Abstract
Genomic instability is often caused by mutations in genes that are involved in DNA repair and/or cell cycle checkpoints, and it plays an important role in tumorigenesis. Poly(ADP-ribose) polymerase (PARP) is a DNA strand break-sensing molecule that is involved in the response to DNA damage and the maintenance of telomere function and genomic stability. We report here that, compared to single-mutant cells, PARP and p53 double-mutant cells exhibit many severe chromosome aberrations, including a high degree of aneuploidy, fragmentations, and end-to-end fusions, which may be attributable to telomere dysfunction. While PARP−/− cells showed telomere shortening and p53−/− cells showed normal telomere length, inactivation of PARP in p53−/− cells surprisingly resulted in very long and heterogeneous telomeres, suggesting a functional interplay between PARP and p53 at the telomeres. Strikingly, PARP deficiency widens the tumor spectrum in mice deficient in p53, resulting in a high frequency of carcinomas in the mammary gland, lung, prostate, and skin, as well as brain tumors, reminiscent of Li-Fraumeni syndrome in humans. The enhanced tumorigenesis is likely to be caused by PARP deficiency, which facilitates the loss of function of tumor suppressor genes as demonstrated by a high rate of loss of heterozygosity at the p53 locus in these tumors. These results indicate that PARP and p53 interact to maintain genome integrity and identify PARP as a cofactor for suppressing tumorigenesis.
Poly(ADP-ribose) polymerase (PARP) (EC 2.4.2.30) is an abundant nuclear protein in mammalian cells. PARP binds rapidly to either single- or double-stranded DNA breaks and catalyzes (ADP-ribose) polymer formation on target proteins following exposure to DNA-damaging agents. The resulting negatively charged PARP is subsequently dissociated from DNA ends, facilitating the DNA repair process (22). This shuttling of PARP on and off the site at DNA strand breaks has led to the hypothesis that PARP and poly(ADP-ribosyl)ation play a multifunctional role in DNA repair, antirecombination, and the maintenance of genomic stability (18, 22). Cells derived from PARP−/− mice exhibit high levels of sister chromatid exchange, chromosomal aberrations (26, 39), and loss or gain of chromosomes (34); in addition, these cells contain shortened telomeres (10), indicative of genomic instability.
p53 is a major genome guardian molecule, the loss of which causes chromosome instability and renders cells susceptible to malignant transformation, most likely by loss of control over cell cycle checkpoints and apoptosis (21). Mutations in the p53 gene were frequently detected in various human malignancies (16) and are mainly responsible for the Li-Fraumeni syndrome (LFS) in humans, which causes the development of various tumor types including soft tissues sarcomas, osteosarcoma, and breast carcinomas as well as brain tumors, leukemia, and lymphomas (24). Inactivation of p53 in mice results in the spontaneous development of tumors which are mainly lymphomas, soft tissue sarcomas, and rare carcinomas (12, 15, 17). The reasons why p53 mutant mice do not perfectly mimic LFS remain elusive. A recent study of LFS patients identified heterozygous germ line mutations in human CHK2, a protein kinase required for DNA damage response and a replication checkpoint (4), suggesting that alteration of DNA repair or damage response and its interaction with p53 play an important role in the etiology of the cancer phenotype in LFS. In addition, although p53 can bind directly to DNA, the function of p53 is believed to be mediated by other DNA break-sensing molecules such as ATM (3, 8).
Biochemical and genetic studies suggest a possible interaction of PARP and p53 in mammalian cells. PARP can bind to specific domains of the p53 protein and modifies its activity by poly(ADP-ribosyl)ation; furthermore, inhibition of PARP leads to abrogation of p21 and mdm2 in response to DNA damage (23, 37). Recent studies have shown that the absence of PARP affects the accumulation or induction of p53 in response to DNA damage (1, 40). All of these in vitro studies suggest an impact of PARP or poly(ADP-ribosyl)ation on the function of p53 and that both proteins could lie in certain common pathways for their biological functions. In this study, we specifically investigated the functional interaction of PARP and p53 in vivo and studied the role of genomic instability induced by PARP deficiency in tumor development using mice deficient in PARP and p53.
MATERIALS AND METHODS
Animal breeding scheme.
PARP−/− mice (38) (129/BL6/CB17) were bred to p53−/− mice (12) (129/Sv; Jackson Laboratory, Bar Harbor, Maine) to generate F1 mice (129/Sv × 129/BL6/CB17)F. These mice were intercrossed for five subsequent generations, and all analyses were carried out using animals from the sixth generation.
Cytogenetic analysis and telomere measurement.
Quantitative fluorescence in situ hybridization (Q-FISH) was applied to determine chromosome aberrations and telomere length (42). Early passages (<5 population doublings) of primary mouse embryonic fibroblasts (MEFs) were isolated from E13.5 embryos derived from intercrosses of PARP+/− p53−/− or PARP−/− p53+/−, according to protocols previously described (38). Thymocytes or thymic lymphoma cells were isolated from adult mice as described previously (39). Q-FISH was performed on chromosome spreads with a Cy-3 labeled (CCCTAA)3 peptide nucleic acid probe and quantified by digital image analyses using TFL-TELO software. One telomere fluorescence unit (TFU) represents ∼1 kb of telomere repeats (42).
Histopathological analysis.
Animals were sacrificed upon decline in health (i.e., weight loss, paralysis, ruffling of fur, or inactivity) by ether anesthesia. Groups of mice from each genotype were monitored for tumor development, and statistical analysis of the numbers of tumor-bearing mice from each genotype was performed using the log rank test. A full autopsy was performed on at least 12 organs from each mouse, which were fixed in 4% neutral-buffered formaldehyde, followed by dehydration and paraffin embedding. Histopathological analyses were carried out on a 3-μm-thick section stained with hematoxylin and eosin.
Mouse genotyping and LOH analysis of tumors.
For genotyping the PARP locus, primers OVL1 (5′-GTT GTG AAC GAC CTT CTG GG-3′), OVL1R (5′-CCT TCC AGA AGC AGG AGA AG-3′), and NEOIIR (5′-GCT TCA GTG ACA ACG TCG AG-3′) were used. For genotyping the p53 locus, primers X7 (5′-TAT ACT CAG AGC CGG CCT-3′), NEO19 (5′-CAT TCA GGA CAT AGC GTT GG-3′), and X6.5 (5′-ACA GCG TGG TGG TAC CTT AT-3′) were used. For Southern blot analysis, genomic DNA was isolated from tumor tissues as well as from normal tissues of the same animal (tail or adjacent tissues) and digested by EcoRI enzymes followed by electrophoresis and blotting. The blot was hybridized with a 32P-labeled p53-specific probe corresponding to exon 11. Loss of heterozygosity (LOH) determinations were based on the quantification of the ratio of wild-type (∼16 kb) and mutant (∼8 kb) bands in autoradiography by PhosphorImager (Molecular Dynamics, Inc., Sunnyvale, Calif.). Ratios between 0 and 30% were regarded as “LOH,” ratios between 70 and 100% were regarded as “no LOH,” and ratios between these two groups were regarded as “partial LOH.”
RESULTS
Severe chromosomal abnormalities in PARP p53 double-mutant cells.
To study the biological significance of the interaction between PARP and p53, PARP−/− mice were crossed with p53 knockout mice, and PARP+/− p53+/− mice were intercrossed to generate different genotypes of double-mutant mice and MEFs. The impact of PARP and p53 interaction on genomic intergrity was first investigated using PARP p53 double-mutant MEFs by Q-FISH. Detailed cytogenetic analysis (Table 1) revealed all kinds of chromosomal abnormalities in double-mutant cells, such as fragmentation, breaks, and end-to-end fusions, including Robertsonian-like configurations and dicentric and ring-like (long arm fusion) chromosomes (Fig. 1A to H). While 19 and 27% aneuploidy were found in PARP−/− and p53−/− cells, respectively, 40% of cells were aneuploid in the double-null cell population. More strikingly, approximately one chromosome fusion per metaphase was detected in PARP−/− p53−/− cells (Table 1). Interestingly, we also observed a high incidence (47%) of end-to-end chromosome fusions in p53−/− MEFs (Fig. 1I). In addition, many chromosomes showed a loss of their telomere signals in PARP−/− cells and p53−/− cells, as well as in PARP p53 double-mutant cells (Fig. 1C and I; Table 1).
TABLE 1.
Impact of PARP and p53 deficiency on chromosomal integrity and telomere function
| Genotype
|
Total no. of metaphases analyzed | % of chromosome aberrationsa
|
No. of undetectable TTAGGG repeatsb | Telomere fluorescence (TFU) (mean ± SE)
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| PARP | p53 | Aneuploid cells | End-to-end fusions | Fragments | p-arm | q-arm | All | ||
| +/+ | +/+ | 16 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 35 ± 0.3 | 48 ± 0.4 | 41 ± 0.2 |
| +/+ | +/− | 16 | 6 (1) | 6 (1R) | 13 (2AC) | 3 (0.1) | 35 ± 0.5 | 49 ± 0.6 | 42 ± 0.4 |
| −/− | +/+ | 16 | 19 (3) | 31 (1RL, 2R, 2DIC) | 44 (5AC, 1CT, 1CH) | 13 (0.5) | 25 ± 0.2 | 31 ± 0.4 | 28 ± 0.2 |
| −/− | +/− | 32 | 28 (9) | 53 (7RL, 5R, 2DIC) | 69 (6C, 12AC, 3CT, 1CH) | 37 (0.7) | 30 ± 0.5 | 55 ± 1.1 | 43 ± 0.6 |
| +/+ | −/− | 59 | 27 (16) | 47 (14RL, 8R, 6DIC) | 68 (11C, 22AC, 4CT, 3CH) | 61 (0.7) | 35 ± 0.5 | 49 ± 0.6 | 42 ± 0.4 |
| +/− | −/− | 33 | 21 (7) | 36 (4RL, 5R, 3DIC) | 21 (1C, 6AC) | 21 (0.4) | 44 ± 0.5 | 62 ± 0.9 | 53 ± 0.6 |
| −/− | −/− | 72 | 40 (29) | 92 (37RL, 14R, 15DIC) | 50 (7C, 18AC, 5CT, 6CH) | 73 (0.8) | 50 ± 0.7 | 79 ± 1.3 | 65 ± 0.8 |
Information in parentheses indicates scored chromosome abnormalities. RL, Robertsonian-like configurations; R, ring-like chromosomes; DIC, dicentric chromosomes; AC, acentric chromosomal fragments; C, centric chromosomal fragments; CT, chromatid breaks; CH, chromosome breaks.
Values in parentheses indicate signal-free ends per total ends.
FIG. 1.
FISH analysis of metaphase chromosomes (DNA stained with 4′,6′-diamidino-2-phenylindole [DAPI] is shown in blue; Cy3-labeled TTAGGG repeats are shown in yellow) of primary MEFs prepared from wild-type (A), PARP−/− p53−/− (B), and PARP+/+ p53−/− (I) embryos. The corresponding DAPI staining of chromosomes in panels C, E, G, and I are shown in panels D, F, H, and J, respectively. Examples of chromosomal abnormalities in PARP−/− p53−/− and PARP+/+ p53−/− MEFs are telomere associations and end-to-end fusions (fu/t), Robertsonian-like configurations (rl), fragmentations (f), dicentrics (d), and ring-like chromosomes (r). Note chromosomes lacking detectable telomere fluorescence (arrows).
PARP p53 double-mutant cells exhibit altered telomeres.
Telomeres play an important role in stabilizing chromosomes. PARP has recently been implicated in telomere function (10), and p53 is also required to maintain chromosome stability. In order to elucidate the role of PARP and p53 in telomere integrity and to determine whether telomere dysfunction could have an impact on the high degree of chromosome instability, we analyzed metaphase spreads prepared from primary MEFs of various littermates of double-mutant mice using Q-FISH. Compared to wild-type controls, PARP−/− cells exhibited shorter telomeres (P < 0.05) whereas p53−/− MEFs did not show significant changes in the mean values of telomere fluorescence (Table 1). However, compared to wild-type cells, the fraction of p53−/− cells containing longer telomeres or loss of telomere signals seemed to be increased (Fig. 2). Surprisingly, telomeres in cells lacking both PARP and p53 were about 45 to 50% longer than in wild-type controls (Table 1), which may be due to an increased proportion of chromosomes having longer telomeres (Fig. 2). In addition, an increased number of chromosomes with low telomere fluorescence values or even loss of telomeric signals in PARP and p53 double-null cells was evident (Fig. 2; Table 1). However, we cannot rule out the possibility that aneuploidy or chromosomal aberrations in these cells could contribute to heterogeneity in telomere fluorescence values. We next examined telomerase activity in PARP and p53 double-mutant cells using the telomere repeat amplification protocol assay and found that levels of telomerase activity in cells from all genotypes tested were similar (data not shown), ruling out the possibility that the observed changes in telomere length are due to an altered telomerase activity.
FIG. 2.
Q-FISH analyses of telomere length in PARP and p53 double-mutant cells. Frequency distributions of telomere fluorescence values (pooled p- and q-arm values) in metaphase chromosomes from representative primary MEFs with the indicated genotype are shown. The x axis depicts the intensity of each signal, expressed in (TFUs), with each unit representing ∼1 kb of telomere repeats (42); the y axis shows the frequency of telomeres of a given length.
Interaction of PARP mutation with p53 homozygous deficiency.
PARP p53 double-mutant mice, obtained at expected frequencies, were fertile and appeared phenotypically normal. However, heterozygous and homozygous mutations of PARP yielded tumors in p53−/− mice as early as 6 weeks, whereas no tumor-related death was observed in PARP+/+ p53−/− mice until 9 weeks of age (Fig. 3A). By 22 weeks, all PARP−/− p53−/ − double-mutant mice had developed tumors, compared to 64% of PARP+/+ p53−/− mice. Heterozygous mutations in PARP resulted in 78% of p53−/− mice developing tumors (Fig. 3A). With the exception of the comparison of the PARP−/− p53−/− and PARP+/− p53−/− groups (P = 0.08), the survival curves for other groups were significantly different. These results demonstrate that PARP mutation accelerates tumorigenesis in p53 null mice. Pathological analysis revealed that the double-mutant mice developed a high rate of lymphomas and sarcomas, including angiosarcoma and some osteosarcoma (Table 2). In addition, a high frequency of carcinomas in the colon, pancreas, liver, skin, and mammary gland was found in p53−/− mice with PARP+/− and PARP−/− backgrounds (16%, 10 out of 61 mice) (Table 2). Immunostaining demonstrated the expression of cytokeratin in these tumors (data not shown), indicative of epithelial origin. Surprisingly, 6 out of 61 (10%) p53−/− mice with PARP+/− and PARP−/− backgrounds developed primitive neuroectodermal tumors (Table 2 and Fig. 3C).
FIG. 3.
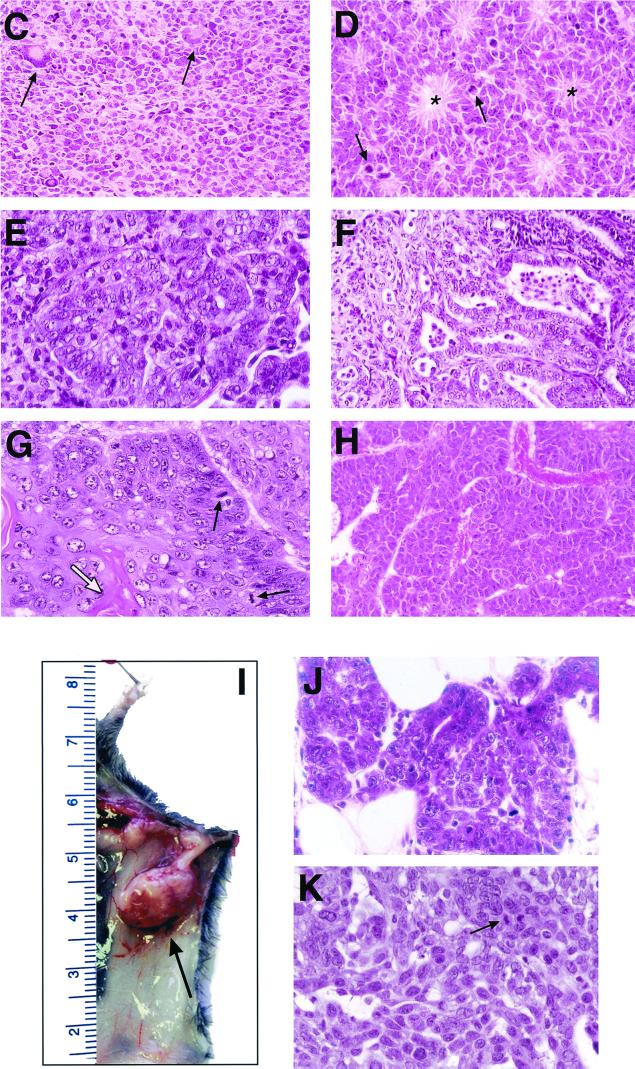
Tumor development in PARP p53 double-mutant mice. (A and B) Tumor incidence in PARP p53 double-mutant mice. PARP-deficient p53−/− animals were monitored over a period of 22 weeks (A), and PARP-deficient p53+/− mice were monitored over a period of 20 months (B). Moribund or tumor-bearing mice were autopsied. (C to K) Histopathological characterization of brain tumors and representative carcinomas from PARP p53 double-mutant mice. (C) Undifferentiated primitive neuroectodermal tumor from a 2.5-month-old PARP−/− p53−/− mouse containing megakaryocytes (arrows) and cells with pleomorphic nuclei and mitotic figures. (D) Ependymoma from a 14.5-month-old PARP−/− p53+/− mouse showing a typical rosette arrangement in the tumor (asterisks), with frequent appearance of mitotic cells (arrows). (E) Lung adenocarcinoma from a 15-month-old PARP−/− p53+/− mouse showing gland-like structure. (F) Prostate carcinoma from a 20-month-old PARP+/− p53+/− mouse showing irregular glandular formation and invasion of the stroma. (G) Keratinizing squamous carcinoma originating from an ear exhibiting numerous keratin pearls (white arrow) and frequent mitotic figures (black arrows). (H) Massive hepatocellular carcinoma showing a trabecular pattern of tumor cells. (I to K), Mammary carcinomas in PARP- and p53-deficient mice. (I) Tumors from the left third pair of mammary glands (arrow) of a 13-month-old PARP−/− p53+/− female mouse. A ductal carcinoma in situ (J) and a solid mammary carcinoma (K) show frequent mitotic figures (arrow).
TABLE 2.
Summary of tumor types in PARP p53 double-mutant mice
| Genotype of mice
|
No. of mice analyzed | No. of tumors analyzed | Tumor typea | No. of tumors | |
|---|---|---|---|---|---|
| PARP | p53 | ||||
| +/+ | −/− | 26 | 31 | Lymphoma | 22 |
| Sarcoma | 7 | ||||
| Teratoma | 2 | ||||
| +/− | −/− | 28 | 39 | Lymphoma | 16 |
| Sarcoma | 16 | ||||
| Primitive neuroectodermal tumor | 1 | ||||
| Colon adenocarcinoma | 3 | ||||
| Hepatocellular carcinoma | 1 | ||||
| Squamous cell carcinoma | 1 | ||||
| Mammary gland carcinoma | 1 | ||||
| −/− | −/− | 33 | 45 | Lymphoma | 21 |
| Sarcoma | 13 | ||||
| Teratoma | 2 | ||||
| Primitive neuroectodermal tumor | 5 | ||||
| Pancreatic carcinomas | 2 | ||||
| Colon adenocarcinoma | 1 | ||||
| Undifferentiated adenocarcinoma | 1 | ||||
| +/+ | +/− | 10 | 12 | Lymphoma | 4 |
| Sarcoma | 7 | ||||
| Ependymoma | 1 | ||||
| +/− | +/− | 35 | 43 | Mammary gland carcinoma | 6 |
| Prostate adenocarcinoma | 5 | ||||
| Squamous cell carcinoma | 2 | ||||
| Lung adenocarcinoma | 2 | ||||
| Hepatocellular carcinoma | 2 | ||||
| Ependymoma | 1 | ||||
| Lymphoma | 7 | ||||
| Sarcoma | 17 | ||||
| Teratoma | 1 | ||||
| −/− | +/− | 33 | 54 | Mammary gland carcinoma | 9 |
| Prostate adenocarcinoma | 3 | ||||
| Lung adenocarcinoma | 6 | ||||
| Squamous cell carcinoma | 4 | ||||
| Ependymoma | 3 | ||||
| Lymphoma | 6 | ||||
| Sarcoma | 23 | ||||
Carcinomas and brain tumors are shown in boldface.
PARP deficiency results in high frequencies of carcinomas in p53+/− mice.
To study the impact of PARP deficiency on tumor susceptibility of p53+/− mice, groups of p53+/− mice with PARP-proficient or -deficient backgrounds were monitored for tumor development. PARP deficiency significantly increased the tumor burden in p53+/− mice. Comparison of tumor-free survival curves indicates that there is a significant difference between PARP−/− p53+/− and PARP+/+ p53+/− mice (P < 0.0001) as well as between PARP−/− p53+/− and PARP+/− p53+/− mice (P < 0.0001) (Fig. 3B). At 20 months of age all PARP−/− p53+/− mice and ∼90% of PARP+/− p53+/− mice developed tumors, whereas only ∼50% of PARP+/+ p53+/− mice suffered from tumors, mainly lymphomas and sarcomas, including angiosarcomas and osteosarcomas. One ependymoma was also found in this group of mice. While lymphomas and soft tissue sarcomas were prominent, a high frequency of osteosarcomas (18%, 12 out of 68 mice analyzed) was found in PARP+/− p53+/− and PARP−/− p53+/− mice. Strikingly, 39 out of 68 (57%) PARP-deficient p53+/− mice developed carcinomas in the mammary gland, lung, liver, prostate, and skin (Table 2 and Fig. 3E to H). Specifically, 8 out of 68 mice (12%) developed lung cancer (Fig. 3E) and 8 out of 27 male mice (30%) developed prostate carcinomas (Fig. 3F). Moreover, 6 out of 24 PARP+/− p53+/− female mice and 9 out of 17 PARP−/− p53+/− female mice developed diverse types of mammary carcinomas (Fig. 3I to K). Whole-mount examination and serial sectioning of mammary glands revealed multiple adenocarcinoma foci in several glands within one mouse. The epithelial origin of these tumors was verified by immunohistochemical staining for cytokeratin (data not shown). Finally, four ependymomas were found in PARP-deficient p53+/− mice (Table 2 and Fig. 3D): they appeared to arise from the ventricle walls of the brain and lacked expression of glial fibrillary acidic protein and synaptophysin (data not shown).
Chromosomal abnormalities and telomere shortening in PARP and p53-deficient tumor cells.
To understand the genetic basis of accelerated tumorigenesis, we analyzed chromosomes and telomeres of primary thymic lymphomas derived from six PARP p53 double-mutant mice and normal thymocytes from the corresponding genotypes. While normal thymocytes contained about 40 chromosomes, all tumor cells were highly aneuploid and exhibited a larger degree of chromosomal aberrations, including end-to-end fusions and fragmentation, than normal thymocyte controls (Table 3). Q-FISH analysis revealed that, similar to MEFs, normal thymocytes derived from PARP-deficient p53−/− mice (B88 and A14) appeared to have overall longer telomeres than their wild-type counterparts; however, all the tumor cells derived from PARP p53 double-mutant mice showed a reduction of telomere length compared to corresponding normal thymocytes (Table 3), suggesting a selective advantage of cells with shorter telomeres during malignant transformation.
TABLE 3.
Summary of cytogenetic changes and telomere fluorescence in normal thymocytes and thymic lymphomas
| Sample | Genotype
|
Chromosome aberrationsa
|
Telomere fluorescence (TFU) (mean ± SE)
|
|||||
|---|---|---|---|---|---|---|---|---|
| PARP | p53 | % of fusionsb | % of fragmentsb | No. of chromosomes/ metaphase (mean ± SE) | p-arm | q-arm | All | |
| Normal thymocytes | ||||||||
| C97 | +/+ | +/+ | 0 (0) | 0 (0) | 40 ± 0.0 | 33 ± 0.4 | 44 ± 0.5 | 38 ± 0.3 |
| B84 | +/− | +/+ | 0 (0) | 0.1 (1) | 40 ± 0.1 | 37 ± 0.4 | 45 ± 0.5 | 41 ± 0.4 |
| B11 | +− | +/− | 0 (0) | 0.1 (1) | 40 ± 0.1 | 39 ± 1.3 | 51 ± 1.8 | 44 ± 1.1 |
| A21 | −/− | +/− | 0.2 (2) | 0.2 (3) | 41 ± 0.8 | 38 ± 1.3 | 45 ± 1.8 | 42 ± 1.1 |
| B88 | +/− | −/− | 0 (0) | 0.2 (1) | 40 ± 0.2 | 53 ± 1.2 | 64 ± 1.6 | 58 ± 1.3 |
| A14 | −/− | −/− | 0.2 (2) | 0.5 (5) | 40 ± 0.4 | 52 ± 1.4 | 65 ± 2.1 | 59 ± 1.3 |
| Thymic lymphomas | ||||||||
| AP35 | −/− | +/− | 1.6 (18) | 1.4 (15) | 59 ± 2.4 | 25 ± 0.6 | 26 ± 0.6 | 25 ± 0.4 |
| TL65 | +/− | −/− | 0.7 (8) | 0.3 (3) | 54 ± 1.4 | 17 ± 0.3 | 22 ± 0.6 | 19 ± 0.2 |
| TL76 | +/− | −/− | 1.2 (12) | 0.4 (4) | 52 ± 3.8 | 21 ± 0.4 | 23 ± 0.4 | 22 ± 0.3 |
| AP6 | +/− | −/− | 0.3 (3) | 0.1 (1) | 66 ± 5.3 | 25 ± 0.5 | 28 ± 0.6 | 26 ± 0.4 |
| AP77 | +/− | −/− | 0.1 (1) | 0.4 (4) | 50 ± 1.6 | 37 ± 0.7 | 43 ± 0.8 | 40 ± 0.6 |
| AP86 | −/− | −/− | 0.3 (3) | 0.0 (0) | 76 ± 2.8 | 45 ± 0.7 | 47 ± 0.8 | 46 ± 0.5 |
At least 10 metaphases from each sample were scored.
Values in parentheses indicate scored chromosome abnormalities.
PARP deficiency promotes loss of the wild-type p53 allele in p53+/− mice.
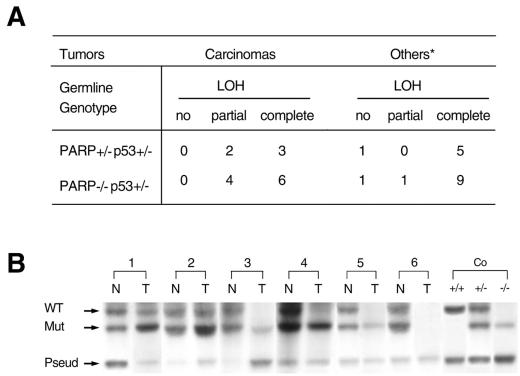
In most cases of LFS, somatic loss of the remaining wild-type p53 allele constitutes the primary initiating event leading to cancer. Heterozygous p53 knockout mice develop spontaneous tumors late in life, partly due to loss of the wild-type p53 allele (12). To investigate whether PARP deficiency-induced genomic instability and telomere dysfunction would promote the loss of function of tumor suppressor genes, we took advantage of p53 heterozygous mice and analyzed for LOH of the wild-type p53 allele in tumors derived from PARP-deficient p53+/− mice. We analyzed 32 tumors by Southern blotting and PCR and found that 30 of them showed loss, or partial loss, of the wild-type p53 allele (Fig. 4A and B). Some tumor samples showed the loss of both mutant and wild-type p53 alleles (Fig. 4B), suggesting chromosome losses or deletions. These data suggest that severe chromosomal instability in mice with a PARP-deficient background accelerates loss of function of tumor suppressor genes.
FIG. 4.
LOH in tumors from PARP-deficient p53+/− mice. (A) Summary of LOH frequency in tumors from PARP-deficient p53+/− mice. ∗, includes lymphoma, sarcomas, and brain tumors. (B) Representative Southern blot analysis showing partial (1T and 2T) and complete (3T, 4T, and 5T) LOH of wild-type p53 in tumors from PARP−/−, p53+/− mice. The loss of both wild-type and mutant p53 bands is also evident in some of the tumors (6T). The wild-type p53 (WT), the mutant allele (Mut), and the p53 pseudogene (Pseud) are indicated. N, normal tissues adjacent to the tumor or tail; T, tumor tissue.
DISCUSSION
We have shown that partial or complete inactivation of the DNA break-sensing molecule PARP renders p53-deficient mice susceptible to various tumor types, including a high frequency of carcinomas in addition to lymphomas and sarcomas. Although p53+/− mice develop breast carcinomas, a tumor type frequently observed in LFS patients (∼25%), at a low frequency (∼2%), many PARP-deficient p53+/− female mice (∼37%) develop this type of tumor. Another prominent tumor type in LFS is the brain tumor; however, it is very rare in p53+/− mice. Here we have shown that PARP deficiency potentiates the formation of brain tumors in p53 mutant mice. Finally, we observed more than one tumor per mouse (∼1.4 tumors/mouse, on average) in both PARP-deficient p53+/− and p53−/− mice (Table 2). Taken together, the tumor spectra, incidence, and multitumor onset observed in these mice demonstrate a close similarity of tumor development observed in PARP p53 double mutant mice and LFS patients (24).
Deficiency in PARP results in a dramatic shift of tumor types to carcinoma formation in p53−/− and p53+/− mutant mice (∼16 and ∼57%, respectively). This observation is striking in light of the fact that p53 knockout mice rarely develop carcinomas and that mice are generally not prone to carcinomas. Although the genetic background can influence the outcome of tumors in p53 mutant mice (13, 19), the shift to carcinomas in PARP p53 double-mutant mice seems to be specifically associated with PARP deficiency as PARP+/+ p53−/− mice and PARP+/+ p53+/− mice develop no carcinomas. However, how PARP deficiency renders the epithelial cells susceptible to transformation is currently unknown. Although other roles are plausible, the ability of PARP to maintain genomic stability and telomere function suggests that it is important in carcinoma development. In this regard, it is interesting to note that telomere shortening has recently been shown to promote carcinoma formation (2). Although this notion seems to be supported by the observation that all tumor cells contained shorter telomeres (Table 3), PARP deficiency results in long and heterogeneous telomeres in mice with a p53 mutant background, which may argue for a different mechanism. However, a common mechanism may be that secondary genome damage, due to either PARP deficiency or telomere loss, shifts the tumor spectrum from within the p53+/− and p53−/− backgrounds.
Why does PARP deficiency induce more tumors in p53+/− mice? A likely explanation is that the role of PARP in DNA break detection and binding and its antirecombinogenic function cooperates with other genome guardian molecules in the maintenance of genomic stability. Loss of these proteins causes a high degree of chromosome instability, leading to mutations in oncogenes and tumor suppressor genes. In support of this hypothesis is the report that the absence of PARP facilitates the loss of tumor suppressor genes, such as Rb, as well as the gain of a chromosome encompassing the c-jun oncogene (34). Consistent with this finding, we have also observed that PARP deficiency results in a high frequency of p53 LOH in tumor cells (Fig. 4) as well as in MEFs during immortalization (our unpublished observation). These results are particularly interesting given the fact that somatic loss of wild-type p53 has been proposed to be a prominent genetic alteration for the initiation of LFS disease. Finally, it is also interesting to note that inactivation of PARP in SCID mice, in which DNA-dependent protein kinase is mutated, induces a high frequency of T-cell lymphoma (27).
Telomeres stabilize the genome and play an important role in tumor development (14). Mice without telomerase and with short telomeres are prone to carcinogenesis (33), and this can be enhanced by deficient checkpoints mediated by p53 (9). PARP deficiency causes telomere shortening in mammalian cells (10), which could be due to either PARP's role at DNA end binding or the lack of PARP activity in modifying telomeric proteins. It is to be noted that another enzyme harboring poly(ADP-ribosyl)ation activity, tankyrase, was found to be involved in telomere regulation (35). Although we did not observe quantitative differences in telomere length in p53 mutant cells, we cannot rule out the possibility that the absence of p53 affects the quality of telomeric DNA repeats. Indeed, a high frequency of end-to-end fusions and telomere loss in p53−/− cells strongly suggests a role for p53 in the maintenance of telomere integrity.
Interestingly, PARP−/− p53−/− double-mutant cells exhibit heterogeneity of telomere length, including very long and very short telomeres or even uncapped chromosomes, suggesting a specific cooperation between p53 and PARP at the telomeres. One explanation for this observation is that the absence of PARP eventually causes telomere elongation via alternative pathways (6), such as the recombination-mediated elongation in yeast (31). However, p53 seems to inhibit this elongation process possibly by its interaction with Rad51 activity in recombination (7). This hypothesis is also supported by the following observations. The absence of PARP elevates sister chromatid exchange (26, 39), believed to be mediated by homologous recombination, in which Rad51 has also been shown to play a role in chicken cells (36). In addition, another DNA damage-sensing complex, Mre11-hRad50-Nbs1, is also involved in recombination (29, 30) and in regulation of telomere function (41). Also, PARP is proposed to be an antirecombinogenic factor (22) and PARP deficiency attenuates the stalled V(D)J recombination in SCID cells (27). Secondly, p53 was shown to harbor 3′ to 5′ exonuclease activity after binding to specific DNA substrates (28), and poly(ADP-ribosyl)ation was shown to negatively regulate the DNA-binding activity of p53 (23). Finally, in apparent consistency with previous observations, telomere elongation in telomerase-negative cells correlates with the loss of p53 expression (32) and p53 null osteosarcoma or carcinoma cells from LFS patients are alternative pathway competent (6). Taken together, these data are in keeping with the recent capping-uncapping model for telomere maintenance (5) and suggest that PARP may act on telomere length in two different pathways, one of which is p53 dependent. However, the mechanism by which p53 regulates telomeres with its interacting molecules requires further investigation.
Although cells with short telomeres and an intact p53 response are culled from the population via apoptosis or cell cycle arrest mediated by p53 and ATM (20), when combined with deficient checkpoints, e.g., those controlled by p53, these telomere alterations result in severe chromosomal abnormalities that facilitate malignant cell growth. Therefore, the enhanced tumor development in PARP and p53 double-mutant mice can be attributed to severe chromosomal instability, which may be caused, at least in part, by telomere dysfunction coupled with loss of function of tumor suppressor genes (11, 25, 34) (Fig. 4).
In summary, the present study has identified PARP as a cofactor, together with p53, for stabilizing the genome and thereby suppressing tumorigenesis in various tissues. Our findings establish mice deficient in both PARP and p53 as models for tumor development in LFS patients, and these mice provide a means to address fundamental aspects of the disease as well as to test therapeutic strategies.
ACKNOWLEDGMENTS
We thank D. Galendo for maintenance of the animal colonies and J. Michelon, M. Laval, and N. Lyandrat for technical assistance. We are also grateful to G. Mollon for the preparation of photographs. Further thanks are due to A. Aguzzi, L. Frappart, P. Kleihues, and H. Ohgaki for help in histopathological examination and to A. Baross, A. Grigoriadis, P. Hainaut, and E. F. Wagner for critical reading of the manuscript.
This research, W.-M.T., and Z.-Q.W. are supported by the Association for International Cancer Research, St. Andrews, Scotland, United Kingdom, and by NIH grant RO1CA79493-01. Research in the laboratory of P.M.L. is supported by NIH grants RO1AI29524 and GM56162 and by a grant from the National Cancer Institute of Canada with funds from The Terry Fox Run.
REFERENCES
- 1.Agarwal M L, Agarwal A, Taylor W R, Wang Z-Q, Wagner E F, Stark G R. Defective induction but normal activation and function of p53 in mouse cells lacking poly-ADP-ribose polymerase. Oncogene. 1997;15:1035–1041. doi: 10.1038/sj.onc.1201274. [DOI] [PubMed] [Google Scholar]
- 2.Artandi S E, Chang S, Lee S L, Alson S, Gottlieb G J, Chin L, DePinho R A. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 3.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 4.Bell D W, Varley J M, Szydlo T E, Kang D H, Wahrer D C, Shanon K E, Lubratovich M, Verselis S J, Issebacher K J, Fraumeni J F, Birch J M, Li F P, Garber J E, Haber D A. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn E H. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 6.Bryan T M, Englezou A, Dalla-Pozza L, Dunham M A, Reddel R R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 7.Buchhop S, Gibson M K, Wang X W, Wagner P, Sturzbecher H W, Harris C C. Interaction of p53 with the human Rad51 protein. Nucleic Acids Res. 1997;25:3868–3874. doi: 10.1093/nar/25.19.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canman C E, Lim D S, Cimprich K A, Taya Y, Tami K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 9.Chin L, Artandi S E, Shen Q, Tam A, Lee S L, Gottlieb G J, Greider C W, DePinho R A. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 10.d'Adda di Fagagna F, Hande M P, Tong W M, Lansdorp P M, Wang Z-Q, Jackson S P. Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat Genet. 1999;23:76–80. doi: 10.1038/12680. [DOI] [PubMed] [Google Scholar]
- 11.de Lange T, Jacks T. For better or worse? Telomerase inhibition and cancer. Cell. 1999;98:273–275. doi: 10.1016/s0092-8674(00)81955-8. [DOI] [PubMed] [Google Scholar]
- 12.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumors. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 13.Donehower L A, Harvey M, Vogel H, McArthur M J, Montgomery C A, Jr, Park S H, Thompson T, Ford R J, Bradley A. Effects of genetic background on tumorigenesis in p53-deficient mice. Mol Carcinog. 1995;14:16–22. doi: 10.1002/mc.2940140105. [DOI] [PubMed] [Google Scholar]
- 14.Greider C W. Telomerase activation. One step on the road to cancer? Trends Genet. 1999;15:109–112. doi: 10.1016/s0168-9525(98)01681-3. [DOI] [PubMed] [Google Scholar]
- 15.Harvey M, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A, Donehower L A. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 16.Hollstein M, Sidransky D, Vogelstein B, Harris C C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 17.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 18.Jeggo P A. DNA repair: PARP—another guardian angel? Curr Biol. 1998;8:R49–R51. doi: 10.1016/s0960-9822(98)70032-6. [DOI] [PubMed] [Google Scholar]
- 19.Jerry D J, Kittrell F S, Kuperwasser C, Laucirica R, Dickinson E S, Bonilla P J, Butel J S, Medina D. A mammary-specific model demonstrates the role of the p53 tumor suppressor gene in tumor development. Oncogene. 2000;19:1052–1058. doi: 10.1038/sj.onc.1203270. [DOI] [PubMed] [Google Scholar]
- 20.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 21.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl T, Satoh M S, Poirier G G, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 23.Malanga M, Pleschke J M, Kleczkowska H E, Althaus F R. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J Biol Chem. 1998;273:11839–11843. doi: 10.1074/jbc.273.19.11839. [DOI] [PubMed] [Google Scholar]
- 24.Malkin D. The Li-Fraumeni syndrome. In: Vogelstein B, Kinzler K W, editors. The genetic basis of human cancer. New York, N.Y: McGraw-Hill; 1998. pp. 393–407. [Google Scholar]
- 25.Martens U M, Zijlmans J M, Poon S S, Dragowska W, Yui J, Chavez E A, Ward R K, Lansdorp P M. Short telomeres on human chromosome 17p. Nat Genet. 1998;18:76–80. doi: 10.1038/ng0198-018. [DOI] [PubMed] [Google Scholar]
- 26.Menessier-de Murcia J, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver F J, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly (ADP-ribose) in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison C, Smith G C M, Stingl L, Jackson S P, Wagner E F, Wang Z-Q. Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nat Genet. 1997;17:479–482. doi: 10.1038/ng1297-479. [DOI] [PubMed] [Google Scholar]
- 28.Mummenbrauer T, Janus F, Muller B, Wiesmuller L, Deppert W, Grosse F. p53 protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85:1089–1099. doi: 10.1016/s0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- 29.Petrini J H J, Walsh M E, Dimare C, Chen X N, Korenberg J R, Weaver D T. Isolation and characterization of the human MRE11 homologue. Genomics. 1995;29:80–88. doi: 10.1006/geno.1995.1217. [DOI] [PubMed] [Google Scholar]
- 30.Petrini J H J. The mammalian Mre11-Rad50-Nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am J Hum Genet. 1999;64:1264–1269. doi: 10.1086/302391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pluta A F, Zakian V A. Recombination occurs during telomere formation in yeast. Nature. 1989;337:429–433. doi: 10.1038/337429a0. [DOI] [PubMed] [Google Scholar]
- 32.Rogan E M, Bryan T M, Hukku B, Maclean K, Chang A C M, Moy E L, Englezou A, Warneford S G, Dalla-Pozza L, Reddel R R. Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li-Fraumeni syndrome fibroblasts. Mol Cell Biol. 1995;15:4745–4753. doi: 10.1128/mcb.15.9.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudolph K L, Chang S, Lee H W, Blasco M, Gottlieb G J, Greider C W, DePinho R A. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 34.Simbulan-Rosenthal C M, Haddad B R, Rosenthal D S, Weaver Z, Coleman A, Luo R, Young H M, Wang Z-Q, Ried T, Smulson M E. Chromosomal aberrations in PARP−/− mice: genome stabilization in immortalized cells by reintroduction of PARP cDNA. Proc Natl Acad Sci USA. 1999;96:13191–13196. doi: 10.1073/pnas.96.23.13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 36.Sonoda E, Sasaki M S, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaziri H, West M D, Allsopp R C, Davison T S, Wu Y S, Arrowsmith C H, Poirier G G, Benchimol S. ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the post-translational activation of p53 protein involving poly(ADP-ribose) polymerase. EMBO J. 1997;16:6018–6033. doi: 10.1093/emboj/16.19.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z-Q, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner E F. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z-Q, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner E F. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wesierska-Gadek J, Wang Z-Q, Schmid G. Reduced stability of regularly spliced but not alternatively spliced p53 protein in PARP-deficient mouse fibroblasts. Cancer Res. 1999;59:28–34. [PubMed] [Google Scholar]
- 41.Zhu X D, Kuester B, Mann M, Petrini J H J, de Lange T. Cell-cycle-regulated association of Rad50/Mre11/Nbs1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 42.Zijlmans J M, Martens U M, Poon S S, Raap A K, Tanke H J, Ward R K, Lansdorp P M. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci USA. 1997;4:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]