Abstract
The relation between the gut microbiota and human health is increasingly recognized. Recently, some evidence suggested that dysbiosis of the oral microbiota may be involved in the development of digestive cancers. A systematic review was conducted according to the PRISMA guidelines to investigate the association between the oral microbiota and digestive cancers. Several databases including Medline, Scopus, and Embase were searched by three independent reviewers, without date restriction. Over a total of 1654 records initially identified, 28 studies (2 prospective cohort studies and 26 case-controls) were selected. They investigated oral microbiota composition in patients with esophageal squamous cell carcinoma (n = 5), gastric cancer (n = 5), colorectal cancer (n = 9), liver carcinoma (n = 2), and pancreatic cancer (n = 7). In most of the studies, oral microbiota composition was found to be different between digestive cancer patients and controls. Particularly, oral microbiota dysbiosis and specific bacteria, such as Fusobacterium nucleatum and Porphyromonas gingivalis, appeared to be associated with colorectal cancers. Current evidence suggests that differences exist in oral microbiota composition between patients with and without digestive cancers. Further studies are required to investigate and validate oral–gut microbial transmission patterns and their role in digestive cancer carcinogenesis.
Keywords: oral microbiota, carcinogenesis, digestive cancer, systematic review
1. Introduction
Digestive cancers include cancers located in the esophagus, stomach, liver, pancreas, colon, and rectum. Their incidence and related mortality are increasing worldwide, but with some characteristic geographical differences [1]. According to the GLOBOCAN, i.e., Cancer Incidence in Five Continents, and the World Health Organization (WHO) mortality databases in 2018, the majority of new cases of digestive cancers (63%) and related deaths (65%) occurred in Asia, followed by Europe and North America. Moreover, esophageal, gastric, and liver cancers appear to be more prevalent in Asia, whereas colorectal and pancreatic cancers are more common in Europe and North America [2]. Most of these cancers are considered sporadic and are influenced by several potentially modifiable environmental factors, such as tobacco smoking, diet, alcohol consumption, physical inactivity, obesity, and immunosuppressive drugs [1]. Some recent evidence suggested a role of the human microbiota in the development of digestive cancers, not only related to the composition and changes in relative abundance of microbes of the gut microbiota [3,4,5] but also linked to a state of dysbiosis of the oral microbiota [6,7]. In this review, we define “dysbiosis” as any change to the composition of resident bacterial communities relative to the community found in healthy individuals [8].
The oral cavity is a reservoir of more 700 species or phylotypes of bacteria, of which approximately 35% have not been cultured yet [9]. The equilibrium of this complex ecosystem is essential for oral health and influences host responses to disease [10]. The disruption in the homeostasis, i.e., dysbiosis, can result in significant metabolic and immunologic effects on the host, ultimately leading to local and systemic diseases [11,12]. These mechanisms are well documented in the pathogenesis of periodontitis, a chronic multifactorial inflammatory disease of the tooth-supporting tissues including the gums, the periodontal ligament, and the alveolar bone [13], but it has also been observed for cardiovascular, neurological, and metabolic disorders as well as digestive cancers and inflammatory bowel diseases [7,13,14,15].
In particular, specific oral bacteria that are typically found in the oral cavity of individuals suffering from periodontal diseases, such as Fusobacterium nucleatum [16] and Porphyromonas gingivalis [17], have been found in significantly high abundance in tumoral tissues and fecal samples of patients affected by colorectal cancer (CRC) [18,19], supporting the ability of these bacteria to migrate through the gastrointestinal tract where they can induce inflammation, alter the host immune response, and create an environment that may eventually favor tumor growth [20,21,22,23]. Specifically, data from a mouse model of intestinal tumorigenesis suggest that fusobacteria generate a proinflammatory microenvironment that is conducive to CRC progression through recruitment of tumor-infiltrating immune cells [24,25,26]. This was confirmed in clinical studies analyzing Fusobacterium nucleatum abundance by quantitative real-time polymerase chain reaction of DNA extracted from colorectal tissue biopsies and surgical resection specimens and observing that Fusobacterium nucleatum was more abundant in stool samples from CRC patients compared with adenomas or controls [27,28,29]. Some authors concluded that Fusobacterium nucleatum could be a novel risk factor for disease progression from adenoma to cancer, possibly affecting patient survival outcomes [27].
In this perspective, recent evidence suggested that certain oral bacteria and in general the characterization of the oral microbiota composition may be used as biomarkers for the detection of certain digestive cancers, with a potential role as a non-invasive screening tool [18,30,31].
The present systemic review aims to provide an updated appraisal of the current literature investigating the association between the oral microbiota and different types of digestive cancers, by (i) describing the observed differences in oral bacterial composition and abundance in cancer patients vs. controls and (ii) suggesting the possible impact of oral microbiota on digestive cancer risk.
2. Materials and Methods
2.1. Protocol Development and Literature Search
The present systematic review was performed according to the Cochrane collaboration-specific protocol [32] and was reported following the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) checklist (Table S1) [33].
Studies that investigated the association between oral microbiota and digestive cancers were searched in the following databases without date restrictions: Medline (through PubMed), Scopus, Embase, Cochrane Library and Google Scholar.
A specific research equation was used for each database, using the following keywords and MeSH terms: oral microbiome, oral microbiota, mouth microbiome, gastrointestinal neoplasms, gastrointestinal cancers, gastrointestinal carcinoma, digestive cancer, digestive neoplasms.
According to the PICOS schema, the following criteria were used to construct the literature search and to select the pertinent articles:
Population: adult patients with a diagnosis of digestive cancer (all types of cancers located in the digestive apparatus).
Intervention: analysis of oral microbiota composition and/or oral microbiome with or without a concomitant oral/periodontal examination. No restriction was applied for the type of microbiological technique used to sample and analyze the oral microbiota/microbiome, which include culture-dependent and genome sequencing methods.
Comparison(s): adult patients without cancer.
Outcome(s): oral microbiota composition (quality and quantity of bacterial species and pathogens related to oral diseases, particularly periodontitis).
Study designs: All types of descriptive (case series, cross-sectional) and analytic studies (cohort, case-control, clinical trial) estimating the magnitude of the association between oral microbiota dysbiosis and digestive cancers.
Results were limited to human clinical studies with review articles, with case reports being excluded. The literature review was completed by an extensive search using the “related articles” function in PubMed. The reference lists of the eligible records and of review articles were double-checked to identify potential additional pertinent articles. Articles were selected and reviewed if written in English only.
The literature search and selection were performed independently and blindly by three reviewers (E.R., G.B. and M.C.C.). Records were removed from the selection if all reviewers excluded the articles at the title/abstract screening levels. Disagreements were resolved with discussion with a third reviewer (N.d.A.).
2.2. Data Extraction
The reviewers performed an independent full-text analysis and data extraction by filling an electronic database. Extracted data included first author name, country, journal, year of publication, number of patients, age of the patients, type of cancer, oral diagnosis, type of microbiota analysis, sample extraction, detection method, and main results.
2.3. Study Quality Assessment and Risk of Bias
The reviewers also carried out a study quality assessment and risk of bias evaluation of the selected articles. According to the type of study design, the Newcastle–Ottawa scale [34] (NOS) was used.
3. Results
3.1. Literature Search and Selection
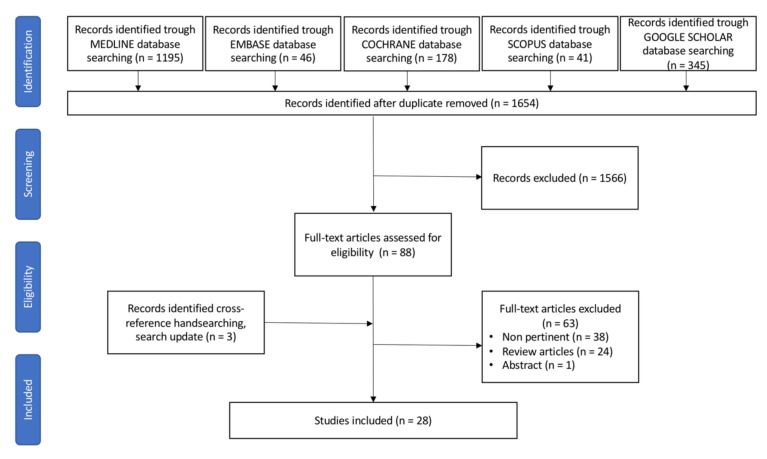
The merged search yielded 1805 results. After removing duplicates, 1654 articles were screened for eligibility based on title and abstract. Twenty-eight articles fulfilled the inclusion criteria and were selected for the present systematic review, as shown in Figure 1.
Figure 1.
PRISMA flow diagram for study search, selection, inclusion, and exclusion. Example of the research strategy: (oral microbiota[Title/Abstract]) OR (oral microbiome[Title/Abstract]) OR (mouth microbiota[Title/Abstract]) OR (mouth microbiome[Title/Abstract]) OR (oral cavity[Title/Abstract]) OR (oral bacteria[Title/Abstract]) AND (digestive cancer[Title/Abstract]) OR (digestive[Title/Abstract]) OR (digestive neoplasm[Title/Abstract]) OR (cancer of digestive system[MeSH Terms]) OR (cancer of the digestive system[MeSH Terms]) OR (cancer, digestive system[MeSH Terms]) OR (cancer of gastrointestinal tract[MeSH Terms]) OR (cancer of the gastrointestinal tract[MeSH Terms]) OR (gastrointestinal neoplasm[Title/Abstract]) OR (gastrointestinal cancer[Title/Abstract]).
3.2. Studies Characteristics
The selected studies were published between 2012 and 2021. There were 2 prospective cohort studies and 26 case-control studies (of which 6 were based on matched case-control groups). The studies were carried out in Europe (n = 3), USA (n = 8), China (n = 14), Japan (n = 1), Turkey (n = 1), and Iran (n = 1). The general characteristics of the studies examined are summarized in Table 1. Concerning the type of cancers, nine studies (31%) investigated oral microbiota composition in patients with colorectal cancer (CRC) [3,16,35,36,37,38,39,40,41], seven on pancreatic cancer (PC) [30,42,43,44,45,46,47], six on gastric cancer (GC) [39,48,49,50,51,52], five on esophageal cancer (EC) [6,53,54,55,56], and two on liver cancer (LC) [57,58].
Table 1.
General characteristics of the selected studies investigating the association between oral microbiota and digestive cancers.
| Reference | Country | Study Design | Study Sample Size (n) | Digestive Cancer Type | Oral Examination/Diagnosis |
|---|---|---|---|---|---|
| Farrell, J. et al. Gut 2012 [30] | USA | Matched case-control | 103 | PC | Not reported |
| Chen, X. et al. PloS ONE 2015 [53] | China | Case-control | 235 | ESCC | Number of teeth; Missing and filled teeth (MFT score), oral hygiene habits |
| Hu, J. et al. Biomed. Res. Int. 2015 [48] | China | Case-control | 146 | GC | No oral disease |
| May, X. et al. J. Periodontol. 2015 [35] | USA | Prospective cohort | 1252 | CRC | Periodontal examination, oral hygiene habits |
| Torres, P. et al. Peer J. 2015 [43] | USA | Case-control | 108 | PC | Not reported |
| Han, S. et al. Int. J. Oncol. 2016 [39] | China | Case-control | 290 | CRC and GC | Tongue examination; No oral disease |
| Kato, I. et al. J. Epidemiol. Res. 2016 [16] | USA | Population-based case-control | 190 | CRC | Not reported |
| Lu, H. et al. Sci. Rep. 2016 [57] | China | Matched case-control | 60 | LC | Full oral examination; No oral disease |
| Peters, B.A. et al. Cancer Res. 2017 [6] | USA | Case control | 316 | ESCC and EAC | Not reported |
| Olson, S. et al. Cancer Causes Control. 2017 [44] | USA | Case-control | 137 | PC, PDAC and IPMN | Number of missing teeth missing; periodontal diseases; number of dental visits in the past 10 years for checkup or cleaning; use of mouthwash at least once a week in the past 5 years |
| Fan, X. et al. Gut 2018 [45] | USA | Population-based nested matched case-control study | 732 | PC | Not reported |
| Flemer, B. et al. Gut 2018 [3] | Ireland | Case-control | 234 | CRC | Not reported |
| Russo, E. et al. Front. Microbiol. 2018 [36] | Italy | Case-control | 20 | CRC | Not reported |
| Sun, J. et al. Oncol. Rep. 2018 [50] | China | Case-control | 50 | GC | Not reported |
| Wu, J. et al. J. Cancer 2018 [51] | China | Case-control | 137 | GC | Not reported |
| Xu, J. et al. Microb. Pathog. 2019 [52] | China | Case-control | 150 | GC | Not reported |
| Lu, H. et al. J. Oral Microbiol. 2019 [46] | China | Case-control | 55 | PC | No oral disease |
| Schmidt, T. et al. eLife 2019 [38] | Europe | Case-control (Multicentric study) |
520 | CRC | Not reported |
| Yang, Y. et al. Int. J. Cancer 2019 [37] | USA | Nested matched case-control study | 693 | CRC | Oral health history |
| Wang, Q. et al. Sci. Rep. 2019 [54] | China | Case-control | 41 | ESCC | Oral health history |
| Guven, D.C. et al. Biomark. Med. 2019 [41] | Turkey | Case-control | 148 | CRC | Oral health history |
| Vogtmann, E. et al. Cancer Med. 2020 [42] | Iran | Case-control | 558 | PC | Not reported |
| Zhao, Q. et al. Front. Cell. Infect. Microbiol. 2020 [55] | China | Case-control | 90 | ESCC | Not reported |
| Zhang, S. et al. Theranostics 2020 [40] | China | Case-control | 253 | CRC | Not reported |
| Kawasaky, M. et al. Cancer 2020 [56] | Japan | Case-control | 122 | ESCC | Not reported |
| Li, D. et al. Microb. Phatog. 2020 [58] | China | Case-control | 24 | LC | Not reported |
| Wei, A.L. et al. World J. Gastroenterol. 2020 [47] | China | Case-control | 114 | PC | Not reported |
| Huang, K. et al. Front. Cell. Infect. Microbiol. 2021 [49] | China | Prospective cohort | 293 | GC | Not reported |
Abbreviations: CRA stands for colorectal adenoma; CRC for colorectal cancer; EAC for esophageal adenocarcinoma; ESCC for esophageal squamous cell carcinoma; GC for gastric cancer; IPMN for intraductal papillary mucinous neoplasms; LC for liver cancer; PC for pancreatic cancer; PDAC for pancreatic ductal adenocarcinoma.
All studies described microbiota composition differences between cases (cancer patients) and controls, and several studies [6,39,40,41,47,49,50,56,57] also analyzed the oral microbiota as being possible biomarkers for cancer screening and early diagnosis. The overall total number of digestive cancer patients considered was 2708, who were compared with 2593 controls. Twenty-six studies used DNA analysis, with 16S rRNA V4 sequencing and PCR as the principal method for microbiota investigation, whereas indirect immunofluorescence was used in one study [36] and shotgun sequence in another one [38]. Study outcomes are displayed in Table 2.
Table 2.
Outcomes of the selected studies investigating the association between oral microbiota and digestive cancers.
| Reference | Study Population Characteristics (n) |
Sampling Method and Type(s) of Microbiological Analysis | Microbiota Associated with Cancer | Bacterial Quantification | Main Finding(s) |
|---|---|---|---|---|---|
| Farrell, J. et al. Gut 2012 [30] |
Discovery cohort:
|
Saliva
|
|
Absolute amount | 31 bacterial species/clusters were increased in the saliva of PC patients, and 25 bacterial species/clusters were decreased in comparison with healthy controls. Salivary microbiota may be a non-invasive biomarker. |
| Chen, X. et al. PloS ONE 2015 [53] |
|
Saliva
|
|
Relative abundance | ESCC patients had a decreased microbial diversity compared with healthy controls and patients with dysplasia. |
| Hu, J. et al. Biomed. Res. Int. 2015 [48] |
|
Tongue coating
|
|
Relative abundance | Thick tongue coatings observed in GC patients presented lower microbial community diversity than thin tongue coatings of healthy controls. |
| May, X. et al. J. Periodontol. 2015 [35] |
Buffalo Osteoporosis and Periodontal Disease Study cohort of postmenopausal females
|
Subgingival plaque
|
|
Relative abundance | No associations were found between the presence of individual subgingival periodontal pathogens and the incident risk of cancer. |
| Torres, P. et al. Peer J. 2015 [43] |
|
Saliva
|
|
Relative abundance | Several bacterial genera differed in abundance between PC patients and controls. Bacteria abundance profiles in saliva may be useful biomarkers. |
| Han, S. et al. Int. J. Oncol. 2016 [39] |
|
Tongue coating
|
|
Relative abundance | Tongue coating is thicker in cancer patients than in healthy controls. Six microorganisms at the species level were significantly different. |
| Kato, I. et al. J. Epidemiol. Res. 2016 [16] |
|
Oral rinse
|
|
Relative abundance | No association between Fusobacterium abundance or presence and CRC. |
| Lu, H. et al. Sci. Rep. 2016 [57] |
|
Tongue coating
|
|
Absolute amount | Significant microbial dysbiosis of tongue coats in LC patients. |
| Peters, B.A. et al. Cancer Res. 2017 [6] |
|
Oral rinse
|
|
Relative abundance | Differences in oral microbiota composition between cases and controls. Possible application for screening purpose. |
| Olson, S. et al. Cancer Causes Control. 2017 [44] |
|
Saliva
|
|
Relative abundance | PDAC cases did not differ in microbiota diversity from controls or IPMN patients. |
| Fan, X. et al. Gut 2018 [45] | From the CPS II and PLCO cohorts
|
Oral rinse
|
|
Relative abundance | Carriage of the periodontal pathogens and decreased relative abundance of Fusobacteria and its genus Leptotrichia are associated with subsequent risk of PC. |
| Flemer, B. et al. Gut 2018 [3] |
|
Oral swabs
|
|
Absolute amount | A classification model of oral swab microbiota distinguishes individuals with CRC or polyps from controls. |
| Russo, E. et al. Front. Microbiol. 2018 [36] |
|
Saliva
|
|
Relative abundance | Bacterial community composition differed significantly between CRC patients and healthy controls. |
| Sun, J. et al. Oncol. Rep. 2018 [50] |
|
Dental plaque and saliva
|
|
Absolute amount | There are differences in the biomass, species richness, and species diversity between GC patients and controls. A microbiome scoring system was designed to be a potential screening method for GC. |
| Wu, J. et al. J. Cancer 2018 [51] |
|
Tongue coating
|
|
Relative abundance | Microbiome in tongue coating may have potential guiding value for early detection and prevention of GC. |
| Xu, J. et al. Microb. Pathog. 2019 [52] |
|
Tongue coating
|
|
Relative abundance | Richness and diversity of microbiome are not related to the variation of the four common types of tongue coating in GC patients. |
| Lu, H. et al. J. Oral. Microbiol. 2019 [46] |
|
Tongue coating
|
|
Absolute amount | PC patients are colonized by remarkably different tongue coating microbiota than healthy controls. |
| Schmidt, T. et al. eLife 2019 [38] |
|
Saliva
|
|
Relative abundance | The oral cavity is an endogenous reservoir for gut microbial strains, with increased levels of transmission in CRC patients. |
| Yang, Y. et al. Int. J. Cancer 2019 [37] |
|
Oral rinse
|
|
Relative abundance | Multiple oral bacterial taxa are associated with CRC risk. |
| Wang, Q. et al. Sci. Rep. 2019 [54] |
|
Saliva
|
|
Relative abundance | Association between oral dysbiosis and risk of ESCC. |
| Guven, D.C. et al. Biomark. Med. 2019 [41] |
|
Saliva
|
|
Absolute amount | Higher amounts of Fusobacterium nucleatum and Streptococcus gallolyticus in CRC patients. |
| Vogtmann, E. et al. Cancer. Med. 2020 [42] |
|
Saliva
|
|
Relative abundance | Increased levels of some oral bacteria and PC, with the overall microbial community different between PC patients and controls. |
| Zhao, Q. et al. Front. Cell. Infect. Microbiol.. 2020 [55] |
|
Saliva
|
|
Relative abundance | Differences in oral microbiota composition between cases and controls. |
| Zhang, S. et al. Theranostics 2020 [40] |
|
Oral swabs
|
|
Relative abundance. | Oral microbial composition and diversity were significantly different among the three groups, and the CRA group had the highest diversity. |
| Kawasaky, M. et al. Cancer 2020 [56] |
|
Dental plaque and saliva
|
|
Relative abundance | Differences in oral microbiota composition between cases and controls. Stronger microbiota association with dental plaque sample. Possible application for screening purpose. |
| Li, D. et al. Microb. Phatog. 2020 [58] |
|
Saliva
|
|
Relative abundance | Difference in oral microbiota composition according to the different grade of disease. |
| Wei, A.L. et al. World J. Gastroenterol. 2020 [47] |
|
Saliva
|
|
Relative abundances | Differences in microbiota composition between cases and controls. |
| Huang, K. et al. Front. Cell. Infect. Microbiol. 2021 [49] |
|
Saliva
|
|
Relative abundance | A distinct salivary microbiota was observed in patients with GC when comparing with SG and AG. Salivary microbiota could be used to predict GC as well as its non-malignant stages. |
Abbreviations: CRA stands for colorectal adenoma; CRC for colorectal cancer; EAC for esophageal adenocarcinoma; ESCC for esophageal squamous cell carcinoma; GC for gastric cancer; IPMN for intraductal papillary mucinous neoplasms; LC for liver cancer; PC for pancreatic cancer; PDAC for pancreatic ductal adenocarcinoma.
3.3. Oral Microbiota and Esophageal Cancer (EC)
Five articles investigated the association between oral microbiota and EC [6,53,54,55,56].
Chen et al. [53] demonstrated that patients with esophageal squamous cell carcinoma (ESCC) have a decreased oral microbiota diversity when compared with non-ESCC controls. The authors described a decreased salivary carriage of genera Lautropia, Bulleidia, Catonella, Corynebacterium, Moryella, Peptococcus, and Cardiobacterium and higher relative abundance of Prevotella, Streptococcus, and Porphyromonas in the ESCC group [53]. Wang et al. [54] showed that Actinomyces and Antopobium were related to a higher risk of ESCC, whereas the healthy group of the study was closely related to Fusobacterium and Porphyromonas. Zhao et al. [55] showed that ESCC is associated with an increased salivary carriage of Firmicutes, Negativicutes, Selenomonadales, Prevotellaceae, Prevotella, and Veillonellaceae and with a decreased taxa of Proteobacteria, Betaproteobacteria, Neisseriales, Neisseriaceae, and Neisseria.
Kawasaky et al. and Peters et al. concluded that differences in microbiota compositions between cases and controls could be used as a possible biomarker for cancer screening [6,56]. Only one study investigated the microbiota in relation to an esophageal adenocarcinoma, showing an association with an increased oral rinse carriage of Tannarella forsythia and a depletion of the commensal genus Neisseria and Streptococcus Pneumoniae [6].
3.4. Oral Microbiota and Liver Cancer (LC)
Only two articles investigated the association between oral microbiota dysbiosis, and LC. Lu et al. [57] identified Oribacterium and Fusobacterium as possible biomarkers to identify LC patients, showing a significant difference in their relative abundance between healthy controls and LC patients [57]. Li et al. compared the oral microbiota of patients with LC with healthy controls and patients with cirrhosis in different stages, finding an association between cancer and Haemophilus, Porphyromonas, and Filifactor.
3.5. Oral Microbiota and Pancreatic Cancer (PC)
In the seven studies dealing with PC, the microbiota analysis was conducted on salivary samples in five studies [30,42,43,44,47], on tongue coating in one study [46], and on oral rinse [45] in the remaining study. Results were heterogeneous. Lu et al. as well as Fan et al. observed an increased risk of cancer linked to Porphyromonas gingivalis, but contrasting findings were reported in relation to Fusobacterium, which was respectively linked to an increased and decreased risk of cancer [45,46]. The remaining studies found different bacteria linked to PC risk, without highlighting the predominance of a specific microbiota.
3.6. Oral Microbiota and Gastric Cancer (GC)
The microbiota analysis was conducted on tongue coating in four studies [39,48,51,52] and on salivary sample in two studies [49,50]. Hu et al. [48], Xu et al. [52], and Han et al. [39] reported that tongue coating characteristics significantly differ between cases and controls, with thicker tongue coating associated with an increased risk of GC. Xu et al. [52] divided the sample of GC patients into five subgroups based on the tongue coating characteristics, reporting different microbial associations in the different groups. However, also for this type of cancer there was no evidence of a specific microbiota being related to the development of GC.
3.7. Oral Microbiota and Colorectal Cancer (CRC)
Microbial samples were collected by oral swab in three studies [3,16,40], oral rinse in one study [45], saliva in three studies [36,38,41], subgingival plaque in one study [35], and tongue coating in one study [39]. Overall, these studies showed the most consistent results. Fusobacterium nucleatum was found to be linked to an increased risk of CRC in five studies [35,36,39,40,41]. Four studies agreed on the association of different species of Prevotella and CRC risk [3,35,37,40], while a borderline association was found in one study [39].
3.8. Study Quality Assessment
Based on the NOS assessment [34], 4 studies had a score of 5/9 [16,38,47,54], 16 studies of 6/9 [3,6,30,36,37,41,42,46,48,49,51,52,53,55,57,58], and 8 studies of 7/9 [35,39,40,43,44,45,50,56] (Table 3). Globally, a high heterogeneity was observed in study designs, study populations, and oral microbiota evaluation methods.
Table 3.
Quality assessment of the selected studies according to the star score of the Newcastle–Ottawa Scale, NOS, based on which * are assigned to three criteria, i.e., selection (with a maximum of 4 stars [****]), comparability (with a maximum of 2 stars [**]), and outcome (with a maximum of 3 stars [***]) for a maximum of 9 stars. Higher scores indicate lower risk of bias.
| Selection | Comparability | Outcome | Total Score | |
|---|---|---|---|---|
| Farrell, J. et al. Gut 2012 [30] | *** | * | ** | 6 |
| Chen, X. et al. PloS ONE 2015 [53] | ** | ** | ** | 6 |
| Hu, J. et al. Biomed. Res. Int. 2015 [48] | ** | ** | ** | 6 |
| May, X. et al. J. Periodontol. 2015 [35] | ** | ** | *** | 7 |
| Torres, P. et al. Peer J. 2015 [43] | **** | * | ** | 7 |
| Han, S. et al. Int. J. Oncol. 2016 [39] | **** | * | ** | 7 |
| Kato, I. et al. J. Epidemiol. Res. 2016 [16] | ** | * | ** | 5 |
| Lu, H. et al. Sci. Rep. 2016 [57] | ** | ** | ** | 6 |
| Peters, B.A. et al. Cancer Res. 2017 [6] | ** | ** | ** | 6 |
| Olson, S. et al. Cancer Causes Control. 2017 [44] | **** | * | ** | 7 |
| Fan, X. et al. Gut 2018 [45] | *** | ** | ** | 7 |
| Flemer, B. et al. Gut 2018 [3] | ** | * | *** | 6 |
| Russo, E. et al. Front. Microbiol. 2018 [36] | ** | * | *** | 6 |
| Sun, J. et al. Oncol. Rep. 2018 [50] | **** | * | ** | 7 |
| Wu, J. et al. J. Cancer 2018 [51] | *** | * | ** | 6 |
| Xu, J. et al. Microb. Pathog. 2019 [52] | ** | ** | ** | 6 |
| Lu, H. et al. J. Oral Microbiol. 2019 [46] | *** | * | ** | 6 |
| Schmidt, T. et al. eLife 2019 [38] | ** | * | ** | 5 |
| Yang, Y. et al. Int. J. Cancer 2019 [37] | ** | ** | ** | 6 |
| Wang, Q. et al. Sci. Rep. 2019 [54] | ** | * | ** | 5 |
| Guven, D.C. et al. Biomark. Med. 2019 [41] | ** | ** | ** | 6 |
| Vogtmann, E. et al. Cancer Med. 2020 [42] | ** | ** | ** | 6 |
| Zhao, Q. et al. Front. Cell. Infect. Microbiol. 2020 [55] | ** | ** | ** | 6 |
| Zhang, S. et al. Theranostics 2020 [40] | *** | ** | ** | 7 |
| Kawasaky, M. et al. Cancer 2020 [56] | *** | ** | ** | 7 |
| Li, D. et al. Microb. Phatog. 2020 [58] | ** | ** | ** | 6 |
| Wei, A.L. et al. World J. Gastroenterol. 2020 [47] | ** | * | ** | 5 |
| Huang, K. et al. Front. Cell. Infect. Microbiol. 2021 [49] | ** | ** | ** | 6 |
4. Discussion
The present systematic review provides a synthesis of studies investigating the association between oral microbiota composition and digestive cancers with a systematic approach. The available evidence suggests that digestive cancer patients present an oral microbiota composition that differs from non-cancer controls and specific oral bacteria may be linked to increased odds for digestive cancers. The predominance and/or relative abundance of these bacteria could have a synergistic effect on digestive cancers’ etiology [3,6,36,37,40,44,45,49,51,55], suggesting a possible role of oral microbiota characterization for screening and risk assessment of some types of cancer [40,47,49,56,59]. However, it is important to highlight that the current body of evidence is still limited, and some digestive cancers are under-investigated. For instance, only two studies were found concerning LC [57,58]. More studies are available concerning the role of oral microbiota in PC, GC, EC, and CRC patients, but further research is needed before advancing any solid conclusion. Notwithstanding this, the overall body of evidence consistently suggests that the oral microbiota is linked to the risk of digestive cancers.
In the digestive tract, the microbiota synthetizes essential amino acids and vitamins taking part in the digestion process. A symbiotic microbiota is normally characterized by a high diversity, and it is able to resist changes that occur during physiological stress [60], and to stimulate an immune response [20]. In recent years, different studies focused on the possible role of intestinal and oral microbiota dysbiosis in disease development, especially in carcinogenesis [7,20,23,53]. The digestive tract microbiota is involved in inflammation, metabolism, and genotoxicity mechanisms taking part in the oncogenic process [60]. Meanwhile, a moderate inflammatory reaction is protective against cancer development, and excessive inflammatory response is correlated with a higher risk of carcinogenesis [59]. Different oral bacteria associated with carcinogenesis, such as Porphyromonas, Prevotella, and Fusobacterium, can contribute to a chronic inflammatory reaction by stimulating the production of inflammatory mediators such as interleukin-1β, interleukin-6, and matrix metalloproteinases [61,62]. Prevotella and Fusobacterium have also been linked to carcinogenesis by the mechanisms of suppressing host immunological functions and promoting the malignant transformation of epithelial cells [28,29,59].
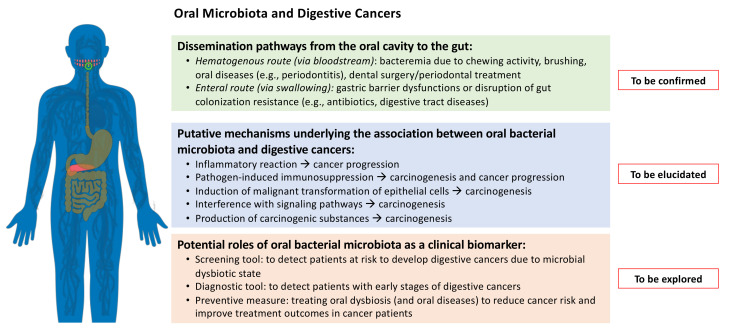
The potential translocation ability of the oral microbiota through the circulation system (hematogenous route) or following the flow of food and fluids into the digestive system (enteral route) could explain its presence in the gut and its role in carcinogenesis [59]. Three possible mechanisms of bacterial translocation in animal models were suggested: (1) disruption of the digestive equilibrium with intestinal bacterial overgrowth, (2) increased permeability of the intestinal mucosal barrier, and (3) deficiencies in host immune defenses. However, the transmission pathway remains questionable, and no definitive reports are currently available on the topic [63]. Zhang et al. [40] conducted an additional functional analysis using PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) and showed that the membrane transport pathway was decreased in CRA and CRC patients compared with healthy controls, with a potential impact on the anti-tumor immune response (such as the response mediated by bacterial outer membrane vesicles). Moreover, the cell motility pathway was found to be overrepresented in cancer patients, promoting tumor invasion and migration [40].
Finding the exact transmission pathways between the oral and the gut microbiota might provide evidence to support new methods for non-invasive cancer screening and could lead to control specific bacteria in their source, i.e., the oral cavity (Figure 2). Schmidt el al. [38] suggested an oral bacteria translocation in patients with CRC cancers. Flemer et al. [3] reported that oral swabs are associated with a sensitivity of 76% and a specificity of 90% in predicting colon adenomas; and a sensitivity of 98% and a specificity of 70% in predicting CRC. Sun et al. [50], found that salivary samples have a sensitivity of 97% and a specificity of 92% in predicting a GC, whereas Lu et al. [46] showed a sensitivity of 77% and a specificity of 78% for tongue coating to predict PC. Overall, the available studies suggest that a single oral bacterium has a limited ability to detect digestive cancer; conversely, multi-bacteria models have better performance and screening accuracy to differentiate patients with PC, EA, ESCC, GC, and CRC and to support the evaluation of the oral microbiota as a potential tool for prediction and prevention of digestive cancers [31]. Some evidence also supports a significant association between specific oral–gut bacteria and tumor stage, cancer-specific survival, and response to treatment [64,65,66,67,68], which, if confirmed and validated, would represent a novel and relevant strategy for reducing digestive cancer risk and improving cancer patients’ outcomes. Indeed, once the putative mechanisms of bacterial carcinogenesis can be elucidated and the role of oral microbiota precisely defined, its detection and characterization could be potentially used as a cancer biomarker and in the treatment of oral dysbiosis (e.g., periodontitis) as a preventive measure.
Figure 2.
Schematic representation of the possible routes of oral bacteria transmigration from the oral cavity to the gut and the possible oral microbiota mechanisms in digestive cancers. The potential role of oral bacterial microbiota characterization for the screening and risk assessment of some types of digestive cancer is also described.
Due to their clinical relevance, these findings need to be validated in future studies with a large population of patients and reproducible protocols for oral microbiota sampling and analysis. The available literature is characterized by a high degree of heterogeneity in the methodology used. Moreover, the role of potential confounders, such as alcohol intake and smoking, which are known to drastically influence oral health and oral microbiota homeostasis [69,70], has been rarely considered. Alcohol abuse and smoking are also related to different cancers [70], but only a few of the studies selected in the present systematic review considered these factors in their analyses of the association between oral microbiota composition and digestive cancers [6,10,40,42,44,45,54,55,56]. Moreover, other confounders, such as oral hygiene habits [71], periodontal health status and severity of periodontitis, presence of other oral diseases, and dietary patterns [72], were disregarded. Tooth loss has been linked to an increased risk of GC development [73] and to an overall increased mortality rate [40]. Poor oral hygiene and tooth loss have also been linked to an increased risk of CRA occurrence [74,75]. Overall, these variables, which may influence oral dysbiosis and cancer development, need to be systematically considered in future research.
Another relevant issue to consider is the sampling method used in the different studies (e.g., saliva, tongue coating, oral rinse, subgingival plaque). The oral cavity consists of different sites (i.e., teeth, tongue, cheeks, hard and soft palates) that create specific niches for microbial colonization characterized by different oxygen levels, nutrient availability, and mechanical stress conditions. Consequently, distinct microbial communities can be found in the oral cavity according to the explored sites and the presence of diseases (e.g., periodontitis) [76]. The type of sample used could therefore affect the results while assessing the link between oral microbiota and cancer. Saliva represents the preferred sampling site to obtain oral microbiota DNA to process since it tends to reflect the microbiota from all oral sites and the associated disease [77]. Ryutaro et al. [78] showed no differences in microbiota diversity in samples of unstimulated saliva, stimulated saliva (through chewing a paraffin gum), and mouth rise water collected in specimen tubes. The authors concluded that mouth rise is a reliable sample for microbiota analysis and that it could be particularly useful in specific subsets of patients, e.g., in elderly patients or in patients with low salivary flow, often providing results comparable with pure saliva samples. Conversely, the study of Gomar-Vecher et al. [79] comparing unstimulated saliva collected on paper points and stimulated saliva collected after chewing paraffin gum showed significant differences in microbiota composition between the two sampling methods. Again, Ryutaro, J. et al. [78] showed a different microbiota composition in tongue coating samples collected by scraping the dorsum of the tongue with a specific specimen compared with saliva and mouth rise water samples. Xu, J. et al. [52] focused on the landmark flora of the four common tongue coatings in GC patients. The samples were collected from the middle section of tongue dorsum using a toothbrush and put into the test tube with saline. By describing the oral microbiota composition, the authors concluded that these data could be useful for future standardization of GC diagnosis based on tongue microbiota. Tongue coating is a common sampling method in Chinese medicine. However, its biological and molecular bases are not completely validated, and thus it is not a widespread sampling method in the Western world [80]. Han, S. et al. [39] showed a relationship between the risk of PC and the tongue coating microbiota collected by scraping the front and middle section of the tongue with a sterile spoon and put into a test tube with saline (repeated three times and centrifuged (2000 r/min) for 5 min). Finally, subgingival plaque can also be used. Theoretically, it reflects the local microbiome composition much more specifically than a salivary sample and should be preferred when simultaneously evaluating periodontal diseases. Nevertheless, this sample collection method is more invasive and requires qualified trained staff to harvest subgingival plaque [56,81]. Moreover, in the presence of periodontitis, great heterogeneity can be found in the microbiota composition of the subgingival plaque sampled in deep vs. shallow periodontal pockets or following periodontal treatments [82]. Kawasaki et al. [56] examined six common periodontal pathogens in the subgingival microbiota of patients with EC and compared them with the salivary microbiota using real-time PCR analysis. A stronger association was found for the microbiota sampled in subgingival plaque. Thus, methodological issues should be considered in the interpretation and generalization of the results [83].
The present systematic review has some limitations related to the type and quality of the pertinent literature considered. Overall, the body of evidence is of low-to-moderate quality, but the results support the need of further studies investigating the role of oral microbiota composition in digestive cancer patients. The qualitative synthesis of the literature highlights the need for standardization in study design and methods to obtain comparable results contributing to the emerging evidence and elucidating the current knowledge. Indeed, the literature is characterized by high heterogeneity related not only to methodological aspects (e.g., sample selection, microbiome preservation, collection methods, taxonomical assignments, DNA extraction methods, and statistical analyses) but also to population characteristics, lifestyle habits, and culture, which may influence oral microbiota composition. Indeed, geography, ethnicity, subsistence-specific variations in human microbiome composition [84], and the genetic risk of cancers [85] could influence the results. Moreover, while most of the studies included healthy controls from comparable populations of those of cancer patients, only two studies reported to have performed endoscopies in controls to rule out the presence of cancer or precancerous lesions. Globally, there is no evidence of external validity so far.
5. Conclusions
Despite the limited evidence available, the literature is consistent in suggesting a significant association between oral microbiota composition and digestive cancers. To date, it is not possible to identify specific bacteria species involved in digestive cancer development, but these findings support further research to characterize the oral microbiota of these patients. Indeed, future studies should analyze the possible microbiota dissemination patterns from the oral cavity and the rest of the digestive tract and the possible role of oral microbiota characterization as a screening tool for digestive cancers.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9122585/s1, Table S1: PRISMA Checklist.
Author Contributions
E.R., M.C.C., G.B. and N.d. contributed to the manuscript the concept, study design, literature search, data interpretation, and article drafting; P.G., F.G., R.M., R.I. and G.L.d. contributed to data interpretation, article drafting, and manuscript critical revision. All authors read and approved the final version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nagai H., Kim Y.H. Cancer prevention from the perspective of global cancer burden patterns. J. Thorac. Dis. 2017;9:448–451. doi: 10.21037/jtd.2017.02.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold M., Abnet C.C., Neale R.E., Vignat J., Giovannucci E.L., McGlynn K.A., Bray F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335–349. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flemer B., Warren R.D., Barrett M.P., Cisek K., Das A., Jeffery I.B., Hurley E., O‘Riordain M., Shanahan F., O’Toole P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2017;67:1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cani P.D. Human gut microbiome: Hopes, threats and promises. Gut. 2018;67:1716–1725. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huttenhower C., Gevers D., Knight R., Abubucker S., Badger J.H., Chinwalla A.T., Creasy H.H., Earl A.M., Fitzgerald M.G., Fulton R.S., et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters B., Wu J., Pei Z., Yang L., Purdue M.P., Freedman N.D., Jacobs E.J., Gapstur S.M., Hayes R., Ahn J. Oral Microbiome Composition Reflects Prospective Risk for Esophageal Cancers. Cancer Res. 2017;77:6777–6787. doi: 10.1158/0008-5472.CAN-17-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauka L., Reitano E., Carra M.C., Gaiani F., Gavriilidis P., Brunetti F., de’Angelis G.L., Sobhani I., de’Angelis N. Role of the intestinal microbiome in colorectal cancer surgery outcomes. World J. Surg. Oncol. 2019;17:1–12. doi: 10.1186/s12957-019-1754-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen C., Round J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aas J.A., Paster B.J., Stokes L.N., Olsen I., Dewhirst F.E. Defining the Normal Bacterial Flora of the Oral Cavity. J. Clin. Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch S.V., Pedersen O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 11.Helmink B.A., Khan M.A.W., Hermann A., Gopalakrishnan V., Wargo J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019;25:377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 12.Lamont R.J., Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 2015;21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jangi S., Gandhi R., Cox L., Li N., Von Glehn F., Yan R., Patel B., Mazzola M.A., Liu S., Glanz B.L., et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novakovic M., Rout A., Kingsley T., Kirchoff R., Singh A., Verma V., Kant R., Chaudhary R. Role of gut microbiota in cardiovascular diseases. World J. Cardiol. 2020;12:110–122. doi: 10.4330/wjc.v12.i4.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duranti S., Gaiani F., Mancabelli L., Milani C., Grandi A., Bolchi A., Santoni A., Lugli G.A., Ferrario C., Mangifesta M., et al. Elucidating the gut microbiome of ulcerative colitis: Bifidobacteria as novel microbial biomarkers. FEMS Microbiol. Ecol. 2016;92:fiw191. doi: 10.1093/femsec/fiw191. [DOI] [PubMed] [Google Scholar]
- 16.Kato I., Vasquez A.A., Moyerbrailean G., Land S., Sun J., Lin H.-S., Ram J.L. Oral microbiome and history of smoking and colorectal cancer. J. Epidemiol. Res. 2015;2:92–101. doi: 10.5430/jer.v2n2p92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y., Luo G.-H. Porphyromonas gingivalisand digestive system cancers. World J. Clin. Cases. 2019;7:819–829. doi: 10.12998/wjcc.v7.i7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S.A., Liu F., Riordan S.M., Lee C.S., Zhang L. Global Investigations of Fusobacterium nucleatum in Human Colorectal Cancer. Front. Oncol. 2019;9:566. doi: 10.3389/fonc.2019.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koliarakis I., Messaritakis I., Nikolouzakis T.K., Hamilos G., Souglakos J., Tsiaoussis J. Oral Bacteria and Intestinal Dysbiosis in Colorectal Cancer. Int. J. Mol. Sci. 2019;20:4146. doi: 10.3390/ijms20174146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J., Li Q., Fu X. Fusobacterium nucleatum Contributes to the Carcinogenesis of Colorectal Cancer by Inducing Inflammation and Suppressing Host Immunity. Transl. Oncol. 2019;12:846–851. doi: 10.1016/j.tranon.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCoy A.N., Araújo-Pérez F., Azcarate-Peril M.A., Yeh J.J., Sandler R.S., Keku T.O. Fusobacterium Is Associated with Colorectal Adenomas. PLoS ONE. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubinstein M.R., Wang X., Liu W., Hao Y., Cai G., Han Y.W. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via its FadA Adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Niu Q., Fan W., Huang F., He H. Oral microbiota and gastrointestinal cancer. OncoTargets Ther. 2019;12:4721–4728. doi: 10.2147/OTT.S194153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M., Clancy T.E., Chung D.C., Lochhead P., Hold G.L., et al. Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., Weng W., Peng J., Hong L., Yang L., Toiyama Y., Gao R., Liu M., Yin M., Pan C., et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor−κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152:851–866.e24. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flynn K.J., Baxter N.T., Schloss P.D. Metabolic and Community Synergy of Oral Bacteria in Colorectal Cancer. mSphere. 2016;1:e00102-16. doi: 10.1128/mSphere.00102-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flanagan L., Schmid J., Ebert M., Soucek P., Kunicka T., Liška V., Bruha J., Neary P., DeZeeuw N., Tommasino M., et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 28.Pascual L.M., Cabrera-Rubio R., Ocon S., Costales P., Parra A., Suarez A., Moris F., Rodrigo L., Mira A., Collado M.C. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J. Gastroenterol. 2015;50:167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- 29.Tahara T., Yamamoto E., Suzuki H., Maruyama R., Chung W., Garriga J., Jelinek J., Yamano H.-O., Sugai T., An B., et al. Fusobacterium in Colonic Flora and Molecular Features of Colorectal Carcinoma. Cancer Res. 2014;74:1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farrell J.J., Zhang L., Zhou H., Chia D., Elashoff D., Akin D., Paster B.J., Joshipura K., Wong D.T.W. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2018;61:582–588. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y., Chen X., Yu H., Zhou H., Xu S. Oral Microbiota as Promising Diagnostic Biomarkers for Gastrointestinal Cancer: A Systematic Review. OncoTargets Ther. 2019;12:11131–11144. doi: 10.2147/OTT.S230262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgins J.P., Thomas J., Chandler J., Cumpston M., Li T., Page M.J., Welch V.A. Cochrane Handbook for Systematic Reviews of Interventions Version 6.0. John Wiley & Sons; New Jersey, NJ, USA: 2019. [Google Scholar]
- 33.Stewart L.A., Clarke M., Rovers M., Riley R.D., Simmonds M., Stewart G., Tierney J.F. Preferred Reporting Items for a Systematic Review and Meta-analysis of Individual Participant Data: The PRISMA-IPD statement. JAMA. 2015;313:1657–1665. doi: 10.1001/jama.2015.3656. [DOI] [PubMed] [Google Scholar]
- 34.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiology. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 35.Mai X., Genco R.J., LaMonte M.J., Hovey K.M., Freudenheim J.L., Andrews C., Wactawski-Wende J. Periodontal Pathogens and Risk of Incident Cancer in Postmenopausal Females: The Buffalo OsteoPerio Study. J. Periodontol. 2016;87:257–267. doi: 10.1902/jop.2015.150433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo E., Bacci G., Chiellini C., Fagorzi C., Niccolai E., Taddei A., Ricci F., Ringressi M.N., Borrelli R., Melli F., et al. Preliminary Comparison of Oral and Intestinal Human Microbiota in Patients with Colorectal Cancer: A Pilot Study. Front. Microbiol. 2018;8:2699. doi: 10.3389/fmicb.2017.02699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y., Cai Q., Shu X., Steinwandel M.D., Blot W.J., Zheng W., Long J. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int. J. Cancer. 2019;144:2381–2389. doi: 10.1002/ijc.31941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt T.S., Hayward M.R., Coelho L.P., Li S.S., Costea P.I., Voigt A.Y., Wirbel J., Maistrenko O.M., Alves R.J., Bergsten E., et al. Extensive transmission of microbes along the gastrointestinal tract. eLife. 2019;8:e42693. doi: 10.7554/eLife.42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han S., Yang X., Qi Q., Pan Y., Chen Y., Shen J., Liao H., Ji Z. Potential screening and early diagnosis method for cancer: Tongue diagnosis. Int. J. Oncol. 2016;48:2257–2264. doi: 10.3892/ijo.2016.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S., Kong C., Yang Y., Cai S., Li X., Cai G., Ma Y. Human oral microbiome dysbiosis as a novel non-invasive biomarker in detection of colorectal cancer. Theranostics. 2020;10:11595–11606. doi: 10.7150/thno.49515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guven D.C., Dizdar O., Alp A., Kittana F.N.A., Karakoc D., Hamaloglu E., Lacin S., Karakas Y., Kilickap S., Hayran M., et al. Analysis of Fusobacterium nucleatum and Streptococcus gallolyticus in saliva of colorectal cancer patients. Biomark. Med. 2019;13:725–735. doi: 10.2217/bmm-2019-0020. [DOI] [PubMed] [Google Scholar]
- 42.Vogtmann E., Han Y., Caporaso J.G., Bokulich N., Mohamadkhani A., MoayyedKazemi A., Hua X., Kamangar F., Wan Y., Suman S., et al. Oral microbial community composition is associated with pancreatic cancer: A case-control study in Iran. Cancer Med. 2019;9:797–806. doi: 10.1002/cam4.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torres P.J., Fletcher E.M., Gibbons S., Bouvet M., Doran K.S., Kelley S.T. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ. 2015;3:e1373. doi: 10.7717/peerj.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson S.H., Satagopan J., Xu Y., Ling L., Leong S., Orlow I., Saldia A., Lilan L., Nunes P., Madonia V., et al. The oral microbiota in patients with pancreatic cancer, patients with IPMNs, and controls: A pilot study. Cancer Causes Control. 2017;28:959–969. doi: 10.1007/s10552-017-0933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan X., Alekseyenko A.V., Wu J., Peters B.A., Jacobs E.J., Gapstur S.M., Purdue M.P., Abnet C.C., Stolzenberg-Solomon R., Miller G., et al. Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut. 2018;67:120–127. doi: 10.1136/gutjnl-2016-312580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu H., Ren Z., Li A., Li J., Xu S., Zhang H., Jiang J., Yang J., Luo Q., Zhou K., et al. Tongue coating microbiome data distinguish patients with pancreatic head cancer from healthy controls. J. Oral Microbiol. 2019;11:1563409. doi: 10.1080/20002297.2018.1563409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei A.-L., Li M., Li G.-Q., Wang X., Hu W.-M., Li Z.-L., Yuan J., Liu H.-Y., Zhou L.-L., Li K., et al. Oral microbiome and pancreatic cancer. World J. Gastroenterol. 2020;26:7679–7692. doi: 10.3748/wjg.v26.i48.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J., Han S., Chen Y., Ji Z. Variations of Tongue Coating Microbiota in Patients with Gastric Cancer. BioMed Res. Int. 2015;2015:173729. doi: 10.1155/2015/173729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang K., Gao X., Wu L., Yan B., Wang Z., Zhang X., Peng L., Yu J., Sun G., Yang Y. Salivary Microbiota for Gastric Cancer Prediction: An Exploratory Study. Front. Cell. Infect. Microbiol. 2021;11:640309. doi: 10.3389/fcimb.2021.640309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun J., Li X., Yin J., Li Y., Hou B., Zhang Z. A screening method for gastric cancer by oral microbiome detection. Oncol. Rep. 2018;39:2217–2224. doi: 10.3892/or.2018.6286. [DOI] [PubMed] [Google Scholar]
- 51.Wu J., Xu S., Xiang C., Cao Q., Li Q., Huang J., Shi L., Zhang J., Zhan Z. Tongue Coating Microbiota Community and Risk Effect on Gastric Cancer. J. Cancer. 2018;9:4039–4048. doi: 10.7150/jca.25280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu J., Xiang C., Zhang C., Xu B., Wu J., Wang R., Yang Y., Shi L., Zhang J., Zhan Z. Microbial biomarkers of common tongue coatings in patients with gastric cancer. Microb. Pathog. 2019;127:97–105. doi: 10.1016/j.micpath.2018.11.051. [DOI] [PubMed] [Google Scholar]
- 53.Chen X., Winckler B., Lu M., Cheng H., Yuan Z., Yang Y., Jin L., Ye W. Oral Microbiota and Risk for Esophageal Squamous Cell Carcinoma in a High-Risk Area of China. PLoS ONE. 2015;10:e0143603. doi: 10.1371/journal.pone.0143603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q., Rao Y., Guo X., Liu N., Liu S., Wen P., Li S., Li Y. Oral Microbiome in Patients with Oesophageal Squamous Cell Carcinoma. Sci. Rep. 2019;9:19055. doi: 10.1038/s41598-019-55667-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Q., Yang T., Yan Y., Zhang Y., Li Z., Wang Y., Yang J., Xia Y., Xiao H., Han H., et al. Alterations of Oral Microbiota in Chinese Patients with Esophageal Cancer. Front. Cell. Infect. Microbiol. 2020;10:541144. doi: 10.3389/fcimb.2020.541144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawasaki M., Ikeda Y., Ikeda E., Takahashi M., Tanaka D., Nakajima Y., Arakawa S., Izumi Y., Miyake S. Oral infectious bacteria in dental plaque and saliva as risk factors in patients with esophageal cancer. Cancer. 2021;127:512–519. doi: 10.1002/cncr.33316. [DOI] [PubMed] [Google Scholar]
- 57.Lu H., Ren Z., Li A., Zhang H., Jiang J., Xu S., Luo Q., Zhou K., Sun X., Zheng S., et al. Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci. Rep. 2016;6:33142. doi: 10.1038/srep33142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li D., Xi W., Zhang Z., Ren L., Deng C., Chen J., Sun C., Zhang N., Xu J. Oral microbial community analysis of the patients in the progression of liver cancer. Microb. Pathog. 2020;149:104479. doi: 10.1016/j.micpath.2020.104479. [DOI] [PubMed] [Google Scholar]
- 59.Sun J., Tang Q., Yu S., Xie M., Xie Y., Chen G., Chen L. Role of the oral microbiota in cancer evolution and progression. Cancer Med. 2020;9:6306–6321. doi: 10.1002/cam4.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwabe R.F., Jobin C. The microbiome and cancer. Nat. Rev. Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Atarashi K., Suda W., Luo C., Kawaguchi T., Motoo I., Narushima S., Kiguchi Y., Yasuma K., Watanabe E., Tanoue T., et al. Ectopic colonization of oral bacteria in the intestine drives T H 1 cell induction and inflammation. Science. 2017;358:359–365. doi: 10.1126/science.aan4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cardoso E.M., Reis C., Manzanares-Céspedes M.C. Chronic periodontitis, inflammatory cytokines, and interrelationship with other chronic diseases. Postgrad. Med. 2018;130:98–104. doi: 10.1080/00325481.2018.1396876. [DOI] [PubMed] [Google Scholar]
- 63.Berg R.D. Bacterial translocation from the gastrointestinal tract. Adv. Exp. Med. Biol. 1999;473:11–30. doi: 10.1007/978-1-4615-4143-1_2. [DOI] [PubMed] [Google Scholar]
- 64.Ito M., Kanno S., Nosho K., Sukawa Y., Mitsuhashi K., Kurihara H., Igarashi H., Takahashi T., Tachibana M., Takahashi H., et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer. 2015;137:1258–1268. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 65.Park C.H., Han D.S., Oh Y.-H., Lee A.-R., Lee Y.-R., Eun C.S. Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis. Sci. Rep. 2016;6:25271. doi: 10.1038/srep25271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamamura K., Baba Y., Nakagawa S., Mima K., Miyake K., Nakamura K., Sawayama H., Kinoshita K., Ishimoto T., Iwatsuki M., et al. Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clin. Cancer Res. 2016;22:5574–5581. doi: 10.1158/1078-0432.CCR-16-1786. [DOI] [PubMed] [Google Scholar]
- 67.Yu T., Guo F., Yu Y., Sun T., Ma D., Han J., Qian Y., Kryczek I., Sun D., Nagarsheth N., et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Serna G., Ruiz-Pace F., Hernando J., Alonso L., Fasani R., Landolfi S., Comas R., Jimenez J., Elez E., Bullman S., et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann. Oncol. 2020;31:1366–1375. doi: 10.1016/j.annonc.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ojima M., Hanioka T., Shimada K., Haresaku S., Yamamoto M., Tanaka K. The role of tobacco use on dental care and oral disease severity within community dental clinics in Japan. Tob. Induc. Dis. 2013;11:13. doi: 10.1186/1617-9625-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fan X., Peters B.A., Jacobs E.J., Gapstur S.M., Purdue M.P., Freedman N.D., Alekseyenko A.V., Wu J., Yang L., Pei Z., et al. Drinking alcohol is associated with variation in the human oral microbiome in a large study of American adults. Microbiome. 2018;6:59. doi: 10.1186/s40168-018-0448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsui M., Chosa N., Shimoyama Y., Minami K., Kimura S., Kishi M. Effects of tongue cleaning on bacterial flora in tongue coating and dental plaque: A crossover study. BMC Oral Health. 2014;14:4. doi: 10.1186/1472-6831-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lassalle F., Spagnoletti M., Fumagalli M., Shaw L., Dyble M., Walker C., Thomas M.G., Migliano A.B., Balloux F. Oral microbiomes from hunter-gatherers and traditional farmers reveal shifts in commensal balance and pathogen load linked to diet. Mol. Ecol. 2018;27:182–195. doi: 10.1111/mec.14435. [DOI] [PubMed] [Google Scholar]
- 73.Ndegwa N., Ploner A., Liu Z., Roosaar A., Axéll T., Ye W. Association between poor oral health and gastric cancer: A prospective cohort study. Int. J. Cancer. 2018;143:2281–2288. doi: 10.1002/ijc.31614. [DOI] [PubMed] [Google Scholar]
- 74.Lee D., Jung K.U., Kim H.O., Kim H., Chun H.-K. Association between oral health and colorectal adenoma in a screening population. Medicine. 2018;97:e12244. doi: 10.1097/MD.0000000000012244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Momen-Heravi F., Babic A., Tworoger S.S., Zhang L., Wu K., Smith-Warner S.A., Ogino S., Chan A.T., Meyerhardt J., Giovannucci E., et al. Periodontal disease, tooth loss and colorectal cancer risk: Results from the Nurses’ Health Study. Int. J. Cancer. 2017;140:646–652. doi: 10.1002/ijc.30486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hall M.W., Singh N., Ng K.F., Lam D.K., Goldberg M.B., Tenenbaum H.C., Neufeld J.D., Beiko R.G., Senadheera D.B. Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. NPJ Biofilms Microbiomes. 2017;3:2. doi: 10.1038/s41522-016-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Slots J., Slots H. Bacterial and viral pathogens in saliva: Disease relationship and infectious risk. Periodontol. 2000. 2010;55:48–69. doi: 10.1111/j.1600-0757.2010.00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jo R., Nishimoto Y., Umezawa K., Yama K., Aita Y., Ichiba Y., Murakami S., Kakizawa Y., Kumagai T., Yamada T., et al. Comparison of oral microbiome profiles in stimulated and unstimulated saliva, tongue, and mouth-rinsed water. Sci. Rep. 2019;9:16124. doi: 10.1038/s41598-019-52445-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gomar-Vercher S., Simon-Soro A., Montiel-Company J.M., Almerich-Silla J.M., Mira A. Stimulated and unstimulated saliva samples have significantly different bacterial profiles. PLoS ONE. 2018;13:e0198021. doi: 10.1371/journal.pone.0198021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liang W., Li X., Li Y., Li C., Gao B., Gan H., Li S., Shen J., Kang J., Ding S., et al. Tongue coating microbiome regulates the changes in tongue texture and coating in patients with post-menopausal osteoporosis of Gan-shen deficiency syndrome type. Int. J. Mol. Med. 2013;32:1069–1076. doi: 10.3892/ijmm.2013.1485. [DOI] [PubMed] [Google Scholar]
- 81.Haririan H., Andrukhov O., Bertl K., Lettner S., Kierstein S., Moritz A., Rausch-Fan X. Microbial Analysis of Subgingival Plaque Samples Compared to that of Whole Saliva in Patients with Periodontitis. J. Periodontol. 2014;85:819–828. doi: 10.1902/jop.2013.130306. [DOI] [PubMed] [Google Scholar]
- 82.Haffajee A.D., Bogren A., Hasturk H., Feres M., Lopez N.J., Socransky S.S. Subgingival microbiota of chronic periodontitis subjects from different geographic locations. J. Clin. Periodontol. 2004;31:996–1002. doi: 10.1111/j.1600-051X.2004.00597.x. [DOI] [PubMed] [Google Scholar]
- 83.Bergsten E., Mestivier D., Sobhani I. The Limits and Avoidance of Biases in Metagenomic Analyses of Human Fecal Microbiota. Microorganisms. 2020;8:1954. doi: 10.3390/microorganisms8121954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gupta V., Paul S., Dutta C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front. Microbiol. 2017;8:1162. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frank S.A. Genetic predisposition to cancer—Insights from population genetics. Nat. Rev. Genet. 2004;5:764–772. doi: 10.1038/nrg1450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available upon request.