Abstract
The Cln3-Cdc28 kinase is required to activate the Swi4-Swi6 transcription complex which induces CLN1 and CLN2 transcription in late G1 and drives the transition to S. Cln3 and Swi4 are both rate limiting for G1 progression, and they are coordinately transcribed to peak at the M/G1 boundary. Early cell cycle box (ECB) elements, which confer M/G1-specific transcription, have been found in both promoters, and elimination of all ECB elements from the CLN3 promoter causes both a loss of periodicity and Cln3-deficient phenotypes, which include an extended G1 interval and increased cell volume. Mutants lacking the ECB elements in both the CLN3 and SWI4 promoters have low and deregulated levels of CLN transcripts, and the G1-to-S transition for these mutants is delayed and highly variable. These observations support the view that the coordinated rise of Cln3 and Swi4 levels mediated by ECB-dependent transcription controls the timing of the G1-to-S phase transition.
Cell cycle-regulated transcription is a motive force for the transitions into and out of G1 in Saccharomyces cerevisiae. The M-to-G1 transition involves elimination of the mitotic cyclins (B-type cyclins [Clbs]). This occurs, in part, through targeted proteolysis of these B-type cyclins by the anaphase-promoting complex (APC) (45). However, even if they are not degraded, the transition to G1 still occurs (1, 35). This is due to the cessation of CLB transcription in late M phase and the burst of M/G1-specific transcription of SIC1 (16), a potent Clb-kinase inhibitor (36). Sic1 and APC activity persist through G1 (1), resulting in the Clb-Cdk-deficient state that is characteristic of G1 and required to set up prereplication complexes on the DNA (29).
Exit from G1 also requires a wave of transcription of CLN1 and CLN2, CLB5, and CLB. Clb5 and Clb6 are initially inactive, but the Cln-Cdk complexes are unaffected by Sic1 and the APC. The latter serve to target Sic1 for degradation (34, 41) and may also inactivate the APC (1), which in turn enables the Clb-Cdk complexes to form and promote the transition into S phase. The third G1 cyclin, Cln3, is required to activate this late G1 wave of cyclin transcription (10, 40, 42). The direct target of Cln3-Cdk activation is unknown, but the G1-specific promoter elements in CLN1 (27) and CLN2 (39) and the two transcription factors (Swi4 and Swi6) that activate these elements have been identified (2, 3, 26). However, neither the timing nor the mechanism by which Cln3-Cdk activity evokes this abrupt transcriptional induction in late G1 has been explained.
Cln3 is unique among the cyclins in that it does not undergo the same radical oscillations in transcript levels as the others do (43). This observation has led many investigators to view the Cln3-Cdk as constitutively active and has left open the question of how a constant kinase activity can trigger the rapid induction of CLN1 and CLN2 transcription in late G1 that is associated with the transition to S phase. Early studies suggested that a threshold level of Cln3-Cdk might initiate CLN1 and CLN2 transcription and thus more Cln-Cdk activity, which would then provide positive feedback and induce more CLN transcription (9, 11). Later, it was shown that Cln1 and Cln2 play no discernible role in their own activation (10, 19, 40), so that model cannot be correct. More recently, investigators have postulated that CLN3 expression is constitutive but that as G1 cells grow their nucleocytoplasmic ratio decreases and their translational capacity increases (5, 12). This could raise the level of Cln3 in the late G1 nucleus above the threshold required to start the cell cycle, but it is difficult to see how such a gradual increase in Cln3-Cdk activity could give rise to the rapid induction of late G1 transcription that is observed.
The activities of the Cln3 protein and the Cln3-Cdc28 kinase have been difficult to measure due to the instability of the Cln3 protein (8, 44), but a modest oscillation of Cln3 protein through the cell cycle is evident from the results of Western analysis of epitope-tagged Cln3 (42). There is also a reproducible three- to fivefold oscillation in CLN3 mRNA in late M/early G1, just before Cln3 is required to activate transcription of its target genes (21). Swi4, a component of the late G1 transcription complex activated by Cln3-Cdk, is also periodically expressed and peaks during the M/G1 interval, and both genes contain M/G1-specific promoter elements called early cell cycle boxes (ECBs). CLN3 and SWI4 heterozygotes delay the G1-to-S transition, indicating that even twofold drops in their gene dosages disrupt normal G1 progression (21). This haploinsufficiency led us to infer that even the modest transcriptional increase observed for CLN3 during late M and throughout early G1 could have a significant impact upon the timing of the transition to S phase.
To test this hypothesis, we eliminated ECB elements from the CLN3 promoter and showed that they are responsible for most of the periodicity of the CLN3 transcript. We then characterized the impact of the loss of ECB-mediated activation of CLN3 and/or SWI4 transcription on the start of the cell cycle. Our data support the view that ECB elements mediate a coordinated increase in the synthesis of these two rate-limiting gene products to a level which triggers the start of the cell cycle.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions.
All strains used were derived from W303 and are listed in Table 1; one-step gene replacements (33), lithium acetate transformations (14), or genetic crosses (37) were used to construct the derivatives. DNA integrations and replacements were confirmed by PCR (23), and site-directed mutations (18) were sequenced before and after transplacement. Unless noted otherwise, cultures were grown at 30°C in YEP (37) medium supplemented with adenine (55 mg/liter) and containing 2% galactose (YEPGal) or glucose (YEPGlu) as the sole carbon source. Synthetic minimal media were as described previously (37). Elutriations from late-log-phase cultures (<5 × 107 cells/ml) were done as described previously (15) with isolated daughter-cell fractions subsequently grown in fresh medium.
TABLE 1.
S. cerevisiae strains generated in this study
| Straina | Genotype |
|---|---|
| BY2125 | aade2-1 his3-11,15 leu2-3,112 trp1-1 ura3 can1-100 ssdl-d |
| BY2270 | aade2-1 his3-11,15 leu2-3,112 trp1-1 ura3 can1-100 ssdl-d Δcln3::URA3 |
| BY2278 | aade2-1 his3-11,15 leu2-3,112 trp1-1 ura3 can1-100 ssd1-d cln3ecb5 |
| BY2679∗ | α ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3 can1-100 ssd1-d |
| BY2680∗ | aade2-1 his3-11,15 leu2-3,112 trp1-1 ura3 can1-100 ssd1-d cln3ecb5 swi4ecb |
| BY2681∗ | aade2-1 his3-11,15 leu2-3,112 trp1-1 ura3 can1-100 ssd1-d cln3ecb5 |
| BY2682∗ | α ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3 can1-100 ssd1-d swi4ecb |
| BY2684 | aade2-1 his3-11,15 leu2-3,112 trp1-1 ura3 can1-100 ssd1-d swi4ecb |
| BY2690 | aade2-1 his3-11,15 leu2-3,112 trp1-1 ura3 can1-100 ssd1-d cln3ecb6 |
| BY2785 | α ade2-1 his3-11,15 leu2-3,112::LEU2 trp1-1 ura3 can1-100 ssd1-d |
| BY2786 | α ade2-1 his3-11,15 leu2-3,112::LEU2 MET3:CLN2 trp1-1 ura3 can1-100 ssd1-d |
| BY2787 | aade2-1 his3-11,15 leu2-3,112::LEU2 trp1-1 ura3 can1-100 ssd1-d cln3ecb5 swi4ecb |
| BY2788 | aade2-1 his3-11,15 leu2-3,112::LEU2 MET3::CLN2 trp1-1 ura3 can1-100 ssd1-d cln3ecb5 swi4ecb |
Strains marked with an asterisk are sister spores from a single tetrad.
The swi4ecb (here and throughout, the suffix -ecb occurring with a genotypic designation indicates the mutation of an ECB element encoded by the gene [in this case, swi4]; if the suffix also includes a number [e.g., -ecb5 or -ecb6], the number indicates how many ECB elements have been mutated) mutant has been described previously (21); the cln3ecb mutants have either five or six putative ECB elements replaced with mutant forms, as shown in Fig. 1. These mutations were introduced into the CLN3 promoter by recombining them into a strain (BY2270) in which the CLN3 promoter (positions −1028 to −414 [positions numbered from the translational start]) was deleted and replaced with the URA3 gene. BY2270 was constructed by introducing an EcoRI site at position −1028 into the CLN3 promoter in pBD1865 to produce pBD2175. This DNA was cut with EcoRI and XhoI (position −414), and a URA3 fragment with like ends from pBD1918 was inserted. The cln3::URA3 DNA construct was used to replace the CLN3 locus of BY2125 and generate BY2270. BY2270 produces no detectable CLN3 mRNA and was used as our cln3 null strain. Replacement with the ECB mutant promoter sequences was carried out by a series of site-directed mutageneses of pBD1865; then these DNAs were used to replace the cln3::URA3 locus of BY2270. Candidates were sequenced. The cln3ecb5 swi4ecb strain was generated by crossing the cln3ecb5 and swi4ecb strains and sequenced to verify that the mutations had been maintained. The plasmid carrying CLN2 under MET3 promoter control (pDS846) was a kind gift from D. Stuart and C. Wittenberg. This plasmid was integrated into BY2679 and BY2680 at leu2 to produce BY2786 and BY2788, respectively. Leu+ control transformants (BY2785 and BY2787) were made by integrating pRS305 (38) at the leu2 locus.
FIG. 1.
CLN3 promoter region showing the positions of the ECB elements (solid boxes) and the glucose response elements (GREs) (open boxes). Below and to the left the sequence alignment of the six potential ECB elements of the CLN3 promoter is shown. Bases identical to the consensus sequence are shaded. To the right the mutations (in italics and lowercase) that have been introduced to prevent Mcm1 binding and to inactivate the ECB elements are shown. The boldface type indicates key residues for Mcm1 binding. uORF, untranslated ORF.
Synchronies and Northern blots
For analysis of cell cycle-regulated transcripts, cells were synchronized either by α-factor treatment (4) or by elutriation (15) and, during outgrowth, samples were removed for isolation of RNA. Aliquots (10 μg) of total RNA were run on 1% agarose gels containing 0.6 M formaldehyde in 40 mM morpholinepropanesulfonic acid (MOPS)-acetate buffer, pH 7.2, transferred to nylon membranes, and hybridized with probes specific for CLN3, SWI4, CLN2, CLN1, HO, and ACT1. Hybridization intensities were quantified with a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.), and the results were normalized to the ACT1 signal.
Cell sizing, fluorescence-activated cell sorter (FACS) analysis, and budding index
Median cell volumes (in femtoliters) were determined for log-phase cultures (5 × 106 to 2 × 107 cells/ml) with a particle counter and size analyzer (model Z2) and AccuComp software (both provided by Coulter Corp., Miami, Fla.). Culture aliquots were washed and resuspended in 2 mM EDTA, sonicated lightly to disperse aggregates, and diluted to 0.5 × 105 to 2.0 × 105 cells/ml into Isoton II diluent (Coulter Corp.) for analysis.
Cell samples for FACS analysis were fixed overnight in 67% ethanol at room temperature, washed with 50 mM Tris-HCl (pH 7.8), incubated in 50 mM Tris-HCl containing 0.2 mg of RNase A/ml at 37°C for 2 to 4 h, and resuspended in a solution containing 200 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 78 mM MgCl2. After addition of propidium iodide (final concentration, 30 μg/ml), the samples were sonicated lightly and analyzed on a FACS Calibur flow cytometer (Becton-Dickinson, San Jose, Calif.). The percentage of G1 cells in the population was determined using MultiCycle software (Phoenix Flow Systems, San Diego, Calif.)
To obtain the budding index, cell growth was arrested with 0.2% sodium azide, the samples were held overnight and sonicated, and then cells were counted microscopically (at least 200 cells were scored per sample). Bud scars on mother cells were visualized by Calcofluor staining (31).
RESULTS
ECB elements confer a coordinated burst of M/G1-specific transcription of CLN3 and SWI4.
ECB elements were first identified in the SWI4 promoter and shown to be sufficient to confer M/G1-specific transcription to a heterologous transcript (21). There are six sequences in the promoter region of the CLN3 gene (Fig. 1) that have various degrees of homology to the 16-bp ECB consensus sequence originally identified. The first four putative ECBs are clustered approximately 1 kb upstream of the translational start. Two more potential ECBs are located about 0.5 kb downstream from these. Previous experiments (21) showed that DNA fragments including sequences from positions −994 to −439 confer M/G1-specific transcription to a heterologous transcript. Moreover, mutations that disrupt three of the clustered sites (sites 1, 3, and 4) eliminate cell cycle regulation of the same reporter construct. This result suggests that sites 2, 5, and 6 are not functional, at least in this heterologous context. However, the activities of individual ECBs have not been investigated in their native locations. Rather, we mutated all six possible ECB elements (Fig. 1) in order to assess their role in the regulation of CLN3 transcription. The CLN3 promoter also contains five A2GA5 sequences (glucose response elements), which have been reported to induce CLN3 transcription in response to glucose (28). CLN3 transcription starts at about position −350, and its untranslated leader sequence contains an upstream open reading frame. There is evidence that this upstream open reading frame imparts an additional level of translational regulation to the CLN3 message (30).
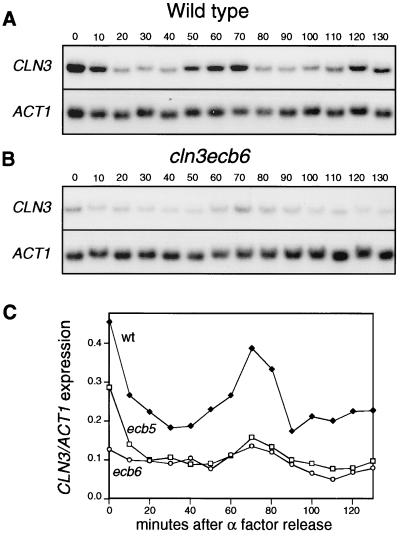
To see if ECB elements are responsible for the M/G1-specific transcription of CLN3, we synchronized wild-type and ECB mutant cells and measured RNA levels through the cell cycle. To avoid glucose-mediated effects on CLN3 transcription, we grew the cells in rich medium containing galactose as the carbon source. Wild-type cells clearly exhibited the same oscillating pattern of CLN3 mRNA levels observed with glucose-grown cells (Fig. 2A), and CLN3 mRNA levels peaked 10 min before the CLN2 mRNA levels did (reference 21 and data not shown). In contrast, strains with mutations in either the first five or all six of the potential ECB elements displayed dramatically reduced transcript levels throughout the cell cycle. These cells retained a modest but reproducible peak of CLN3 mRNA coincident with the ECB-induced peak, which suggests that there may be another unrecognized promoter element or a component of posttranscriptional control which makes a minor contribution to CLN3 expression. However, this non-ECB-mediated induction never exceeded the trough level of CLN3 transcript from a wild-type cell, so its significance is unclear.
FIG. 2.
ECB elements are important for cell cycle regulation of CLN3. (A and B) Cycling of CLN3 mRNA in wild type (BY2125) and cln3ecb6 mutant (BY2690), respectively. Cells were grown in YEPGal and synchronized with α-factor. Cells were released into fresh medium at time point 0 and samples were taken every 10 min, as indicated by the numbers above the lanes. The expression of CLN3 and ACT1 was analyzed by Northern blot hybridizations. The blots were hybridized at the same time to the same probes, and exposure times were identical. (C) CLN3 transcript levels in the wild-type strain (BY2125) (closed squares) and cln3ecb5 (BY2278) (open squares) and cln3ecb6 (BY2690) (open circles) mutant strains grown in parallel were measured throughout the cell cycle, and the results of Northern blotting were quantitated with a PhosphorImager.
We also noted that mutation of the sixth potential ECB had no impact on the transcription pattern through the cell cycle. This provides further evidence that this poor match to the consensus is not an active ECB. Thus, all subsequent experiments were carried out with a strain carrying mutations in the first five putative ECBs, and this multiple mutant is referred to as cln3 ecb.
ECB-regulated transcription of SWI4 and CLN3 affects the length of G1 and size control.
Since Cln3 and Swi4 are rate-limiting activators of the transition to S phase (7, 21, 24) and their transcription is coordinately regulated by ECB elements, we wished to know how ECB elements contribute to the regulation of G1 progression. To address this issue, we measured the length of G1 and the sizes of cells carrying mutations in the CLN3 ECB elements and/or the SWI4 ECB. Table 2 shows that cells lacking ECB-activated transcription of either SWI4 or CLN3 or both spend a larger proportion of their cell cycle in G1 than wild-type cells. Perhaps due to the glucose induction of CLN3 transcription, the effects of these mutations were reduced when YEPGlu was used, but they are qualitatively similar to those observed when YEPGal was used. Loss of the ECB elements from both SWI4 and CLN3 promoters caused a G1 delay roughly equivalent to that of a cln3 null strain, which we found to maintain the level of cells in G1 at 43% in YEPGal medium. The swi4 deletion strain is lethal in this background so that comparison cannot be made.
TABLE 2.
Percentage of cells in G1a
| Yeast strain | % of cells in G1 (n)
|
|
|---|---|---|
| Glucose | Galactose | |
| SWI4 CLN3 | 19.1 ± 1.2 (8) | 29.8 ± 1.8 (14) |
| swi4ecb | 22.5 ± 1.3 (5) | 33.1 ± 1.7 (10) |
| cln3ecb | 23.2 ± 0.2 (4) | 37.8 ± 0.6 (4) |
| cln3ecb swi4ecb | 24.8 ± 1.3 (6) | 41.6 ± 1.3 (12) |
Cells were grown exponentially to densities between 4 × 106 and 2.4 × 107 cells per ml in YEP medium supplemented with adenine and either 2% glucose or 2% galactose. The cells were fixed and prepared for FACS analysis as described in Materials and Methods. The percentages of cells in G1 were determined from the FACS profiles using MultiCycle, and the values shown here are means ± standard deviations. n, the number of independent measurements (given for each genotype).
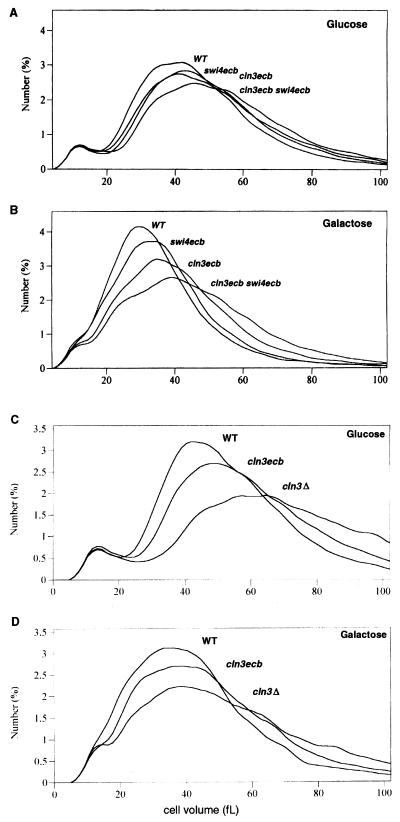
In addition to prolonging G1, elimination of ECB-mediated activation of CLN3 and SWI4 expression also caused a concomitant increase in cell volume (Fig. 3A and B). The data presented in Fig. 3 indicate that the M/G1-specific transcription of these two genes makes a substantial and additive contribution to the control of the G1-to S-transition, especially for cells grown in the absence of glucose. Figure 3C and D show a direct comparison of the size distribution of cln3ecb cells with that of cells with no Cln3 at all. Consistent with the FACS analysis data, the absence of Cln3 results in the most severe phenotype and loss of the ECB-dependent transcription of CLN3 has an intermediate phenotype. Thus, both basal and cell cycle-regulated transcription of CLN3 contributed to size control under the conditions tested.
FIG. 3.
Loss of ECB-mediated transcription of key cell cycle regulators leads to increased cell volume. Exponential-phase cultures of strains BY2679, BY2680, BY2681, and BY2682 grown in YEPGlu (A) or YEPGal (B) and of strains BY2125, BY2270, and BY2278 grown in YEPGlu (C) or YEPGal (D) were analyzed for cell size profile as described in Materials and Methods. The relevant genotypes are indicated.
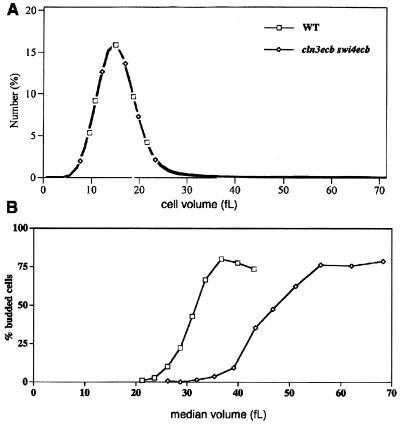
To further characterize G1 progression and size heterogeneity in cells lacking ECB function in their SWI4 and/or CLN3 promoters, we purified small G1 daughters of each genotype and compared their behavior to that of wild-type cells. Several size fractions of G1 daughter cells of each genotype were inoculated into fresh medium, and their growth, budding kinetics, and DNA profiles were monitored. As has been previously shown (20), wild-type cells show remarkable uniformity in that they shift from 0 to 50% budded as the cells enlarge from a volume of 26 to 31 fl (Fig. 4A). This concerted transition reflects a uniform response to the signal to start the cell cycle which is tightly correlated with cell size. Cells that lack the ECB-mediated burst of either CLN3 or SWI4 expression (Fig. 4A and B, respectively) bud at larger cell sizes and those lacking ECB activation of both CLN3 and SWI4 are even larger and more heterogeneous at budding (Fig. 4C). In contrast to the uniform behavior observed with wild-type cells, the cln3ecb swi4ecb cultures nearly doubled in volume as they went from 0% to 50% budding. Figure 4D shows wild-type and cln3ecb swi4ecb mutant cells at the time point when 50% of the cells had budded. It is clear that the wild-type cells were of uniform size, as were their buds, indicating that bud initiation occurred at about the same time in the growth of this population. However, budding occurred more slowly and heterogeneously in the double mutant, leading to cells with buds of various sizes. This effect was not due to the size heterogeneity of the starting population, because when we started with cells with precisely the same size distribution, buds accumulated in the double-mutant population much more gradually than in the wild type population (Fig. 5.) This heterogeneity of response also was not due to contamination of the double-mutant population with petite mutants, as measured by growth of the starting population on glycerol, or to the presence of mother cells, as measured by Calcofluor staining for bud scars (data not shown.)
FIG. 4.
ECB elements confer rapid, synchronous progression through G1 in daughter-cell fractions isolated by elutriation. Late-exponential-phase cultures (ca. 5 × 107 cells/ml) grown in YEPGal were fractionated by elutriation. Fractions containing <5% budded cells were diluted into fresh YEPGal at densities between 2.4 × 106 and 2 × 107 cells/ml, grown at 30°C, and sampled at 15-min intervals for determinations of size and budding indexes. The median cell volumes (in femtoliters) were determined by sizing on a Coulter model Z2 analyzer), as described in Materials and Methods. (A) Budding profiles of cells lacking the ECB-mediated burst of CLN3 expression. Open symbols, wild-type [BY2679(CLN3 SWI)] (starting fractions had median volumes of from 13.0 to 15.8 fl); closed circles, cln3ecb mutant (BY2681) (starting fractions had median volumes of from 22.7 to 25.9 fl). (B) Budding profiles of cells lacking the ECB-mediated burst of SWI4 expression. Circles, swi4ecb (BY2682) (starting fractions had median volumes of from 14.8 to 17.2 fl); open squares, wild type (profile shown for comparison). (C) Budding profiles of cells lacking ECB activation of both CLN3 and SWI4. Circles, cln3ecb swi4ecb (BY2680) (starting fractions had median volumes of from 15.0 and 28.4 fl); open squares, wild type (profile shown for comparison). (D) Phase-contrast photomicrographs of fractions of wild-type (initial median volume = 16.9 fl) and cln3ecb swi4ecb (initial median volume = 18.8 fl) elutriated cells after outgrowth to ca. 50% budded cells. The images shown are at the same magnification.
FIG. 5.
Wild-type (BY2679) and cln3ecb swi4ecb (BY2680) daughter cells exhibit very different budding kinetics in spite of their identical initial size distributions. Elutriated fractions of wild-type and cln3ecb swi4ecb daughter cells with identical size distributions (A) and starting median volumes of 15.1 and 15.0 fl, respectively, were sampled during outgrowth and monitored for budding and median cell size (B).
As expected, the double-mutant cells were more sensitive to α-factor than wild-type cells are. In addition, we found that their transit to S phase after release from the arrest was slower and more heterogeneous than that of wild-type cells. Figure 6 shows the FACS profiles for wild-type and cln3ecb swi4ecb mutant cells obtained as they were released from α-factor arrest. Not only was initiation of the first S phase at least 15 min slower for the cln3ecb swi4ecb cells but the synchrony of the culture was lost within the first cell cycle. The delay of S phase, as well as the size heterogeneity and FACS profiles observed in unperturbed, growing populations of cln3ecb swi4ecb cells, leads us to conclude that the defects in G1 progression manifested by the ECB mutants are not artifacts of recovery from elutriation, nor are they specific to daughter cells. Rather, they reflect an inefficient and deregulated G1-to-S transition. However, the heterogeneity between different elutriated fractions that was especially evident with the double mutant (Fig. 4C) may have resulted in part from the elutriation treatment. We see a general tendency for the larger daughter-cell fractions to bud at larger cell volumes, suggesting that the double mutant is particularly sensitive to disruptions in the growth cycle in late G1.
FIG. 6.
Coupling between the G1-to-S phase transition and initiation of budding is maintained in cln3ecb swi4ecb mutants. Unbudded daughter-cell fractions from elutriation were sampled at 15-min intervals during outgrowth for budding (squares) and initiation of DNA synthesis (circles). The percentage of cells in each sample that had exited G1 was determined by subtracting the percentage of cells in G1 (determined by analysis of the FACS profiles with MultiCycle software) from 100. Open symbols, wild-type (BY2679); closed symbols, cln3ecb swi4ecb (BY2680).
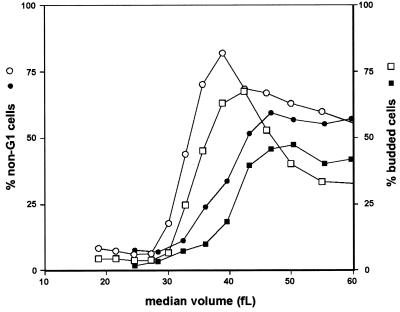
Budding is typically coordinated with the transition to S phase, so we also monitored exit from G1 in wild-type and cln3ecb swi4ecb cells by FACS analysis. As indicated in Fig. 7, the percentage of cells that exited G1 increased smoothly and in parallel to the budding profile, which lagged by about the same amount in both strains. Thus, the coupling between budding and the transition to S phase was not altered in the double mutant.
FIG. 7.
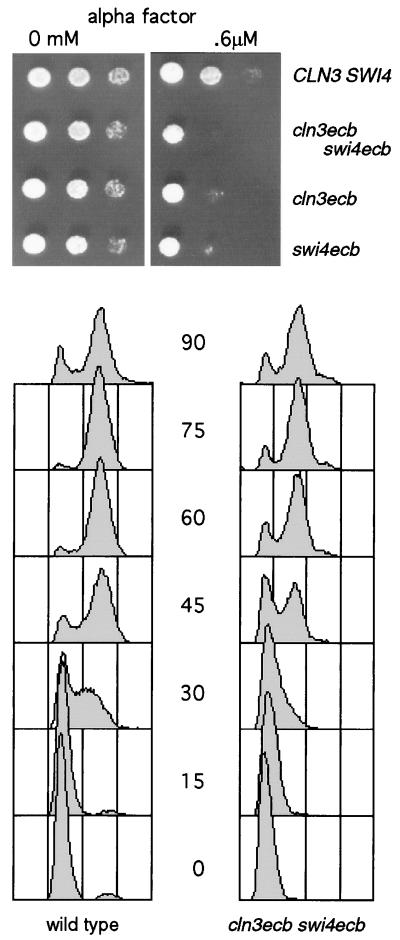
ECB mutants are more sensitive to α-factor and recover from α-factor arrest more slowly than wild-type cells. The upper portion of the figure shows 3,000, 1,000 and 300 cells of the genotypes indicated (strains BY2125, BY2680, BY2681, and BY2684) that were spotted on YEPglu plates containing 0 or 0.6 μM α-factor and allowed to grow for 2 days at 30°C. The lower portion of the figure shows FACS profiles of wild-type (BY2125) and cln3ecb swi4ecb (BY2680) cells that were grown in YEPGal. Growth was arrested by treatment with 5 μg of α-factor/ml for 135 min, and then the cells were released into the same medium and sampled at 15-min intervals.
CLN1 and CLN2 are critical targets of CLN3 and SWI4.
The long G1 phase and heterogeneous cell size of cln3ecb swi4ecb cells suggest that their ability to induce CLN1 and CLN2 transcription is highly impaired. To see if this is the case, we carried out Northern blot analysis of mRNAs from elutriated daughter cells harvested at intervals as they progressed through the cell cycle with the wild-type and double mutant cells. The starting population of wild-type G1 cells had high levels of CLN3 and SWI4 mRNAs which continued to rise throughout G1 and then fell as cells began to bud. The pattern of CLN1 and CLN2 transcription paralleled the pattern of ECB-mediated transcription but showed a slight lag. However, in the double-mutant population, both CLN3 and SWI4 transcripts remained at a low constitutive level (Fig. 8). CLN1 and CLN2 mRNA levels also remained low but rose gradually, as they did in a cln3 mutant population (10, 40). HO, another target gene which is not expressed in daughter cells (25), peaked only in the second cycle.
FIG. 8.
ECB elements are required for transcriptional regulation of CLN3 and SWI4 and their target genes (CLN1, CLN2, and HO) through the cell cycle. Unbudded daughter-cell fractions from elutriation were grown in YEPGal, and samples were taken at 15-min intervals for isolation of total RNA, as well as for size determination, FACS analysis, and budding index. Data shown are from a single Northern blot iteratively hybridized with probes for the indicated transcripts; multiple blotting experiments (data not shown) were carried out to confirm the validity of the cycling pattern shown. Initial cell volumes of these preparative elutriation fractions were 18.4 fl (wild-type strain [BY2679]) (open squares) and 23.4 fl (cln3ecb swi4ecb strain [BY2680]) (filled squares). The graph at the upper left shows budding kinetics, and the other five graphs show quantitated hybridization signals for the transcripts indicated (data normalized to the ACT1 signal and plotted with the lowest point in the wild-type samples set at 1.0). Below, hybridization patterns for each transcript scanned by a PhosphorImager are shown.
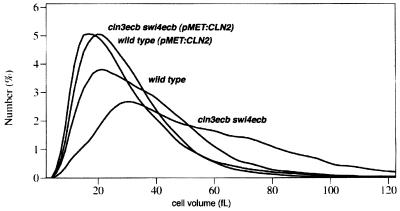
From these results one would predict that enhanced expression of CLN2 in these cells would effectively suppress these phenotypes. As shown in Fig. 9, ectopic expression of CLN2 from the MET3 promoter in cln3ecb swi4ecb cells shifted their size distribution to a uniform and smaller cell volume comparable to that observed for wild-type cells. This decrease in cell size was accompanied by a concomitant decrease in the percentage of G1 cells in the population (data not shown).
FIG. 9.
Ectopic expression of CLN2 suppresses the increased cell size and heterogeneity of cln3ecb swi4ecb cells. A DNA construct with CLN2 under the control of the MET3 promoter or a control vector was integrated at leu2 in the cln3ecb swi4ecb strain and the comparable wild-type strain. For analysis, cultures were grown in synthetic minimal glucose medium lacking methionine to derepress the promoter, and size profiles were determined on a Coulter model Z2 analyzer.
DISCUSSION
ECB elements, which are sufficient to promote M/G1-specific transcription, have been identified in both the SWI4 and CLN3 promoters. In a previous study, it was shown that loss of the ECB element from SWI4 caused a 10-min delay of its transcription and a change of start site, but SWI4 expression was still periodic owing to the MluI cell cycle box elements that also reside in the SWI4 promoter. This 10-min delay led to a lengthening of G1 and an enlarged cell size (21). By introducing inactivating mutations in all six of the potential ECB elements in the CLN3 promoter, we have shown that these elements are responsible for most of the cell cycle regulation of the CLN3 transcript. The resulting strain also suffers a lengthening of G1 and an increase in cell size, indicating that ECB-mediated transcription of CLN3 is required for efficient transit through G1 and into S phase. Thus, elimination of the modest oscillation of CLN3 transcription or decoupling the peak of CLN3 transcription from that of SWI4 by 10 min has a negative effect upon G1 progression.
Loss of the ECB elements from both promoters has a more dramatic effect, consistent with our previous findings that Swi4 and Cln3 make independent, rate-limiting contributions to G1 progression (21). The tight correlation between budding and the transition to S phase is not perturbed in the double mutant. This is not surprising because CLN1 and CLN2, the critical targets of Swi4 and Cln3, initiate both the budding cycle and the transition to S, so both should be equivalently delayed, as they are in the absence of Cln3 (10). However, the mechanism which triggers the late G1 transcription responsible for these events at a specific time and at a specific cell size is absent from these cells.
With a clonal population of wild-type daughter cells under constant environmental conditions, the balance between biosynthetic capacity and the activity of key regulators causes cells to progress through G1 at a particular pace and achieve a characteristic and uniform cell size before budding and the transition to S phase is triggered. In contrast, cells lacking ECB-mediated transcription of CLN3 and SWI4 exit G1 in a delayed and highly asynchronous manner, which is reflected in the diverse sizes these cells attain. Thus, instead of the highly regulated and sharp transition observed with wild-type cells, the cln3ecb swi4ecb cells behave as though the triggering mechanism has been lost and commitment to start the cell cycle has become a stochastic process, subject to random fluctuations in the concentration of key regulators from one cell to another. These observations show that the ECB elements, which coordinate the rise of both SWI4 and CLN3 mRNA levels, are important components of the mechanism which controls the timing of the G1-to-S phase transition in daughter cells.
Due to the asymmetric growth pattern of budding yeast, daughters are typically smaller than their mothers and spend more time in their first G1 before achieving a characteristic cell size and committing to another round of division (for a review, see reference 32). This homeostatic mechanism has long been thought to involve accumulation of an unstable activator (22), and experimental evidence has shown that Cln3 could be that activator (7, 24). Our data demonstrate a clear role for ECB-mediated transcription of CLN3 and SWI4 in determining the timing of the G1-to-S transition in daughter cells. There is an ECB-mediated, early G1 peak of both CLN3 and SWI4 transcription in elutriated daughters which is necessary for the normal induction of late G1 transcripts. In the absence of this transcriptional control, the transition to S phase is delayed and highly heterogeneous. We also observed heterogeneous cell sizes and G1 delays in exponentially growing mixtures of cln3ecb swi4ecb mothers and daughters, so we expect that G1 progression is also ECB-mediated in mother cells, but the specific pattern of ECB-mediated transcription could differ in mother and daughter cells.
Our initial observations showed that CLN3 mRNA levels peak in very late M phase and remain high throughout G1 in mixed populations of mothers and daughters (21). In this paper, we show that elutriated G1 daughters start with high levels of CLN3 and SWI4 mRNA which continue to rise as cells transit through G1 and peak just minutes before the peak of their target genes. This is consistent with the possibility that ECB activity peaks later in daughter cells than in mothers. This is an interesting possibility, as it could explain how G1 is prolonged in daughter cells. However, the transcript pattern we observed in these daughters could also be an artifact of elutriation.
The discovery of the importance of transcriptional regulation of CLN3 and SWI4 by ECB elements provides a simple model for how the timing of the G1-to-S transition is controlled. Cln3 and Swi4 are unstable proteins (44; L. L. Breeden, unpublished data). This does not favor models in which either protein must accumulate to some threshold level to trigger start. However, if their stability were constant through the cell cycle, the transcriptional oscillation mediated by the ECB elements would result in a parallel oscillation in the protein levels, enabling the concentration of both proteins to rise coordinately and peak in early G1. The coordinated increase in the levels of these two rate-limiting regulators would promote the efficient formation and activation of the Swi4-Swi6-DNA complex and result in a burst of transcription of the next wave of cyclins that are responsible for an efficient transition to S phase. In vivo binding studies indicate that Swi4-Swi6 complexes are detectable on the DNA throughout G1, during the interval of maximum SWI4 and CLN3 transcription, but activation of the complex occurs in late G1 (6, 13, 17). This lag between the formation of the complex and its activation suggests that there may be other steps involved in the activation process.
ACKNOWLEDGMENTS
We gratefully acknowledge helpful discussions with and technical assistance from other members of the lab, particularly J. Sidorova, S. Plante, C. McInerny, and J. Partridge.
This work was supported by a grant from the National Institutes of Health (GM41073) to L.L.B.
REFERENCES
- 1.Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 2.Andrews B J, Herskowitz I. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell. 1989;57:21–29. doi: 10.1016/0092-8674(89)90168-2. [DOI] [PubMed] [Google Scholar]
- 3.Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 4.Breeden L L. Alpha factor synchronization of budding yeast. Methods Enzymol. 1997;283:332–341. doi: 10.1016/s0076-6879(97)83027-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen K C, Csikasz-Nagy A, Gyorffy B, Val J, Novak B, Tyson J J. Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol Biol Cell. 2000;11:369–391. doi: 10.1091/mbc.11.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally-regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 7.Cross F R. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross F R, Blake C M. The yeast Cln3 protein is an unstable activator of Cdc28. Mol Cell Biol. 1993;13:3266–3271. doi: 10.1128/mcb.13.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cross F R, Tinkelenberg A H. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 10.Dirick L, Bohm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- 12.Futcher B. Cyclins and the wiring of the yeast cell cycle. Yeast. 1996;12:1635–1646. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1635::AID-YEA83%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 13.Harrington L A, Andrews B J. Binding to the yeast Swi4,6-dependent cell cycle box, CACGAAA, is cell cycle regulated in vivo. Nucleic Acids Res. 1996;24:558–565. doi: 10.1093/nar/24.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston L H, Johnson A L. Elutriation of budding yeast. Methods Enzymol. 1997;283:342–350. doi: 10.1016/s0076-6879(97)83028-5. [DOI] [PubMed] [Google Scholar]
- 16.Knapp D, Bhoite L, Stillman D J, Nasmyth K. The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol Cell Biol. 1996;16:5701–5707. doi: 10.1128/mcb.16.10.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at Start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–141. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- 18.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine K, Huang K, Cross F R. Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol Cell Biol. 1996;16:6794–6803. doi: 10.1128/mcb.16.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linskens M, Tyers M, Futcher B. CLN3 functions in both daughter and mother cells of S. cerevisiae. Cell. 1993;72:487–489. doi: 10.1016/0092-8674(93)90067-z. [DOI] [PubMed] [Google Scholar]
- 21.McInerny C J, Partridge J F, Mikesell G E, Creemer D P, Breeden L L. A novel Mcm1-dependent promoter element in the SWI4, CLN3, CDC6 and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- 22.Moore S A. Kinetic evidence for a critical rate of protein synthesis in the Saccharomyces cerevisiae yeast cell cycle. J Biol Chem. 1988;263:9674–9681. [PubMed] [Google Scholar]
- 23.Mullis K, Faloona F. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 24.Nash R, Tokiwa G, Anand S, Erickson K, Futcher A B. The WHI1 gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasmyth K. Molecular analysis of cell lineage. Nature. 1983;302:670–676. doi: 10.1038/302670a0. [DOI] [PubMed] [Google Scholar]
- 26.Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- 27.Partridge J F, Mikesell G E, Breeden L L. Cell cycle-dependent transcription of CLN1 involves Swi4 binding to MCB-like elements. J Biol Chem. 1997;272:9071–9077. doi: 10.1074/jbc.272.14.9071. [DOI] [PubMed] [Google Scholar]
- 28.Parviz F, Hall D D, Markwardt D D, Heideman W. Transcriptional regulation of CLN3 expression by glucose in Saccharomyces cerevisiae. J Bacteriol. 1998;180:4508–4515. doi: 10.1128/jb.180.17.4508-4515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piatti S, Bohm T, Cocker J H, Diffley J F X, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 30.Polymenis M A, Schmidt E V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pringle J R. Staining of bud scars and other cell wall chitin with Calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- 32.Pringle J R, Hartwell L H. The Saccharomyces cerevisiae cell cycle. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1981. pp. 97–142. [Google Scholar]
- 33.Rothstein R. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 34.Schneider B L, Yang Q-H, Futcher A B. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 35.Schwab M, Lutum A S, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 36.Schwob E, Bohm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 37.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. 1st ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 38.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuart D, Wittenberg C. Cell cycle-dependent transcription of CLN2 is conferred by multiple distinct cis-acting regulatory elements. Mol Cell Biol. 1994;14:4788–4801. doi: 10.1128/mcb.14.7.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1996;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 41.Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wittenberg C, Sugimoto K, Reed S I. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- 44.Yaglom Y, Linskens M H K, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]