Western analysis of the levels of endogenous p53 in the samples prepared in the results shown in panel A. These data indicate that transfection with Sin3 and transfection with p14ARF both significantly stabilize endogenous p53, compared to transfection with equal amounts, in micrograms, of GFP vector alone. Equal protein loading was confirmed by Western analysis for β-actin.
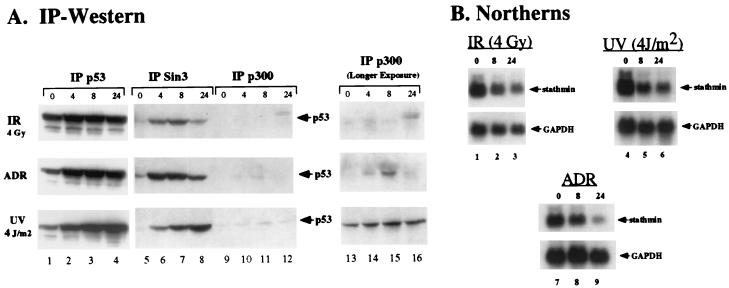
The Sin3-p53 complex appears for prolonged periods after DNA damage.
Taken together, the above data indicate that Sin3 stabilizes p53 and that this stabilization requires an interaction between these two proteins. These data raise the interesting hypothesis that repression complexes containing p53 and Sin3 exist more stably in the cell than transactivation complexes containing p53 and p300, which are targeted by MDM2 for degradation (16). As an examination of this hypothesis, we analyzed the abundance of p53-Sin3 immunocomplexes in cells following genotoxic stress and compared this to p53-p300 complexes. For these studies, p53, Sin3, and p300 were immunoprecipitated from MCF-7 cells treated with γ irradiation, the DNA-damaging agent doxorubicin (adriamycin), and UV radiation; treated cells were harvested after 0, 4, 8, and 24 h. These immunoprecipitates were analyzed by Western analysis for the presence of p53. As expected, these studies revealed that stabilization of p53 protein occurred as early as 4 h in response to all genotoxic stresses tested (Fig. 8A, lane 2). The Sin3-p53 immunocomplex appeared at the earliest time point (4 h) and persisted in abundance throughout the time course, though with slightly different kinetics for the three genotoxic stresses. Notably, the formation and persistence of this complex paralleled the down-regulation of stathmin, a p53-repressed gene (1) (Fig. 8B).
FIG. 8.
The p53-Sin3 complex is immunologically detectable for prolonged periods following genotoxic stress, compared to the p53-p300 complex. (A) IP-Western analysis of MCF-7 cells treated with 4 Gy of irradiation (IR), 0.5 μg of adriamycin (ADR)/ml, or 4 J of UV radiation/m2. Cells were harvested at the indicated time points following treatment (0, 4, 8, and 24 h), and lysates were immunoprecipitated with polyclonal antisera to p53, Sin3 (AK-11; Santa Cruz Biotechnology), and p300 (Ab-1; Calbiochem), followed by Western analysis for p53 with a mouse monoclonal antibody (Ab-6; Calbiochem). Longer exposures for the p300 IPs are shown on the right, and shorter p53 exposures are shown on the left. The data depicted are representative of three independent experiments. (B) Northern analysis of the p53-repressed gene stathmin in cells treated identically to those shown in panel A. For all stresses tested, down-regulation of stathmin is evident by 8 h and increases at 24 h, concomitant with the appearance of the p53-Sin3 complex. The glyceraldehyde-3-phosphate dehydrogenase housekeeping gene (GAPDH) is included as a control for RNA loading and integrity.
The p300-p53 complex was much less detectable than the Sin3-p53 complex, possibly because p300 targets p53 for MDM2-mediated degradation (16). Further, in the case of doxorubicin-treated cells, this complex consistently appeared to be transient, peaking at 4 or 8 h and becoming undetectable by 24 h (Fig. 8A). A similarly transient nature was observed for the MDM2-p53 complex (data not shown). Identical results were obtained with three different antibodies to Sin3 and p300 (data not shown). These in vivo data support the physiological relevance of our finding that interaction with Sin3 leads to stabilization of p53. These data also indicate that different pools of p53 may have different half-lives in the cell and that regulation of p53 stability may have a direct impact on p53 function (for example, transactivation versus transrepression).
DISCUSSION
In the present study, we demonstrate that ectopic expression of the corepressor protein Sin3 leads to stabilization of both transfected and endogenous p53. That this effect is a direct impact of interaction of Sin3 with p53 is supported by our finding that a 15-amino-acid deletion mutant of p53 that is incapable of interacting with Sin3 also fails to be stabilized by this protein. Similarly, mutation of proline 71 of p53 to arginine or leucine significantly impairs Sin3 binding and Sin3-mediated stabilization. These data support a tight correlation between Sin3 binding and stabilization of p53. Our data also indicate that stabilization by Sin3 is likely the result of inhibition of proteasome-mediated degradation of p53. Interestingly, unlike p14ARF, MDMX, and pRB, Sin3 does not require the presence of MDM2 for this effect. Therefore, these findings point to the existence of a potentially novel pathway for p53 stabilization that does not involve inhibition of MDM2 function.
At least two proteins are implicated in p53 degradation in a manner that is independent of MDM2. These are the human papillomavirus type 16 or 18 (HPV-16 or -18) E6 protein (coupled with the accessory protein E6-AP) and JNK. As the E6–E6-AP complex and JNK both target p53 for degradation in an MDM2-independent manner (2, 12, 13), it is formally possible that Sin3 stabilizes p53 by inhibiting the action of these proteins. We have found that Sin3 also stabilizes the Δ92–112 mutant of p53 (J. T. Zilfou and M. Murphy, unpublished data); this deletion overlaps with the JNK binding site, indicating that it is unlikely that Sin3 stabilizes p53 by inhibiting JNK binding and/or destabilization. The contribution of the E6/E6-AP pathway to p53 degradation in normal (non-HPV-infected) cells remains controversial. Two groups found no evidence for a role for E6-AP protein in p53 degradation in normal cells (5, 47). In contrast, the E6-AP knockout mouse shows increased p53 levels in certain cell types, indicating that this degradative pathway may play a cell-type-specific role in p53 degradation in normal cells (24). It is notable that we have mapped the Sin3-binding domain of p53 to a region that is required for degradation by the HPV-16–HPV-18 E6 protein (34). The possibility that Sin3 interferes with E6–E6-AP-mediated degradation of p53 is currently being tested in the laboratory.
The combined data implicate the Sin3-binding domain of p53, from amino acids 61 to 75, as a region that normally confers destabilization to this protein. This region of p53 has been previously implicated as a destabilization domain for this protein; specifically, removal of amino acids 62 to 96 has been shown to stabilize p53 in certain cell types (19). Interestingly, this region also overlaps with the proline-rich domain of p53 (amino acids 64 to 91), which is absolutely essential for apoptosis induction by this protein (40, 49, 50). A smaller deletion in this region, retaining only the Sin3-binding domain (amino acids 62 to 73), also retains the ability to induce apoptosis (53) and points to the importance of the p53-Sin3 interaction for the ability of p53 to induce apoptosis. Significantly, the proline-rich domain has also been shown to be necessary for transcriptional repression by p53 (49, 50). Therefore, Sin3 is not only one of the first proteins found to interact with the proline-rich domain of p53, it is also the first protein implicated in transcriptional repression found to interact there.
It should be noted that our data do not imply that interaction with Sin3 is the exclusive, or even the major, mechanism whereby p53 is stabilized following DNA damage. In fact, our data indicate that the amount of p53 associated with Sin3 following DNA damage is rather small, approaching only 10% of total p53 (Fig. 8 and data not shown). Additionally, data from several other groups support the notion that decreased association with MDM2 is the major mechanism whereby p53 is stabilized in the cell in response to genotoxic stress (6–8, 26, 38). Therefore, the ability of Sin3 to stabilize p53 is relevant only for that pool of p53 that is bound to Sin3; this stabilization may even occur exclusively at the promoters of p53-repressed genes.
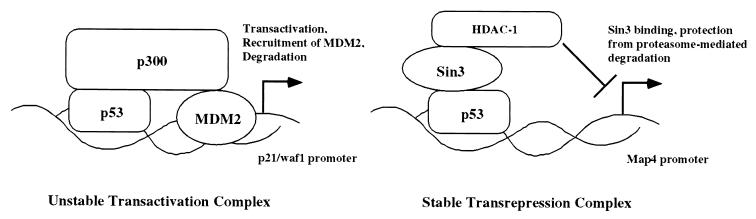
The identification of Sin3 as a stabilizer of p53 appears logical when the mechanism of action of the p53-Sin3 complex is taken into consideration. Transactivation of target genes by p53 can proceed effectively in a transient manner; this transient nature may even be facilitated by the coactivator p300, which serves as a scaffold for MDM2-mediated degradation of p53 (16). In contrast, it could be argued that transcriptional repression is effective only if repression complexes exist stably on the promoters of repressed genes for prolonged periods of time. By protecting p53 from degradation, the corepressor Sin3 therefore would be predicted to enhance the efficacy of p53 as a transcriptional repressor (Fig. 9). In an analogous manner, E2F-1 and c-Myc, both of which are unstable transcriptional activators, have been shown to be protected from proteasome-mediated degradation by their respective transcriptional repression partners, pRB and miz-1 (20, 41). Our in vivo data, indicating that the p53-Sin3 complex exists for sustained periods following exposure to DNA damage, support this hypothesis. In sum, the data presented in this paper support a novel mechanism for the control of p53 stability and activity and point to a physiologically relevant mechanism for the control of p53 degradation, mediated by the Sin3-binding domain of p53.
FIG. 9.
Model for the differential stability of p53-dependent transactivation and transrepression complexes. p53-dependent transactivation of p53-induced genes such as p21 utilizes p300 as a coactivator; p300 serves as a scaffold for interaction between p53 and MDM2, thus targeting p53 for degradation and prohibiting prolonged transactivation. In contrast, the p53 protein present at the promoters of p53-repressed genes like map4 would be bound to the corepressor Sin3 (shown here bound to histone deacetylase 1 [HDAC-1]) and is predicted to be protected from proteasome-mediated degradation. Such protection would facilitate prolonged and efficient transcriptional repression.
ACKNOWLEDGMENTS
We thank Peter Adams, Warren Davis, Margret Einarson, Geraldine O'Neill, and Rebecca Raftogianis for critical reading of the manuscript. We also thank Thanos Halazonetis for the p53TZ construct, Jonathan Boyd for confocal expertise, and Pearl Huang and Siham Biade for helpful discussions throughout the course of this work. We thank Stephanie Stehman for technical assistance for some of these studies.
This work was supported by NIH grants CA80854 (M.M.) and CA66741 (D.L.G.), as well as by a generous grant from the W. W. Smith Charitable Trust (M.M).
REFERENCES
- 1.Ahn J, Murphy M, Kratowicz S, Wang A, Levine A J, George D L. Down-regulation of the stathmin/Op18 and FKBP25 genes following p53 induction. Oncogene. 1999;18:5954–5958. doi: 10.1038/sj.onc.1202986. [DOI] [PubMed] [Google Scholar]
- 2.Alarcon R, Koumenis C, Geyer R K, Maki C G, Giaccia A J. Hypoxia induces p53 accumulation through MDM2 down-regulation and inhibition of E6-mediated degradation. Cancer Res. 1999;59:6046–6051. [PubMed] [Google Scholar]
- 3.Ayer D E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 4.Bartek J, Iggo R, Gannon J, Lane D P. Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene. 1990;5:893–899. [PubMed] [Google Scholar]
- 5.Beer-Romero P, Glass S, Rolfe M. Antisense targeting of E6AP elevates p53 in HPV-infected cells but not in normal cells. Oncogene. 1997;14:595–602. doi: 10.1038/sj.onc.1200872. [DOI] [PubMed] [Google Scholar]
- 6.Bottger A, Bottger V, Sparks A, Liu W, Howard S F, Lane D P. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- 7.Chehab N H, Malikzay A, Stavridi E S, Halazonetis T D. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci USA. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Agrawal S, Zhou W, Zhang R, Chen J. Synergistic activation of p53 by inhibition of MDM2 expression and DNA damage. Proc Natl Acad Sci USA. 1998;95:195–200. doi: 10.1073/pnas.95.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Ko L J, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 10.Chowdary D R, Dermody J J, Jha K K, Ozer H L. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol Cell Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damalas A, Ben-Ze'ev A, Simcha I, Shtutman M, Fernando J, Leal M, Zhurinsky J, Geiger B, Oren M. Excess β-catenin promotes accumulation of transcriptionally active p53. EMBO J. 1999;18:3054–3063. doi: 10.1093/emboj/18.11.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freedman D A, Levine A J. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs S Y, Adler V, Buschmann T, Yin Z, Wu X, Jones S N, Ronai Z. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 1998;12:2658–2663. doi: 10.1101/gad.12.17.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geyer R K, Yu Z K, Maki C G. The MDM2 RING finger is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- 15.Gottlieb T M, Oren M. p53 in growth control and neoplasia. Biochem Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 16.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 17.Gu J, Chen D, Rosenblum J, Rubin R M, Yuan Z-M. Identification of a sequence element from p53 that signals for MDM2-targeted degradation. Mol Cell Biol. 2000;20:1243–1253. doi: 10.1128/mcb.20.4.1243-1253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 19.Hengstermann A, Whitaker N J, Zimmer D, Zentgraf H, Scheffner M. Characterization of sequence elements involved in p53 stability regulation reveals cell type dependence for p53 degradation. Oncogene. 1998;17:2933–2941. doi: 10.1038/sj.onc.1202282. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann F, Martelli F, Livingston D M, Wang Z. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 21.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 22.Honda R, Yasuda H. Association of p19ARF with MDM2 inhibits ubiquitin ligase activity of MDM2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh J, Chan F S G, O'Connor D J, Mittnacht S, Zhong S, Lu X. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell. 1999;3:181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y H, Armstrong D, Albrecht U, Atkins C M, Noebels J L, Eichele G, Sweatt J D, Beaudet A L. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 25.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Functional and physical interactions of the ARF tumor suppressor protein with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci USA. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 28.Kubbutat M H, Vousden K H. Proteolytic cleavage of human p53 by calpain: a potential regulator of p53 stability. Mol Cell Biol. 1997;17:460–468. doi: 10.1128/mcb.17.1.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubbutat M H, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 30.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 31.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 32.Maki C G, Huibregtse J M, Howley P M. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 33.Maki C G. Oligomerization is required for p53 to be efficiently ubiquitinated by MDM2. J Biol Chem. 1999;274:16531–16535. doi: 10.1074/jbc.274.23.16531. [DOI] [PubMed] [Google Scholar]
- 34.Mansur C P, Marcus B, Dalal S, Androphy E J. The domain of p53 required for binding HPV 16 E6 is separable from the degradation domain. Oncogene. 1995;10:457–465. [PubMed] [Google Scholar]
- 35.Murphy M, Hinman A, Levine A J. Wild-type p53 negatively regulates the expression of a microtubule associated protein. Genes Dev. 1996;10:2971–2980. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- 36.Murphy M, Ahn J, Walker K K, Hoffman W H, Evans R M, Levine A J, George D L. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oren M. Regulation of the p53 tumor suppressor protein. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 38.Prives C. Signaling to p53: breaking the MDM2-p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 39.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakamuro D, Sabbatini P, White E, Prendergast G C. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene. 1997;15:887–898. doi: 10.1038/sj.onc.1201263. [DOI] [PubMed] [Google Scholar]
- 41.Salghetti S E, Kim S Y, Tansey W P. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharp D A, Kratowicz S A, Sank M J, George D L. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- 43.Shieh S Y, Taya Y, Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spataro V, Norbury C, Harris A L. The ubiquitin-proteasome pathway in cancer. Br J Cancer. 1998;77:448–455. doi: 10.1038/bjc.1998.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stott F J, Bates S, James M C, McConnell B B, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden K H, Peters G. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talis A L, Huibregtse J M, Howley P M. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem. 1998;273:6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 48.Tao W, Levine A J. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998;17:4668–4679. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker K K, Levine A J. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waterman M J F, Waterman J L F, Halazonetis T D. An engineered four-stranded coiled coil substitutes for the tetramerization domain of wild-type p53 and alleviates transdominant inhibition by tumor-derived p53 mutants. Cancer Res. 1996;56:158–163. [PubMed] [Google Scholar]
- 52.Zhang Y, Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]
- 53.Zhu J, Jiang J, Zhou W, Zhu K, Chen X. Differential regulation of cellular target genes by p53 devoid of the PXXP motifs with impaired apoptotic activity. Oncogene. 1999;18:2149–2155. doi: 10.1038/sj.onc.1202533. [DOI] [PubMed] [Google Scholar]