Abstract
An increasing number of studies have demonstrated the antiviral nature of polyphenols, and many polyphenols have been proposed to inhibit SARS-CoV or SARS-CoV-2. Our previous study revealed the inhibitory mechanisms of polyphenols against DNA polymerase α and HIV reverse transcriptase to show that polyphenols can block DNA elongation by competing with the incoming NTPs. Here we applied computational approaches to examine if some polyphenols can also inhibit RNA polymerase (RdRp) in SARS-CoV-2, and we identified some better candidates than remdesivir, the FDA-approved drug against RdRp, in terms of estimated binding affinities. The proposed compounds will be further examined to develop new treatments for COVID-19.
Keywords: COVID-19, polyphenol, natural product, antiviral, molecular docking, MD simulation, remdesivir, drug discovery, virtual screening
1. Introduction
Polyphenols, commonly found in plants, fruits, and tea, are antioxidants with many health benefits [1,2]. Previous studies have shown polyphenols hold antiviral properties against various viruses [3,4,5,6], especially those targeting the respiratory tract [7,8,9,10]. Moreover, many polyphenols were found to inhibit SARS-CoV or its target proteins [11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26]. The anti-inflammatory properties of polyphenols are strong enough that they have been suggested as a supplement for obese and elderly COVID-19 patients [1]. Considerations like these have prompted researchers to investigate polyphenols’ ability to inhibit SARS-CoV-2 proteins. In an in silico analysis performed by Singh et al., TF3, TF2b, TF1, TF2a, and hesperidin all had better binding scores compared to remdesivir [27]. Though not better than remdesivir, gallic acid had a large binding energy when docked to RdRp in a study of plant polyphenols by Nourhan et al., showing strong inhibitory potential [28]. Despite the promising results, these studies sampled a very small variety of polyphenols, mainly tea extracts. Therefore, it is necessary to conduct large-scale virtual screening for a larger number of polyphenols.
Since it was first identified in late 2019, SARS-CoV-2 has devastated the world, infecting more than 200 million people and killing more than 4.7 million [29]. In December 2020, the FDA approved the Pfizer-BioNTech vaccine for emergency use, resulting in more than 100 million vaccinations in the U.S. alone [30]. Despite the success of the vaccines, COVID-19 cases continue to increase, making the discovery of safe antiviral drugs a significant concern.
SARS-CoV-2 is an enveloped, positive-sense RNA virus closely related to SARS-CoV, the cause of the SARS pandemic in 2002–2003 [31,32]. The virus’ genome consists of 14 ORFs, with ORF 1a and 1b being the most important for RNA replication [33,34]. After entry into a cell, ORF 1a and 1b are translated into two polyproteins that are further broken down into 16 non-structural proteins (nsps) [33]. These proteins assemble to form a replication complex that transcribes and replicates the virus’ RNA [35]. This RNA is ultimately packaged to make new SARS-CoV-2 viruses.
As the enzyme complex responsible for generating RNA, RNA-dependent RNA polymerase (RdRp) is central to viral replication [32]. The main protein in the complex is nsp12 bound by two accessory subunits, nsp 7 and nsp8 [36]. Given that human cells do not use RdRp and that its role in the viral life cycle is critical, RdRp is a popular drug target [37,38]. Inhibitors of RdRp fall into two categories: nucleoside inhibitors (NI) and non-nucleoside inhibitors (NNI). After being incorporated, nucleoside inhibitors block new nucleotides from entering the RNA chain, so synthesis stops [36]. In an in vitro study using a newly developed CoV-RdRp-Gluc reporter assay, remdesivir and molupiravir, both NIs, inhibited RdRp with EC50 values of 11.11 and 0.22 µM, respectively [32]. NNIs inhibit RdRp by changing its shape after binding to allosteric sites on the protein [36]. In a molecular docking study by Zijing et al., tegobuvir, an NNI drug in development for hepatitis C, bonded to the nsp12-7 and nsp12-8 sections of RdRp with docking scores ranging from −7.8 to −8.4 kcal/mol, showing an inhibitory effect [39]. Compared to NIs, which are vulnerable to exonuclease (nsp14) cleavage and competition with nucleotides present in the cell, NNIs have the potential to be highly potent drugs against SARS-CoV-2 [32,36].
Thus, to find potential therapeutic agents against COVID-19, this study screened 480 polyphenols to bind to SARS-CoV-2 RdRp. The structures of the 480 polyphenols were obtained from the Phenol-Explorer 3.6 database [40,41,42], and molecular docking was conducted using Maestro. Molecular mechanics with generalized Born and surface area solvation (MM-GBSA) scores were collected to quantify the affinity of the molecules for the proteins, and then absorption, distribution, metabolism, and excretion (ADME) and drug-likeness properties were analyzed for further screening. This study identifies three polyphenols with extremely low binding affinities to the SARS-CoV-2 RdRp as potential natural products for COVID-19 treatment.
2. Results
2.1. Docking Analysis of Polyphenols against SARS-CoV-2 RdRp
To develop effective inhibitors from polyphenols against SARS-CoV-2 RdRp, 480 polyphenols were docked onto SARS-CoV-2 RdRp (PDB ID: 7BV2). Based on docking poses, the binding energies were calculated using Prime MM-GBSA in Maestro. As a result, the top three protein–ligand complexes, namely RdRp–cyanidin 3-O-rutinoside (−107.68 kcal/mol), RdRp–petunidin 3,5-O-diglucoside (−99.18 kcal/mol), and RdRp–delphinidin 3-O-rutinoside (−90.70 kcal/mol), have better estimated binding energies (Table 1). These three polyphenols belong to the group of anthocyanins [43,44,45]. Meanwhile, we selected four compounds—remdesivir-TP (−55.00 kcal/mol), theaflavin 3,3′-digallate (TF3) (−77.89 kcal/mol), swertiapuniside (−39.42 kcal/mol), and ATP (−57.83 kcal/mol)—as a control group. Here, remdesivir-TP is the best drug candidate, which was identified in our previous study [46]. TF3 and swertiapuniside were proposed as the top-ranked inhibitors of RdRp in the studies from Singh et al. [27] and Koulgi et al. [47], respectively. From Table 1, we find that the top three polyphenols show better binding energies than remdesivir-TP, TF3, and swertiapuniside. This result indicates that the top three polyphenols possess the potential to inhibit RdRp. Furthermore, the binding energies of the top three polyphenols are better than that of ATP, which also suggests that the top three polyphenols might exert strong competitiveness at the ATP binding site.
Table 1.
The estimated binding energies of the top three polyphenols and the control compounds.
| Compound | Binding Energy (kcal/mol) |
|---|---|
| Cyanidin 3-O-rutinoside | −107.68 |
| Petunidin 3,5-O-diglucoside | −99.18 |
| Delphinidin 3-O-rutinoside | −90.70 |
| Remdesivir-TP 1 | −55.00 |
| TF3 2 | −77.89 |
| Swertiapuniside 3 | −39.42 |
| ATP 4 | −57.83 |
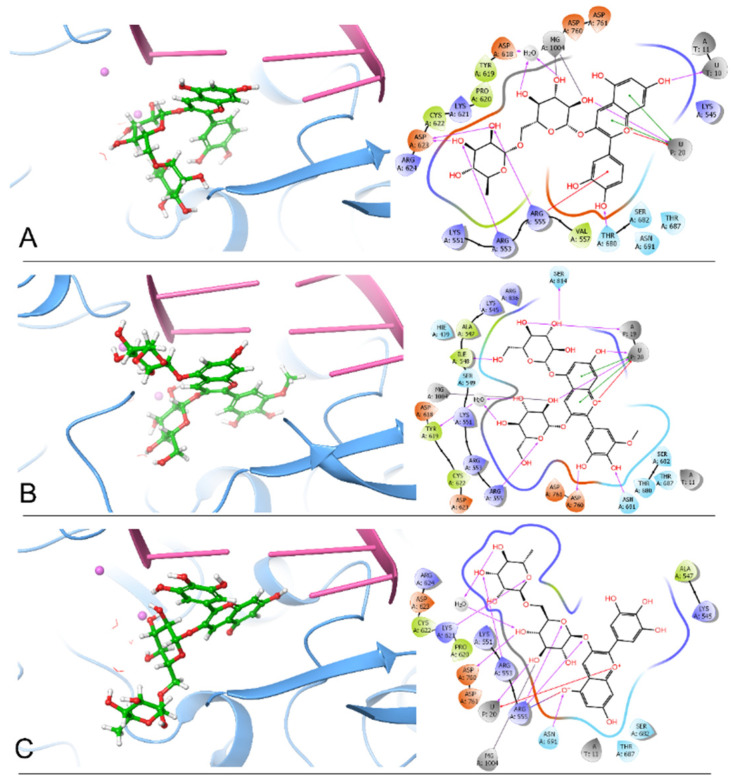
By comparing the 2D ligand–protein interactions of the top three polyphenols bound to RdRp (Figure 1), we find that they all interact with U20, the terminal nucleotide that interacts with incoming ATP. Cyanidin 3-O-rutinoside interacts with U20 by forming one hydrogen bond, two π-π stackings, and one π–cation interaction. By forming two hydrogen bonds, two π-π stackings, and one π–cation interaction, petunidin 3,5-O-diglucoside interacts with U20. Meanwhile, delphinidin 3-O-rutinoside interacts with U20 by forming one hydrogen bond and one π–cation interaction. These interactions between the top three polyphenols and U20 suggest that these polyphenols might strongly bind to RNA and compete with incoming ATP for the binding site so that viral RNA elongation is blocked.
Figure 1.
The docking poses and 2D ligand–protein interaction diagrams of 7BV2 and the top three ligands: (A), Cyanidin 3-O-rutinoside; (B), Petunidin 3,5-O-diglucoside; (C), Delphinidin 3-O-rutinoside. The pink spheres represent Mg2+ ions. The purple arrows indicate the hydrogen bonds; the green line represents π-π stacking; the red line represents π–cation interaction; the blue-red line represents the salt bridge; the gray line represents metal coordination.
Additionally, we summarized the number of hydrogen bonds between the top three polyphenols and the essential residues in Table 2. From Table 2, we find that these three polyphenols all interact with Arg555 by forming hydrogen bonds: one hydrogen bond for cyanidin 3-O-rutinoside, one hydrogen bond for petunidin 3,5-O-diglucoside, and two hydrogen bonds for delphinidin 3-O-rutinoside. Moreover, cyanidin 3-O-rutinoside interacts with Arg555 by forming a π–cation interaction, and delphinidin 3-O-rutinoside interacts with Agr555 through a salt bridge (Figure 1A,C). This result suggests that Arg555 is a critical residue in the binding pocket. Furthermore, cyanidin 3-O-rutinoside and delphinidin 3-O-rutinoside both interact with Arg553 by forming one hydrogen bond each. In forming one hydrogen bond each, petunidin 3,5-O-diglucoside and delphinidin 3-O-rutinoside both interact with Asn691 and Asp760. Notably, the ligand delphinidin 3-O-rutinoside has a phenoxide ion interacting with Arg555 and Asn691 (Figure 1C). Accordingly, we propose that cyanidin 3-O-rutinoside, petunidin 3,5-O-diglucoside, and delphinidin 3-O-rutinoside are the three best inhibitor candidates of RdRp among all 480 polyphenols based on these docking results.
Table 2.
The number of hydrogen bonds formed between the top three polyphenols and essential residues of SARS-CoV-2 RdRp.
| Cyanidin 3-O-rutinoside | Petunidin 3,5-O-diglucoside | Delphinidin 3-O-rutinoside | |
|---|---|---|---|
| Ile548 | 1 | ||
| Arg553 | 1 | 1 | |
| Arg555 | 1 | 1 | 2 |
| Lys621 | 2 | ||
| Asp623 | 2 | ||
| Thr680 | 1 | ||
| Asn691 | 1 | 1 | |
| Asp760 | 1 | 1 | |
| Ser814 | 1 |
2.2. Physicochemical Properties Prediction
To explore the physicochemical properties of the top three polyphenols, we predicted the ADME and drug-likeness properties using Qikprop in Maestro. The results are shown in Table 3. Meanwhile, the physicochemical properties of three control compounds are also predicted and listed in Table 3. First, the molecular weight of the top three polyphenols is within the recommended range of 130.0 to 725.0, whereas the control compound TF3 falls out of the recommended range. QPlogS is another important property to predict aqueous solubility, and its recommended range is from −6.5 to 0.5. All the compounds in Table 3 are in that range. In addition, the Lipinski’s rule of five (RO5) and the Jorgensen’s rule of three (RO3) are essential descriptors for evaluating drug-likeness. The top three polyphenols all pass the RO5 and RO3, which further suggests the drug-likeness of these polyphenols. In summary, Qikprop predictions indicate that cyanidin 3-O-rutinoside, petunidin 3,5-O-diglucoside, and delphinidin 3-O-rutinoside can be considered as viable drug candidates worthy of further research.
Table 3.
Selected Qikprop descriptors of the top three polyphenols and the control compounds.
| Compound | mol_MW 1 | QPlogS 2 | RO5 3 | RO3 4 |
|---|---|---|---|---|
| Cyanidin 3-O-rutinoside | 596.541 | −2.112 | 3 | 2 |
| Petunidin 3,5-O-diglucoside | 642.566 | −1.187 | 3 | 2 |
| Delphinidin 3-O-rutinoside | 612.540 | −2.663 | 3 | 2 |
| Remdesivir-TP | 531.205 | −1.742 | 3 | 1 |
| TF3 | 868.714 | −4.852 | 3 | 2 |
| Swertiapuniside | 531.205 | −1.742 | 3 | 1 |
1 mol_MW represents molecular weight of the molecule. The recommended range is 130.0–725.0. 2 QPlogS is the predicted aqueous solubility. The recommended range is −6.5~0.5. 3 RO5: number of violations of Lipinski’s rule of five [48]. The recommended range: maximum is 4. 4 RO3: Number of violations of Jorgensen’s rule of three [49]. The recommended range: maximum is 3.
2.3. Molecular Dynamics (MD) Simulation Analysis
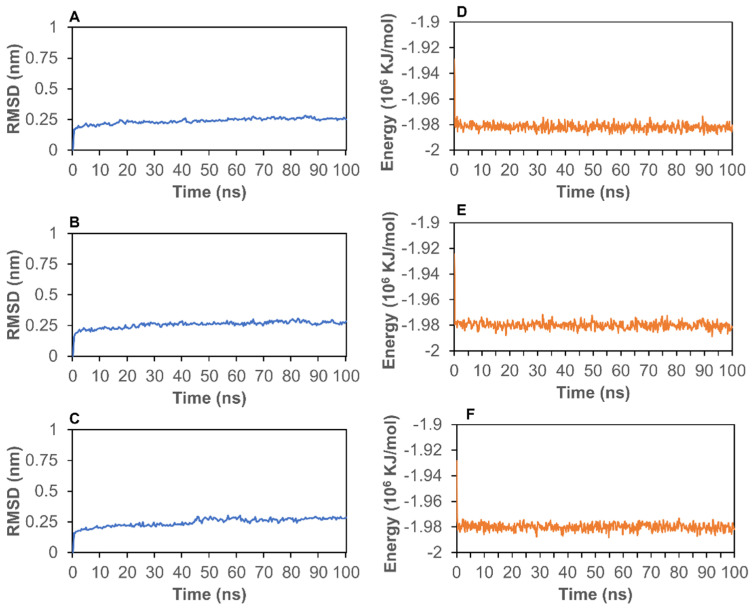
To further analyze the stability of the complexes, we conducted MD simulations to calculate RMSD and energy for the top three protein–ligand complexes. First, RMSD can be used to assess the stability of a protein–ligand complex. As shown in Figure 2A–C, the RMSD for the 7BV2–ligand complexes stabilized at around 0.25 nm after 5 ns, which suggests that the protein–ligand complexes are stable during the simulation process. Moreover, the total energies of these three complexes are shown in Figure 2D–F. The energies of 7BV2–ligand complexes stabilized at around −1.98 × 106 kJ/mol after 2 ns, further revealing the good stabilities of these three systems.
Figure 2.
The RMSD and energy of protein–ligand complexes. (A), the RMSD of 7BV2–cyanidin 3-O-rutinoside; (B), the RMSD of 7BV2–petunidin 3,5-O-diglucoside; (C), the RMSD of 7BV2–delphinidin 3-O-rutinoside; (D), the energy of 7BV2–cyanidin 3-O-rutinoside; (E), the energy of 7BV2–petunidin 3,5-O-diglucoside; (F), the energy of 7BV2–delphinidin 3-O-rutinoside.
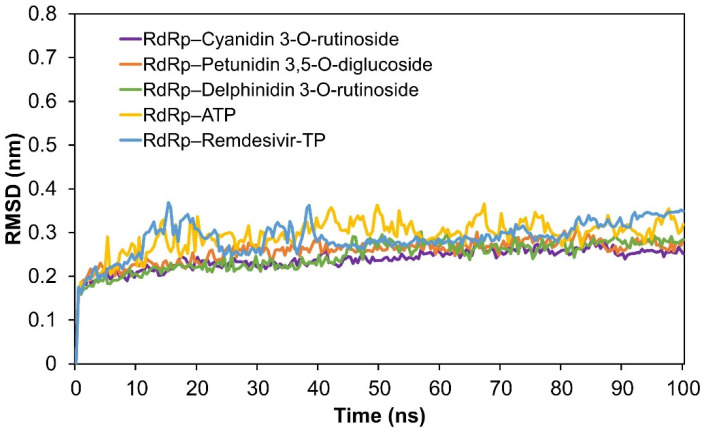
To compare the stability of RdRp with the three best polyphenols and the control group, we conducted MD simulations on the control complexes: RdRp–remdesivir-TP and RdRp–ATP. The RMSD values were then calculated and shown in Figure 3. As a result, the RMSD values of RdRp–remdesivir-TP and RdRp–ATP were higher than those of the complexes of RdRp with the three best ligands, which suggests that these three top-scored polyphenols, bound to RdRp, show better stability. Figure S1 shows that the RMSD values of 7BV2 before and after docking all stabilize at around 0.25 nm from 5 ns to 100 ns. Interestingly, the RMSD value of 7BV2 before ligand docking is slightly higher than the others, which indicates that the protein structure of RdRp becomes more stable after ligand–protein docking. Therefore, we conclude that the top three polyphenols bound to RdRp promote the stability of the protein structure. After MD simulations, we created three 2D interaction diagrams between RdRp and the three top-scored polyphenols (Figure S2). Compared to the docking results, the number of interactions (hydrogen bonds and π–π stacking) was reduced (Figure 1 vs. Figure S2A–C) because the fluctuation of the structures is the nature of MD simulations. However, we also found that the most significant interactions between the ligands and the protein remained stable (Figure S2). For example, the hydrogen bond, π–cation, and π–π interactions between the top-scored polyphenols and Arg555 and U20, the terminal nucleotide, remained stable during the MD simulations. As the control, the complex of RdRp and remdesivir-TP had fewer interactions after the MD simulation, while remdesivir-TP still interacted with U20 but not with Arg555 (Figure S3).
Figure 3.
The comparison of the RMSD of protein–ligand complexes. The purple, red, green, yellow, and blue lines represent the RMSD of complexes of RdRps binding to cyanidin 3-O-rutinoside, petunidin 3,5-O-diglucoside, delphinidin 3-O-rutinoside, ATP and remdesivir, respectively.
3. Discussion
Polyphenols possess a wide range of health benefits and biological activities, including antioxidant [50], antitumoral [51], anti-inflammatory [52], and antiviral properties [53], the latter of which suggests that polyphenols may be useful in the battle against multiple viruses, even SARS-CoV-2 [27,54,55,56,57,58,59,60,61,62]. In fact, numerous studies have shown that polyphenols exhibit antiviral effects by inhibiting DNA/RNA synthesis [63,64,65,66,67]. Our previous study also demonstrated that miquelianin, a flavonol glucuronide, can compete for the binding site of dATP on HIV-1 reverse transcriptase, inhibiting the viral DNA synthesis by interacting with the last nucleotide of the RNA chain and the binding residues of DNA polymerase [68]. Competing for the binding site of the incoming dNTP on the 3′ end of the elongating chain of DNA or RNA on the DNA/RNA polymerases is a significant mechanism through which polyphenols or other artificial compounds such as remdesivir exert their inhibitory effects. Therefore, to virtually screen potential inhibitors against DNA/RNA polymerases, the DNA or RNA molecule has to be considered and located precisely in the docking box. However, compounds were docked onto the empty active site of RdRp of SARS-CoV-2 without the RNA chain involved in many previous studies. This should be a reason why many drug candidates identified in in silico experiments are false positives.
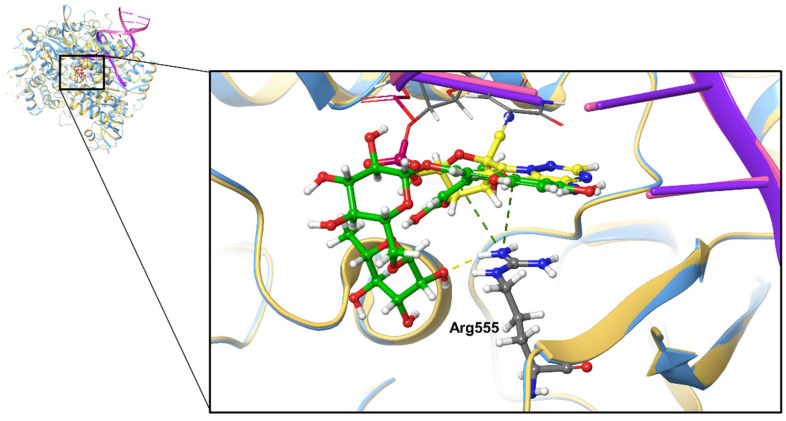
In this study, we found that the binding affinities of the top-ranked polyphenols all showed much better results than remdesivir in terms of binding affinity. To further discuss the docking pose of the best drug candidate, we overlapped the docking pose of 7BV2–cyanidin 3-O-rutinoside with the original structure of 7BV2 (Figure 4). We found that cyanidin 3-O-rutinoside occupies the position of the original ligand remdesivir-MP, but that it also interacts with Arg555 and the last nucleotide of the RNA chain in addition to the interactions between RdRp and the original ligand remdesivir-MP. Cyanidin 3-O-rutinoside interacts with Arg555 by forming one hydrogen bond and one π–cation interaction; however, remdesivir-MP interacts with Arg555 only through one π–cation interaction. Cyanidin 3-O-rutinoside interacts with U20, the last nucleotide, through a π–π interaction as well. These findings indicate that cyanidin 3-O-rutinoside, the polyphenol with the best scores, should compete for the binding site to block viral RNA synthesis.
Figure 4.
Superposition of cyanidin 3-O-rutinoside docked onto SARS-CoV-2 RdRp (7BV2) and cryo-EM structure of SARS-CoV-2 RdRp with remdesivir-MP (7BV2). The yellow-colored compound represents remdesivir-MP on 7BV2 (yellow-colored); the green-colored compound represents cyanidin 3-O-rutinoside docked on 7BV2 (blue-colored). The yellow dash line represents a hydrogen bond, and the green dash line represents a π–cation interaction.
4. Materials and Methods
4.1. Ligand Preparation
The structures of 480 tested polyphenols were retrieved from Phenol-Explorer 3.6 (http://phenol-explorer.eu/) (accessed on 1 December 2020). All the tested compounds were prepared using Ligprep in Maestro 12.4 (Schrödinger). The process for Ligprep includes adding hydrogens, computing correct partial charges, and generating possible conformations. The force field is OPLS3e by default [69].
4.2. Protein Preparation
The protein structures of RNA-dependent RNA polymerase (PDB ID: 7BV2) from RCSB’s Protein Data Bank (https://www.rcsb.org/) were prepared for use by Maestro in three steps: preprocessing, optimization, and minimization [70,71]. The OPLS3e force field was applied in both the optimization and the minimization steps [69].
4.3. Ligand–Protein Docking
To estimate the interactions between target proteins and polyphenols, we conducted ligand–protein docking by using the Ligand Docking panel in Maestro. Before running docking jobs, a receptor grid box was generated based on the existing ligand remdesivir in the protein structure. The size of the receptor grid box was set as default (20 Å). Ligand–protein docking was performed in extra-precision (XP) mode.
4.4. MM-GBSA Calculation
To predict the binding energies of polyphenols bound to RdRp, we performed Prime MM-GBSA (molecular mechanics generalized Born surface area) in Maestro. In the MM-GBSA panel, the pose viewer files of the docked complex were uploaded into the MM-GBSA panel. The force field was OPLS3e [69].
4.5. ADME and Drug-Likeness Properties Prediction
Qikprop module in Maestro was applied to predict the absorption, distribution, metabolism, and excretion (ADME) and drug-likeness properties for further screening [72]. For Qikprop, the top-ranked polyphenols were prepared by using Ligprep. Finally, descriptors such as RuleOfFive (RO5) and RuleOfThree (RO3) were applied to analyze the candidates.
4.6. Molecular Dynamics Simulation
To further investigate the dynamic interactions between RdRp and the top three polyphenols, we conducted molecular dynamics (MD) simulations by using GROMACS version 2018.1 and CHARMM36 force field [73]. The starting coordinates of the protein–ligand complex were obtained from a ligand–protein docking study. Then, we used CHARMM-GUI to build the MD simulation solution box, a cubic box with a length of 125 Å, which was then filled with water [74,75,76]. Next, the minimized structures were equilibrated using an NVT ensemble (constant Number of particles, Volume, and Temperature) and NPT ensemble (the Number of particles, Pressure, and Temperature). The target equilibration temperature was 300 K. Finally, MD simulations were performed for 100 ns. After the MD simulations, we calculated the root-mean-square deviation (RMSD) and the potential energies.
5. Conclusions
In summary, this study identified three polyphenols out of 480 as the best drug candidates for COVID-19 treatment. They all showed better estimated binding affinities than control compounds such as remdesivir. This can offer inspiration for new drug development.
Acknowledgments
We would like to acknowledge the Georgia Advanced Computing Resource Center (GACRC) and the College of Engineering’s IT department from the UGA for technical support. This work used the Extreme Science and Engineering Discovery Environment (XSEDE) Bridges GPU at the Pittsburgh Supercomputing Center through allocation TGDPP180005.
Supplementary Materials
The following are available online, Figure S1: The RMSD of 7BV2 before and after ligand-protein docking. Figure S2: 2D ligand-protein interaction diagrams of RdRp and the top three ligands after MD simulations: A, Cyanidin 3-O-rutinoside; B, Petunidin 3,5-O-diglucoside; C, Delphinidin 3-O-rutinoside. Figure S3: 2D ligand-protein interaction diagrams of RdRp-remdesivir-TP before (A) and after (B) MD simulation.
Author Contributions
Conceptualization, K.Y.C. and Z.-R.X.; virtual screening, Y.W., M.C.H., C.B., E.D., L.N.M. and E.B.S.; MD simulation, L.L.; data curation, Y.W., D.C. and S.D.P.; writing—original draft preparation, Y.W., M.C.H., E.D. and L.N.M.; writing—review and editing, D.C., S.D.P., K.Y.C. and Z.-R.X.; visualization and validation, Y.W. and D.C.; funding acquisition, K.Y.C. and Z.-R.X. All authors have read and agreed to the published version of the manuscript.
Funding
In this research, Y.W., L.L. and Z.-R.X. were supported by a faculty seed grant, 2231464F22, from the University of Georgia, and K.Y.C. was supported by the Ministry of Science and Technology, R.O.C. under MOST-108-2221-E-019-052.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Santos J.C., Ribeiro M.L., Gambero A. The Impact of Polyphenols-Based Diet on the Inflammatory Profile in COVID-19 Elderly and Obese Patients. Front. Physiol. 2021;11:612268. doi: 10.3389/fphys.2020.612268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pham A.H., Phillips M.A., Xie Z.-R. Resveratrol: State-of-the-Art Science and Health Applications. World Scientific; Singapore: 2018. Resveratrol and Advancing Virtual Drug Discovery; pp. 275–308. [DOI] [Google Scholar]
- 3.De Leo A., Arena G., Lacanna E., Oliviero G., Colavita F., Mattia E. Resveratrol inhibits Epstein Barr Virus lytic cycle in Burkitt’s lymphoma cells by affecting multiple molecular targets. Antivir. Res. 2012;96:196–202. doi: 10.1016/j.antiviral.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Yiu C.-Y., Chen S.-Y., Chang L.-K., Chiu Y.-F., Lin T.-P. Inhibitory Effects of Resveratrol on the Epstein-Barr Virus Lytic Cycle. Molecule. 2010;15:7115–7124. doi: 10.3390/molecules15107115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Annunziata G., Maisto M., Schisano C., Ciampaglia R., Narciso V., Tenore G.C., Novellino E. Resveratrol as a Novel Anti-Herpes Simplex Virus Nutraceutical Agent: An Overview. Viruses. 2018;10:473. doi: 10.3390/v10090473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faith S.A., Sweet T.J., Bailey E., Booth T., Docherty J.J. Resveratrol suppresses nuclear factor-κB in herpes simplex virus infected cells. Antivir. Res. 2006;72:242–251. doi: 10.1016/j.antiviral.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Lin C.J., Lin H.J., Chen T.H., Hsu Y.A., Liu C.S., Hwang G.Y., Wan L. Polygonum cuspidatum and its active components inhibit replication of the influenza virus through Toll-like receptor 9-induced interferon beta expression. PLoS ONE. 2015;10:e0117602. doi: 10.1371/journal.pone.0117602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zang N., Xie X., Deng Y., Wu S., Wang L., Peng C., Li S., Ni K., Luo Y., Liu E. Resveratrol-Mediated Gamma Interferon Reduction Prevents Airway In-flammation and Airway Hyperresponsiveness in Respiratory Syncytial Virus-Infected Immunocompromised Mice. J. Virol. 2011;85:13061–13068. doi: 10.1128/JVI.05869-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu T., Zang N., Zhou N., Li W., Xie X., Deng Y., Ren L., Long X., Li S., Zhou L., et al. Resveratrol Inhibits the TRIF-Dependent Pathway by Upregulating Sterile Alpha and Armadillo Motif Protein, Contributing to Anti-Inflammatory Effects after Respiratory Syncytial Virus Infection. J. Virol. 2014;88:4229–4236. doi: 10.1128/JVI.03637-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mastromarino P., Capobianco D., Cannata F., Nardis C., Mattia E., De Leo A., Restignoli R., Francioso A., Mosca L. Resveratrol inhibits rhinovirus replication and expression of inflammatory mediators in nasal epithelia. Antivir. Res. 2015;123:15–21. doi: 10.1016/j.antiviral.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Cho J.K., Curtis-Long M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J., Park K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013;21:3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H.-Y., Eo E.-Y., Park H., Kim Y.-C., Park S., Shin H.-J., Kim K. Medicinal herbal extracts of Sophorae radix, Acanthopanacis cortex, Sanguisorbae radix and Torilis fructus inhibit coronavirus replication in vitro. Antivir. Ther. 2010;15:697–709. doi: 10.3851/IMP1615. [DOI] [PubMed] [Google Scholar]
- 13.Chen C.N., Lin C.P.C., Huang K.K., Chen W.C., Hsieh H.P., Liang P.H., Hsu J.T.-A. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3,3′- digallate (TF3) Evid.-Based Complement Altern. Med. 2005;2:209–215. doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin L.-T., Hsu W.-C., Lin C.-C. Antiviral Natural Products and Herbal Medicines. J. Tradit. Complement. Med. 2014;4:24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.-Y., Kim D., Naguyen T.T.H., Park S.-J., Chang J.S., Park K.H., et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng P.-W., Ng L.-T., Chiang L.-C., Lin C.-C. Antiviral Effects Of Saikosaponins On Human Coronavirus 229E In Vitro. Clin. Exp. Pharmacol. Physiol. 2006;33:612–616. doi: 10.1111/j.1440-1681.2006.04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin C.-W., Tsai F.-J., Tsai C.-H., Lai C.-C., Wan L., Ho T.-Y., Hsieh C.-C., Chao P.-D.L. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 2005;68:36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau K.-M., Lee K.-M., Koon C.-M., Cheung C.S.-F., Lau C.-P., Ho H.-M., Lee M.Y.-H., Au S.W.-N., Cheng C.H.-K., Lau C., et al. Immunomodulatory and anti-SARS activities of Houttuynia cordata. J. Ethnopharmacol. 2008;118:79–85. doi: 10.1016/j.jep.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S.-Y., Chen C., Zhang H.-Q., Guo H.-Y., Wang H., Wang L., Zhang X., Hua S.-N., Yu J., Xiao P.-G., et al. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005;67:18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J.-Y., Kim J.H., Kwon J.M., Kwon H.-J., Jeong H.J., Kim Y.M., Kim D., Lee W.S., Ryu Y.B. Dieckol, a SARS-CoV 3CL(pro) inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg. Med. Chem. 2013;21:3730–3737. doi: 10.1016/j.bmc.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park H.R., Yoon H., Kim M.K., Lee S.D., Chong Y. Synthesis and antiviral evaluation of 7-O-arylmethylquercetin derivatives against SARS-associated coronavirus (SCV) and hepatitis C virus (HCV) Arch. Pharmacal. Res. 2012;35:77–85. doi: 10.1007/s12272-012-0108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C., Lee J.M., Lee N.-R., Kim D.-E., Jeong Y.-J., Chong Y. Investigation of the pharmacophore space of Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) NTPase/helicase by dihydroxychromone derivatives. Bioorganic Med. Chem. Lett. 2009;19:4538–4541. doi: 10.1016/j.bmcl.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C., Lee J.M., Lee N.-R., Jin B.-S., Jang K.J., Kim D.-E., Jeong Y.-J., Chong Y. Aryl diketoacids (ADK) selectively inhibit duplex DNA-unwinding activity of SARS coronavirus NTPase/helicase. Bioorganic Med. Chem. Lett. 2009;19:1636–1638. doi: 10.1016/j.bmcl.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J.Y., Yuk H.J., Ryu H.W., Lim S.H., Kim K.S., Park K.H., Ryu Y.B., Lee W.S. Evaluation of polyphenols from Broussonetia papyrifera as coro-navirus protease inhibitors. J. Enzym. Inhib. Med. Chem. 2017;32:504–512. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho T.-Y., Wu S.-L., Chen J.-C., Li C.-C., Hsiang C.-Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2007;74:92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G., Zhang H., Luo H., Zhu L., Jiang P., et al. Small Molecules Blocking the Entry of Severe Acute Respiratory Syndrome Coronavirus into Host Cells. J. Virol. 2004;78:11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh S., Sk M.F., Sonawane A., Kar P., Sadhukhan S. Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 via RNA-dependent RNA polymerase (RdRp) inhibition: An in-silico analysis. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1796810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Sohaimy S., Abdo N., Shehata M.G., Moheyeldin O. Inhibition of COVID-19 RNA-Dependent RNA Polymerase by Natural Bioactive Compounds: Molecular Docking Analysis. Egypt. J. Chem. 2021;64:1989–2001. doi: 10.21608/ejchem.2021.45739.2947. [DOI] [Google Scholar]
- 29.World Health Organisation WHO Coronavirus. 2020; April: 18–19. [(accessed on 16 September 2021)]. Available online: https://www.who.int/health-topics/coronavirus.
- 30.Centers for Disease Control and Prevention COVID-19 Vaccinations in the United States (as of 22 May 2021). Centers for Disease Control and Prevention. 2021:30. [(accessed on 16 September 2021)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/vaccines/index.html.
- 31.Fehr A.R., Perlman S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J., Guo S., Yi D., Li Q., Ma L., Zhang Y., Wang J., Li X., Guo F., Lin R., et al. A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase. Antivir. Res. 2021;190:105078. doi: 10.1016/j.antiviral.2021.105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chand G.B., Banerjee A., Azad G.K. Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure. PeerJ. 2020;8:e9492. doi: 10.7717/peerj.9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aftab S.O., Ghouri M.Z., Masood M.U., Haider Z., Khan Z., Ahmad A., Munawar N. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J. Transl. Med. 2020;18:275. doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yadav R., Chaudhary J., Jain N., Chaudhary P., Khanra S., Dhamija P., Sharma A., Kumar A., Handu S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells. 2021;10:821. doi: 10.3390/cells10040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian L., Qiang T., Liang C., Ren X., Jia M., Zhang J., Li J., Wan M., YuWen X., Li H., et al. RNA-dependent RNA polymerase (RdRp) inhibitors: The current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 2021;213:113201. doi: 10.1016/j.ejmech.2021.113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vicenti I., Zazzi M., Saladini F. SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19. Expert Opin Ther Pat. 2021;31:325–337. doi: 10.1080/13543776.2021.1880568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu W., Chen C.Z., Gorshkov K., Xu M., Lo D.C., Zheng W. RNA-Dependent RNA Polymerase as a Target for COVID-19 Drug Discovery. SLAS Discov. Adv. Life Sci. R&D. 2020;25:1141–1151. doi: 10.1177/2472555220942123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruan Z., Liu C., Guo Y., He Z., Huang X., Jia X., Yang T. SARS-CoV-2 and SARS-CoV: Virtual screening of potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12) J. Med. Virol. 2021;93:389–400. doi: 10.1002/jmv.26222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neveu V., Perez-Jiménez J., Vos F., Crespy V., du Chaffaut L., Mennen L., Knox C., Eisner R., Cruz J., Wishart D. Phenol-Explorer: An online comprehensive database on polyphenol contents in foods. Database. 2010;2010:bap024. doi: 10.1093/database/bap024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothwell J.A., Urpi-Sarda M., Boto-Ordoñez M., Knox C., Llorach R., Eisner R., Cruz J., Neveu V., Wishart D., Manach C. Phenol-Explorer 2.0: A major update of the Phenol-Explorer database integrating data on polyphenol metabolism and pharmacokinetics in humans and experimental animals. Database. 2012;2012:bas031. doi: 10.1093/database/bas031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothwell J.A., Perez-Jimenez J., Neveu V., Medina-Remon A., M’hiri N., García-Lobato P., Manach C., Knox C., Eisner R., Wishart D.S. Phenol-Explorer 3.0: A major update of the Phenol-Explorer database to incorporate data on the effects of food processing on polyphenol content. Database. 2013;2013:bat070. doi: 10.1093/database/bat070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Artemio Z., Tulio J., Reese R.N., Wyzgoski F.J., Rinaldi P.L., Fu R., Scheerens J.C., Miller A.R. Cyanidin 3-Rutinoside and Cyanidin 3-Xylosylrutinoside as Primary Phenolic Antioxidants in Black Raspberry. J Agric. Food Chem. 2008;56:1880–1888. doi: 10.1021/jf072313k. [DOI] [PubMed] [Google Scholar]
- 44.León-González A.J., Sharif T., Kayali A., Abbas M., Dandache I., Etienne-Selloum N., Kevers C., Pincemail J., Auger C., Chabert P., et al. Delphinidin-3-O-glucoside and delphinidin-3-O-rutinoside mediate the redox-sensitive caspase 3-related pro-apoptotic effect of blackcurrant juice on leukaemia Jurkat cells. J. Funct. Foods. 2015;17:847–856. doi: 10.1016/j.jff.2015.06.043. [DOI] [Google Scholar]
- 45.Anari Z., Mai C., Sengupta A., Howard L., Brownmiller C., Wickramasinghe S.R. Combined Osmotic and Membrane Distillation for Concentration of Anthocyanin from Muscadine Pomace. J. Food Sci. 2019;84:2199–2208. doi: 10.1111/1750-3841.14717. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y., Chang K.Y., Lou L., Edwards L.G., Doma B.K., Xie Z.-R. In silico identification of drug candidates against COVID-19. Inform. Med. Unlocked. 2020;21:100461. doi: 10.1016/j.imu.2020.100461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koulgi S., Jani V., Uppuladinne V.N.M., Sonavane U., Joshi R. Natural plant products as potential inhibitors of RNA dependent RNA polymerase of Severe Acute Respiratory Syndrome Coronavirus-2. PLoS ONE. 2021;16:e0251801. doi: 10.1371/journal.pone.0251801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 49.Jorgensen W.L., Duffy E.M. Prediction of drug solubility from structure. Adv. Drug Deliv. Rev. 2002;54:355–366. doi: 10.1016/S0169-409X(02)00008-X. [DOI] [PubMed] [Google Scholar]
- 50.Pandey K.B., Rizvi S.I. Plant Polyphenols as Dietary Antioxidants in Human Health and Disease. Oxid. Med. Cell. Longev. 2009;2:270–278. doi: 10.4161/oxim.2.5.9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fantini M., Benvenuto M., Masuelli L., Frajese G.V., Tresoldi I., Modesti A., Bei R. In Vitro and in Vivo Antitumoral Effects of Combinations of Polyphenols, or Polyphenols and Anticancer Drugs: Perspectives on Cancer Treatment. Int. J. Mol. Sci. 2015;16:9236–9282. doi: 10.3390/ijms16059236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yahfoufi N., Alsadi N., Jambi M., Matar C. The Immunomodulatory and Anti-Inflammatory Role of Polyphenols. Nutrients. 2018;10:1618. doi: 10.3390/nu10111618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Musarra-Pizzo M., Ginestra G., Smeriglio A., Pennisi R., Sciortino M.T., Mandalari G. The Antimicrobial and Antiviral Activity of Polyphenols from Almond (Prunus dulcis L.) Skin. Nutrients. 2019;11:2355. doi: 10.3390/nu11102355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaihatsu K., Yamabe M., Ebara Y. Antiviral Mechanism of Action of Epigallocatechin-3-O-gallate and Its Fatty Acid Esters. Molecules. 2018;23:2475. doi: 10.3390/molecules23102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Y., Pegan S.D., Crich D., Desrochers E., Starling E.B., Hansen M.C., Booth C., Mullininx L.N., Lou L., Chang K.Y., et al. Polyphenols as alternative treatments of COVID-19. Comput. Struct. Biotechnol. J. 2021;19:5371–5380. doi: 10.1016/j.csbj.2021.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mani J.S., Johnson J., Steel J.C., Broszczak D.A., Neilsen P.M., Walsh K.B., Naiker M. Natural product-derived phytochemicals as potential agents against coronaviruses: A review. Virus Res. 2020;284:197989. doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galanakis C.M., Aldawoud T.M., Rizou M., Rowan N.J., Ibrahim S.A. Food Ingredients and Active Compounds against the Coronavirus Disease (COVID-19) Pandemic: A Comprehensive Review. Foods. 2020;9:1701. doi: 10.3390/foods9111701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alkhatib A. Antiviral Functional Foods and Exercise Lifestyle Prevention of Coronavirus. Nutrients. 2020;12:2633. doi: 10.3390/nu12092633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russo M., Moccia S., Spagnuolo C., Tedesco I., Russo G.L. Roles of flavonoids against coronavirus infection. Chem. Interact. 2020;328:109211. doi: 10.1016/j.cbi.2020.109211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chojnacka K., Witek-Krowiak A., Skrzypczak D., Mikula K., Młynarz P. Phytochemicals containing biologically active polyphenols as an effective agent against Covid-19-inducing coronavirus. J. Funct. Foods. 2020;73:104146. doi: 10.1016/j.jff.2020.104146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keflie T.S., Biesalski H.K. Micronutrients and bioactive substances: Their potential roles in combating COVID-19. Nutrients. 2021;84:111103. doi: 10.1016/j.nut.2020.111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zahedipour F., Hosseini S.A., Sathyapalan T., Majeed M., Jamialahmadi T., Al-Rasadi K., Banach M., Sahebkar A. Potential effects of curcumin in the treatment of COVID -19 infection. Phytother. Res. 2020;34:2911–2920. doi: 10.1002/ptr.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ono K., Nakane H. Mechanisms of Inhibition of Various Cellular DNA and RNA Polymerases by Several Flavonoids. J. Biochem. 1990;108:609–613. doi: 10.1093/oxfordjournals.jbchem.a123251. [DOI] [PubMed] [Google Scholar]
- 64.Smith D., Dou Q. Green tea polyphenol epigallocatechin inhibits DNA replication and consequently induces leukemia cell apoptosis. Int. J. Mol. Med. 2001;7:645–652. doi: 10.3892/ijmm.7.6.645. [DOI] [PubMed] [Google Scholar]
- 65.Lee J.-H., Bae S.Y., Oh M., Seok J.H., Kim S., Chung Y.B., Gowda K.G., Mun J.Y., Chung M.S., Kim K.H. Antiviral effects of black raspberry (Rubus coreanus) seed extract and its polyphenolic compounds on norovirus surrogates. Biosci. Biotechnol. Biochem. 2016;80:1196–1204. doi: 10.1080/09168451.2016.1151337. [DOI] [PubMed] [Google Scholar]
- 66.Rabie A.M. Potent toxic effects of Taroxaz-104 on the replication of SARS-CoV-2 particles. Chem. Interact. 2021;343:109480. doi: 10.1016/j.cbi.2021.109480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tabari M.K., Iranpanah A., Bahramsoltani R., Rahimi R. Flavonoids as Promising Antiviral Agents against SARS-CoV-2 Infection: A Mechanistic Review. Molecules. 2021;26:3900. doi: 10.3390/molecules26133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Y., Hsieh T.-C., Wu J.M., Wang X., Christopher J.S., Pham A.H., Swaby J.D.-L., Lou L., Xie Z.-R. Elucidating the Inhibitory Effect of Resveratrol and Its Structural Analogs on Selected Nucleotide- Related Enzymes. Biomolecules. 2020;10:1223. doi: 10.3390/biom10091223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roos K., Wu C., Damm W., Reboul M., Stevenson J.M., Lu C., Dahlgren M.K., Mondal S., Chen W., Wang L., et al. OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput. 2019;15:1863–1874. doi: 10.1021/acs.jctc.8b01026. [DOI] [PubMed] [Google Scholar]
- 70.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schrödinger Release 2021-3: QikProp. Schrödinger, LLC.; New York, NY, USA: 2021. [Google Scholar]
- 73.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., De Groot B.L., Grubmüller H., MacKerell A.D. CHARMM36: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Biophys. J. 2017;112:175a–176a. doi: 10.1016/j.bpj.2016.11.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 75.Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J., Wei S., Buckner J., Jeong J.C., Qi Y., et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brooks B.R., Brooks C.L., Mackerell A.D., Nilsson L., Petrella R.J., Roux B. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.