Abstract
Apricot (Prunus armeniaca L.) is an important temperate fruit crop worldwide. The availability of wild apricot germplasm and its characterization through genomic studies can guide us towards its conservation, increasing productivity and nutritional composition. Therefore, in this study, we carried out the genomic characterization of 50 phenotypically variable accessions by using SSR markers in the erstwhile States of Jammu and Kashmir to reveal genetic variability among accessions and their genetic associations. The genetic parameter results revealed that the number of alleles per locus (Na) ranged from 1 to 6 with a mean Na value of 3.89 and the mean effective number of alleles (Ne) per locus 1.882 with a range of 1.22 to 2. Similarly, the polymorphic information content (PIC) values ranged from 0.464 to 0.104. The observed heterozygosity (Ho) (0.547) was found to have higher than expected heterozygosity (He) (0.453) with average heterozygosity of 0.4483. The dendrogram clustered genotypes into three main clades based on their pedigree. The population structure revealed IV sub-populations with all admixtures except the III sub-population, which was mainly formed of exotic cultivars. The average expected heterozygosity (He) and population differentiation within four sub-populations was 1.78 and 0.04, respectively, and explained 95.0% of the total genetic variance in the population. The results revealed that the SSR marker studies could easily decrypt the genetic variability present within the germplasm, which may form the base for the establishment of good gene banks by reducing redundancy of germplasm, selection of parents for any breeding program.
Keywords: diversity, apricot, polymorphism, genotype, structure, population
1. Introduction
Apricot (Prunus armeniaca L) is one of the influential fruits of the Rosaceae family that is mostly produced in temperate climates. Apricots have been divided into seven eco-geographical groups based on their origin [1]. Among all groups, the Central Asian cultivars are the most diverse and the oldest of all. These cultivars form two main gene pools such as Central Asia and Eastern Asia [2] and are characterized by high chilling requirements. Major regions for the cultivation of apricot accessions are Central Asia and China, from Kashmir to Tien-shan [3]. The Central Asian geographical region shows the richest variability [4]. Regions such as Turkey, Italy, Spain, the USA and France are widely known for producing apricot in colossal amounts [5]. In Asia, apricot is majorly produced in North-Western Himalayan regions, where it has been reported to grow wild in desert areas of Tibet that mostly remain cold, Southern regions of China and in some Northern parts of India, which include the temperate areas of Himachal Pradesh, Jammu and Kashmir and Uttarakhand that fall in the elevation range of 2000 to 2500 m above sea level. Jammu and Kashmir has rich genetic diversity of apricot, possessing both indigenous accessions cultivated through seeds and some exotic collections propagated by grafting traditional local cultivars. The exotic apricot cultivars such as ‘Harcot’, ‘Hartlay’, and ‘New-Castle’ have been introduced in different temperate areas including Jammu and Kashmir because of their highly productive nature, self-compatibility, resistance to diseases and long shelf life [1,6]. Cultivation of only a few of these commercial cultivars having commercial importance in place of diverse indigenous local cultivars may lead to genetic erosion because of a decrease in genetic diversity [7]. However, the presence of diverse plant genetic diversity as found in Jammu and Kashmir is important for increasing crop productivity and development of new cultivars. Therefore, evaluating these potential plant genetic resources is essential for future plant breeding and maintaining natural populations as viable evolutionary units in genetic resource management [8,9]. In addition, such studies help to determine the extent of genetic deviation among and within populations and reveal the processes that support these variations. Different scientists have used different morphological characters to assess the degree of diversity. However, the diversity assessment through morphological characterization is expensive, lengthy and is influenced by the environment. Therefore the DNA markers are used for plant diversity evaluation [10,11]. Various primers were significantly used in apricot to explore diversity such as amplified fragment length polymorphism (AFLP), restriction fragment length polymorphisms (RFLP) and randomly amplified polymorphic DNAs (RAPD) and ISSR [12,13,14,15,16,17,18]. Repeated DNA sequences or microsatellites are the markers that are intermittently used for diversity studies because of their even distribution throughout the genome, codominance and highly polymorphic nature [19,20]. Earlier investigations have acknowledged the considerable extent of molecular variation in Prunus genotypes [21,22,23,24] through microsatellites [25,26,27,28,29,30] utilized specific marker pairs devised primarily for other Prunus species and stone fruits [31,32,33,34,35,36]. The numbers of primers have been fabricated recently by utilizing sequence information of the apricot genome [37,38]. For diversity analysis, SSR markers have been adapted in Turkey, China, Morocco and other diverse eco-geography groups [2,3,11,39,40,41,42,43]. These studies have not only helped us to understand the molecular genetic variability and population structure of the local population but have helped researchers to advance biological research and the development of future breeding programs in Prunus armeniaca L. In this part of India, no such type of study has been carried out so far to evaluate the genetic diversity of the Prunus armeniaca L from the whole temperate areas of Jammu and Kashmir through SSR markers. Therefore, this study was undertaken to examine the genetic diversity and population structure between 50 apricot genotypes (local and exotic cultivars) and to evaluate the degree of variation among and within eco-geographical groups and subgroups of apricot germplasm taken into consideration.
2. Results
2.1. SSR Genotyping and Genetic Diversity Analysis
Genetic diversity was assessed among 50 apricot genotypes by using 46 SSR markers. The genetic parameters are shown in Table 1.
Table 1.
Genetic parameters of 46 SSR markers evaluated on 50 Prunus armeniaca L. accessions.
| Marker | No. of Alleles | PIC Value | Obs Hom |
Obs Het |
Exp Hom |
Exp Het | Ave Het | Ne | I | Fst |
|---|---|---|---|---|---|---|---|---|---|---|
| RPPG1-017 | 6 | 0.374 | 0.8333 | 0.1667 | 0.7193 | 0.2807 | 0.2778 | 1.3846 | 0.4506 | 0.05 |
| RPPG1-026 | 6 | 0.350 | 0.5833 | 0.4167 | 0.5088 | 0.4912 | 0.4861 | 1.9459 | 0.6792 | 0.02 |
| RPPG1-032 | 5 | 0.362 | 0.5417 | 0.4583 | 0.5026 | 0.4974 | 0.4922 | 1.9692 | 0.6853 | 0.04 |
| RPPG1-037 | 3 | 0.283 | 0.5208 | 0.4792 | 0.5055 | 0.4945 | 0.4894 | 1.9584 | 0.6825 | 0.07 |
| RPPG1-041 | 2 | 0.196 | 0.7083 | 0.2917 | 0.5509 | 0.4491 | 0.4444 | 1.8 | 0.6365 | 0.02 |
| RPPG2-011 | 1 | 0.104 | 0.4167 | 0.5833 | 0.5825 | 0.4175 | 0.4132 | 1.7041 | 0.6036 | 0 |
| RPPG2-022 | 1 | 0.104 | 0.8542 | 0.1458 | 0.8634 | 0.1366 | 0.1352 | 1.1563 | 0.2611 | 0.08 |
| RPPG3-026 | 2 | 0.186 | 0.2917 | 0.7083 | 0.4982 | 0.5018 | 0.4965 | 1.9862 | 0.6897 | 0.01 |
| RPPG4-059 | 4 | 0.350 | 0.6458 | 0.3542 | 0.5125 | 0.4875 | 0.4824 | 1.9321 | 0.6755 | 0.01 |
| RPPG4-067 | 2 | 0.203 | 0.9375 | 0.0625 | 0.5318 | 0.4682 | 0.4633 | 1.8633 | 0.656 | 0.04 |
| RPPG4-077 | 3 | 0.284 | 0.2917 | 0.7083 | 0.5377 | 0.4623 | 0.4575 | 1.8432 | 0.65 | 0 |
| RPPG4-084 | 2 | 0.194 | 0.7083 | 0.2917 | 0.5263 | 0.4737 | 0.4688 | 1.8824 | 0.6616 | 0.06 |
| RPPG4-091 | 6 | 0.462 | 0.2917 | 0.7083 | 0.4982 | 0.5018 | 0.4965 | 1.9862 | 0.6897 | 0 |
| RPPG5-018 | 4 | 0.326 | 0.4167 | 0.5833 | 0.5825 | 0.4175 | 0.4132 | 1.7041 | 0.6036 | 0 |
| RPPG5-022 | 3 | 0.284 | 0.4792 | 0.5208 | 0.495 | 0.505 | 0.4998 | 1.9991 | 0.6929 | 0.05 |
| RPPG5-023 | 2 | 0.188 | 0.5208 | 0.4792 | 0.5055 | 0.4945 | 0.4894 | 1.9584 | 0.6825 | 0 |
| RPPG5-025 | 1 | 0.105 | 0.7292 | 0.2708 | 0.5055 | 0.4945 | 0.4894 | 1.9584 | 0.6825 | 0.03 |
| RPPG5-030 | 4 | 0.354 | 0.000 | 1.000 | 0.4947 | 0.5053 | 0.500 | 2.000 | 0.6931 | 0.01 |
| RPPG6-009 | 4 | 0.351 | 0.5 | 0.5 | 0.5167 | 0.4833 | 0.4783 | 1.9168 | 0.6713 | 0.02 |
| RPPG6-032 | 4 | 0.354 | 0.5625 | 0.4375 | 0.495 | 0.505 | 0.4998 | 1.9991 | 0.6929 | 0 |
| RPPG6-033 | 2 | 0.190 | 0.7083 | 0.2917 | 0.4947 | 0.5053 | 0.500 | 2.000 | 0.6931 | 0.05 |
| RPPG7-015 | 4 | 0.353 | 0.2708 | 0.7292 | 0.5055 | 0.4945 | 0.4894 | 1.9584 | 0.6825 | 0.02 |
| RPPG7-026 | 4 | 0.354 | 0.5417 | 0.4583 | 0.643 | 0.357 | 0.3533 | 1.5463 | 0.5383 | 0.04 |
| RPPG7-032 | 3 | 0.279 | 0.875 | 0.125 | 0.4956 | 0.5044 | 0.4991 | 1.9965 | 0.6923 | 0 |
| RPPG8-007 | 1 | 0.107 | 0.875 | 0.125 | 0.5026 | 0.4974 | 0.4922 | 1.9692 | 0.6853 | 0.03 |
| RPPG8-028 | 3 | 0.260 | 0.1875 | 0.8125 | 0.5125 | 0.4875 | 0.4824 | 1.9321 | 0.6755 | 0.01 |
| Aprigms18 | 5 | 0.412 | 0.3542 | 0.6458 | 0.495 | 0.505 | 0.4998 | 1.9991 | 0.6929 | 0 |
| UDP98-405 | 6 | 0.447 | 0.5833 | 0.4167 | 0.5825 | 0.4175 | 0.4132 | 1.7041 | 0.6036 | 0 |
| UDP98-406 | 6 | 0.462 | 0.375 | 0.625 | 0.5026 | 0.4974 | 0.4922 | 1.9692 | 0.6853 | 0.02 |
| UDP98-409 | 4 | 0.337 | 0.125 | 0.875 | 0.5026 | 0.4974 | 0.4922 | 1.9692 | 0.6853 | 0.05 |
| UDP98-411 | 6 | 0.464 | 0.0833 | 0.9167 | 0.4982 | 0.5018 | 0.4965 | 1.9862 | 0.6897 | 0.05 |
| Pchgms4 | 6 | 0.463 | 0.8333 | 0.1667 | 0.7789 | 0.2211 | 0.2188 | 1.2800 | 0.3768 | 0.01 |
| Pchgms5 | 5 | 0.414 | 1.000 | 0.000 | 0.8114 | 0.1886 | 0.1866 | 1.2295 | 0.3341 | 0.06 |
| Bppct007 | 6 | 0.464 | 0.5833 | 0.4167 | 0.5509 | 0.4491 | 0.4444 | 1.800 | 0.6365 | 0.01 |
| Bppct025 | 2 | 0.193 | 0.375 | 0.625 | 0.5658 | 0.4342 | 0.4297 | 1.7534 | 0.6211 | 0 |
| Bppct030 | 6 | 0.462 | 0.3542 | 0.6458 | 0.5441 | 0.4559 | 0.4512 | 1.8221 | 0.6435 | 0.05 |
| PacA10 | 6 | 0.462 | 0.000 | 1.000 | 0.4947 | 0.5053 | 0.5000 | 2.000 | 0.6931 | 0 |
| PacA18 | 6 | 0.464 | 0.2083 | 0.7917 | 0.4956 | 0.5044 | 0.4991 | 1.9965 | 0.6923 | 0.04 |
| PacA33 | 6 | 0.463 | 0.2292 | 0.7708 | 0.5213 | 0.4787 | 0.4737 | 1.9002 | 0.6667 | 0.01 |
| PacA22 | 6 | 0.460 | 0.125 | 0.875 | 0.4947 | 0.5053 | 0.500 | 2.000 | 0.6931 | 0 |
| PacA26 | 6 | 0.462 | 0.375 | 0.625 | 0.4947 | 0.5053 | 0.500 | 2.000 | 0.6931 | 0.02 |
| PacA35 | 4 | 0.35 | 0.500 | 0.500 | 0.6211 | 0.3789 | 0.375 | 1.600 | 0.5623 | 0.01 |
| PacC3 | 2 | 0.20 | 0.4167 | 0.5833 | 0.5088 | 0.4912 | 0.4861 | 1.9459 | 0.6792 | 0.02 |
| PacC25 | 3 | 0.27 | 0.4792 | 0.5208 | 0.5739 | 0.4261 | 0.4217 | 1.7291 | 0.6126 | 0.02 |
| PacA58 | 2 | 0.190 | 0.3125 | 0.6875 | 0.495 | 0.505 | 0.4998 | 1.9991 | 0.6929 | 0 |
| PdavW3 | 4 | 0.352 | 0.3542 | 0.6458 | 0.5441 | 0.4559 | 0.4512 | 1.8221 | 0.6435 | 0.02 |
| Mean | 3.89 | 0.320 | 0.4774 | 0.5226 | 0.5470 | 0.4530 | 0.4483 | 1.8221 | 0.6371 | 0.0228 |
Legend: (Exp Ho) Expected homozygosity, (Exp He) heterozygosity, (Ob He) observed heterozygosity, (Ob Ho) homozygosity, (Ave Het) Average Heterozygosity, Ne = Effective number of alleles, I = Shannon’s information index, Fst = Genetic differentiation.
Using 46 SSR markers on 50 apricot genotypes, a total of 179 alleles were detected, and the number of alleles (Na) ranged from 1 to 6, with an average value of 3.89. Among the 46 markers, the highest number of alleles, 6 per locus, was realized with 14 markers, and the highest number of effective alleles (Ne) was observed, 2 per locus with five markers such as RPPG5-030, RPPG6-033, PacA10, PacA22 and PacA26. The PIC value varied in range from 0.104 to 0.464, with an average value of 0.320. Furthermore, the number of effective alleles (Ne) ranged from 1.1563 to 2 with an average value of 1.8821. The average observed homozygosity (Ho) was 0.4774, and varied from 0 to 1, whereas the average expected homozygosity was 0.5470 which ranged from 0.4947 to 0.8634. Similarly, the observed heterozygosity ranged from 0 to 1 with an average value of 0.5226, and expected heterozygosity (He) ranged from 0.1366 to 0.5053 and produced an average value of 0.4530. The overall average heterozygosity was 0.4483 and ranged from 0.1352 to 0.5. The Shannon’s diversity index (I) ranged from 0.2611 to 0.6931 with an average value of 0.6371, and the genetic differentiation (Fst) ranged from 0 to 0.08 with an average value of 0.0228. Different parameters showed a lot of variabilities indicating high genetic diversity.
2.2. Cluster and PCoA Analysis
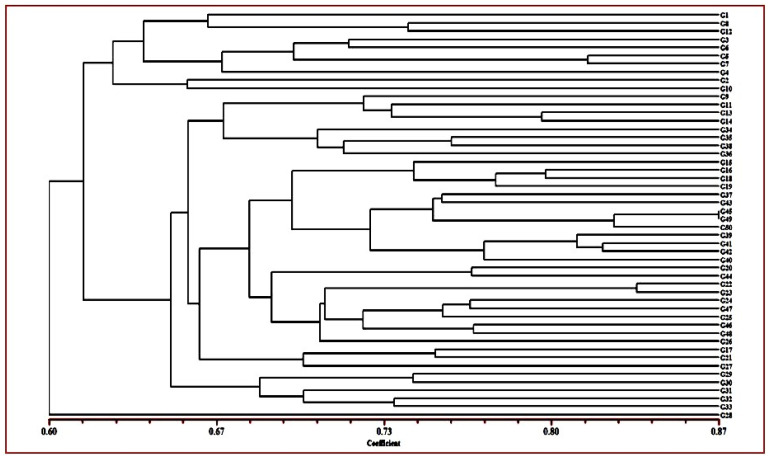
The Jaccard’s similarity coefficients between the germplasms were calculated for UPGMA clustering. The diverse group of 50 germplasms were divided into three primary groups based on their genetic similarity at a distance of 0.614 as cluster I, cluster II and cluster III (Figure 1).
Figure 1.
UPGMA dendrogram showing clustering of 50 Apricot genotypes based on Jaccard’s Similarity Coefficient.
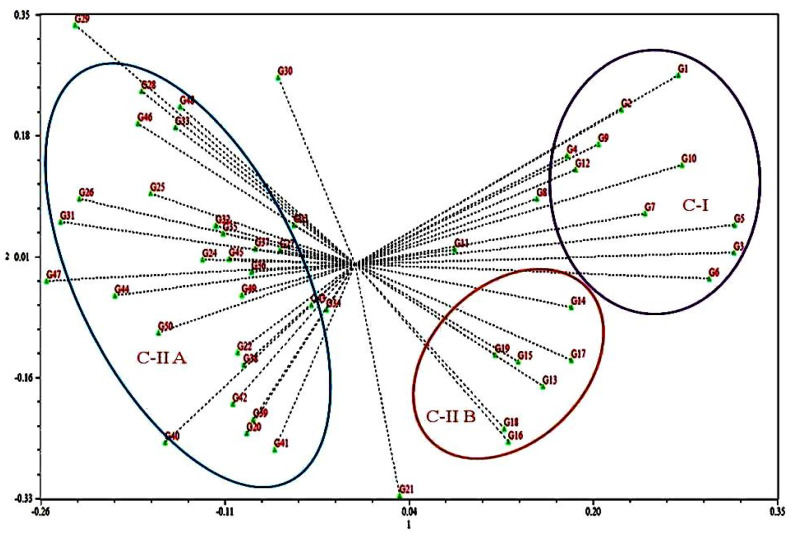
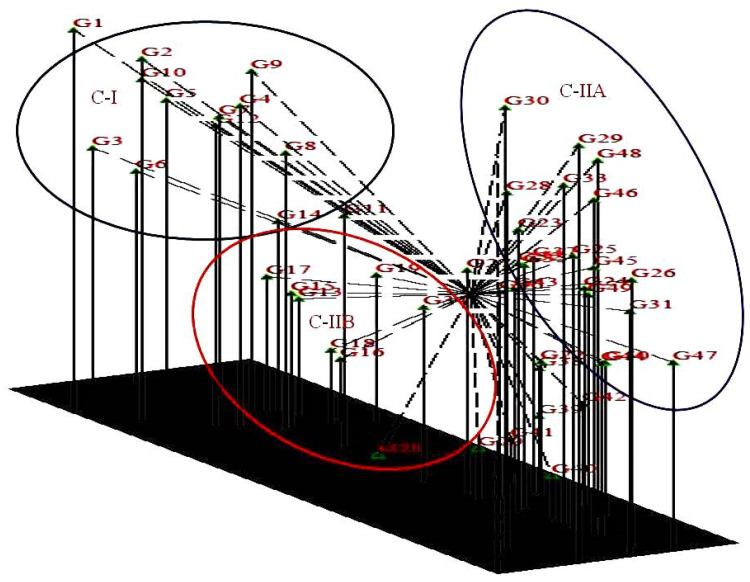
Cluster I consisted of 10 exotic genotypes divided further into two sub-clusters IA which included 8 genotypes (G1, G3, G4, G5, G6, G7, G8 and G12) and IB, which contained two genotypes (G2 and G10). Cluster II was the largest and contained 39 genotypes that are mainly indigenous to Jammu and Kashmir and form two sub-clusters: cluster IIA and cluster IIB. Cluster IIA is further divided into two sub-clusters and contained 8 genotypes in the first cluster (G9, G11, G13, G14, G34, G35, G36 and G38) and 26 genotypes (G15, G16, G18, G19, G37, G43, G45, G49, G50, G39, G41, G42, G40, G20, G44, G22, G23, G24, G47, G25, G46, G48, G26, G17, G21 and G27). Cluster IIB contained 5 genotypes (G29, G30, G31, G32 and G33) and cluster III contained single genotype G28. The Jaccard’s similarity among the genotypes ranged from 0.508 to 0.867. The highest similarity 0.867 was observed between indigenous accessions G45 and G49, which are accessions from Banihal and Doda regions of Jammu province of J&K and the lowest similarity 0.508 was observed between exotic cultivar Hartley and indigenous accession G31 from Malapora area of Baramullah. A 3D clustering plot revealed that 50 accessions produce three clusters C-I and C-IIA and C-IIB. In addition, the results from 2D PCoA clustering (Figure 2) were consistent with the results of 3DPCoA (Figure 3).
Figure 2.
A biplot of the first two principal components of 50 apricot genotypes using 46 microsat−ellite markers.
Figure 3.
Three-dimensional principal coordinates analysis (PCoA) of 50 apricot genotypes using 46 microsatellite markers.
The clustering pattern of apricot accessions from 2D and 3D PCoA plots and the UPGMA clustering graph were highly consistent. The UPGMA clustering tree graph provides abundant information and categorizes the accessions into different groups. The information produced by the 2D PCoA plot, although not sufficient, produced a flat and direct view of the relationship between different accessions as compared to 3D PCoA which provides sufficient information in different layers and directions. The combined result analysis of population structure through genetic similarity and PCoA provides valuable information to understand the genetic structure of the accessions.
2.3. Population Structure
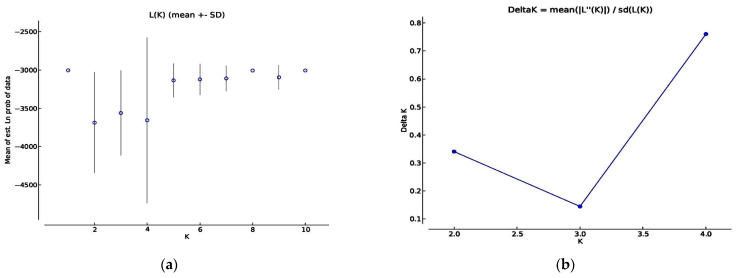
An investigation carried out for population structure utilizing marker data assisted in recognizing four (K = 4) genetically different sub-populations in 50 diverse apricot genotypes. Initially, we were unable to estimate the number of subpopulations as the LnP (K) values decreased from 1 to K = 2 and then increased at K = 3 and then again decreased at K = 4 before subsequently increasing at K = 5 to K = 8 and started again declined at K = 9, and finally at K = 10 increased (Figure 4a). Thus, no comprehensive outcome emerged regarding the probable number of subpopulations using LnP (K) values. Accordingly, to deduce the accurate number of all subpopulations in our population of 50 apricot accessions, the 1K approach developed by Evanno et al. [44] was utilized. The 1K approach calculates the rate of change of the mean probability values (LnP) of all subpopulations. According to this approach, the proportion of change was higher (1830.5) at K = 4 (Figure 4b).
Figure 4.
The figures show different methods of calculation of sub-populations (a) Non−parametric test showing a probable number of subpopulations using LnP (K) values. (b) Delta K showing peak value at K = 4 calculated by Evano et al. (2005) method.
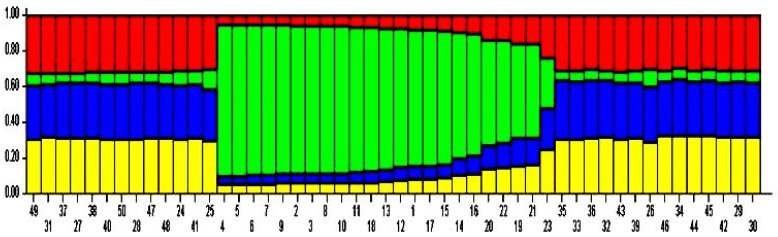
Hence, in our population of 50 apricot accessions, we found 4 subpopulations. Subpopulations 1, 2, 4 contained 36 genotypes that were all indigenous, while as in the 3rd subpopulation, only one indigenous genotype was spotted, the remaining 13 of the genotypes found were exotic. This arrangement pattern was also revealed in the structure graph (Figure 5), depicting the distribution of local (indigenous genotypes) vs. exotic genotypes separately. Further, in the 3rdsubpopulation, all the genotypes had affiliation likelihood more significant than 80%, and hence in this subpopulation, no apricot genotype was displayed as admixture (Table 2). The genotypes in sub-population 1, 2 and 4 have affiliation probability <80%, hence all individuals in these sub-populations were admixtures. The expected heterozygosity was calculated to estimate individuals’ mean distance among and within clusters/subpopulations. The expected heterozygosity, which calculates the likelihood that two randomly selected individuals would be heterozygous at a particular locus, ranged from 1.81 in the third sub-population to 1.77 in the other three sub-populations, with a mean of 1.78. Similarly, population differentiation measurements (Fst) ranged from 0.143 (in the second sub-population) to 0.05 (in the first sub-population), with an average of 0.04 (Table 3).
Figure 5.
Each column in the figure shows an individual and the X coordinate represents the name of the sample. The length of color represents the proportion of ancestors in the individual genome.
Table 2.
Distribution of individuals to sub-populations (K) on the basis of genetic ancestry.
| Code | Genotype | K1 | K2 | K3 | K4 | Sub-Population |
|---|---|---|---|---|---|---|
| 1 | G1 | 0.049 | 0.045 | 0.861 | 0.045 | 3 |
| 2 | G2 | 0.029 | 0.025 | 0.923 | 0.024 | 3 |
| 3 | G3 | 0.028 | 0.025 | 0.92 | 0.027 | 3 |
| 4 | G4 | 0.022 | 0.023 | 0.935 | 0.021 | 3 |
| 5 | G5 | 0.021 | 0.02 | 0.937 | 0.022 | 3 |
| 6 | G6 | 0.024 | 0.025 | 0.928 | 0.024 | 3 |
| 7 | G7 | 0.024 | 0.024 | 0.928 | 0.024 | 3 |
| 8 | G8 | 0.032 | 0.028 | 0.915 | 0.026 | 3 |
| 9 | G9 | 0.035 | 0.027 | 0.911 | 0.027 | 3 |
| 10 | G10 | 0.029 | 0.026 | 0.918 | 0.027 | 3 |
| 11 | G11 | 0.033 | 0.03 | 0.903 | 0.034 | 3 |
| 12 | G12 | 0.084 | 0.057 | 0.807 | 0.051 | 3 |
| 13 | G13 | 0.075 | 0.047 | 0.833 | 0.046 | 3 |
| 14 | G14 | 0.101 | 0.077 | 0.751 | 0.071 | Admixture of 1,2,3,4 |
| 15 | G15 | 0.101 | 0.069 | 0.76 | 0.07 | Admixture of 1,2,3,4 |
| 16 | G16 | 0.116 | 0.081 | 0.719 | 0.084 | Admixture of 1,2,3,4 |
| 17 | G17 | 0.094 | 0.062 | 0.783 | 0.061 | Admixture of 1,2,3,4 |
| 18 | G18 | 0.036 | 0.034 | 0.897 | 0.033 | 3 |
| 19 | G19 | 0.178 | 0.146 | 0.53 | 0.146 | Admixture of 1,2,3,4 |
| 20 | G20 | 0.16 | 0.123 | 0.595 | 0.122 | Admixture of 1,2,3,4 |
| 21 | G21 | 0.168 | 0.156 | 0.516 | 0.16 | Admixture of 1,2,3,4 |
| 22 | G22 | 0.15 | 0.136 | 0.579 | 0.134 | Admixture of 1,2,3,4 |
| 23 | G23 | 0.248 | 0.239 | 0.287 | 0.227 | Admixture of 1,2,3,4 |
| 24 | G24 | 0.3 | 0.328 | 0.047 | 0.325 | Admixture of 1,2,3,4 |
| 25 | G25 | 0.296 | 0.317 | 0.078 | 0.309 | Admixture of 1,2,3,4 |
| 26 | G26 | 0.312 | 0.326 | 0.057 | 0.306 | Admixture of 1,2,3,4 |
| 27 | G27 | 0.323 | 0.318 | 0.024 | 0.335 | Admixture of 1,2,3,4 |
| 28 | G28 | 0.329 | 0.311 | 0.028 | 0.333 | Admixture of 1,2,3,4 |
| 29 | G29 | 0.329 | 0.313 | 0.027 | 0.331 | Admixture of 1,2,3,4 |
| 30 | G30 | 0.298 | 0.339 | 0.03 | 0.333 | Admixture of 1,2,3,4 |
| 31 | G31 | 0.318 | 0.321 | 0.024 | 0.337 | Admixture of 1,2,3,4 |
| 32 | G32 | 0.324 | 0.319 | 0.02 | 0.337 | Admixture of 1,2,3,4 |
| 33 | G33 | 0.315 | 0.332 | 0.03 | 0.322 | Admixture of 1,2,3,4 |
| 34 | G34 | 0.307 | 0.327 | 0.028 | 0.338 | Admixture of 1,2,3,4 |
| 35 | G35 | 0.315 | 0.335 | 0.022 | 0.328 | Admixture of 1,2,3,4 |
| 36 | G36 | 0.311 | 0.321 | 0.025 | 0.342 | Admixture of 1,2,3,4 |
| 37 | G37 | 0.314 | 0.317 | 0.024 | 0.345 | Admixture of 1,2,3,4 |
| 38 | G38 | 0.308 | 0.337 | 0.025 | 0.329 | Admixture of 1,2,3,4 |
| 39 | G39 | 0.323 | 0.32 | 0.032 | 0.325 | Admixture of 1,2,3,4 |
| 40 | G40 | 0.308 | 0.337 | 0.033 | 0.322 | Admixture of 1,2,3,4 |
| 41 | G41 | 0.316 | 0.33 | 0.038 | 0.316 | Admixture of 1,2,3,4 |
| 42 | G42 | 0.319 | 0.323 | 0.029 | 0.329 | Admixture of 1,2,3,4 |
| 43 | G43 | 0.318 | 0.328 | 0.025 | 0.329 | Admixture of 1,2,3,4 |
| 44 | G44 | 0.33 | 0.32 | 0.029 | 0.322 | Admixture of 1,2,3,4 |
| 45 | G45 | 0.299 | 0.316 | 0.029 | 0.356 | Admixture of 1,2,3,4 |
| 46 | G46 | 0.312 | 0.327 | 0.033 | 0.328 | Admixture of 1,2,3,4 |
| 47 | G47 | 0.314 | 0.326 | 0.026 | 0.334 | Admixture of 1,2,3,4 |
| 48 | G48 | 0.305 | 0.34 | 0.035 | 0.32 | Admixture of 1,2,3,4 |
| 49 | G49 | 0.314 | 0.328 | 0.031 | 0.327 | Admixture of 1,2,3,4 |
| 50 | G50 | 0.296 | 0.327 | 0.028 | 0.349 | Admixture of 1,2,3,4 |
Table 3.
Heterozygosity and Fst value of four sub-populations of the apricot.
| S. No | Sub-Population | Exp Het | Fst |
|---|---|---|---|
| 01 | 1 | 1.77 | 0.005 |
| 02 | 2 | 1.81 | 0.143 |
| 03 | 3 | 1.77 | 0.006 |
| 04 | 4 | 1.77 | 0.006 |
| Average | 1.78 | 0.04 | |
2.4. AMOVA
The purpose of the analysis of molecular variance was to see if there was any genetic variation across populations as well as within populations. According to our results, 95% of the variance was observed within the population, whereas only 5% of the overall genetic diversity was identified between populations (Table 4).
Table 4.
Summary AMOVA table.
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among populations | 1 | 32.385 | 32.385 | 0.557 | 5% |
| Within populations | 98 | 973.665 | 9.935 | 9.935 | 95% |
| Total | 99 | 1006.050 | 10.492 | 100% |
3. Discussion
3.1. SSR Genotyping and Genetic Diversity Analysis
Microsatellite markers have been successfully employed by several studies to identify molecular genetic variation in apricot genotype collections and populations [2,3,22,45,46,47,48,49,50,51]. In this study, we found most of the amplification bands size range between 90–280 bp, similar size range was observed in cultivated apricot [22,23,24] and peach [52,53]. The large range of allele sizes found revealed a significant amount of genetic distance and diversity among the germplasms examined, which is usually as a result of Russian Botanist Vavilov [54], who considered this zone a rich area of diversity. The diversity indices Na, Ne, Ho, and He were evaluated to assess the degree of genetic variation among wild native apricots and exotic genotypes. The average number of alleles (Na) indicates the richness of alleles in the population and the degree of variability it has [36] and the effective number of alleles (Ne) reflects gene frequency in a population [42]. The observed number of alleles (Na) varied from one to six per locus, and the total number of alleles amplified was 179. Bourguiba et al. [2] reported 609 alleles among 890 worldwide accessions. Among the 46 primers, the highest number of alleles, 6 per locus, was realized with 14 markers, and the highest number of effective alleles (Ne) was identified, two per locus with five markers. In their study, Vilanova et al. [55] reported that the Na ranged from two to seven in apricot accessions. In another study, Zhebentyayeva et al. [24] revealed a higher range of Na 2 to 13 alleles per locus in very diverse germplasm. The lower sample size in our study may have resulted in a lesser number of alleles. The mean Na 3.89 per locus found by us is less than 23.00 found in wild apricot 16.75 [11], Decroocq et al. [3] 6.50 found in landraces [56], 4.00 reported for traditional cultivars [48], 4.27 found in apricot germplasm [7], 4.62 found in common apricot [57], 7.64 reported in endemic apricot cultivars [24] and 15.14 realized in 94 Prunus genotypes [26]. The number of alleles was, however, greater than that recorded by Romero et al. [21] in different cultivars (3.1) and almost similar to 3.9 reported by Sanchez-Perez et al. [23]. The average number of effective alleles (Ne) was 1.8821, with a range of 1.1563 to 2. Expected heterozygosity (He) or gene diversity in our investigation varied from 0.13 to 0.50, with an average of 0.45, which was lower than the observed heterozygosity (Ho) of 0.5470. The He range observed by us was narrower in range than 0.4607 to 0.8339 reported by Pedryc et al. [4], 0.37–0.82 by Vilanova et al. [55] and 0.5949–0.8487 by Maghuly et al. [46]. Bourguiba et al. [19] observed that the expected heterozygosity (He) for particular loci differed from 0.04 to 0.82, with a mean value of 0.56 among Tunisian Apricot cultivars. Furthermore, Bourguiba et al. [40] investigated the genetic variability of the apricots grown in Algeria, Morocco and Tunisia and showed expected heterozygosity of 0.593, greater than the average expected 0.45 in this study. Zhang et al. [26] also observed a higher average He of 0.792 in China, Wang et al. [36] observed a He of 0.731 in 150 core samples of Chinese apricot germplasms, Bourguibaet al. [19] revealed that the expected heterozygosity (He) with a mean value of 0.56 among Tunisian Apricot cultivars. The observed heterozygosity ranged from 0 to 1 with a mean value of 0.5226. These values were comparable with 0.51, 0.52, 0.52 reported by Hormaza [22], Raji et al. [24] and Zhebentyayeva et al. [51], respectively, whereas the He value was lesser than 0.58,0.63,0.65,0.68 and 0.72 reported by Ruthner et al. [34], Maghuly et al. [46], Liu et al. [47], Gurcan et al. [58] and Akpinar et al. [59]. The PIC value varied in range from 0.104 to 0.464, with a mean value of 0.320. The average values for PIC in our investigation are less than 0.81 reported by Dehkordi et al. [60]. The Microsatellite sites are the most illuminating ones, those with a greater number of alleles can be utilized directly as DNA fingerprints for apricot cultivar genotype/variety identification. The Shannon information index (I), which estimates diversity, ranged from 0.00 to 0.69 with a mean value of 0.63. Bourgiba et al. [2] found a wider range of I 0.840 to 2.516 with an average value of 2.516. The FST value varied from 0.000 to 0.08, with a mean of 0.022, which was lower than the 0.14, 0.32, 0.38, and 0.5768 reported by Martin et al. [7], Tian-Ming et al. [11], Romero et al. [21], Maghuly et al. [46] and Batnini et al. [50], respectively, in apricot specifying a comparatively low genetic differentiation between genotypes.
3.2. Cluster and PCoA Analysis
All genotypes were divided into three main clusters, cluster I, cluster II, and cluster III, with varying degrees of sub-clustering based on the dendrogram. Cluster I comprised ten accessions, the majority of which were exotic genotypes. Cluster I was subdivided into two sub-clusters, IA and IB, which contained eight and two genotypes. Cluster II contained 40 genotypes that are mainly indigenous to Jammu and Kashmir. The grouping of genotypes revealed by the principal coordinate analysis (PCoA), biplot and cluster dendrogram is similar and shows consistency of the results of the grouping of genotypes based on the geographic areas of the sample collection. The first two coordinates of PCoA contributed 68.43% of total genetic variability and the maximum share of this genetic variation is contributed by cluster first (C-I) and cluster IIB (C-IIB). Previously, apricot accessions were arranged using molecular markers according to their geographic origins [12,22,24]. Romero et al. [21] investigated 40 apricot accessions using SSR markers and showed that the accessions were distinguished according to their ecological and geographical origin. According to Zhang et al. [26] and Herrera et al. [56] SSR markers may easily identify natural germplasm or landraces from breeding releases or cultivars. These results also confirm the different genetic nature of exotic and indigenously grown genotypes. These results also show that the members of cluster I had a significant genetic relationship to each other and are genetically distant from other clusters. The similarity coefficient indicated that the highest similarity 0.867 was observed between indigenous accessions 45 (cluster II) and 49 (cluster II), which are from Banihal and Doda areas of Jammu and Kashmir, respectively, and the lowest similarity was observed between exotic cultivar and indigenous accession 2 (cluster I) and 31 (cluster II) (0.508). The highest similarity among the indigenous accessions may be due to the geographical closeness of these genotypes, and the lowest similarity among exotic and indigenous is due to the difference in the genetic makeup of these genotypes.
3.3. Population Structure
STRUCTURE analysis of the population is a convincing approach to examine genetic relationships and ancestry of individuals within gene banks [61]. The STRUCTURE revealed four sub-populations and sub-population 1, 2, 4 contained only indigenous accessions and sub-population 3rd contained mostly exotic populations. This arrangement pattern is following the cluster dendrogram, 2D PCoA and 3D PCoA plots depicting the separation of local (indigenous genotypes) vs. exotic genotypes separately. Further, the exotic genotypes in the 3rdsubpopulation show no admixture and each genotype in this sub-population can be considered as genetically pure. The genetically pure nature of these cultivars may be due to the recent inclusion of these genotypes for cultivation in this area. The genotypes in sub-population 1, 2 and 4 were all admixtures. The admixture nature of these genotypes may be due to long periods of gene flow among the genotypes without any geographical barrier. Four genetic subpopulations in our study were also identified as per the accession’s geographical location by [19,49,62]. Zehdi et al. [63] in date palm and Haouane et al. [64] in olive found a similar association between the genetic structure and the geographic origin of the plant material. The expected heterozygosity and population differentiation between and within populations reflected that genetic variations within populations were more substantial than differences among populations and that gene flow among populations was rare [65]. Using the software program STRUCTURE, the allele-frequency deviation between populations (Net nucleotide distance) was calculated by applying point estimation of P. The distance between the two identified subpopulations was found to be 0.2119.
3.4. AMOVA
AMOVA revealed a 95% variation within populations and 5.0% of the total molecular variability between populations. Gomez et al. [66] and Vendramin et al. [67], in their findings, also observed an immense amount of genetic deviation occurred within populations of wild apricot (86.3% and 83.6%, respectively). The presence of high variance within the population shows high allelic diversity within populations. This may be due to easy gene flow within individuals of the population than among populations.
4. Materials and Methods
4.1. DNA Extraction and Amplification
Fresh young tender leaves during preflowering season from each accession were taken in a plastic bag from the field and flash frozen in liquid nitrogen to keep them at −80 °C until DNA extraction. The geo-referenced data, name and exact location of apricot leaf sample collection from the field are shown in Table 5. The germplasm of exotic apricot genotypes were preserved and grown at Central Institute of Temperate Horticulture (CITH), Srinagar. The other indigenous genotypes were grown by farmers in their fields in different districts of Jammu and Kashmir. Furthermore, all the sampled genotypes were phenotypically different showing diverse nature of the experimental study material.
Table 5.
Geographical coordinates and location of Apricot accessions evaluated in this study.
| S.NO | Genotype Name | Code | Location | District | Latitude | Longitude | Origin |
|---|---|---|---|---|---|---|---|
| 1 | ‘Harcot’ | G1 | CITH | Budgam | 33.9749° N | 74.7895° E | Exotic |
| 2 | ‘Hartlay’ | G2 | CITH | Budgam | 33.9741° N | 74.7889° E | Exotic |
| 3 | ‘Irani’ | G3 | CITH | Budgam | 33.9739° N | 74.7884°E | Exotic |
| 4 | ‘Communis-Holi’ | G4 | CITH | Budgam | 33.9725° N | 74.7882° E | Exotic |
| 5 | ‘Tilton’ | G5 | CITH | Budgam | 33.9719° N | 74.7875° E | Exotic |
| 6 | ‘Rival’ | G6 | CITH | Budgam | 33.9716° N | 74.7872° E | Exotic |
| 7 | ‘Tokpopanimu’ | G7 | CITH | Budgam | 33.9711° N | 74.7863° E | Exotic |
| 8 | ‘Fair medister’ | G8 | CITH | Budgam | 33.9706° N | 74.7858° E | Exotic |
| 9 | ‘Viva Gold’ | G9 | CITH | Budgam | 33.9701° N | 74.7850° E | Exotic |
| 10 | ‘Cummins’ | G10 | CITH | Budgam | 33.9721° N | 74.7877° E | Exotic |
| 11 | ‘Turkey’ | G11 | CITH | Budgam | 33.9714° N | 74.7867° E | Exotic |
| 12 | ‘New-Castle’ | G12 | CITH | Budgam | 33.9731° N | 74.7881°E | Exotic |
| 13 | ‘Chinese Apricot’ | G13 | CITH | Budgam | 33.9752° N | 74.7497° E | Exotic |
| 14 | Unknown | G14 | Hardas | Ladakh | 34.6061° N | 76.0981° E | Indigenous |
| 15 | Unknown | G15 | Ushkara | Baramulla | 34.2504° N | 74.3788° E | Indigenous |
| 16 | Unknown | G16 | Chardari | Baramulla | 34.1852° N | 74.3634° E | Indigenous |
| 17 | Unknown | G17 | Kantibag | Baramulla | 34.2406° N | 74.3674° E | Indigenous |
| 18 | Unknown | G18 | Uri | Baramulla | 34.0831° N | 74.0543° E | Indigenous |
| 19 | Unknown | G19 | Rangwar | Baramulla | 34.2343° N | 74.3676° E | Indigenous |
| 20 | Unknown | G20 | Beerwah | Budgam | 34.0128° N | 74.5956° E | Indigenous |
| 21 | Unknown | G21 | Katiyawali | Baramulla | 34.1754° N | 74.3531° E | Indigenous |
| 22 | unknown | G22 | Gatha Baderwah | Doda | 32.9973° N | 75.7007° E | Indigenous |
| 23 | unknown | G23 | Khanpora | Baramulla | 34.2086° N | 74.3275° E | Indigenous |
| 24 | unknown | G24 | Brazllo | Kulgam | 33.6467° N | 75.0589° E | Indigenous |
| 25 | unknown | G25 | Shiva | Baramulla | 34.3521° N | 74.4748° E | Indigenous |
| 26 | unknown | G26 | Dogar | Baramulla | 33.1829° N | 74.3619° E | Indigenous |
| 27 | unknown | G27 | Narapora | Shopian | 34.7611° N | 74.8019° E | Indigenous |
| 28 | unknown | G28 | Buniyar | Baramulla | 34.1009° N | 74.2004° E | Indigenous |
| 29 | unknown | G29 | Gozahama | Ganderbal | 34.1934° N | 74.6755° E | Indigenous |
| 30 | unknown | G30 | Kokarnag | Anantnag | 33.6801° N | 75.3895° E | Indigenous |
| 31 | unknown | G31 | Malpora | Baramulla | 34.3528° N | 74.4732° E | Indigenous |
| 32 | unknown | G32 | Dangerpora | Pulwama | 33.8756° N | 74.9793° E | Indigenous |
| 33 | unknown | G33 | Sopore | Baramulla | 34.2604° N | 74.4681° E | Indigenous |
| 34 | unknown | G34 | Duroo | Baramulla | 34.3516° N | 74.4633° E | Indigenous |
| 35 | unknown | G35 | Pazelpora | Baramulla | 34.3587° N | 74.4831° E | Indigenous |
| 36 | unknown | G36 | Kanispora | Baramulla | 34.2184° N | 74.3998° E | Indigenous |
| 37 | unknown | G37 | Darpora | Baramulla | 34.3570° N | 74.4323° E | Indigenous |
| 38 | unknown | G38 | Goripora | Baramulla | 34.3465° N | 74.4212° E | Indigenous |
| 39 | unknown | G39 | Mundji | Baramulla | 34.3607° N | 74.4738° E | Indigenous |
| 40 | unknown | G40 | Handwara | Kupwara | 34.4043° N | 74.2831° E | Indigenous |
| 41 | unknown | G41 | Brath Kalan | Baramulla | 34.3446° N | 74.4065° E | Indigenous |
| 42 | unknown | G42 | Wadura | Baramulla | 34.3528° N | 74.4018° E | Indigenous |
| 43 | unknown | G43 | Badwenchak | Qazigund | 33.5927° N | 75.1658° E | Indigenous |
| 44 | unknown | G44 | Sheeri | Baramulla | 34.1107° N | 74.1837° E | Indigenous |
| 45 | unknown | G45 | Krawah | Banihal | 33.2518° N | 75.1048° E | Indigenous |
| 46 | unknown | G46 | Chadoora | Budgam | 33.9453° N | 75.7967° E | Indigenous |
| 47 | unknown | G47 | Kralpora | Budgam | 34.4997° N | 74.1177° E | Indigenous |
| 48 | unknown | G48 | Bhangra | Doda | 32.9831° N | 75.7116° E | Indigenous |
| 49 | unknown | G49 | KapraBaderwah | Doda | 32.9833° N | 75.7112° E | Indigenous |
| 50 | unknown | G50 | Rawalpora | Srinagar | 34.0042° N | 74.4676° E | Indigenous |
The procedure described by Doyle and Doyle [68] was used to extract genomic DNA. The presence of genomic DNA isolated from 50 genotypes was examined by agarose gel electrophoresis using 1% agarose gel. The purity and amount were examined using a nano-drop spectrophotometer (Thermo Scientific, Waltham, MA, USA). The extracted DNA of each sample was stored at −20 °C after normalization of DNA quantity of each sample to 50 ng/μL for PCR amplification. Forty-six microsatellite markers were used to determine the genetic diversity of apricot samples [69,70,71]. The primers were selected by screening the recent literature related to SSR genetic diversity in apricot and related species, and finally, those primers were selected which have been found polymorphic by evaluating genetic parameters of each primer (Table 6).
Table 6.
List of evaluated SSR markers screened with their primer sequence and allele size range calculated in apricot genotypes studied.
| SSR Marker | Primer Sequence 5′→3′ | Reference | Size Range (bp) |
|---|---|---|---|
| RPPG1-017 | F:GCTCATCAAAACTCTCAACCA R:CCCTTTCTTCAATCCCATC |
Dettori et al., 2015 | 90–220 |
| RPPG1-026 | F:CTTCTGGCACTCTTCCATTT R:GTTCCCAAGTTTTCCTCTCA |
Dettori et al., 2015 | 90–220 |
| RPPG1-032 | F:ATGGCAGAGAGCACAACAA R:TTGAGAGGTAACAGCGAGAA |
Dettori et al., 2015 | 90–250 |
| RPPG1-037 | F:GTCTCTGATCCAAGCCAACT R:ACGCTGCCATTGTTTCTATT |
Dettori et al., 2015 | 100–250 |
| RPPG1-041 | F:TGTTGTAATGGATGGTGTCTTC R:CTTGGTCTTGGTTTCATTCA |
Dettori et al., 2015 | 120–220 |
| RPPG2-011 | F:TTTACAGGTGCCTCAACAAA R:GTACAGCCGATGGAGAGAAA |
Dettori et al., 2015 | 180 |
| RPPG2-022 | F:CTGCTGCGTCTGATGATG R:ACAGGACAGGACCACTTTCT |
Dettori et al., 2015 | 200 |
| RPPG3-026 | F:AGAACGCTATTCCCCTGTAA R:TCATCCTCTCCAAATGTCAA |
Dettori et al., 2015 | 90–200 |
| RPPG4-059 | F:GACGGCTGTTTATTTGCATT R:TGCATTTGTGATCTCGTTTC |
Dettoriet al., 2015 | 100–180 |
| RPPG4-067 | F:AGAAGGGAGGGTGAGAGAAG R:CACGAAGGAAGAAACGAAGT |
Dettori et al., 2015 | 100–210 |
| RPPG4-077 | F:CCTCGTCTTCAGTCTTTTCTG R:CTGTCCCTTCTGTGTTCCTAA |
Dettori et al., 2015 | 90–150 |
| RPPG4-084 | F:TCCTCAAAAGTTACCCCAAG R:CTTGCTGTGGAAGAAGAACC |
Dettori et al., 2015 | 120–200 |
| RPPG4-091 | F:GGAGGGTAGAGAACAGAGCA R:CGGAAGATGTGATTGTGAGA |
Dettori et al., 2015 | 90–220 |
| RPPG5-018 | F:GCATGAAATTGACCCATACA R:TAATTGCTTTGGGGAGGAC |
Dettori et al., 2015 | 90–200 |
| RPPG5-022 | F:CTTGTGAACTGGCATCTGTC R:AGTTGTATGGGCATGTTGTG |
Dettori et al., 2015 | 90–180 |
| RPPG5-023 | F:TTGTTTGCACTAGGCTTTGA R:TTCTTCTTGCATGTCCTTGA |
Dettori et al., 2015 | 90–150 |
| RPPG5-025 | F:GTGTCTCCTCCTCAAAGCAA R:TACGGCAACCAAGAACATC |
Dettori et al., 2015 | 120 |
| RPPG5-030 | F:AAGGCAAGGAATTGGGTAGT R:TGGTTTGTCGTAAGAGTCCA |
Dettori et al., 2015 | 90–280 |
| RPPG6-009 | F:GGGCTTGGCTGATAAAATAA R:TGGTAAAATAGAAGAGCGAGAAG |
Dettori et al., 2015 | 100–120 |
| RPPG6-032 | F:TCCTATGGCAAAAACAAAATC R:TGAAGAGATGGAGTGGAAGAG |
Dettori et al., 2015 | 90–150 |
| RPPG6-033 | F:CATTATCAAACCACGACCAA R:AAAGCTCAACAGCGACTTCT |
Dettori et al., 2015 | 100–200 |
| RPPG7-015 | F:TCTTGGTGGTGGTGAAGTAA R:GAGAGATGGAGGAGGCTGA |
Dettori et al., 2015 | 90–180 |
| RPPG7-026 | F:TTTGGTGAGTGGGCTCTATT R:CTATCGTTCGCTGGTCTTCT |
Dettori et al., 2015 | 90–180 |
| RPPG7-032 | F:AAGGGAGGAGGATTGTGAA R:TGGTAGACGGGTAGATGTTG |
Dettori et al., 2015 | 90–180 |
| RPPG8-007 | F:ACCACCACCTCTTCCAATC R:ACCTCAAAGTGTCCCAGAAA |
Dettori et al., 2015 | 150 |
| RPPG8-028 | F:AAGGAGCCGACATCAGAAC R:TGACCAGAAGCCAAATACATC |
Dettori et al., 2015 | 120–180 |
| Aprigms18 | F:TCTGAGTTCAGTGGGTAGCA R:ACAGAATGTGCGTTGCTTTA |
Liu et al., 2015 | 90–200 |
| UDP98-405 | F:ACGTCATGAACTGACACCCA R:GAGTCTTTGCTCTGCCATCC |
Liu et al., 2015 | 90–120 |
| UDP98-406 | F:TCGGAAACTGGTAGTATGAACAGA R:ATGGGTCGTATGCACAGTCA |
Liu et al., 2015 | 90–120 |
| UDP98-409 | F:GCTGATGGGTTTTATGGTTTTC R:CGGACTCTTATCCTCTATCAACA |
Liu et al., 2015 | 90–150 |
| UDP98-411 | F:AAGCCATCCACTCAGCACTC R:CCAAAAACCAAAACCAAAGG |
Liu et al., 2015 | 90–180 |
| Pchgms4 | F:ATCTTCACAACCCTAATGTC R:GTTGAGGCAAAAGACTTCAAT |
Liu et al., 2015 | 90–280 |
| Pchgms5 | F:CGCCCATGACAAACTTA R:GTCAAGAGGTACACCAG |
Liu et al., 2015 | 150–280 |
| Bppct007 | F:TCATTGCTCGTCATCAGC R:CAGATTTCTGAAGTTAGCGGTA |
Liu et al., 2015 | 150–420 |
| Bppct025 | F:TCCTGCGTAGAAGAAGGTAGC R:CGACATAAAGTCCAAATGGC |
Liu et al., 2015 | 90–150 |
| Bppct030 | F:AATTGTACTTGCCAATGCTATGA R:CTGCCTTCTGCTCACACC |
Liu et al., 2015 | 90–180 |
| PacA10 | F:TGAGCATAATTGGGGCAG R:GCCAGAGAAGCCATTTCAGT |
Lambert et al., 2004 | 120–250 |
| PacA18 | F:TCCAAACCTACCGTTTCTCAT R:CAACAGCACAAACAGAACCAC |
Lambert et al., 2004 | 180–250 |
| PacA33 | F:TCAGTCTCATCCTGCATACG R:CATGTGGCTCAAGGATCAAA |
Lambert et al., 2004 | 90–250 |
| PacA22 | F:AACCAGTTGCCTCTAGATTTTG R:AGCTGAAAGTCAATTCAGAGTAGTT |
Lambert et al., 2004 | 100–180 |
| PacA26 | F:CCAATCATGAAAATCATAAAAGCAA R:TGGGATGTCCTATTGTTTTCA |
Lambert et al., 2004 | 100–200 |
| PacA35 | F:ATTGCGATTTCGGTCTGTT R:CCATCCCAAATTGCTTACTT |
Lambert et al., 2004 | 120–180 |
| PacC3 | F:TGACTTGATCAGACTCGACA R:TTGCATTTGCATTTACAATAGA |
Lambert et al., 2004 | 90–200 |
| PacC25 | F:GTGTTTTGACAAGAAATGAATTG R:TCCATTCGCAGTAAAATTAAAC |
Lambert et al., 2004 | 100–200 |
| PacA58 | F:GACATTGCGATTTCGGTC R:TCCATCCCAAATTGCTTACT |
Lambert et al., 2004 | 100–180 |
| PdavW3 | F:GAGGGCTGGATCATGACG R:AACCCAGTGGCACAATCGTA |
Lambert et al., 2004 | 90–200 |
PCR reaction was carried out in a 20 μL reaction mixture with 50 ng/µL DNA templates, 10X PCR buffer, 2.5 mM MgCl2, 10 mM dNTPs, 1U Taq DNA polymerase, and both primer pairs. A thermal cycler (Takara Thermal Cycler Dice, TD 600, Shiga, Japan) was used for amplification. The PCR amplification steps were executed as initial denaturation for 5 min at 94 °C followed by 35 cycles of 60 s at 94 °C denaturation, 49 to 58 °C for 60 s for optimal annealing temperature for different primers, 90 s at 72 °C for extension and final extension for 10 min at 72 °C followed by cooling at 4 °C. The procedure was performed three consecutive times with the same primers and genotypes to check out the reproducibility. The PCR amplification products and the 100 bp DNA marker were separated on 3% agarose gel with 0.5× TBE buffers using Ethidium bromide (EtBr) as a staining agent on the gel. The banding pattern of the amplified bands was examined under a gel documentation imaging system.
4.2. Data Analysis
For all accessions, the composition of alleles and each microsatellite locus were used to calculate the total number of alleles. Indices of molecular characterization were statistically evaluated, including the expected heterozygosity (He), the observed heterozygosity (Ho), the effective number of alleles (Ne), Shannon’s information index (I), the coefficient of gene differentiation (Fst) by applying the POPGENE 1.32 [72,73,74]. In addition to this, based on Jaccard’s similarity coefficient, the Unweighted pair-group method with arithmetic means (UPGMA) hierarchical clustering tree was designed for distinct apricot cultivar groups [75]. STRUCTURE 2.3.4 software was used to analyze ancestral population structure based on Bayesian clustering [76]. STRUCTURE was run ten times, with each run consisting of 100,000-steps followed by 500,000 Markov Chain Monte Carlo (MCMC) iterations, presuming an admixture framework with correlated allelic and several clusters (K) ranging from 1 to 10. The Pritchard et al. [76] criteria and the 1K approach, defined by Evanno et al. [44] and implemented in the STRUCTURE HARVESTER v2.3.4. Websites were used to determine the precise number of populations (K) [77]. CLUMPP v1.1 software [78] was utilized using optimistic algorithms, 10,000 random input orders, and 10,000 repeats to estimate the mean pairwise similarity of runs and produce optimum alignment of independent runs. To graphically display the results, the output files of CLUMPP were used as input files for DISTRUCT v1.1 software, the output of CLUMPP was immediately fed into DISTRUCT v1.1 [79]. The probability membership of each accession was ascertained, they were allocated to the appropriate cluster if their affiliation was higher than 80%; otherwise, they were labeled admixture. For estimation of genetic differentiation among and within populations, AMOVA analysis was done in software GenAlEx v6.503 [80].
5. Conclusions
In conclusion, our investigation has dispensed a broader context on genetic variability and core structure among apricot accessions in Jammu and Kashmir. The results revealed that the SSR marker studies could easily decrypt the genetic variability present within the germplasm. This was the first kind of study carried out in this area to distinguish exotic genotypes from indigenous genotypes via molecular markers and showed a high level of polymorphism. Genetic variability between exotic and indigenous genotypes can provide an excellent opportunity for new cultivar development through hybridization and advanced genetic tools such as molecular markers. These diversity analysis tools could be utilized for the establishment and collection of gene banks and core collections by reducing redundancy of germplasm, selection of parents for any breeding program and genome-wide association studies for mapping of different traits.
Acknowledgments
This research was funded by Institutional Fund Projects grant no. (IFPRP:915-130-1442). Therefore, authors gratefully acknowledge technical and financial support from the Ministry of Education and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
Author Contributions
Conceptualization, V.S.; Data curation, Z.N.S., V.S., R.A.S. and J.I.M.; Formal analysis, Z.N.S., V.S., R.A.S., K.R.H. and J.I.M.; Investigation, Z.N.S.; Methodology, Z.N.S.; Software, R.A.S., K.R.H., M.A., N.A. and Z.N.S.; Supervision, V.S. and J.I.M.; Validation, Z.N.S., K.R.H., V.S., M.A., N.A. and J.I.M.; Funding, K.R.H., M.A. and N.A.; Visualization, V.S., Writing—original draft, Z.N.S.; Writing—review & editing, Z.N.S., V.S., R.A.S., K.R.H., M.A., N.A. and S.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Institutional Fund Projects grant no. (IFPRP:915-130-1442). Therefore, authors gratefully acknowledge technical and financial support from the Ministry of Education and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Herrera S., Lora J., Hormaza J., Herrero M., Rodrigo J. Optimizing production in the new generation of Apricot cultivars: Self-incompatibility, S-RNase allele identification, and incompatibility group assignment. Front. Plant Sci. 2018;27:527. doi: 10.3389/fpls.2018.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourguiba H., Scotti I., Sauvage C., Zhebentyayeva T., Ledbetter C., Krska B., Remay A., Donofrio C., Iketani H., Christen D., et al. Genetic structure of a worldwide germplasm collection of Prunus armeniaca L. reveals three major diffusion routes for varieties coming from the species’ center of origin. Front. Plant Sci. 2020;25:638. doi: 10.3389/fpls.2020.00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decroocq S., Cornille A., Tricon D., Babayeva S., Chague A., Eyquard P., Karychev R., Dolgikh S., Kostritsyna T., Liu S., et al. New insights into the history of domesticated and wild apricots and its contribution to plum pox virus resistance. Mol. Ecol. 2016;25:2712–4729. doi: 10.1111/mec.13772. [DOI] [PubMed] [Google Scholar]
- 4.Pedryc A., Ruthner S., Herman R., Krska B., Hegedus A., Halasz J. Genetic diversity of apricot revealed by a set of SSR markers from linkage group G1. Sci. Hortic. 2009;121:19–26. doi: 10.1016/j.scienta.2009.01.014. [DOI] [Google Scholar]
- 5.Faostat. 2013. [(accessed on 4 February 2015)]. Available online: http://apps.fao.org.
- 6.Zhebentyayeva T., Ledbetter C., Burgos L., Llacer G. Infruit Breeding. Springer; Boston, MA, USA: 2012. Apricot; pp. 415–458. [DOI] [Google Scholar]
- 7.Martín C., Herrero M., Hormaza J. Molecular characterization of apricot germplasm from an old stone collection. PLoS ONE. 2011;25:e23979. doi: 10.1371/journal.pone.0023979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Y., Zhou H., Lin-Wang K., Vimolmangkang S., Espley V., Wang L., Allan C., Han Y. Transcriptome analysis and transient transformation suggest an ancient duplicated MYB transcription factor as a candidate gene for leaf red coloration in peach. BMC Plant Biol. 2014;14:388. doi: 10.1186/s12870-014-0388-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah R.A., Bakshi P., Sharma N., Jasrotia A., Itoo H., Gupta R., Singh A. Diversity assessment and selection of superior Persian walnut (Juglans regia L.) trees of seedling origin from North-Western Himalayan region. Resour. Environ. Sustain. 2021;1:100015. doi: 10.1016/j.resenv.2021.100015. [DOI] [Google Scholar]
- 10.Corrado G., Forlani M., Rao R., Basile B. Diversity and Relationships among Neglected Apricot (Prunus armeniaca L.) Landraces Using Morphological Traits and SSR Markers: Implications for Agro-Biodiversity Conservation. Plants. 2021;10:1341. doi: 10.3390/plants10071341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian-Ming H., Xue-Sen C., Zheng X., Jiang-Sheng G., Pei-Jun L., Wen L., Qing L., Yan W. Using SSR markers to determine the population genetic structure of wild apricot (Prunus armeniaca Prunus armeniaca L.) in the Ily Valley of West China. Genet. Resour. Crop Evol. 2007;54:563–572. doi: 10.1007/s10722-006-0013-5. [DOI] [Google Scholar]
- 12.Hagen L., Khadari B., Lambert P., Audergon J.M. Genetic diversity in apricot revealed by AFLP markers: Species and cultivar comparisons. Theor. Appl. Genet. 2002;105:298–305. doi: 10.1007/s00122-002-0910-8. [DOI] [PubMed] [Google Scholar]
- 13.Geuna F., Toschi M., Bassi D. The use of AFLP markers for cultivar identification in apricot. Plant Breed. 2003;122:526–531. doi: 10.1111/j.1439-0523.2003.00897.x. [DOI] [Google Scholar]
- 14.Kumar M., Mishra G.P., Singh R., Kumar J., Naik P.K., Singh S.B. Correspondence of ISSR and RAPD markers for comparative analysis of genetic diversity among different apricot genotypes from cold arid deserts of trans-Himalayas. Physiol. Mol. Biol. Plants. 2009;15:225–236. doi: 10.1007/s12298-009-0026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krichen L., Martins J.M., Lambert P., Daaloul A., Trifi-Farah N., Marrakchi M., Audergon J.M. Using AFLP markers for the analysis of the genetic diversity of apricot cultivars in Tunisia. J. Am. Soc. Hortic. 2008;133:204–212. doi: 10.21273/JASHS.133.2.204. [DOI] [Google Scholar]
- 16.Fang J., Tao J., Chao C.T. Genetic diversity in fruiting-mei, apricot, plum and peach revealed by AFLP analysis. J. Hortic. Sci. Biotechnol. 2006;81:898–902. doi: 10.1080/14620316.2006.11512156. [DOI] [Google Scholar]
- 17.Zargar S.A., Wani A.A., Saggoo M.I. Analysis of phenotypic diversity of apricot (Prunus armeniaca L.) accessions from Jammu and Kashmir, India. Plant Genet. Resour. 2021;19:203–215. doi: 10.1017/S1479262121000241. [DOI] [Google Scholar]
- 18.Sheikh Z.N., Sharma V., Shah R.A., Sharma N., Summuna B., Al-Misned F.A., El-Serehy H.A., Mir J.I. Genetic diversity analysis and population structure in apricot (Prunus armeniaca L.) grown under north-western himalayas using ISSR markers. Saudi J. Biol. Sci. 2021;28:5989–5995. doi: 10.1016/j.sjbs.2021.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourguiba H., Krichen L., Audergon J.M., Khadari B., Trifi-Farah N. Impact of mapped SSR markers on the genetic diversity of apricot (Prunus armeniaca L.) in Tunisia. Plant Mol. Biol. Rep. 2010;28:578–587. doi: 10.1007/s11105-010-0189-x. [DOI] [Google Scholar]
- 20.Shah R.A., Baksi P., Jasrotia A., Bhat D.J., Gupta R., Bakshi M. Genetic diversity of walnut (Juglans regia L.) seedlings through SSR markers in north-western Himalayan region of Jammu. Bangladesh J. Bot. 2020;49:1003–1012. doi: 10.3329/bjb.v49i4.52517. [DOI] [Google Scholar]
- 21.Romero C., Pedryc A., Munoz V., Llacer G., Badenes M.L. Genetic diversity of different apricot geographical groups determined by SSR markers. Genome. 2003;46:244–252. doi: 10.1139/g02-128. [DOI] [PubMed] [Google Scholar]
- 22.Hormaza J.I. Molecular characterization and similarity relationships among apricot (Prunus armeniaca L.) genotypes using simple sequence repeats. Theor. Appl. Genet. 2002;104:321–328. doi: 10.1007/s001220100684. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez-Perez R., Martinez-Gomez P., Dicenta F., Egea J., Ruiz D. Level and transmission of genetic heterozygosity in apricot (Prunus armeniaca L.) explored using simple sequence repeat markers. Genet. Resour. Crop. Evol. 2006;53:763–770. doi: 10.1007/s10722-004-4636-0. [DOI] [Google Scholar]
- 24.Zhebentyayeva T., Reighard G., Gorina V., Abbott A. Simple sequence repeat (SSR) analysis for assessment of genetic variability in apricot germplasm. Theor. Appl. Genet. 2003;106:435–444. doi: 10.1007/s00122-002-1069-z. [DOI] [PubMed] [Google Scholar]
- 25.Li M., Zheng P., Ni B., Hu X., Miao X., Zhao Z. Genetic diversity analysis of apricot cultivars grown in China based on SSR markers. Eur. J. Hortic. Sci. 2018;83:18–27. doi: 10.17660/eJHS.2018/83.1.3. [DOI] [Google Scholar]
- 26.Zhang Q.P., Liu D.C., Liu S., Liu N., Wei X., Zhang A.M., Liu W.S. Genetic diversity and relationships of common apricot (Prunus armeniaca L.) in China based on simple sequence repeat (SSR) markers. Genet. Resour. Crop. Evol. 2014;61:357–368. doi: 10.1007/s10722-013-0039-4. [DOI] [Google Scholar]
- 27.Hu X., Zheng P., Ni B., Miao X., Zhao Z., Li M. Population genetic diversity and structure analysis of wild apricot (Prunus armeniaca L.) revealed by SSR markers in the Tien-Shan mountains of China. Pak. J. Bot. 2018;50:609–615. [Google Scholar]
- 28.Bakir M., Dumanoglu H., Erdogan V., Ernim C., Macit T. Characterization of wild apricot (Prunus armeniaca L.) genotypes selected from Cappadocia region (Nevşehir-Turkey) by SSR markers. J. Agric. Sci. 2019;25:498–507. doi: 10.15832/ankutbd.457850. [DOI] [Google Scholar]
- 29.Yuan W.Z., Bai Y. Analysis of Genetic Diversity in Prunus sibirica L. in Inner Mongolia Using SCoT Molecular Markers. Res. Square. 2021:1–17. doi: 10.21203/rs.3.rs-652651/v1. [DOI] [Google Scholar]
- 30.Wang L. Genetic Diversity and Population Genetic Structure of Armeniaca Sibirica Clones. Shenyang Agricultural University; Shenyang, China: 2019. [Google Scholar]
- 31.Dirlewanger E., Cosson P., Tavaud M., Aranzana M., Poizat C., Zanetto A., Arus P., Laigret F. Development of microsatellite markers in peach Prunus persica L. Batsch and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.) Theor. Appl. Genet. 2002;105:127–138. doi: 10.1007/s00122-002-0867-7. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Zhang J., Sun H., Ning N., Yang L. Construction and evaluation of a primary core collection of apricot germplasm in China. Sci. Hortic. 2011;128:311–319. doi: 10.1016/j.scienta.2011.01.025. [DOI] [Google Scholar]
- 33.Aranzana M.J., Garcia-Mas J., Carbo J., Arus P. Development and variability analysis of microsatellite markers in peach. Plant Breed. 2002;121:87–92. doi: 10.1046/j.1439-0523.2002.00656.x. [DOI] [Google Scholar]
- 34.Ruthner S., Pedryc A., Kriska B., Romero C. Molecular characterization of apricot (Prunus armenica L.) cultivars using cross species SSR amplification with peach primers. Int. J. Hortic. Sci. 2006;12:53–57. doi: 10.31421/IJHS/12/3/659. [DOI] [Google Scholar]
- 35.Dondini L., Lain O., Geuna F., Banfi R., Gaiotti F., Tartarini S., Bassi D., Testolin R. Development of a new SSR-based linkage map in apricot and analysis of synteny with existing Prunus maps. Tree Genet. Genomes. 2007;3:239–249. doi: 10.1007/s11295-006-0059-8. [DOI] [Google Scholar]
- 36.Wang Y.J., Li X.Y., Han J., Fang W.M., Li X.D., Wang S.S., Fang J.G. Analysis of genetic relationships and identification of flowering-mei cultivars using EST-SSR markers developed from apricot and fruiting-mei. Sci. Hortic. 2011;132:12–17. doi: 10.1016/j.scienta.2011.09.013. [DOI] [Google Scholar]
- 37.Hagen L.S., Chaïb J., Fady B., Decroocq V., Bouchet J.P., Lambert P., Audergon J.M. Genomic and cDNA microsatellites from apricot (Prunus armeniaca L.) Mol. Ecol. Notes. 2004;4:742–745. doi: 10.1111/j.1471-8286.2004.00802.x. [DOI] [Google Scholar]
- 38.Messina R., Lain O., Marrazzo M.T., Cipriani G., Testolin R. New set of microsatellite loci isolated in apricot. Mol. Ecol. Notes. 2004;3:432–434. doi: 10.1111/j.1471-8286.2004.00674.x. [DOI] [Google Scholar]
- 39.Xie R., Li X., Chai M., Song L., Jia H., Wu D., Gao Z. Evaluation of the genetic diversity of Asian peach accessions using a selected set of SSR markers. Sci. Hortic. 2010;4:622–629. doi: 10.1016/j.scienta.2010.05.015. [DOI] [Google Scholar]
- 40.Bourguiba H., Audergon J.M., Krichen L., Trifi-Farah N., Mamouni A., Trabelsi S., D’Onofrio C., Asma B.M., Santoni S., Khadari B. Loss of genetic diversity as a signature of apricot domestication and diffusion into the Mediterranean Basin. BMC Plant Biol. 2012;12:49. doi: 10.1186/1471-2229-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yilmaz K.U., Paydas-Kargi S.E., Dogan Y., Kafkas S.A. Genetic diversity analysis based on ISSR, RAPD and SSR among Turkish apricot germplasms in Iran caucasian eco-geographical group. Sci. Hortic. 2012;138:138–143. doi: 10.1016/j.scienta.2012.02.017. [DOI] [Google Scholar]
- 42.Chen J., Dong S., Zhang X., Wu Y., Zhang H., Sun Y., Zhang J. Genetic diversity of Prunus sibirica L. superior accessions based on the SSR markers developed using restriction-site associated DNA sequencing. Genet. Resour. Crop. Evol. 2021;68:615–628. doi: 10.1007/s10722-020-01011-5. [DOI] [Google Scholar]
- 43.Rezaei M., Rahmati M., Kavand A., Hemati M., Kazemi S.R. Screening of some Iranian Commercial Apricot Cultivars by SSR Markers for identification of Synonyms. Int. J. Hortic. Sci. 2021;35 doi: 10.22067/jhs.2021.61722.0. [DOI] [Google Scholar]
- 44.Evanno G., Regnaut S., Goudet J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecolo. 2005;8:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 45.Groppi A., Liu S., Cornille A., Decroocq S., Bui Q.T., Tricon D., Cruaud C., Arribat S., Belser C., Marande W., et al. Population genomics of apricots unravels domestication history and adaptive events. Nat. Commun. 2021;12:3956. doi: 10.1038/s41467-021-24283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maghuly F., Fernandez E.B., Ruthner S., Pedryc A., Laimer M. Microsatellite variability in apricots (Prunus armeniaca L.) reflects their geographic origin and breeding history. Tree Genet. Genomes. 2005;4:151–165. doi: 10.1007/s11295-005-0018-9. [DOI] [Google Scholar]
- 47.Liu S., Cornille A., Decroocq S., Tricon D., Chague A., Eyquard J.P., Liu W.S., Giraud T., Decroocq V. The complex evolutionary history of apricots: Species divergence, gene flow and multiple domestication events. Mol. Ecol. 2019;24:5299–5314. doi: 10.1111/mec.15296. [DOI] [PubMed] [Google Scholar]
- 48.Martinez Mora C., Rodríguez J., Cenis J.L., Ruiz García L. Genetic variability among local apricots (Prunus armeniaca L.) from the Southeast of Spain. Span. J. Agric. 2009;7:855–868. doi: 10.5424/sjar/2009074-1099. [DOI] [Google Scholar]
- 49.Bourguiba H., Khadari B., Krichen L., Trifi-Farah N., Mamouni A., Trabelsi S., Audergon J.M. Genetic relationships between local North African apricot (Prunus armeniaca L.) germplasm and recently introduced varieties. Sci. Hortic. 2013;152:61–69. doi: 10.1016/j.scienta.2013.01.012. [DOI] [Google Scholar]
- 50.Batnini M.A., Krichen L., Bourguiba H., Trifi-Farah N., González D.R., Gómez P.M., Rubio M. Comparative analysis of traditional and modern apricot breeding programs: A case of study with Spanish and Tunisian apricot breeding germplasm. Span. J. Agric. 2016;14:14. doi: 10.5424/sjar/2016143-8638. [DOI] [Google Scholar]
- 51.Raji R., Jannatizadeh A., Fattahi R., Esfahlani M.A. Investigation of variability of apricot (Prunus armeniaca L.) using morphological traits and microsatellite markers. Sci. Hortic. 2014;176:225–231. doi: 10.1016/j.scienta.2014.06.033. [DOI] [Google Scholar]
- 52.Sosinski B., Gannavarapu M., Hager L.D., Beck L.E., King G.J., Ryder C.D., Rajapakse S., Baird W.V., Ballard R.E., Abbott A.G. Characterization of microsatellite markers in peach (Prunus persica L.) Batsch. Theor. Appl. Genet. 2000;101:421–428. doi: 10.1007/s001220051499. [DOI] [Google Scholar]
- 53.Testolin R., Marrazzo T., Cipriani G., Quarta R., Verde I., Dettori M.T., Pancaldi M., Sansavini S. Microsatellite DNA in peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome. 2000;43:512–520. doi: 10.1139/g00-010. [DOI] [PubMed] [Google Scholar]
- 54.Vavilov N.I. The origin, variation, immunity and breeding of cultivated plants. LWW. Soil Sci. 1951;72:482. doi: 10.1097/00010694-195112000-00018. [DOI] [Google Scholar]
- 55.Vilanova S., Soriano J.M., Lalli D.A., Romero C., Abbott A.G., Llacer G., Badenes M.L. Development of SSR markers located in the G1 linkage group of apricot (Prunus armeniaca L.) using a bacterial artificial chromosome library. Mol. Ecol. Notes. 2006;3:789–791. doi: 10.1111/j.1471-8286.2006.01346.x. [DOI] [Google Scholar]
- 56.Herrera S., Rodrigo J., Hormaza J.I., Lora J. Identification of self-incompatibility alleles by specific PCR analysis and S-RNase sequencing in apricot. Int. J. Mol. Sci. 2018;11:3612. doi: 10.3390/ijms19113612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khadivi-Khub A., Jafari H.R., Zamani Z. Phenotypic and genotypic variation in Iranian sour and duke cherries. Trees. 2013;27:1455–1466. doi: 10.1007/s00468-013-0892-y. [DOI] [Google Scholar]
- 58.Gurcan K., Ocal N., Yılmaz K.U., Ullah S., Erdogan A., Zengin Y. Evaluation of Turkish apricot germplasm using SSR markers: Genetic diversity assessment and search for Plum pox virus resistance alleles. Sci. Hortic. 2015;193:155–164. doi: 10.1016/j.scienta.2015.07.012. [DOI] [Google Scholar]
- 59.Akpınar A.E., Koçal H., Ergül A., Kazan K., Şelli M.E., Bakır M., Aslantaş Ş., Kaymak S., Sarıbaş R. SSR-based molecular analysis of economically important Turkish apricot cultivars. Genet. Mol. Res. 2010;9:324–332. doi: 10.4238/vol9-1gmr727. [DOI] [PubMed] [Google Scholar]
- 60.Dehkordi M., Beigzadeh T., Sorkheh K. Novel in silico EST-SSR markers and bioinformatic approaches to detect genetic variation among peach (Prunus persica L.) germplasm. J. For. Res. 2020;4:1359–1370. doi: 10.1007/s11676-019-00922-z. [DOI] [Google Scholar]
- 61.Song Y., Fan L., Chen H., Zhang M., Ma Q., Zhang S., Wu J. Identifying genetic diversity and a preliminary core collection of Pyrus pyrifolia cultivars by a genome-wide set of SSR markers. Sci. Hortic. 2014;167:5–16. doi: 10.1016/j.scienta.2013.12.005. [DOI] [Google Scholar]
- 62.Krichen H.B., Jean-Marc A., Neila T.F. Comparative analysis of genetic diversity in Tunisian apricot germplasm using AFLP and SSR markers. Sci. Hortic. 2010;127:54–63. doi: 10.1016/j.scienta.2010.09.012. [DOI] [Google Scholar]
- 63.Zehdi-Azouzi S., Cherif E., Moussouni S., Gros-Balthazard M., Abbas N.S., Ludena B., Castillo K., Chabrillange N., Bouguedoura N., Bennaceur M., et al. Genetic structure of the date palm (Phoenix dactylifera) in the old world reveals a strong differentiation between eastern and western populations. Ann. Bot. 2015;116:101–112. doi: 10.1093/aob/mcv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haouane H., Bakkali A., Moukhli A., Tollon C., Santoni S., Oukabli A., El Modafar C., Khadari B. Genetic structure and core collection of the world olive germplasm bank of marrakech towards the optimised management and use of mediterranean olive genetic resources. Genetica. 2011;139:1083–1094. doi: 10.1007/s10709-011-9608-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pereira-Dias L., Vilanova S., Fita A., Prohens J., Rodríguez-Burruezo A. Genetic diversity, population structure, and relationships in a collection of pepper (Capsicum spp.) landraces from the Spanish centre of diversity revealed by genotyping-by-sequencing (GBS) Hortic. Res. 2019;6:54. doi: 10.1038/s41438-019-0132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martinez-Gomez P., Arulsekar S., Potter D., Gradziel T.M. Relationships among peach, almond, and related species as detected by simple sequence repeat markers. J. Am. Soc. Hortic. 2003;128:667–671. doi: 10.21273/JASHS.128.5.0667. [DOI] [Google Scholar]
- 67.Vendramin G.G., Anzidei M., Madaghiele A., Bucci G. Distribution of genetic diversity in Pinus pinaster Ait. as revealed by chloroplast microsatellites. Theor. Appl. Genet. 1998;97:456–463. doi: 10.1007/s001220050917. [DOI] [Google Scholar]
- 68.Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 69.Dettori M.T., Micali S., Giovinazzi J., Scalabrin S., Verde I., Cipriani G. Mining microsatellites in the peach genome: Development of new long-core SSR markers for genetic analyses in five Prunus species. Springer Plus. 2015;4:337. doi: 10.1186/s40064-015-1098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu M.P., Du H.Y., Zhu G.P., Fu D.L., Tana W.Y. Genetic diversity analysis of sweet kernel apricot in China based on SSR and ISSR markers. Genet. Mol. Res. 2015;14:9722–9729. doi: 10.4238/2015.August.19.4. [DOI] [PubMed] [Google Scholar]
- 71.Lambert P., Hagen L.S., Arus P., Audergon J.M. Genetic linkage maps of two apricot cultivars (Prunus armeniaca L.) compared with the almond Texas×peach Earlygold reference map for Prunus. Theor. Appl. Genet. 2004;108:1120–1130. doi: 10.1007/s00122-003-1526-3. [DOI] [PubMed] [Google Scholar]
- 72.Nei M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shannon C.E. A mathematical theory of communication. ACM SIGMOBILE Mob. Comput. Commun. Rev. 2001;5:3–55. doi: 10.1145/584091.584093. [DOI] [Google Scholar]
- 74.Yeh F.C., Yang R.C., Boyle T.B., Ye Z.H., Mao J.X. POPGENE, the User-Friendly Shareware for Population Genetic Analysis. Volume 10. Molecular Biology and Biotechnology Centre, University of Alberta; Edmonton, AB, Canada: 1997. pp. 295–301. [Google Scholar]
- 75.Rohlf F.J. NTSYS-pc Version. 2.02i Numerical Taxonomy and Multivariate Analysis System. Applied Biostatistics Inc.; Setauket, NY, USA: 1997. [Google Scholar]
- 76.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Earl D.A. Structure harvester: A website and program for visualizing STRUCTURE output and implementing the evanno method. Conserv. Genet. Resour. 2012;2:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 78.Jakobsson M., Rosenberg N.A. CLUMPP: Cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 79.Rosenberg N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes. 2004;1:137–148. doi: 10.1046/j.1471-8286.2003.00566.x. [DOI] [Google Scholar]
- 80.Peakall R.O., Smouse P.E. Genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes. 2006;1:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are contained within the article.