Abstract
The Salmonella Enterica subsp. Enterica serovar Enteritidis is one of main serovars isolated from human patients with food poisoning and poultry without clinical signs. Consumption of poultry products contaminated with Salmonella Enteritidis is a common source of human salmonellosis; 82 Salmonella spp. were isolated from 291 samples of retail chicken meat, 201 one-day-old chicks, 30 internal organs of chickens, 156 chicken eggs, 100 duck eggs, 38 straw bedding samples, 18 samples of retail duck meat, and 19 swab samples from slaughterhouses in 2019 and 2020. An antibiotic susceptibility test was performed for all isolates, revealing 33 multidrug-resistant (MDR) strains. The whole genome of 33 MDR strains isolated in 2019 and 2020 and 10 strains isolated in 2011, 2012, and 2017 was sequenced using the MinION sequencing protocol. Within these 43 samples, 5 serovars were identified: S. Enteritidis, S. Agona, S. Virchow, S. Albany, and S. Bareilly. The most common serovar was S. Enteritidis (26/43), which showed the highest resistance to ampicillin (100%), followed by nalidixic acid (90%) and colistin (83%). Core genome multilocus sequence typing analysis showed that the S. Enteritidis strains isolated from different sources and in different years were clustered together. In addition, the S. Enteritidis strains isolated since 2011 consistently harbored the same antibiotic resistance patterns.
Keywords: whole-genome sequencing, Salmonella Enteritidis, multidrug resistance, virulence plasmid, salmonellosis
1. Introduction
Non-typhoidal S. Enterica causes foodborne salmonellosis and has become a global health threat [1]. Salmonella Enteritidis is frequently isolated from human patients with salmonellosis caused by the consumption of contaminated chicken meat and chicken products, such as eggs [1]. Fever, abdominal cramps, and diarrhea are common clinical symptoms of Salmonella Enteritidis infection in humans, which appear 12 to 72 h after consuming the contaminated food [2]. In most cases, the symptoms last 4 to 7 days and resolve on their own without the need for antibiotics. However, in the elderly, infants, and people with weakened immune systems, the diarrhea symptoms can be severe, and septicemia, and even death, can occur [3]. No clinical symptoms are observed in birds infected with Salmonella Enteritidis [4].
In Korea, antibiotic usage is not regulated in the poultry industry, to which about 1000 tons of antibiotics were sold between 2011 and 2017 [5]. In 2012, 720,000 tons of chicken meat were produced, which represents the second-largest protein source in Korea [6]. It is noteworthy that the prevalence of antibiotic-resistant Salmonella is high in retail chicken in Korea [5,7].
Recently, the isolation of multidrug-resistant (MDR) Salmonella, which is resistant not only to traditional first-line antibiotics such as ampicillin, chloramphenicol, and trimethoprim but also to currently recommended antibiotics such as fluoroquinolones and extended-spectrum cephalosporins, has increased dramatically [8,9]. MDR Salmonella with the same antibiotic resistance phenotype has been isolated from humans and poultry [10]. The spread of antibiotic resistance genes from animal bacteria to human populations is a major public health concern [11]. Investigation and epidemiological analysis of the MDR Salmonella in foods are necessary to prevent the development of the spread of MDR strains [12].
Epidemiological sources of Salmonella outbreaks have been investigated using pulsed-field gel electrophoresis (PFGE) and multiple-locus variable-number tandem repeats (MLVA) analysis [13]. In previous studies, these approaches have successfully detected the genetic relationship between Salmonella Enteritidis strains isolated from human patients or poultry sources in Korea [14,15]. However, the discrimination power of these methods for genetically closely related Salmonella Enteritidis strains is limited [16,17].
Whole-genome sequencing (WGS) has improved the resolution of genome analyses; thus, the sources of Salmonella outbreaks can now be traced [18,19,20]. Two analytic methods are commonly used for WGS-based genome analysis: single-nucleotide polymorphism (SNP) analysis and multilocus sequence typing (MLST) [21]. SNPs are identified by mapping sequence data of isolates to a reference genome and then recording the nucleotides that differ within the datasets [22]. The MLST method explores the allelic difference in a predefined set of gene loci [23]. In order to improve the resolution power of the MLST, the number of genes included in the scheme was increased [24]. Core genome MLST (cgMLST) balances the number of loci included in a scheme by considering those loci present in the majority of the isolates (ranging from 95% to 99%) in a given species [22,25].
Recently, several studies have used WGS to investigate the molecular relationship of Salmonella isolated from various sources [26,27,28,29]. However, only a small number of studies using WGS for the epidemiological analysis of Salmonella spp. have been reported in Korea [30,31,32].
Here, we have isolated 82 MDR Salmonella strains from 853 poultry sources in Korea and sequenced their genomes using the Oxford Nanopore approach. In order to investigate the relationships between the Salmonella Enteritidis strains, the whole genome sequences of the MDR isolates were compared using cgMLST and whole-genome SNP (wgSNP).
2. Materials and Methods
2.1. Sample Collection
Between 1 April 2019 to 11 May 2020, a total of 853 samples (291 samples of retail chicken meat, 201 one-day-old chicks, 30 internal organs of chickens for pet food, 156 chicken eggs, 100 duck eggs, 38 straw bedding samples, 18 samples of retail duck meat, and 19 swab samples from slaughterhouses) were collected. Retail chicken meats were purchased in two local supermarkets, three traditional markets, and from the internet. From each sample, 2–10 pieces of packed chicken meat were collected. Chicken eggs were purchased in one local supermarket. Retail duck meat samples were purchased in two local supermarkets and one traditional market. Duck eggs were purchased in one local supermarket and from online stores. Straw bedding samples from geographically separated multiple poultry farms were collected. Swab samples positive for Salmonella, as determined by polymerase chain reaction (PCR), from eight geographically separated slaughterhouses were collected. In addition, 11 Salmonella Enteritidis strains isolated in 2011, 2012, and 2017 were kindly provided by the Avian Disease Laboratory, College of Veterinary Medicine, Konkuk University, Korea.
2.2. Salmonella Isolation
For retail chicken, duck meat, and internal organs of chicken, each sample was aseptically placed in a sterile plastic bag containing 400 mL of buffered peptone water broth (BPW, Difco, Detroit, MI, USA) and shaken for 2 min. The rinsed material (20 mL) was vortex-mixed in 20 mL of BPW for 15 s, and then incubated at 37 °C for 24 h. For straw bedding and swab samples, each sample was aseptically placed in a sterile plastic bag containing 20 mL of BPW and shaken for 15 s. The rinsed material (0.1 mL) was incubated at 37 °C for 24 h. Chicken and duck egg samples were incubated in the egg incubator for 21 and 28 days, respectively. Liver, spleen, and cecal tonsil were collected from the egg embryo. The organs were placed into a sterile plastic bag containing 20 mL of BPW and homogenized using the stomacher for 2 min. The homogenized sample (0.1 mL) was vortex-mixed in 10 mL of Rappaport-Vassiliadis broth (RV, Difco, Detroit, MI, USA) and incubated at 37 °C for 24 h. Incubated BPW (100 µL) was vortex-mixed for 15 s in 10 mL of RV and then incubated at 41.5 °C for 20 h. The presence of Salmonella spp. in the incubated RV was analyzed by PCR, as described previously [33]. Samples that yielded positive results were streaked onto Salmonella ChromoSelect agar (Sigma-Aldrich, St. Louis, MO, USA), followed by incubation at 37 °C for 24 h. Pink colonies of Salmonella spp. on the agar were validated by PCR, and positive colonies were stored at −80 °C in glycerol.
2.3. Antibiotic Susceptibility Test
Antibiotic susceptibility was determined using the Sensititre panel (KRCDC2F; Thermo Fisher Scientific, Waltham, MA, USA) with the following antibiotics: ciprofloxacin (CIP, 0.03–0.5 µg), nalidixic acid (NAL, 2–128 µg), imipenem (IMI, 1–8 µg), colistin (COL, 2–16 µg), ampicillin (AMP, 2–64 µg), tetracycline (TET, 2–128 µg), chloramphenicol (CHL, 2–32 µg), azithromycin (AZI, 2–32 µg), gentamicin (GEN, 1–64 µg), streptomycin (STR, 2–128 µg), amikacin (AMI, 4–64 µg), trimethoprim/sulfamethoxazole (SXT, 1/19–16/304), cefotaxime (FOT, 1–32 µg), ceftriaxone (AXO, 1–32 µg), cefoxitin (FOX, 4–32 µg), and ceftazidime (TAZ, 1–16 µg), according to the Clinical and Laboratory Standards Institute guidelines (Wayne, PA, USA) [34]. Briefly, 10 µL portions of Salmonella spp. strains (1 × 105 cfu/mL) cultured overnight were thoroughly mixed with 11 mL of Muller Hinton Broth with N-Tris (hydroxymethyl) methyl-2-aminoethanesulfonic acid; 50 µL portions were placed in the wells of the Sensititre panel. The panel was sealed with film, and the results were assessed manually after 24 h incubation at 37 °C. The minimum inhibitory concentration (MIC) was recorded as the lowest concentration of antibiotic that inhibited visible growth, identified as a turbidity or deposit of cells at the bottom of a wall. Escherichia coli (ATCC25922) was used as the quality control standard. Salmonella spp. resistant to more than three classes and more than one antibiotic in a single class were designated as an MDR strain.
2.4. Extraction and WGS of MDR Salmonella Genomic DNA
Genomic DNA was extracted from overnight cultured MDR Salmonella spp. using a MagAttract kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The purity and concentration of the extracted DNA were measured with a NanoDrop spectrophotometer (Thermo Fisher Scientific) and a Quantus fluorometer (Promega, Madison, WI, USA), respectively. A library was prepared for sequencing using native barcoding genomic DNA kits, and WGS sequencing was performed using the MinION system (Oxford Nanopore Technologies, Oxford, UK), as described by the respective manufacturers. The library was loaded onto FLO-MIN106 R9.4.1 flow cells and sequenced for 48 h. Data were base-called using Albacore (Oxford Nanopore Technologies). A library prepared using a TrueSeq Nano DNA instrument (Illumina, San Diego, CA, USA) was also sequenced using the HiSeq4000 system (Illumina, San Diego, CA, USA) for error correction of the nanopore sequencing results.
2.5. Assembly, Polishing, and Annotation of MDR Salmonella DNA
Reads generated from nanopore sequencing were downsampled to generate ~100× coverage depth of the Salmonella genome (4.9 Mb) using seqtk (https://github.com/lh3/seqtk; accessed on 22 September 2021). Downsampled reads were de novo assembled using the Flye algorithm [35] with default parameters. The assembled contigs were polished using unicycler_polish [36] with the Illumina fastq reads with default parameters. The assembled Salmonella genome was annotated using Prokka [37].
2.6. Data Analysis
MDR Salmonella serovar was predicted using SeqSero from assembled contigs [38]. CgMLST was determined using SeqSero. The minimum spanning tree of core genome MLST was visualized using GrapeTree [39]. Antibiotic resistance genes were identified using Resfinder [40]. SNPs between the whole genomes of the sequenced Salmonella Enteritidis strains in this study and those of the Korean Salmonella Enteritidis strains deposited in the public database were identified and aligned using kSNP3.0 [41] with the optimum kmer size 19. The genomic sequence of the Salmonella Enteritidis P125109 strain (GenBank no. NC011294) was used as the reference genome for SNP calling. A whole-genome SNP tree was constructed based on the pan SNPs generated by kSNP3.0 using RAxML, with the General Time Reversible gamma substitution model and 1000 bootstrap replicates. The phylogenetic tree with antibiotic resistance genes was visualized using the interactive Tree of Life version 5 (iTOLv5) (http://itol.embl.de/; accessed on 22 September 2021).
3. Results
3.1. Prevalence of Salmonella spp.
In total, 82 Salmonella spp. were isolated from 853 samples: 100% from swab samples from the slaughterhouses (19/19), 61% from retail duck meat (11/18), 26.7% from internal organs of chickens (8/30), 14.1% from retail chicken meat (41/291), 5.3% from straw bedding samples (2/38), and 0.5% from one-day-old chicks (1/201). No Salmonella spp. was detected from chicken and duck eggs.
3.2. Antibiotic Resistance Profiles of the Isolated Salmonella spp.
The antibiotic resistance profiles of the Salmonella spp. isolates are shown in Table 1. Among the 82 Salmonella spp. tested, 40% isolates were identified as MDR strains (33/82). All 10 Salmonella Enteritidis strains isolated in 2011, 2012, and 2017 were identified as MDR. The highest resistance rate was to ampicillin (100%, 43/43), followed by nalidixic acid (76.74%, 33/43), tetracycline (74.42, 32/43), and colistin (53.49%, 23/43). All MDR isolates were susceptible to imipenem, azithromycin, and amikacin.
Table 1.
Antibiotic susceptibility profiles of MDR Salmonella spp.
| Strain | CIP | NAL | IMI | COL | AMP | TET | CHL | AZI | GEN | STR | AMI | SXT | FOT | AXO | FOX | TAZ | Number of Resistance | Number of Antibiotics Classes | Isolation Year | Source of Isolation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Z0719SL0002 | I | R | S | R | R | R | S | S | R | S | S | S | R | R | S | R | 8 | 6 | 2019 | Internal organs of chicken |
| Z0719SL0007 | I | R | S | R | R | R | S | S | R | S | S | S | R | R | S | R | 8 | 6 | 2019 | Straw bedding samples |
| Z0719SL0011 | I | R | S | R | R | R | S | S | R | S | S | S | R | R | S | R | 8 | 6 | 2019 | Retail chicken meat |
| Z0719SL0012 | I | R | S | R | R | R | S | S | R | S | S | S | R | R | S | R | 8 | 6 | 2019 | Retail chicken meat |
| Z0719SL0013 | I | R | S | R | R | R | S | S | R | S | S | S | R | R | S | R | 8 | 6 | 2019 | Internal organs of chicken |
| Z0719SL0014 | I | R | S | R | R | R | S | S | R | S | S | S | R | R | S | R | 8 | 6 | 2019 | Retail chicken meat |
| Z0719SL0018 | I | R | S | R | R | R | S | S | R | S | S | S | R | R | S | R | 8 | 6 | 2019 | Internal organs of chicken |
| Z0720SL0031 | I | R | S | R | R | R | S | S | R | S | S | S | R | R | S | R | 8 | 6 | 2019 | Retail chicken meat |
| Z0719SL0004 | I | R | S | S | R | S | S | S | S | S | S | S | R | R | R | R | 6 | 3 | 2019 | Retail chicken meat |
| Z0719SL0001 | I | R | S | R | R | R | S | S | S | R | S | S | S | S | S | S | 5 | 5 | 2019 | Internal organs of chicken |
| Z0720SL0023 | I | R | S | R | R | R | S | S | S | R | S | S | S | S | S | S | 5 | 5 | 2019 | Internal organs of chicken |
| Z0719SL0008 | R | R | S | S | R | S | R | S | S | S | S | S | S | S | R | S | 5 | 4 | 2019 | Straw bedding samples |
| Z0719SL0009 | I | R | S | S | R | R | R | S | S | S | S | R | S | S | S | S | 5 | 5 | 2019 | Slaughterhouse |
| Z0719SL0010 | I | R | S | S | R | R | R | S | S | S | S | R | S | S | S | S | 5 | 5 | 2019 | Slaughterhouse |
| Z0719SL0021 | I | R | S | S | R | R | R | S | S | S | S | R | S | S | S | S | 5 | 5 | 2019 | Slaughterhouse |
| Z0720SL0026 | I | R | S | S | R | R | R | S | S | S | S | R | S | S | S | S | 5 | 5 | 2019 | Slaughterhouse |
| Z0719SL0003 | I | R | S | R | R | S | S | S | S | S | S | R | S | S | S | S | 4 | 4 | 2019 | Retail chicken meat |
| Z0719SL0005 | I | R | S | R | R | S | S | S | S | R | S | S | S | S | S | S | 4 | 4 | 2019 | Retail chicken meat |
| Z0719SL0017 | R | S | S | S | R | R | I | S | S | R | S | S | S | S | S | S | 4 | 4 | 2019 | Slaughterhouse |
| Z0719SL0022 | R | S | S | S | R | R | I | S | S | R | S | S | S | S | S | S | 4 | 4 | 2019 | Slaughterhouse |
| Z0720SL0029 | I | R | S | S | R | S | S | S | S | S | S | S | R | R | I | I | 4 | 3 | 2019 | Retail chicken meat |
| Z0719SL0006 | I | R | S | S | R | S | S | S | S | R | S | S | S | S | S | S | 3 | 3 | 2019 | Retail chicken meat |
| Z0719SL0015 | I | S | S | S | R | R | S | S | S | R | S | S | S | S | S | S | 3 | 3 | 2019 | Slaughterhouse |
| Z0719SL0016 | I | S | S | S | R | R | S | S | S | R | S | S | S | S | S | S | 3 | 3 | 2019 | Slaughterhouse |
| Z0719SL0019 | I | S | S | S | R | R | S | S | S | R | S | S | S | S | S | S | 3 | 3 | 2019 | Internal organs of chicken |
| Z0719SL0020 | I | S | S | S | R | R | S | S | S | R | S | S | S | S | S | S | 3 | 3 | 2019 | Slaughterhouse |
| Z0720SL0025 | I | S | S | S | R | R | S | S | S | R | S | S | S | S | S | S | 3 | 3 | 2019 | Slaughterhouse |
| Z0720SL0027 | I | S | S | S | R | R | S | S | S | R | S | S | I | S | S | S | 3 | 3 | 2019 | Slaughterhouse |
| Z0720SL0030 | I | R | S | R | R | S | S | S | S | S | S | S | S | S | S | S | 3 | 3 | 2019 | Internal organs of chicken |
| Z0720SL0028 | I | S | S | S | R | R | S | S | S | R | S | S | S | S | S | S | 3 | 3 | 2019 | Slaughterhouse |
| Z0720SL0033 | I | R | S | R | R | S | S | S | S | R | S | R | S | S | S | S | 5 | 5 | 2020 | Retail chicken meat |
| Z0720SL0034 | R | R | S | S | R | R | I | S | S | R | S | S | S | S | S | S | 5 | 4 | 2020 | Retail chicken meat |
| Z0720SL0032 | I | R | S | S | R | S | S | S | S | S | S | R | S | S | S | S | 3 | 3 | 2020 | Retail chicken meat |
| Z0720SL0035 | I | R | S | R | R | R | S | S | S | R | S | S | S | S | S | S | 5 | 5 | 2012 | Slaughterhouse |
| Z0720SL0037 | I | R | S | R | R | R | S | S | R | R | S | S | R | R | S | R | 9 | 6 | 2012 | Truck |
| Z0720SL0038 | S | S | S | S | R | R | R | S | S | R | S | R | S | S | S | S | 5 | 5 | 2012 | Truck |
| Z0720SL0039 | I | R | S | R | R | R | S | S | S | S | S | R | S | S | S | S | 5 | 5 | 2017 | Environment |
| Z0720SL0040 | I | R | S | R | R | R | S | S | S | R | S | S | S | S | S | S | 5 | 5 | 2017 | Retail chicken meat |
| Z0720SL0041 | I | R | S | R | R | S | R | S | S | R | S | S | S | S | S | S | 5 | 5 | 2017 | Environment |
| Z0720SL0042 | I | R | S | R | R | R | S | S | R | R | S | S | R | R | S | R | 9 | 6 | 2011 | Retail chicken meat |
| Z0720SL0043 | R | R | S | R | R | R | S | S | R | R | S | S | R | R | S | R | 10 | 6 | 2011 | Retail chicken meat |
| Z0720SL0044 | I | R | S | R | R | R | S | S | R | R | S | S | R | R | S | R | 9 | 6 | 2011 | Retail chicken meat |
| Z0720SL0045 | I | R | S | R | R | S | R | S | S | R | S | S | S | S | S | S | 5 | 5 | 2011 | Retail chicken meat |
Ciprofloxacin (CIP), nalidixic acid (NAL), imipenem (IMI), colistin (COL), ampicillin (AMP), tetracycline (TET), chloramphenicol (CHL), azithromycin (AZI), gentamicin (GEN), streptomycin (STR), amikacin (AMI), trimethoprim/sulfamethoxazole (SXT), cefotaxime (FOT), ceftriaxone (AXO), cefoxitin (FOX), ceftazidime (TAZ); R, resistant; S, susceptible; I, intermediate.
3.3. Results of Whole-Genome Sequencing and In-Silico Serotyping of MDR Salmonella spp.
The genomic features of the MDR Salmonella spp. are shown in Table 2. The sequence data for all MDR isolates yielded a depth of greater than 100, except for one sample with a depth of 87.5. The genome assembly generated 1–5 contigs. The size of the chromosome was 4,547,043–4,878,409 bp. MDR isolates were assigned to five serovars. Salmonella Enteritidis was the most prevalent serovar (57.78%, 26/43) followed by Salmonella Agona (15.6%, 7/43), Salmonella Virchow (13.3%, 6/43), Salmonella Albany (8.89%, 4/43), and Salmonella Bareilly (2.2%, 1/43).
Table 2.
The genomic features of the MDR Salmonella spp.
| Sample Name | Data Output (gb) |
Fold Coverage (X) | Chromosome Size (bp) | Number of Plasmid | Serovar |
|---|---|---|---|---|---|
| Z0720SL0023 | 1.8 | 375.0 | 4,783,705 | 1 | Enteritidis |
| Z0719SL0001 | 5.2 | 1083.3 | 4,783,876 | 1 | Enteritidis |
| Z0719SL0004 | 1.8 | 375.0 | 4,673,348 | 0 | Virchow |
| Z0719SL0003 | 6.4 | 1333.3 | 4,679,604 | 3 | Enteritidis |
| Z0719SL0002 | 5.9 | 1229.2 | 4,680,702 | 1 | Enteritidis |
| Z0719SL0005 | 1.75 | 364.6 | 4,779,036 | 1 | Enteritidis |
| Z0719SL0006 | 5.9 | 1229.2 | 4,779,850 | 0 | Enteritidis |
| Z0719SL0007 | 2.8 | 583.3 | 4,681,486 | 1 | Enteritidis |
| Z0719SL0008 | 2.5 | 520.8 | 4,670,331 | 2 | Virchow |
| Z0719SL0011 | 4.5 | 937.5 | 4,681,460 | 1 | Enteritidis |
| Z0719SL0012 | 4.7 | 979.2 | 4,681,475 | 1 | Enteritidis |
| Z0719SL0009 | 2.1 | 437.5 | 4,809,470 | 1 | Albany |
| Z0719SL0010 | 2.1 | 437.5 | 4,809,485 | 2 | Albany |
| Z0719SL0013 | 4.8 | 1000.0 | 4,683,147 | 1 | Enteritidis |
| Z0719SL0014 | 5.3 | 1104.2 | 4,678,918 | 1 | Enteritidis |
| Z0719SL0018 | 1 | 208.3 | 4,681,459 | 1 | Enteritidis |
| Z0719SL0019 | 7 | 1458.3 | 4,843,579 | 1 | Agona |
| Z0720SL0025 | 0.62 | 129.2 | 4,878,409 | 1 | Agona |
| Z0719SL0021 | 2.85 | 593.8 | 4,844,531 | 0 | Albany |
| Z0720SL0026 | 0.42 | 87.5 | 4,844,485 | 0 | Albany |
| Z0720SL0027 | 0.6 | 125.0 | 4,593,080 | 1 | Virchow |
| Z0720SL0028 | 2.09 | 435.4 | 4,677,146 | 2 | Virchow |
| Z0719SL0015 | 2.7 | 562.5 | 4,843,592 | 1 | Agona |
| Z0719SL0016 | 5 | 1041.7 | 4,843,581 | 1 | Agona |
| Z0719SL0017 | 4.9 | 1020.8 | 4,877,928 | 1 | Agona |
| Z0719SL0022 | 2.05 | 427.1 | 4,877,150 | 1 | Agona |
| Z0719SL0020 | 2.05 | 427.1 | 4,878,418 | 1 | Agona |
| Z0720SL0030 | 0.51 | 106.3 | 4,547,043 | 0 | Enteritidis |
| Z0720SL0031 | 0.54 | 112.5 | 4,681,359 | 4 | Enteritidis |
| Z0720SL0032 | 3.3 | 687.5 | 4,679,600 | 4 | Enteritidis |
| Z0720SL0033 | 1.05 | 218.8 | 4,679,611 | 4 | Enteritidis |
| Z0720SL0034 | 1.1 | 229.2 | 4,670,318 | 0 | Virchow |
| Z0720SL0035 | 1.31 | 272.9 | 4,680,380 | 1 | Enteritidis |
| Z0720SL0037 | 1.55 | 322.9 | 4,680,192 | 3 | Enteritidis |
| Z0720SL0038 | 1.56 | 325.0 | 4,680,091 | 2 | Enteritidis |
| Z0720SL0039 | 1.53 | 318.8 | 4,807,544 | 2 | Enteritidis |
| Z0720SL0040 | 2.55 | 531.3 | 4,782,444 | 1 | Enteritidis |
| Z0720SL0041 | 1.32 | 275.0 | 4,783,583 | 1 | Enteritidis |
| Z0720SL0042 | 1.29 | 268.8 | 4,678,693 | 1 | Enteritidis |
| Z0720SL0043 | 1.47 | 306.3 | 4,664,874 | 1 | Enteritidis |
| Z0720SL0044 | 1.36 | 283.3 | 4,680,669 | 1 | Enteritidis |
| Z0720SL0045 | 1.49 | 310.4 | 4,679,466 | 1 | Enteritidis |
3.4. Antibiotic Resistance Profiles of MDR Salmonella Enteritidis
The highest resistance observed in MDR Salmonella Enteritidis strains was to ampicillin (100%, 26/26), followed by nalidixic acid (96.15%, 25/26), colistin (88.46%, 23/26), and tetracycline (69.23%, 18/26). Resistance to third-generation cephalosporins (cefotaxime, ceftriaxone, and ceftazidime) was observed in 16 isolates (46.15%). These isolates were also resistant to nalidixic acid, colistin, ampicillin, tetracycline, and gentamicin. Isolation sources of these isolates were internal organs of chicken, retail chicken meat, straw for bedding, and farm environment. MDR Salmonella Enteritidis strains that were resistant to colistin were also resistant to ampicillin and nalidixic acid. All the sequenced Salmonella Enteritidis strains harbored antibiotic resistance genes that coincided with antibiotic resistance phenotypes except colistin resistance (Table 3). All MDR Salmonella Enteritidis strains carried the aac(6)-Iaa gene. The mobile colistin resistance (mcr) gene and chromosomal mutations related to colistin resistance were not found.
Table 3.
Antibiotic resistance patterns and antibiotic resistance genes in MDR Salmonella Enteritidis strains.
| Antibiotic Resistance | Sources of Isolation | Antibiotic Resistance Gene | No. of Isolates | No. of Antibiotics | No. of Classes |
|---|---|---|---|---|---|
| NAL-COL-AMP-TET-GEN-FOT-AXO-TAZ | Retail chicken meat | aac(6′)-Iaa_1 aac(3)-IId_1 blaCTX-M-15_1 tet(A)_6 |
3 | 8 | 6 |
| Straw for bedding | 1 | ||||
| Internal organs of chicken | 3 | ||||
| NAL-COL-AMP-TET-GEN-STR-FOT-AXO-TAZ | Retail chicken meat | aac(6′)-Iaa_1 sul2_3 aph(3″)-Ib_5 aph(6)-Id_1 aph(3′)-Ia_1 aac(3)-IId_1 blaCTX-M-15_1 tet(A)_6 |
2 | 9 | 6 |
| Truck | 1 | ||||
| CIP-NAL-COL-AMP-TET-GEN-STR-FOT-AXO-TAZ | Retail chicken meat | aac(6′)-Iaa_1 aac(3)-IId_1 sul2_3 aph(3″)-Ib_5 aph(6)-Id_1 aph(3′)-Ia_1 tet(A)_6 blaCTX-M-15_1 |
1 | 10 | 6 |
| NAL-COL-AMP-TET-STR | Retail chicken meat | aac(6′)-Iaa_1 blaTEM-1B_1 aph(6)-Id_1 aph(3″)-Ib_5 tet(A)_6 aph(6)-Id_1 aph(3″)-Ib_5 sul2_2 |
1 | 5 | 5 |
| Environment | 1 | ||||
| Internal organs of chicken | 2 | ||||
| NAL-COL-AMP-STR-SXT | Retail chicken meat | aac(6′)-Iaa_1 blaTEM-1B_1 dfrA1_10 |
1 | 5 | 5 |
| NAL-COL-AMP-CHL-STR | Slaughterhouse | aac(6′)-Iaa_1 blaTEM-1B_1 aph(6)-Id_1 aph(3′)-Ib_5 tet(A)_6 aph(6)-Id_1 aph(3″)-Ib_5 sul2_2 |
1 | 5 | 5 |
| Retail chicken meat | 1 | ||||
| NAL-COL-AMP-TET-SXT | Truck | aac(6′)-Iaa_1 blaTEM-1B_1 aph(6)-Id_1 aph(3″)-Ib_5 tet(A)_6 aph(6)-Id_1 aph(3″)-Ib_5 sul2_2 |
1 | 5 | 5 |
| AMP-TET-CHL-STR-SXT | Environment | aac(6′)-Iaa_1 catA2_1 sul2_2 aph(3″)-Ib_5 aph(6)-Id_1 blaTEM-1B_1 |
1 | 5 | 5 |
| NAL-COL-AMP-SXT | Retail chicken meat | aac(6′)-Iaa_1 blaTEM-1B_1 dfrA1_10 |
2 | 4 | 4 |
| NAL-COL-AMP-STR | Retail chicken meat | sul2_2 aph(3″)-Ib_5 aph(6)-Id_1 blaTEM-1B_1 aac(6′)-Iaa_1 |
1 | 4 | 4 |
| NAL-AMP-STR | Retail chicken meat | sul2_2 aph(3″)-Ib_5 aph(6)-Id_1 blaTEM-1B_1 aac(6′)-Iaa_1 |
1 | 3 | 3 |
| NAL-COL-AMP | Internal organs of chicken | aac(6′)-Iaa_1 | 1 | 3 | 3 |
| NAL-AMP-SXT | Retail chicken meat | dfrA1_10 blaTEM-1B_1 aac(6′)-Iaa_1 |
1 | 3 | 3 |
3.5. CgMLST
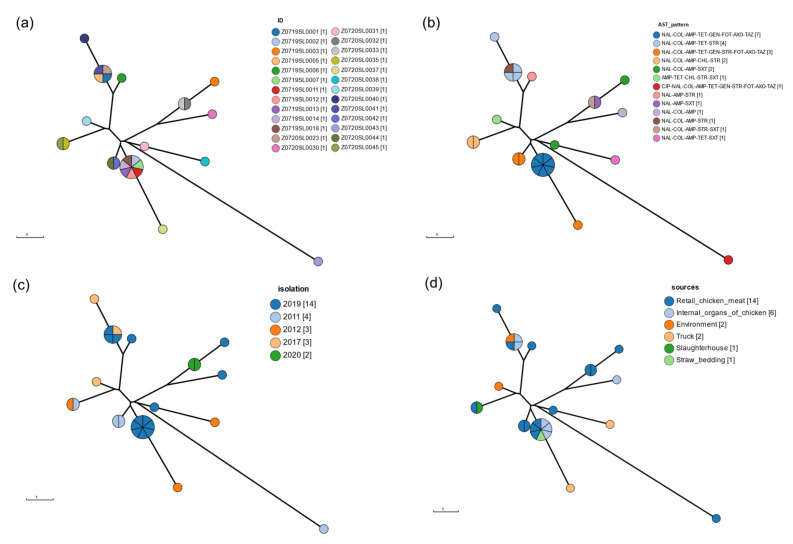
The results of CgMLST analysis showed that the MDR Salmonella Enteritidis strains isolated from different sources and years were clustered together (Figure 1). MDR Salmonella Enteritidis strains isolated in 2019 with antibiotic resistance patterns NAL-COL-AMP-TET-GEN-FOT-AXO-TAX clustered with Salmonella Enteritidis strains isolated in 2011 and 2012 with antibiotic resistance patterns NAL-COL-AMP-TET-GEN-STR-FOT-AXO-TAX. These strains were isolated from retail chicken meat, internal organs of chicken, trucks, and straw for bedding.
Figure 1.
Minimum spanning tree of CgMLST of MDR Salmonella Enteritidis. The color of the node indicates (a) isolates, (b) year of isolation, (c) source of isolation, and (d) antibiotic resistance profiles.
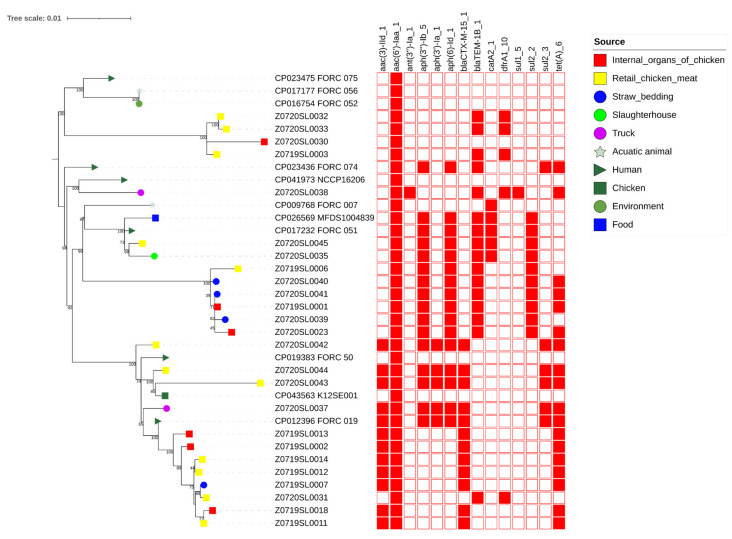
3.6. wgSNP Phylogenetic Analysis of the Salmonella Enteritidis Strains
The Salmonella Enteritidis genomes were clustered into five different groups (I to VII) with one singleton genome (Figure 2). The Salmonella Enteritidis strains in the same cluster had similar antibiotic resistance genes. Strains from different sources and different years were grouped with monophyletic clusters. In Cluster I, isolates from humans, the environment, and aquatic animals were clustered together. Four isolates from chicken meat (n = 3) and internal organs of chicken (n = 1) were grouped in Cluster II. Cluster III contained two isolates from truck and human. Cluster IV included four isolates from food, humans, a slaughterhouse, and chicken meat. Six isolates from straw bedding (n = 3), internal organs of chicken (n = 2), and chicken meat (n = 1) were clustered in Cluster V. In Cluster VI, 15 isolates from chicken meat (n = 7), chicken (n = 1), internal organs of chicken (n = 3), humans (n = 2), straw bedding (n = 1), and trucks (n = 1) were included. The Salmonella Enteritidis in Clusters II and IV carried blaTEM-1B in a plasmid, and those in Cluster V carried blaTEM-1B in a chromosome. In Cluster VI, the Salmonella Enteritidis strains carrying blaCTX-M-15 were isolated from chicken meat, internal organs of chicken, straw bedding, humans, and trucks. The Z0720SL0042, Z0720SL0043, and Z0720SL0044 strains were isolated in 2011. The Z0720SL0037 and FORC_019 strains were isolated in 2012 and 2015, respectively. The Z0719SL0002, Z0719SL0007, Z0719SL0011, Z0719SL0012, Z0719SL0013, Z0719SL0014, and Z0719SL0018 strains were isolated in 2019.
Figure 2.
wgSNPs phylogenetic tree of MDR Salmonella Enteritidis. Phylogeny was rooted at the midpoint. The presence of antibiotic resistance genes is highlighted in red. The source of isolation of each Salmonella Enteritidis is indicated.
4. Discussion
To the best of our knowledge, this is the first study that uses WGS to determine the genetic relationship among Salmonella Enteritidis strains isolated from humans and poultry sources in Korea. Previous findings obtained by PFGE and MLVA analysis of Salmonella Enteritidis strains isolated from humans and poultry sources suggest that Salmonella Enteritidis strains have already been transmitted from poultry sources to humans in Korea [14,15]. In this study, results of wgSNP analysis between Salmonella Enteritidis strains isolated from humans and poultry sources were consistent with those of the previous PFGE and MLVA studies. The wgSNP phylogenetic analysis revealed a monophyletic relationship, with the support of 100 bootstrap replicates between Salmonella Enteritidis isolated from different sources. The Salmonella Enteritidis strains in the same cluster had identical antimicrobial resistance gene patterns, indicating that vertical clonal expansion occurred rather than horizontal transmission of the antimicrobial resistance gene.
Salmonella is the most common zoonotic foodborne pathogen responsible for gastroenteritis in humans [1]. The rapid emergence of antibiotic resistance in Salmonella has been a serious public health problem worldwide [42]. The isolation of MDR Salmonella, which is resistant to not only traditional first-line antibiotics such as ampicillin, chloramphenicol, and trimethoprim but also to currently recommended antibiotics, including fluoroquinolones and extended-spectrum cephalosporins, has recently dramatically increased [8,9]. Genotypic and phenotypic antibiotic resistance of the Salmonella observed in this study was consistent with the previous findings that MDR Salmonella Enteritidis strains isolated showed a high resistance rate to antibiotics commonly used in the Korean poultry industry [5]. Because of the risk of human transmission, the high prevalence of MDR Salmonella Enteritidis in the Korean poultry food chain is concerning. MDR Salmonella isolated from poultry sources has also been reported in other countries [43,44]. Nevertheless, the prevalence of MDR strains reported herein and in a previous study [5], 50.5% and 50.9, respectively, were higher than that in Spain (9.7%) [45] and China (24.3%) [46].
Colistin was recently used as a last-resort therapeutic option for the therapy of intestinal infections in humans [47]. It is critical to monitor resistance to this agent in isolates from food-producing animals worldwide. Colistin, in turn, has been widely used in the food-animal industry in several countries for the purpose of therapeutic, prophylactic, and growth promotion [48,49]. Colistin was included in poultry formula feeds in Korea prior to 2009. Resistance to colistin can be conferred by various mechanisms, including chromosomal mutation and transmissible genetic mobile elements carrying the colistin resistance gene [50]. Lipid A modification, mediated by mutations in the pmrHFIJKLM operon, have been shown to confer resistance to colistin in Enterobacteriaceae [51]. Mutations in pmrAB and pmrLM, as well as the AcrAB efflux pump, have been shown to confer resistance in S. Typhymurium [52]. The MDR isolates in this study had no mutations in any of these genes. mcr in the plasmid confers resistance by reducing the anionic changes of lipid A, resulting in a lower binding affinity to colistin [51]. To date, 10 variants of mcr have been described [53]. The MDR isolates in this study had no mcr gene. The absence of colistin resistance, conferring mutations and plasmid-mediated colistin resistance genes in Salmonella Enteritidis, has been previously reported [54], suggesting the presence of a novel mechanism for colistin resistance.
Extended-spectrum cephalosporins (ESCs) are the first-line antibiotics for treating salmonellosis and other bacterial infections [55]. Salmonella isolates resistant to ciprofloxacin and ESC have increased in recent years [56,57]. Herein, 12 isolates, which were resistant to three third-generation cephalosporins, were clustered together in Cluster VII. Those isolates showed similar antibiotic resistance patterns (NAL-COL-AMP-TET-GEN-FOT-AXO-TAZ). Isolates collected before 2015 showed additional resistance to STR, and one isolate from 2011 showed resistance to both STR and CIP. Most of the antibiotic-resistant genes in strains within Cluster VII were located in the plasmid. MDR Salmonella Enteritidis strains in this clade were isolated from various sources and different years. This is a serious public health problem because the vertical clonal expansion of those strains has occurred and been transferred to humans through contaminated food sources.
In conclusion, this study reveals the high prevalence of MDR Salmonella in poultry sources in Korea. Considering the location of the antibiotic resistance genes (mainly in the plasmid), analysis of how these plasmids evolve is still warranted to further elucidate the epidemiological emergence of MDR Salmonella spp.
Author Contributions
Conceptualization, T.-M.L.; methodology, H.-J.L. and T.K.; software, T.-M.L.; validation, T.-M.L.; formal analysis, T.-M.L.; investigation, J.-B.L., S.-Y.P., I.-S.C., S.-W.L.; writing—original draft preparation, T.-M.L.; writing—review and editing, J.-B.L., S.-Y.P., I.-S.C., S.-W.L.; visualization, T.-M.L.; supervision, J.-B.L., S.-Y.P., I.-S.C., S.-W.L.; funding acquisition, S.-W.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded (2019ER540501) by the Korea Centers for Disease Control and Prevention.
Institutional Review Board Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Informed Consent Statement
Not applicable.
Data Availability Statement
The complete genomes of Salmonella spp. in this study are available from the GenBank under BioProjects PRJNA658425 and PRJNA780385.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dewey-Mattia D., Manikonda K., Hall A.J., Wise M.E., Crowe S.J. Surveillance for Foodborne Disease Outbreaks—United States, 2009–2015. MMWR Surveill. Summ. 2018;67:1–11. doi: 10.15585/mmwr.ss6710a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eng S.-K., Pusparajah P., Mutalib N.-S.A., Ser H.-L., Chan K.-G., Lee L.-H. Salmonella: A Review on Pathogenesis, Epidemiology and Antibiotic Resistance. Front. Life Sci. 2015;8:284–293. doi: 10.1080/21553769.2015.1051243. [DOI] [Google Scholar]
- 3.Omwandho C.O.A., Kubota T. Salmonella Enterica Serovar Enteritidis: A Mini-Review of Contamination Routes and Limitations to Effective Control. Jpn. Agric. Res. Q. 2010;44:7–16. doi: 10.6090/jarq.44.7. [DOI] [Google Scholar]
- 4.Hogue A., White P., Guard-Petter J., Schlosser W., Gast R., Ebel E., Farrar J., Gomez T., Madden J., Madison M., et al. Epidemiology and Control of Egg-Associated Salmonella Enteriditis in the United States of America. Rev. Sci. Tech. 1997;16:542–553. doi: 10.20506/rst.16.2.1045. [DOI] [PubMed] [Google Scholar]
- 5.Sin M., Yoon S., Kim Y.B., Noh E.B., Seo K.W., Lee Y.J. Molecular Characteristics of Antimicrobial Resistance Determinants and Integrons in Salmonella Isolated from Chicken Meat in Korea. J. Appl. Poult. Res. 2020;29:502–514. doi: 10.1016/j.japr.2019.12.010. [DOI] [Google Scholar]
- 6.Choi S.-W., Ha J.-S., Kim B.-Y., Lee D.-H., Park J.-K., Youn H.-N., Hong Y.-H., Lee S.-B., Lee J.-B., Park S.-Y., et al. Prevalence and Characterization of Salmonella Species in Entire Steps of a Single Integrated Broiler Supply Chain in Korea. Poult. Sci. 2014;93:1251–1257. doi: 10.3382/ps.2013-03558. [DOI] [PubMed] [Google Scholar]
- 7.Seo K.W., Lee Y.J. Prevalence and Characterization of β-Lactamases Genes and Class 1 Integrons in Multidrug-Resistant Escherichia coli Isolates from Chicken Meat in Korea. Microb. Drug Resist. 2018;24:1599–1606. doi: 10.1089/mdr.2018.0019. [DOI] [PubMed] [Google Scholar]
- 8.Yan M., Li X., Liao Q., Li F., Zhang J., Kan B. The Emergence and Outbreak of Multidrug-Resistant Typhoid Fever in China. Emerg. Microbes Infect. 2019;5:1–6. doi: 10.1038/emi.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crump J.A., Mintz E.D. Global Trends in Typhoid and Paratyphoid Fever. Clin. Infect. Dis. 2010;50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamang M.D., Nam H.-M., Kim T.-S., Jang G.-C., Jung S.-C., Lim S.-K. Emergence of Extended-Spectrum β-Lactamase (CTX-M-15 and CTX-M-14)-Producing Nontyphoid Salmonella with Reduced Susceptibility to Ciprofloxacin among Food Animals and Humans in Korea. J. Clin. Microbiol. 2011;49:2671–2675. doi: 10.1128/JCM.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dessie H.K., Bae D.H., Lee Y.J. Characterization of Integrons and Their Cassettes in Escherichia coli and Salmonella Isolates from Poultry in Korea. Poult. Sci. 2013;92:3036–3043. doi: 10.3382/ps.2013-03312. [DOI] [PubMed] [Google Scholar]
- 12.Hyeon J.-Y., Chon J.-W., Hwang I.-G., Kwak H.-S., Kim M.-S., Kim S.-K., Choi I.-S., Song C.-S., Park C., Seo K.-H. Prevalence, Antibiotic Resistance, and Molecular Characterization of Salmonella Serovars in Retail Meat Products. J. Food Prot. 2016;74:161–166. doi: 10.4315/0362-028X.JFP-10-327. [DOI] [PubMed] [Google Scholar]
- 13.Boxrud D., Pederson-Gulrud K., Wotton J., Medus C., Lyszkowicz E., Besser J., Bartkus J.M. Comparison of Multiple-Locus Variable-Number Tandem Repeat Analysis, Pulsed-Field Gel Electrophoresis, and Phage Typing for Subtype Analysis of Salmonella Enterica Serotype Enteritidis. J. Clin. Microbiol. 2006;45:536–543. doi: 10.1128/JCM.01595-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee D.-H., Hyeon J.-Y., Kim J., Kim J.S., Kim S.J., Jeon S.-E., Choi S.-W., Hong W.-T., Song C.-S., Lee S.-W. Close Genetic Relationship between Salmonella Enterica Serovar Enteritidis Isolated from Patients with Diarrhoea and Poultry in the Republic of Korea. Clin. Microbiol. Infect. 2015;21:e68–e70. doi: 10.1016/j.cmi.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 15.Kang M.-S., Oh J.-Y., Kwon Y.-K., Lee D.-Y., Jeong O.-M., Choi B.-K., Youn S.-Y., Jeon B.-W., Lee H.-J., Lee H.-S. Public Health Significance of Major Genotypes of Salmonella Enterica Serovar Enteritidis Present in Both Human and Chicken Isolates in Korea. Res. Vet. Sci. 2017;112:125–131. doi: 10.1016/j.rvsc.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Allard M.W., Luo Y., Strain E., Pettengill J., Timme R., Wang C., Li C., Keys C.E., Zheng J., Stones R., et al. On the Evolutionary History, Population Genetics and Diversity among Isolates of Salmonella Enteritidis PFGE Pattern JEGX01.0004. PLoS ONE. 2013;8:e55254. doi: 10.1371/journal.pone.0055254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saeed A.M., Walk S.T., Arshad M., Whittam T.S. Clonal Structure and Variation in Virulence of Salmonella Enteritidis Isolated from Mice, Chickens, and Humans. J. AOAC Int. 2006;89:504–511. doi: 10.1093/jaoac/89.2.504. [DOI] [PubMed] [Google Scholar]
- 18.Taylor A.J., Lappi V., Wolfgang W.J., Lapierre P., Palumbo M.J., Medus C., Boxrud D. Characterization of Foodborne Outbreaks of Salmonella Enterica Serovar Enteritidis with Whole-Genome Sequencing Single Nucleotide Polymorphism-Based Analysis for Surveillance and Outbreak Detection. J. Clin. Microbiol. 2015;53:3334–3340. doi: 10.1128/JCM.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson B.R., Tarr C., Strain E., Jackson K.A., Conrad A., Carleton H., Katz L.S., Stroika S., Gould L.H., Mody R.K., et al. Implementation of Nationwide Real-Time Whole-Genome Sequencing to Enhance Listeriosis Outbreak Detection and Investigation. Clin. Infect. Dis. 2016;63:380–386. doi: 10.1093/cid/ciw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Den Bakker H.C., Allard M.W., Bopp D., Brown E.W., Fontana J., Iqbal Z., Kinney A., Limberger R., Musser K.A., Shudt M., et al. Rapid Whole-Genome Sequencing for Surveillance of Salmonella Enterica Serovar Enteritidis. Emerg. Infect. Dis. 2014;20:1306–1314. doi: 10.3201/eid2008.131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown E., Dessai U., McGarry S., Gerner-Smidt P. Use of Whole-Genome Sequencing for Food Safety and Public Health in the United States. Foodborne Pathog. Dis. 2019;16:441–450. doi: 10.1089/fpd.2019.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Den Bakker H.C., Switt A.I.M., Cummings C.A., Hoelzer K., Degoricija L., Rodriguez-Rivera L.D., Wright E.M., Fang R., Davis M., Root T., et al. A Whole-Genome Single Nucleotide Polymorphism-Based Approach to Trace and Identify Outbreaks Linked to a Common Salmonella Enterica Subsp. Enterica Serovar Montevideo Pulsed-Field Gel Electrophoresis Type. Appl. Environ. Microbiol. 2011;77:8648–8655. doi: 10.1128/AEM.06538-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiden M.C.J., Bygraves J.A., Feil E., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., et al. Multilocus Sequence Typing: A Portable Approach to the Identification of Clones within Populations of Pathogenic Microorganisms. Proc. Natl. Acad. Sci. USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann M., Zhao S., Pettengill J., Luo Y., Monday S.R., Abbott J., Ayers S.L., Cinar H.N., Muruvanda T., Li C., et al. Comparative Genomic Analysis and Virulence Differences in Closely Related Salmonella Enterica Serotype Heidelberg Isolates from Humans, Retail Meats, and Animals. Genome Biol. Evol. 2014;6:1046–1068. doi: 10.1093/gbe/evu079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bratcher H.B., Corton C., Jolley K.A., Parkhill J., Maiden M.C. A Gene-by-Gene Population Genomics Platform: De Novo Assembly, Annotation and Genealogical Analysis of 108 Representative Neisseria meningitidis Genomes. BMC Genom. 2014;15:1138. doi: 10.1186/1471-2164-15-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leekitcharoenphon P., Hendriksen R.S., Hello S.L., Weill F.-X., Baggesen D.L., Jun S.-R., Ussery D.W., Lund O., Crook D.W., Wilson D.J., et al. Global Genomic Epidemiology of Salmonella Enterica Serovar Typhimurium DT104. Appl. Environ. Microbiol. 2016;82:2516–2526. doi: 10.1128/AEM.03821-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng X., Desai P.T., den Bakker H.C., Mikoleit M., Tolar B., Trees E., Hendriksen R.S., Frye J.G., Porwollik S., Weimer B.C., et al. Genomic Epidemiology of Salmonella Enterica Serotype Enteritidis Based on Population Structure of Prevalent Lineages. Emerg. Infect. Dis. 2014;20:1481–1489. doi: 10.3201/eid2009.131095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toro M., Retamal P., Ayers S., Barreto M., Allard M., Brown E.W., Gonzalez-Escalona N. Whole-Genome Sequencing Analysis of Salmonella Enterica Serovar Enteritidis Isolates in Chile Provides Insights into Possible Transmission between Gulls, Poultry, and Humans. Appl. Environ. Microbiol. 2016;82:6223–6232. doi: 10.1128/AEM.01760-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearce M.E., Alikhan N.-F., Dallman T.J., Zhou Z., Grant K., Maiden M.C.J. Comparative Analysis of Core Genome MLST and SNP Typing within a European Salmonella Serovar Enteritidis Outbreak. Int. J. Food Microbiol. 2018;274:1–11. doi: 10.1016/j.ijfoodmicro.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park A.K., Shin E., Kim S., Park J., Jeong H.J., Chun J.-H., Hwang K.J., Kim J. Traveller-Associated High-Level Ciprofloxacin-Resistant Salmonella Enterica Serovar Kentucky in the Republic of Korea. J. Glob. Antimicrob. Resist. 2020;22:190–194. doi: 10.1016/j.jgar.2019.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Kim S., Kim E., Park S., Hahn T.-W., Yoon H. Genomic Approaches for Understanding the Characteristics of Salmonella Enterica subsp. Enterica Serovar Typhimurium ST1120, Isolated from Swine Feces in Korea. J. Microbiol. Biotechnol. 2017;27:1983–1993. doi: 10.4014/jmb.1708.08027. [DOI] [PubMed] [Google Scholar]
- 32.Hyeon J.-Y., Li S., Mann D.A., Zhang S., Kim K.-J., Lee D.-H., Deng X., Song C.-S. Whole-Genome Sequencing Analysis of Salmonella Enterica Serotype Enteritidis Isolated from Poultry Sources in South Korea, 2010–2017. Pathogens. 2021;10:45. doi: 10.3390/pathogens10010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halatsi K., Oikonomou I., Lambiri M., Mandilara G., Vatopoulos A., Kyriacou A. PCR Detection of Salmonella spp. Using Primers Targeting the Quorum Sensing Gene sdiA. FEMS Microbiol. Lett. 2006;259:201–207. doi: 10.1111/j.1574-6968.2006.00266.x. [DOI] [PubMed] [Google Scholar]
- 34.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2019. CLSI Document M100-S29. [Google Scholar]
- 35.Kolmogorov M., Yuan J., Lin Y., Pevzner P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 36.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seemann T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 38.Zhang S., Yin Y., Jones M.B., Zhang Z., Kaiser B.L.D., Dinsmore B.A., Fitzgerald C., Fields P.I., Deng X. Salmonella Serotype Determination Utilizing High-Throughput Genome Sequencing Data. J. Clin. Microbiol. 2015;53:1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z., Alikhan N.-F., Sergeant M.J., Luhmann N., Vaz C., Francisco A.P., Carriço J.A., Achtman M. GrapeTree: Visualization of Core Genomic Relationships among 100,000 Bacterial Pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardner S.N., Slezak T., Hall B.G. KSNP3.0: SNP Detection and Phylogenetic Analysis of Genomes without Genome Alignment or Reference Genome. Bioinformatics. 2015;31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 42.Su L.-H., Chiu C.-H., Chu C., Ou J.T. Antimicrobial Resistance in Nontyphoid Salmonella Serotypes: A Global Challenge. Clin. Infect. Dis. 2004;39:546–551. doi: 10.1086/422726. [DOI] [PubMed] [Google Scholar]
- 43.De Oliveira S.D., Flores F.S., dos Santos L.R., Brandelli A. Antimicrobial Resistance in Salmonella Enteritidis Strains Isolated from Broiler Carcasses, Food, Human and Poultry-Related Samples. Int. J. Food Microbiol. 2005;97:297–305. doi: 10.1016/j.ijfoodmicro.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 44.Thung T.Y., Mahyudin N.A., Basri D.F., Radzi C.W.J.W.M., Nakaguchi Y., Nishibuchi M., Radu S. Prevalence and Antibiotic Resistance of Salmonella Enteritidis and Salmonella Typhimurium in Raw Chicken Meat at Retail Markets in Malaysia. Poult. Sci. 2016;95:1888–1893. doi: 10.3382/ps/pew144. [DOI] [PubMed] [Google Scholar]
- 45.Lamas A., Fernandez-No I.C., Miranda J.M., Vázquez B., Cepeda A., Franco C.M. Prevalence, Molecular Characterization and Antimicrobial Resistance of Salmonella Serovars Isolated from Northwestern Spanish Broiler Flocks (2011–2015) Poult. Sci. 2016;95:2097–2105. doi: 10.3382/ps/pew150. [DOI] [PubMed] [Google Scholar]
- 46.Cui M., Xie M., Qu Z., Zhao S., Wang J., Wang Y., He T., Wang H., Zuo Z., Wu C. Prevalence and Antimicrobial Resistance of Salmonella Isolated from an Integrated Broiler Chicken Supply Chain in Qingdao, China. Food Control. 2016;62:270–276. doi: 10.1016/j.foodcont.2015.10.036. [DOI] [Google Scholar]
- 47.Ahmed M.A.E.-G.E.-S., Zhong L.-L., Shen C., Yang Y., Doi Y., Tian G.-B. Colistin and Its Role in the Era of Antibiotic Resistance: An Extended Review (2000–2019) Emerg. Microbes Infect. 2020;9:868–885. doi: 10.1080/22221751.2020.1754133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhouma M., Beaudry F., Thériault W., Letellier A. Colistin in Pig Production: Chemistry, Mechanism of Antibacterial Action, Microbial Resistance Emergence, and One Health Perspectives. Front. Microbiol. 2016;7:1789. doi: 10.3389/fmicb.2016.01789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poirel L., Jayol A., Nordmann P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun S., Negrea A., Rhen M., Andersson D.I. Genetic Analysis of Colistin Resistance in Salmonella Enterica Serovar Typhimurium. Antimicrob. Agents Chemother. 2009;53:2298–2305. doi: 10.1128/AAC.01016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekyere J.O., Govinden U., Bester L.A., Essack S.Y. Colistin and Tigecycline Resistance in Carbapenemase-producing Gram-negative Bacteria: Emerging Resistance Mechanisms and Detection Methods. J. Appl. Microbiol. 2016;121:601–617. doi: 10.1111/jam.13169. [DOI] [PubMed] [Google Scholar]
- 52.Morales A.S., de Araújo J.F., de Moura Gomes V.T., Costa A.T.R., dos Prazeres Rodrigues D., Ferreira T.S.P., de Lima Filsner P.H.N., Felizardo M.R., Moreno A.M. Colistin Resistance in Escherichia coli and Salmonella Enterica Strains Isolated from Swine in Brazil. Sci. World J. 2012;2012:109795. doi: 10.1100/2012/109795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C., Feng Y., Liu L., Wei L., Kang M., Zong Z. Identification of Novel Mobile Colistin Resistance Gene mcr-10. Emerg. Microbes Infect. 2020;9:508–516. doi: 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rule R., Mbelle N., Sekyere J.O., Kock M., Hoosen A., Said M. A Rare Case of Colistin-Resistant Salmonella Enteritidis Meningitis in an HIV-Seropositive Patient. BMC Infect. Dis. 2019;19:806. doi: 10.1186/s12879-019-4391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White D.G., Zhao S., Sudler R., Ayers S., Friedman S., Chen S., McDermott P.F., McDermott S., Wagner D.D., Meng J. The Isolation of Antibiotic-Resistant Salmonella from Retail Ground Meats. N. Engl. J. Med. 2001;345:1147–1154. doi: 10.1056/NEJMoa010315. [DOI] [PubMed] [Google Scholar]
- 56.Williamson D.A., Lane C.R., Easton M., Valcanis M., Strachan J., Veitch M.G., Kirk M.D., Howden B.P. Increasing Antimicrobial Resistance in Nontyphoidal Salmonella Isolates in Australia from 1979 to 2015. Antimicrob. Agents Chemother. 2018;62:e02012-17. doi: 10.1128/AAC.02012-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trongjit S., Angkititrakul S., Tuttle R.E., Poungseree J., Padungtod P., Chuanchuen R. Prevalence and Antimicrobial Resistance in Salmonella Enterica Isolated from Broiler Chickens, Pigs and Meat Products in Thailand-Cambodia Border Provinces. Microbiol. Immunol. 2017;61:23–33. doi: 10.1111/1348-0421.12462. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genomes of Salmonella spp. in this study are available from the GenBank under BioProjects PRJNA658425 and PRJNA780385.