Abstract
Plasmids containing oriP, the latent origin of replication for Epstein-Barr virus, support efficient replication in selected cell clones when the viral protein EBNA-1 is provided, being lost at a rate of 2 to 4% per cell generation after removal of selection (A. L. Kirchmaier and B. Sugden, J. Virol. 69:1280–1283, 1995; B. Sugden and N. Warren, Mol. Biol. Med. 5:85–94, 1988). We refer to these plasmids as established replicons in that they support efficient DNA synthesis and partitioning each cell cycle. Unexpectedly, we have found that upon introduction of oriP plasmids into a population of EBNA-1-positive cells, oriP plasmids replicate but are lost precipitously from cells during 2 weeks posttransfection (>25% rate of loss per cell generation). Upon investigation of these disparate observations, we have found that only 1 to 10% of cells transfected with an oriP plasmid expressing EBNA-1 and hygromycin phosphotransferase give rise to drug-resistant clones in which the oriP replicon is established. A hereditable alteration in these drug-resistant cell clones, manifested at the genetic or epigenetic level, does not underlie the establishment of oriP, as newly introduced oriP plasmids replicate but are also lost rapidly from these cells. In addition, a genetic alteration in the oriP plasmid is not responsible for establishment, as oriP plasmids isolated from an established cell clone, propagated in Escherichia coli, and reintroduced into EBNA-1-positive cells are likewise established inefficiently. Our findings demonstrate that oriP replicons are not intrinsically stable in EBNA-1-positive cell lines. Rather, the establishment of an oriP replicon is conferred upon the replicon by a stochastic, epigenetic event that occurs infrequently and, therefore, is detected in only a minority of cells.
It is essential for cells and latently infecting viruses to duplicate their genomes and transmit their genetic information to daughter cells during each cell cycle, processes that we condense into the term replication. The plasmid replicon of Epstein-Barr virus (EBV) serves as a model for studying replication in that, akin to the genome of its host cell, this replicon is synthesized only once per cell cycle during S phase (1, 51) and is efficiently partitioned to daughter cells (20, 25, 45, 47). Only two viral components are required for the replication of EBV's genome, the latent origin oriP and its binding protein Epstein-Barr-associated nuclear antigen 1 (EBNA-1); all else is contributed by the cell (26, 49, 52). Plasmids containing oriP support efficient replication in EBNA-1-positive cells selected to maintain them, being lost at a rate of 2 to 4% per cell generation after removal of selection (20, 47), a rate of loss which resembles that of ARS/CEN plasmids in Saccharomyces cerevisiae (23). We define these plasmids as established replicons in that they support efficient DNA synthesis and partitioning each cell cycle.
What events ensure efficient replication of oriP plasmids? The answer to this question largely remains enigmatic but should provide insights into the replication of EBV and perhaps into the replication of mammalian genomes. Genetic and biochemical studies of the eukaryotes S. cerevisiae and Schizosaccharomyces pombe have unveiled three regulatory steps required for high-fidelity replication which occur at distinct time points: during the initiation of DNA synthesis, during the completion of DNA synthesis, and during the partitioning of daughter molecules at mitosis. The mechanisms underlying these checkpoints may provide a framework for understanding the efficient replication of EBV's plasmid replicon.
Efficient initiation of DNA synthesis in eukaryotes depends upon genetic and epigenetic parameters. cis-acting elements known as origins must effectively recruit components of the prereplication complex (pre-RC), including ORC and MCM proteins (36). These specific initiation sites are defined by nuclear structure as demonstrated by studies of the dihydrofolate reductase origin in Xenopus egg extracts (10, 13), and chromatin, a component of nuclear structure, affects the timing of DNA synthesis at these origins as well (43). Epigenetic factors not only can define origin usage and timing but also can modulate origin activity as illustrated by experiments in which the activity of ARS1, an origin of DNA synthesis in S. cerevisiae, is compromised when located within a nucleosome but restored upon the positioning of the nucleosome outside the origin (41).
After initiation of DNA synthesis within the eukaryotic cell, several checkpoints are imposed to ensure completion of genome duplication prior to entry into mitosis and to prevent additional rounds of DNA synthesis in the absence of intervening mitoses. The former of these checkpoints involves components of the replication fork, such as DNA polymerase epsilon in S. cerevisiae, which is thought to sense single-stranded DNA accumulated during an S-phase block and transduce a Rad-dependent signaling cascade which halts mitosis (6, 31). When the genome is completely duplicated, additional rounds of DNA synthesis must be inhibited. The M-phase kinase Cdk1/cyclin B regulates this process by inhibiting pre-RC formation until mitosis is complete (8, 32). Deletion of the mitotic B cyclin p56cdc13 in S. pombe has disastrous results, with cells accumulating up to 32 copies of their genome due to the lack of intervening mitoses (16).
Upon completion of a single round of DNA synthesis within the eukaryotic cell, a single sister chromatid must be partitioned to each daughter cell. The fidelity of this process is ensured in part by the assembly of functional centromeres. In S. pombe, an epigenetic event involving maintenance of underacetylated centromeric chromatin is essential for centromere function (11, 14, 42). Upon association of the centromere-bound kinetochore with the mitotic spindle, a MAD2 checkpoint is imposed in which MAD1, MAD2, MAD3, and BUB1 inhibit the activity of the anaphase-promoting complex activator Cdc20 (APCcdc20), thereby blocking sister chromatid separation (4, 7). Anaphase can ensue upon attachment of these chromatids to spindle fibers emanating from opposing poles.
Eukaryotes have orchestrated multiple regulatory events to ensure high-fidelity replication of their genomes, which EBV is likely to use. EBV, in contrast to latently infecting viruses such as bovine papilloma virus, does not override the cellular synthetic machinery to maintain its genome extrachromosomally but rather appears to insinuate itself into this machinery. For example, initiation of DNA synthesis within EBV and oriP plasmids occurs at or near the dyad symmetry element (DS) of oriP (12), an origin that is licensed analogously to origins in eukaryotic cells (39). Likewise, partitioning of oriP plasmids is nonrandom, akin to eukaryotic genomes, in that cells containing, on average, two oriP plasmids lose these plasmids at the same rate as do cells having 10 or more oriP plasmids (20). The events that underlie the establishment of oriP plasmids therefore likely mirror those of eukaryotic cells.
Our studies on the replication of oriP plasmids have provided insights into an event required for the establishment of a replicon. While oriP plasmids support efficient replication in selected cell clones, we have found that upon introduction of oriP plasmids into a population of EBNA-1-positive cells, the oriP plasmids support replication but are lost precipitously from these cells during 2 weeks posttransfection. Investigation of these disparate findings has revealed that only 1 to 10% of those cells transfected with an oriP plasmid expressing EBNA-1 and hygromycin phosphotransferase gave rise to a drug-resistant clone in which an oriP replicon was established. A hereditable alteration in these cell clones was not responsible for the establishment of oriP, as newly introduced oriP plasmids supported replication but were also lost precipitously in these cells. In addition, establishment is not dependent upon genetic alteration of the oriP plasmid, as oriP plasmids which were rescued from an established cell clone, propagated in Escherichia coli, and reintroduced into EBNA-1-positive cells were likewise established inefficiently. Our experiments demonstrate that the stable replication of an oriP plasmid is not intrinsic to the plasmid but is conferred upon the replicon by a stochastic, epigenetic event that occurs infrequently and therefore is detected in a minority of cells. This epigenetic event must allow efficient completion of one or more of the regulatory steps described above. Defining this epigenetic event will provide insights into the cellular and viral contributors to the establishment of oriP and perhaps to that of other cellular and viral replicons.
MATERIALS AND METHODS
Plasmids.
2278, the oriP test plasmid, and 2276, the prokaryotic backbone plasmid, are derived from 2275, a plasmid containing a ColE1 origin and supF marker for propagation in E. coli and a neomycin phosphotransferase gene driven by the thymidine kinase promoter of herpes simplex virus. 2278 was constructed by inserting oriP between the HpaI and NsiI sites of 2275. 2276 was constructed by replacing the neomycin phosphotransferase gene with the firefly luciferase gene, thereby generating a length polymorphism upon amplification with primers 1588 and 86, as described below. 1728 contains oriP and encodes hygromycin B phosphotransferase and a derivative of EBNA-1 containing only five copies of the Gly-Gly-Ala repeat (2). 2048 is pcDNA3 (Invitrogen) in which the neomycin phosphotransferase gene was deleted by digestion with EcoRV and Bst1107I and religated. 2145 encodes enhanced green fluorescent protein (EGFP), whose expression is driven by the cytomegalovirus promoter, and lacks the neomycin phosphotransferase cassette. 2264, the competitor DNA, contains a fragment of DNA composed of pBR322 and LEU2 flanked by primer 1588 and primer 86 binding sites, such that amplification by PCR yields a 656-bp product. 2446 contains the luciferase gene from pGL2 Basic (Stratagene) cloned in place of EGFP in a plasmid construction (1783) containing oriP, and a cytomegalovirus promoter-driven IRES-EGFP cassette.
p220 was constructed by addition of the SmaI/HaeIII polylinker from pUC12 into the NarI site of p201 (52). This plasmid contains oriP, the beta-lactamase gene and origin of replication from pBR322, and encodes hygromycin B phosphotransferase and EBNA-1. The DS of oriP was removed from p220 by digestion with EcoRV and HpaI and religated, resulting in p220ΔDS (plasmid 2368, a kind gift from Jun Komano). pHEBo is the parental plasmid of p220 and lacks EBNA-1-encoding sequences (46). Low-molecular weight DNA from an established 293 cell clone harboring p220 (293/p220#2) was isolated (a kind gift from Jun Komano) and introduced into STBL2 E. coli (Gibco BRL). Two pools of approximately 1,000 colonies each were propagated on a large scale and plasmid DNA was purified (37), resulting in plasmids 2798 and 2799. The establishment efficiency of these plasmids (collectively referred to as “rescued p220”) in 293 cells was monitored as described for plasmid 1728 (see below).
Plasmid constructions were confirmed by sequencing and/or restriction enzyme digestions. Our plasmid database is accessible at: http://mcardle.oncology.wisc.edu/sugden/.
Cell lines and transfections.
The cell lines used for the replication assays include (i) H1299, a p53-null human lung carcinoma cell line (ATCC CRL-5803); (ii) 293, a human embryonic kidney cell line (ATCC CRL-1573); (iii) 143B, a human osteosarcoma cell line (ATCC CRL-8303); (iv) 293/EBNA-1, a 293 cell line which stably expresses EBNA-1 and neomycin phosphotransferase (ATCC CRL-10852); and (v) C33A/EBNA-1, a human cervical carcinoma cell line (ATCC HTB-31) into which the plasmid 1553, which expresses EBNA-1 and hygromycin B phosphotransferase, was integrated (2). Cell lines were grown in Dulbecco's modified Eagle's medium–high glucose supplemented with 10% fetal bovine serum, 200 U of penicillin per ml and 200 μg of streptomycin sulfate per ml. 143B cells were grown in medium containing calf serum instead of fetal bovine serum. 293/EBNA-1 and C33A/EBNA-1 cells were also grown in the presence of 200 μg of G418 sulfate per ml and 100 μg of hygromycin B per ml, respectively. Cells were grown at 37°C in a humidified 5% CO2 atmosphere. The doubling time of these cell lines is 22 to 24 h.
For the time course experiments, calcium phosphate precipitates containing equimolar amounts of oriP test plasmid (10 μg) and prokaryotic backbone plasmid (6.5 μg), 5 μg of 2145, (an expression vector for EGFP), and 10 μg of 1728 (an oriP-based EBNA-1 expression plasmid) or 10 μg of 2048 (an empty expression plasmid) were prepared and placed onto 15-cm dishes containing adherent cell lines (37). Plasmids 1728 and 2048 were not introduced into 293/EBNA-1 and C33A/EBNA-1 cells. H1299 cells were suspended by trypsinization, mixed with the precipitate, and plated onto a 15-cm dish. The medium was changed 5 to 8 h after addition of the precipitate. DNAs were introduced into 143B cells by electroporation (22). At 2 days posttransfection, cells were harvested and the percentage of EGFP-positive cells was determined as a measure of the transfection efficiency. (The transfection efficiency of oriP plasmids should be equivalent to or greater than that of the EGFP expression vector, as the FR of oriP promotes plasmid retention [29, 24].) Cells were expanded on 15-cm dishes and harvested at 4 to 6 days posttransfection, at which time a dilution of cells was replated such that the dishes were near confluence at the next time point, and the remaining cells were prepared by Hirt extraction, as described below.
Quantitative competitive PCR assay. (i) Hirt extraction and digestion of low-molecular-weight DNAs.
At the indicated time points posttransfection, cells were harvested and low-molecular-weight DNA was extracted by the method of Hirt (17). The supernatant was transferred to a new tube, incubated with 100 μg of RNase A per ml at 37°C for 2 h, and subsequently treated with 200 μg of proteinase K per ml overnight at 37°C. Samples were extracted with phenol-chloroform and the DNA was precipitated with isopropanol. The DNA pellet was rinsed with 70% ethanol, dried, and resuspended at 5 × 105 cell equivalents per μl in Tris-EDTA (TE) (10 mM Tris, pH 7.0, 1 mM EDTA). Fifty million cell equivalents were digested in a volume of 500 μl overnight at 37°C with 200 U of DpnI, to cleave the input fully methylated DNAs and hemimethylated DNAs, and 100 U of XhoI, to linearize the oriP test and prokaryotic backbone plasmids. The next day, an additional 100 U of DpnI was added and incubated overnight. Samples were extracted with phenol-chloroform, ethanol precipitated, and resuspended at 5 × 104 cell equivalents per μl in TE for analysis by quantitative competitive PCR or resuspended at 5 × 105 cell equivalents per μl in TE for analysis by Southern blotting.
(ii) Preparation of competitor DNA.
Twenty micrograms of 2264 was linearized with MluI for 2 h at 37°C, extracted with phenol-chloroform, ethanol precipitated, and resuspended in TE. This DNA was resolved on an agarose gel using 0.5× TBE buffer (0.45 M Tris-borate, 0.5 mM EDTA) containing 100 ng of ethidium bromide per ml. Images were captured with a charge-coupled device camera (IS-1000 digital imaging system; Alpha Innotech Corporation) and analyzed with ImageQuant software (Molecular Dynamics). The amount of linearized 2264 was determined by comparison to known quantities of lambda HindIII and phi X HaeIII DNA markers run in parallel.
(iii) PCR assay and data analysis.
A modified quantitative competitive PCR assay (19) was used to measure the amount of replicated, DpnI-resistant plasmid DNA present at various times posttransfection. Five PCRs were performed per sample using decreasing amounts of competitor DNA: 9 pg (corresponding to approximately 3.12 × 106 molecules), 3 pg, 600 fg, 120 fg and 24 fg. One hundred thousand cell equivalents of digested, Hirt-extracted DNA was added to a tube containing competitor DNA, 1× Taq buffer (Roche), 0.2 mM each deoxynucleoside triphosphate, 10 pmol of each primer (0.17 μM each), and 1.5 U of Taq DNA polymerase (Roche) in a total volume of 60 μl. DNA was amplified by a touch-down protocol in a Hybaid Omn-E Thermocycler for 22 cycles using the following conditions: two cycles of 94°C for 60 s, 60°C for 30 s and 72°C for 75 s; in the remaining cycles, DNA was denatured at 94°C for only 30 s and the annealing temperature was decreased by 1°C every second cycle until it reached 55°C. The primers used included 5′-GATCAAGAGACAGGATGAGGATCG-3′ (primer 1588), which lies in the herpes simplex virus thymidine kinase promoter region, and the previously described primer 5′-ACGATTCCGAAGCCCAACCTTTCA-3′ (primer 86) (19). The sizes of the amplified products generated from the primers are 656 bp for the competitor DNA, 931 bp for the oriP test plasmid, and 1,164 bp for the prokaryotic backbone plasmid.
One-third of the PCR mixture was electrophoresed through a 1.2% agarose gel using 0.5× TBE buffer containing 100 ng of ethidium bromide per ml. Signals were captured with a charge-coupled device camera (IS-1000 digital imaging system; Alpha Innotech Corporation) and analyzed with ImageQuant software (Molecular Dynamics). The number of molecules of replicated oriP test plasmid and prokaryotic backbone plasmid were determined by interpolation between known quantities of competitor DNA. This number was divided by the number of transfected cells analyzed to give the average number of molecules per transfected cell. Bromodeoxyuridine-labeling studies demonstrate that the oriP test plasmid is synthesized only once per cell cycle (Leight and Sugden, unpublished observations).
Southern blotting.
Approximately 107 cell equivalents of linearized, DpnI-digested DNA were loaded per lane on a 0.7% agarose gel and electrophoresed in 0.5× TBE buffer containing 100 ng of ethidium bromide per ml. The DNA was denatured and transferred to Gene Screen Plus hybridization membrane (NEN Life Sciences) (37). A probe was prepared by nick translation of 1728 (Amersham Pharmacia Biotech nick translation kit N 5000) and hybridized to the membrane. Signals were captured by a PhosphorImager (Molecular Dynamics) and analyzed using ImageQuant software (Molecular Dynamics).
Measurement of cell growth rates.
2446, an oriP plasmid expressing luciferase, and 2145, a plasmid expressing EGFP, were cotransfected into 293/EBNA-1 cells by calcium phosphate precipitation (37). At 2 days posttransfection, cells were harvested, the transfection efficiency was determined (as described above), and cells were plated at two cells, one cell, and one-half cell per well onto two 24-well dishes. The cells were allowed to grow in the absence of selection for 14 days, after which the number of wells containing cells was determined. For dishes plated at one cell per well and one-half cell per well, 24 of the 48 wells and 17 of the 48 wells contained cells, respectively. Using the Poisson distribution, we calculated that the colonies present on the dishes plated at one cell per well and one-half cell per well had a 69 and 80% probability, respectively, of arising from a single cell. The colonies on these dishes were triturated and allowed to grow an additional 3 days (to ensure even cell growth on the dish), after which the number of cells present per well and the luciferase activity present per well were determined as previously described (30).
Measurement of establishment efficiency and isolation of oriP-positive cell clones.
1728, an oriP plasmid expressing EBNA-1 and hygromycin phosphotransferase, and 2145, a plasmid expressing EGFP, were cotransfected into H1299, 293, and 143B cells as described above. At 2 days posttransfection, the percentage of EGFP-positive cells was determined and cells were plated at 105, 104, and 103 cells per 15-cm dish in media containing hygromycin B (H1299, 300 μg/ml; 293, 200 μg/ml; 143B, 150 μg/ml). Selection was applied for 3 to 4 weeks, at which time the drug-resistant colonies were enumerated. Several of these colonies were isolated and expanded in the presence of selection, giving rise to the H1299/1728#3, 293/1728#1, and 293/1728#5 cell clones. oriP test and prokaryotic backbone plasmids were then introduced into these cell clones, and replication was monitored over 2 weeks posttransfection as described above.
The cloning efficiency refers to the percentage of cells that are competent to plate and form a colony in the absence of selection and was determined by plating one cell per well onto a 96-well dish and enumerating those wells without cell growth after approximately 2 weeks in the absence of selection. The cloning efficiency was calculated using the Poisson distribution, where f(0), the probability of no cell growth in a well, is equal to e−m (m is the number of viable cells). For example, if 73 of 96 wells plated contain no cell growth, f(0) = 0.76, and m therefore equals 0.27. Given that only one cell was plated per well, the cloning efficiency (number of viable cells/number of cells plated) is merely m. These data were compiled to determine the establishment efficiency, which is defined as (no. of drug-resistant colonies/no. of transfected, clonable cells plated) × 100%.
RESULTS
Replicated oriP plasmids are lost precipitously from cells during 2 weeks posttransfection.
To further our understanding of the replication of oriP plasmids soon after their introduction into human cells, we assayed the pool of replicated oriP plasmids in the absence and presence of EBNA-1 in various cell lines during approximately 2 weeks posttransfection. Equimolar amounts of oriP test plasmid and prokaryotic backbone plasmid (which serves as an internal negative control) were introduced into H1299, 293, and 143B cells with or without an expression plasmid for EBNA-1. At different time points posttransfection, plasmid DNA was isolated by Hirt extraction and digested exhaustively with DpnI, and the level of replicated, DpnI-resistant DNA was determined by quantitative competitive PCR (19). Under our conditions, DpnI cleaves input methylated plasmid prepared from dam+ E. coli and hemimethylated plasmid which has undergone one round of DNA synthesis in mammalian cells (3). At 4 days posttransfection in a representative experiment with H1299 cells, the level of replicated oriP test plasmid detected in the presence of EBNA-1 was 28-fold greater than the level detected in the absence of EBNA-1 (Fig. 1A). This replicated oriP test plasmid was lost precipitously during the subsequent 10 days, such that the level of replicated oriP test plasmid detected at 14 days posttransfection was less than 1% of the level detected at 4 days posttransfection. That is, the oriP test plasmid replicated but was rapidly lost from the majority of cells in the H1299 cell line during 2 weeks posttransfection. Clearly, this loss could reflect inefficient DNA synthesis, inefficient partitioning, or both.
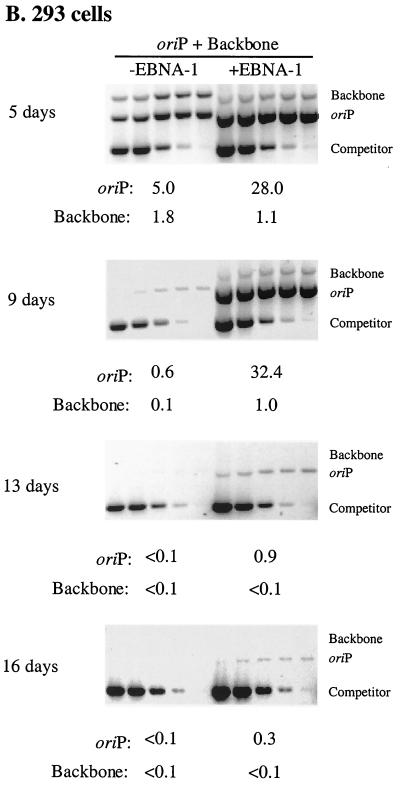
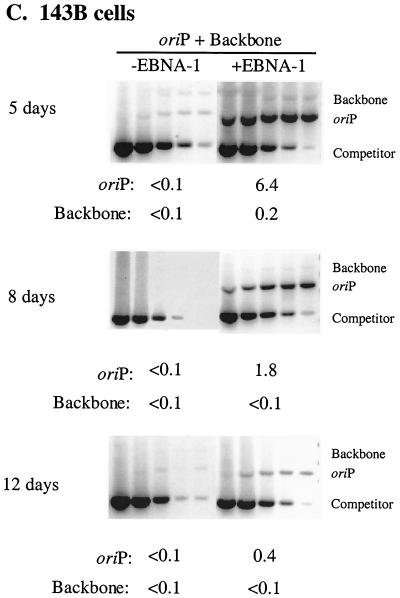
FIG. 1.
Newly introduced oriP plasmids replicate within EBNA-1-positive cells but are lost precipitously during 2 weeks posttransfection. Equimolar amounts of oriP test plasmid (oriP) and prokaryotic backbone plasmid (Backbone) were introduced into H1299 (A), 293 (B), and 143B (C) cells with (+EBNA-1) or without (−EBNA-1) an expression plasmid for EBNA-1 and into 293/EBNA-1 cells (D) which stably express EBNA-1. At the indicated time points posttransfection, plasmid DNA was isolated by Hirt extraction and digested with XhoI and DpnI, and the level of replicated, DpnI-resistant DNA was determined by quantitative competitive PCR. PCRs were performed using 105 cell equivalents and a competitor DNA standard curve (9 pg, 3 pg, 600 fg, 120 fg, and 24 fg). Numbers below gels refer to the average number of molecules present per transfected cell. The 143B time course was conducted in duplicate, and gels were rearranged to present a single experiment. The amount of replicated oriP test plasmid present at 6 days posttransfection in 293/EBNA-1 cells was quantified from a separate gel in which 3.3 × 103 cell equivalents were assayed.
To determine if the precipitous loss of oriP plasmids from EBNA-1-expressing cells during 2 weeks posttransfection was peculiar to the H1299 cell line, we monitored the fate of oriP test plasmids in the 293 and 143B human cell lines which historically have been used to monitor replication of oriP plasmids (35, 52). In a representative experiment with 293 cells at 5 days posttransfection, the replication efficiency of the oriP test plasmid increased sixfold when EBNA-1 was provided in trans. (Replication efficiency refers to the average number of DpnI-resistant molecules present per transfected cell and encompasses both DNA synthetic and partitioning events.) This augmentation by EBNA-1 was not as dramatic as that detected in H1299 and 143B cells (compare Fig. 1 A, B, and C at 4 to 5 days posttransfection) and reflects the high permissivity of 293 cells for replication of any introduced DNAs in the absence of EBNA-1 at early time points posttransfection (see also reference 50). As in H1299 cells, in 293 cells the replicated oriP test plasmid detected in the presence of EBNA-1 was lost at such a precipitous rate that the level of replicated oriP test plasmid detected at 16 days posttransfection was 1% of the level detected at 5 days posttransfection (Fig. 1B). Furthermore, in a representative experiment with 143B cells, the level of replicated oriP test plasmid detected at 5 days posttransfection in the presence of EBNA-1 was 64-fold greater than the level detected in the absence of EBNA-1. However, this replicated oriP test plasmid decreased by a factor of sixteen between 5 and 12 days posttransfection even though EBNA-1 was provided in trans (Fig. 1C). These experiments demonstrate that in multiple human cell lines an oriP plasmid is competent to support replication; however, this replicated DNA is lost rapidly from the majority of the cell population even though EBNA-1 is provided in trans.
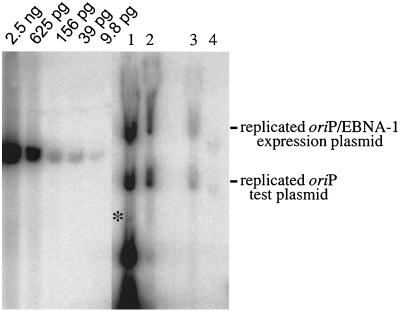
In the experiments presented in Fig. 1 A, B, and C, EBNA-1 was expressed from an oriP plasmid provided in trans (1728, an oriP/EBNA-1 expression plasmid). We wanted to determine if this plasmid was also lost rapidly, as the loss of this plasmid would result in a loss of EBNA-1 expression that could affect the observations for the oriP test plasmid. Equimolar amounts of the oriP test plasmid and prokaryotic backbone plasmid were introduced into 293 cells together with the oriP/EBNA-1 expression plasmid, and the level of replicated plasmid present at 5, 9, 13 and 16 days posttransfection was determined by Southern blotting. The oriP/EBNA-1 expression plasmid supported replication but was lost precipitously from the population of cells during the time course, as was the oriP test plasmid (Fig. 2). The replicated oriP/EBNA-1 expression plasmid was lost at the same rate as the replicated oriP test plasmid, indicating that declining levels of EBNA-1 could contribute to the loss of these replicated plasmids.
FIG. 2.
The oriP/EBNA-1 expression plasmid supports replication but is lost precipitously during 2 weeks posttransfection, as is the oriP test plasmid. Equimolar amounts of oriP test plasmid and prokaryotic backbone plasmid were introduced into 293 cells together with 1728, an oriP-based expression plasmid for EBNA-1 (oriP/EBNA-1 expression plasmid). At 5, 9, 13, and 16 days posttransfection (lanes 1 to 4, respectively), plasmid DNA was isolated by Hirt extraction and digested with XhoI and DpnI and the level of replicated, DpnI-resistant DNA was determined by Southern blotting. Approximately 107 cell equivalents were loaded per lane and electrophoresed beside a standard curve of linearized 1728 DNA. The Hirt DNAs ran aberrantly in the gel due to the presence of contaminating chromosomal and mitochondrial DNA. Approximately 5% of chromosomal DNA is present in the Hirt extract (3 μg per 107 cells). The replicated backbone plasmid (indicated by an asterisk) cannot be detected clearly because it migrates with the DpnI-sensitive material near the bottom of the gel.
We therefore investigated whether newly introduced oriP plasmids were lost precipitiously in cell lines which expressed EBNA-1 stably. The oriP test plasmid and prokaryotic backbone plasmid were introduced into 293/EBNA-1 cells, a cell line which expresses EBNA-1 stably, and the level of replicated plasmid was monitored over 16 days. At 6 days posttransfection in a representative experiment, the level of replicated oriP test plasmid was 34-fold greater than the level of replicated prokaryotic backbone plasmid. This replicated oriP test plasmid was rapidly lost over the subsequent 10 days such that the level of replicated oriP test plasmid present at 16 days posttransfection was 1% of the level detected at 6 days posttransfection (Fig. 1D). This oriP test plasmid was also lost rapidly in C33A/EBNA-1 cells, which express EBNA-1 stably, such that the level of replicated plasmid detected at 16 days posttransfection was less than 7% of the level detected at 6 days posttransfection (data not shown). That is, even when EBNA-1 is expressed stably in cells, oriP plasmids undergo replication but still are lost precipitously from the majority of the cell population.
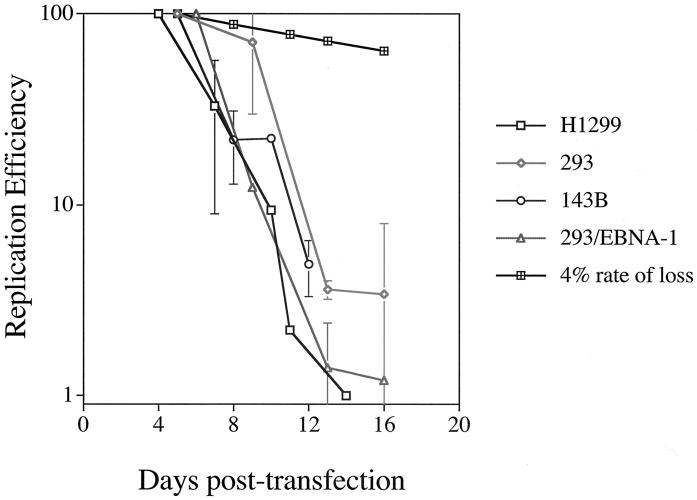
A graphical representation of multiple, independent time course experiments conducted with H1299, 293, and 143B cells, in which EBNA-1 was provided in trans, and 293/EBNA-1 cells, in which EBNA-1 was expressed stably, is presented in Fig. 3. For each of these cell lines, the loss of oriP plasmids from the population of cells over 2 weeks posttransfection varied dramatically from the loss of oriP plasmids in an established oriP-positive cell clone. That is, in established oriP-positive cell clones, oriP plasmids are lost at a rate of 2 to 4% per cell generation after removal of selection (20, 47), whereas during 2 weeks posttransfection, oriP plasmids are lost at an average rate of 26 to 37% per cell generation from the majority of the cell population. Why then are oriP plasmids stable in the established oriP-positive cell clones yet unstable upon introduction into a population of EBNA-1-expressing cells? We examined two general explanations for these disparate experimental observations. First, the precipitous loss of replicated oriP plasmids could result from a selective growth advantage of the untransfected cells relative to the transfected cells, such that in the absence of selection for the oriP plasmid, the transfected oriP-positive cells are overgrown by untransfected cells. Alternatively, given no selective growth advantage of the untransfected cells, our observations would suggest that the establishment of an oriP replicon is inefficient such that oriP plasmids are replicated efficiently in only a minority of transfected cells.
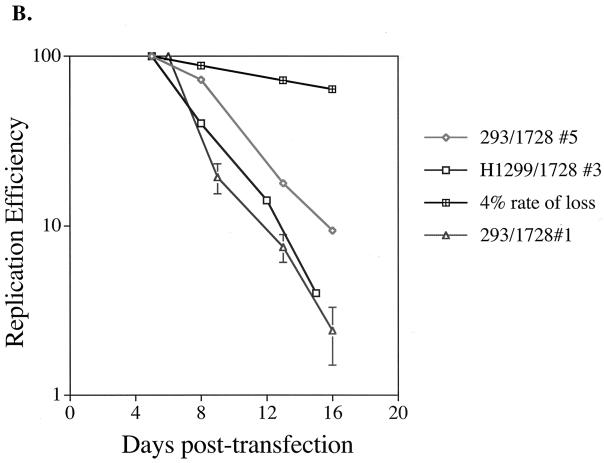
FIG. 3.
The loss of oriP plasmids from a population of cells during 2 weeks posttransfection varies dramatically from an established oriP-positive cell clone. A graphical representation of multiple, independent time course experiments with H1299 (open boxes), 293 (diamonds), and 143B (circles) cells in which EBNA-1 is provided in trans and 293/EBNA-1 cells (triangles) in which EBNA-1 is expressed stably is shown. Experiments were conducted as described in the legend to Fig. 1. The replication efficiency is plotted versus the days posttransfection. For each independent experiment, the level of replicated oriP test plasmid detected at the first time point posttransfection was set to 100% and the replication efficiency of this plasmid at later time points was set relative to this initial time point. Data for H1299 cells represent the average ± standard deviation of two experiments, except for the 10-day time point, which represents a single experiment, and the 11- and 14-day time points, which represent a second independent experiment. For 293 cells, data represent the average ± standard deviation of three experiments, except for the 13-day time point, which represents two experiments. Data for 143B cells represent the average ± standard deviation of two experiments for the 8- and 12-day time points and a single experiment for the 10- and 16-day time points (16-day data were not plotted but were less than 7% of the replicated oriP test plasmid detected at the 5-day time point). For 293/EBNA-1 cells, data represent the average ± standard deviation of two experiments, except for the 16-day time point, which represents one experiment. A 4% rate of loss, measured for selected cell clones, is depicted by cross-hatched boxes.
Precipitous loss of oriP plasmids does not result from a selective growth advantage of the untransfected cells relative to the transfected cells.
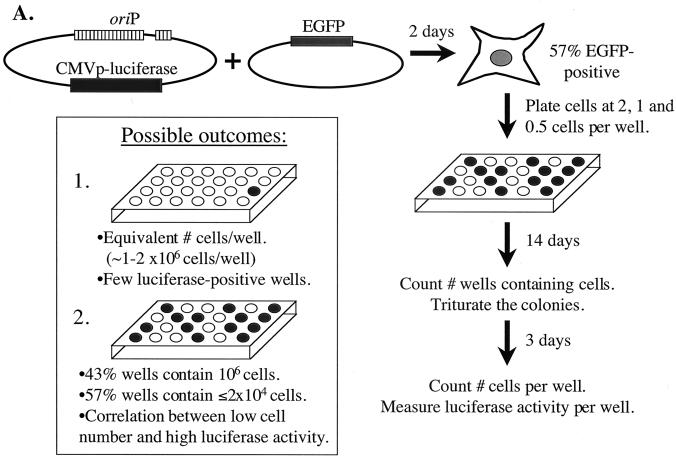
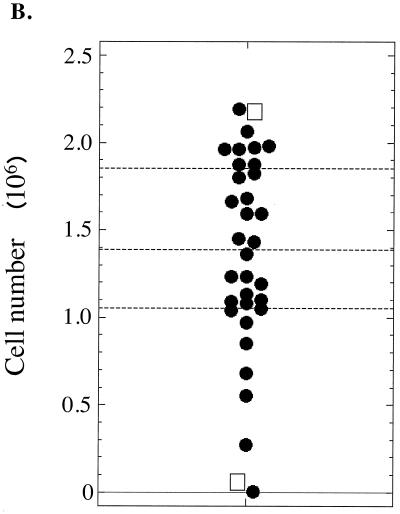
To test the first scenario of varying growth advantages, we measured the proliferation of transfected and untransfected cells as depicted in Fig. 4A. An oriP plasmid expressing luciferase and a plasmid expressing EGFP were introduced into 293/EBNA-1 cells, and the transfection efficiency was determined at 2 days posttransfection by calculating the percentage of EGFP-positive cells. Cells were then plated at two cells, one cell, and one-half cell per well onto 24-well dishes and allowed to grow in the absence of selection for 14 days, at which time the number of wells containing cells was enumerated. Based on the Poisson distribution, the colonies present on the dishes plated at one and one-half cell per well had a 69 and 80% probability, respectively, of arising from a single cell. The colonies on these dishes were triturated and allowed to grow an additional 3 days (to allow even growth in the wells), after which the cells present in each well were enumerated and the average luciferase activity present per cell was measured. Luciferase activity serves as an indicator for the presence of oriP plasmids. During 17 days, 293/EBNA-1 cells are expected to undergo 20 to 21 doublings, giving rise to 1 × 106 to 2 × 106 cells per well. If the untransfected and transfected cell populations have similar growth kinetics, 1 × 106 to 2 × 106 cells should be present per well and few wells should exhibit high luciferase activity, indicating a loss of oriP plasmids from the cell population. Alternatively, if the untransfected cell population has a selective growth advantage, only those untransfected cells should give rise to 1 × 106 to 2 × 106 cells per well after 17 days. In addition, the transfected cells (57% of the population) should give rise to 1 to 2% the number of cells as the untransfected population (based on the 50- to 100-fold decrease in replicated oriP plasmids during 2 weeks posttransfection as shown in Fig. 3) and should exhibit high luciferase activity per cell. Our experimental observations are consistent with the former of these two outcomes. The numbers of transfected and untransfected cells per well which accumulated after 17 days were similar, with a variance of only 43% (average of 1.35 × 106 ± 5.9 × 105 cells) (Fig. 4B). With respect to luciferase activity, at 2 days posttransfection, an average of 3 × 103 relative light units (RLU) per transfected cell was detected, while 10−4 RLU per untransfected cell was measured. After 17 days, <10−3 RLU/cell was detected in 80% of the wells, between 10−1 and 10−2 RLU/cell was detected in 17% of the wells, and 2 RLU/cell was detected in the well possessing the highest luciferase activity. That is, the luciferase activity of the transfected cells decreased rapidly during the time course, indicating loss of the oriP plasmid. Most importantly, there was no correlation between low cell number and high luciferase activity according to Kendall's rank correlation test (P [two sided] = 0.79). These findings demonstrate that the rapid loss of replicated oriP plasmids does not result from a selective growth advantage of the untransfected cells relative to the transfected cells.
FIG. 4.
The precipitous loss of newly introduced oriP plasmids does not result from a selective growth advantage of the untransfected cell population relative to the transfected cell population. (A) The experiment depicted was conducted as described in Results. Given the 57% transfection efficiency, 14 of every 24 wells (plated at one cell/well) should contain the oriP plasmid expressing luciferase, as indicated by dark wells. The inset illustrates two possible outcomes. For outcome 1, with no selective growth advantage of the untransfected cell population, the cells would be expected to undergo 20 to 21 doublings, resulting in 1 × 106 to 2 × 106 cells per well, with approximately 1% of the wells exhibiting high luciferase activity, indicating the loss of oriP plasmids from the majority of the cell population. In outcome 2, the untransfected cell population has a selective growth advantage, so only those untransfected cells (43% of the population) would give rise to 1 × 106 to 2 × 106 cells per well after 17 days. In this scenario, the transfected cells (57% of the population) would give rise to ≤2% of the number of cells of the untransfected population (i.e., ≤2 × 104 cells/well) and would exhibit high luciferase activity per cell. (B) Shown is a dot plot depicting the number of cells which accumulated after 17 days from 34 separate wells. The quartiles are indicated by dashed lines. The transfected and untransfected cells which accumulated after 17 days were similar, with a variance of only 43% (average of 1.35 × 106 ± 5.9 × 105 cells). Two wells contained few cells, with only 3 × 103 and 5 × 104 cells present. The luciferase activity from each well was normalized to the number of cells present in the well. The two wells containing the highest luciferase activity are indicated by boxes (0.4 RLU/cell for the well containing 2.2 × 106 cells; 2.3 RLU/cell for the well containing 5.3 × 104 cells). According to Kendall's rank correlation test, there is no correlation between low cell number and high luciferase activity (P [two-sided] = 0.79).
Inefficient establishment of oriP plasmids in multiple cell lines.
That the rapid loss of replicated oriP plasmids is not the result of a selective growth disadvantage indicates that the majority of transiently transfected cells support oriP's replication inefficiently. Yet oriP-positive cell clones in which oriP plasmids are replicated (i.e., synthesized and partitioned faithfully) in 96 to 98% of cell cycles can be selected (20, 47). Given these disparate results, we predicted that the establishment of an oriP replicon in human cell lines would be inefficient. To test this prediction, an oriP plasmid expressing EBNA-1 and hygromycin phosphotransferase and a plasmid expressing EGFP were cotransfected into H1299, 293, and 143B cells. At 2 days posttransfection, the percentage of EGFP-positive cells was determined and cells were plated at 105, 104, and 103 cells per 15-cm dish in media containing hygromycin B. Selection was applied for 3 to 4 weeks, after which the drug-resistant colonies were enumerated. In addition, the cloning efficiency of each cell line was determined by plating one cell per well onto a 96-well dish and counting those wells with and without cell growth after approximately 2 weeks in the absence of selection. Compilation of these data allowed calculation of the establishment efficiency, which is defined as follows: (no. of drug-resistant colonies/no. of transfected, clonable cells plated) × 100%. For H1299 and 293 cells, approximately 1% of clonable cells transfected with the oriP plasmid established a drug-resistant colony which supported oriP's efficient replication (Table 1 and Fig. 5). In 143B cells, only 6 to 10% of transfected, clonable cells gave rise to drug-resistant colonies. These experiments demonstrate that the establishment of an oriP replicon is inefficient in multiple human cell lines. This inefficient establishment of EBV's oriP replicon correlates with the precipitous loss of oriP plasmids during 2 weeks posttransfection. Together these observations support a model in which replicated oriP plasmids are lost rapidly from the majority of EBNA-1-expressing cells but a minority of transfected cells supports efficient replication of oriP plasmids and gives rise to established, drug-resistant colonies.
TABLE 1.
Establishment of an oriP replicon in multiple cell lines
| Cell line | Avg no. of drug-resistant coloniesa | No. of transfected cells plated (103)b | Cloning efficiencyc(%) | Establishment efficiencyd(%) |
|---|---|---|---|---|
| H1299 | 76 | 5.0 | 79 | 1.9 |
| 93 | 13 | 0.9 | ||
| 293 | 12.5 | 5.8 | 31 | 0.7 |
| 16.5 | 4.6 | 1.2 | ||
| 18e | 5.6 | 1.1 | ||
| 143B | 132 | 2.2 | 60 | 9.9 |
| 56.5 | 1.5 | 6.2 |
Numbers reflect the average of 2 dilutions of cells plated.
Transfection efficiency is based upon the percentage of EGFP-positive cells present at 2 days posttransfection.
Cloning efficiency refers to the percentage of cells which are competent to plate and form a colony in the absence of selection and is defined by the variable m in the Poisson distribution, as described in Materials and Methods. Percentages are based on two independent experiments.
Establishment efficiency refers to the percentage of cells which take up the oriP plasmid and support its stable replication, thereby generating a drug-resistant colony. The establishment efficiency is defined by the following equation: (no. of drug-resistant colonies/no. of transfected, clonable cells plated) × 100%.
Number reflects a single dilution of cells plated.
FIG. 5.
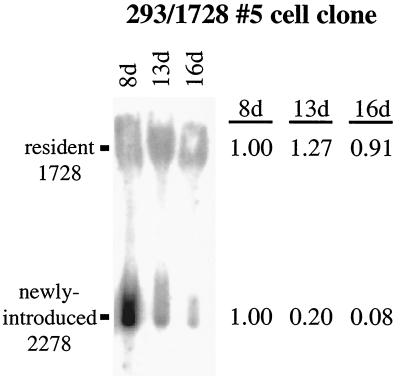
Replicated, newly introduced oriP plasmids are lost precipitously from an established, oriP-positive cell clone, while resident oriP plasmids are stable. Equimolar amounts of oriP test plasmid (2278) and prokaryotic backbone plasmid (2276) were introduced into the established oriP-positive cell clone 293/1728#5, which harbors the 1728 oriP/EBNA-1 expression plasmid. Plasmid DNA was isolated by Hirt extraction at the indicated time points posttransfection and digested with XhoI and DpnI. The levels of replicated, DpnI-resistant 1728 (resident 1728) and 2278 (newly introduced 2278) were determined by Southern blotting. The level of replicated DNA detected at 8 days posttransfection was set to 1 for each of the two plasmids. The Hirt DNAs ran aberrantly in the gel due to the presence of contaminating chromosomal and mitochondrial DNA. Approximately 5% of chromosomal DNA is present in the Hirt extract (3 μg per 107 cells).
Newly introduced oriP plasmids replicate but are also lost precipitously in established oriP-positive cell clones.
Why is a minority of the cell population competent to support efficient replication of oriP plasmids but the majority rapidly loses replicated oriP plasmids? A hereditable alteration in the minority of the cell population, manifested at the genetic or epigenetic level, may ensure faithful replication of oriP plasmids. Alternatively, a hereditable alteration in the newly introduced oriP plasmid may be required for its establishment. To distinguish between these possibilities, we determined the fate of newly introduced oriP plasmids in drug-resistant cell clones which harbor established oriP replicons. If a hereditable alteration within a cell clone is responsible for the establishment of oriP plasmids, newly introduced oriP plasmids should be efficiently replicated within them. However, if modification of the newly introduced oriP plasmid is required for its establishment, newly introduced oriP plasmids should replicate but should be lost from the majority of cells as they are from other recipients. For this experiment, an oriP plasmid expressing EBNA-1 and hygromycin phosphotransferase was introduced into the H1299 and 293 cell lines and hygromycin-resistant cell clones, referred to as H1299/1728#3, 293/1728#1, and 293/1728#5, were isolated. These cell clones maintained 1728 as a plasmid and expressed EBNA-1 functionally (data not shown). Equimolar amounts of oriP test plasmid and prokaryotic backbone plasmid were introduced into these cell clones, the clones were grown in the absence of selection for the resident oriP plasmid (1728), and the fate of the plasmids was monitored over 2 weeks posttransfection by Southern blotting or quantitative competitive PCR.
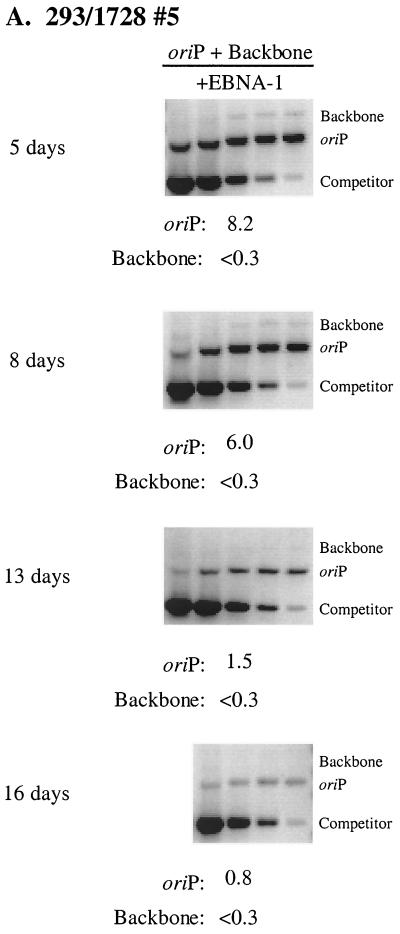
A Southern blot analysis was conducted to monitor the fate of the resident 1728 plasmid in comparison to the newly introduced oriP test plasmid in the 293/1728#5 cell clone. The level of replicated, resident plasmid 1728 remained stable over the time course, whereas the level of replicated, newly introduced plasmid 2278 decreased by a factor of 12 over the 8-day time period (Fig. 5). In parallel with the Southern blot analysis, the levels of replicated, newly introduced oriP test plasmid and prokaryotic backbone plasmid were determined by quantitative competitive PCR. At 5 days posttransfection in the 293/1728#5 cell clone, the level of replicated oriP test plasmid was at least 27-fold greater than the level of replicated backbone plasmid (Fig. 6A). As also seen by Southern analysis, the replicated, newly introduced oriP test plasmid was rapidly lost such that the level of replicated oriP test plasmid present at 16 days posttransfection was 10% of the level present at the 5-day time point. The replicated, newly introduced oriP test plasmid similarly was lost in the H1299/1728#3 and 293/1728#1 cell clones such that the level of replicated plasmid decreased by factors of 25 and 42, respectively, during a period of 10 days (Fig. 6B), while the resident plasmid 1728 remained stable (data not shown). That is, in multiple established oriP-positive cell clones, the newly introduced oriP test plasmids supported replication but were lost precipitously while the resident oriP plasmids remained stable. These experiments demonstrate that the establishment of oriP plasmids within selected oriP-positive cell clones is not due to hereditable alteration(s) in the host cell clones.
FIG. 6.
Newly introduced oriP plasmids support replication but are lost precipitously in oriP-positive cell clones. (A) The experiment was conducted as described in the legend to Fig. 5. The level of replicated, DpnI-resistant oriP test plasmid (oriP) and prokaryotic backbone plasmid (Backbone) was determined by quantitative competitive PCR. PCRs were performed using 105 cell equivalents and a competitor DNA standard curve (9 pg, 3 pg, 600 fg, 120 fg, 24 fg). Numbers below each gel refer to the average number of molecules present per transfected cell. Note that for the 16-day time point, a PCR mixture containing 9 pg of competitor was not analyzed. (B) Graphical representation of time course experiments in the selected, oriP-positive cell clones 293/1728#5 (diamonds), H1299/1728#3 (boxes), and 293/1728#1 (triangles). Experiments were conducted as described above. For each independent experiment, the level of replicated oriP test plasmid detected at the first time point posttransfection was set to 100%. Data represent a single experiment for the 293/1728#5 and H1299/1728#3 cell clones and three independent experiments for the 293/1728#1 cell clone. A 4% rate of loss is depicted by black cross-hatched boxes.
Establishment of an oriP replicon is dependent upon its epigenetic modification.
Given that establishment of an oriP replicon is not due to a hereditable alteration in the drug-resistant cell clones, a hereditable alteration in the newly introduced oriP plasmid must be required for its establishment. Such a hereditable modification may be manifested at the genetic or epigenetic level. We therefore investigated whether established cell clones harbor intact oriP plasmids in which the DS is of wild-type sequence. (The DS is the site at which DNA synthesis initiates within oriP [12].) The DS of oriP with flanking sequences was PCR amplified from three separate Hirt extractions of the 293/1728#5 cell clone and one Hirt extraction of the H1299/1728#3 cell clone and sequenced. In these cell clones, 1728 is present as an extrachromosomal element at approximately one copy per cell as determined by Southern blotting (data not shown). These 500 nucleotides of oriP were wild type in sequence (data not shown). These results are consistent with previous studies in which plasmids containing two copies of oriP were rescued from established cell clones and introduced into E. coli. Both copies of oriP were shown to be intact by restriction endonuclease digestion, and each functioned to support long-term replication (47).
While oriP is intact in established cell clones, genetic alteration(s) in the plasmid backbone may promote the replicon's establishment. Hence, we addressed whether oriP plasmids isolated from an established cell clone functionally behave as do native oriP plasmids. If genetic modification of an oriP plasmid were required for its establishment, the plasmids rescued from a drug-resistant cell clone should be established efficiently when reintroduced into mammalian cells. The 293/p220#2 drug-resistant cell clone was utilized for this experiment because it harbors 20 to 50 established p220 replicons per cell (Jun Komano, personal communication). p220 is a derivative of p201, a plasmid used historically to identify EBNA-1 as the sole viral trans-acting factor required for the stable replication of oriP plasmids (52). p220 contains oriP, the beta-lactamase gene and origin of replication from pBR322, and encodes hygromycin B phosphotransferase and EBNA-1. p220 was rescued from the 293/p220#2 established cell clone and introduced into E. coli. Plasmid DNA was isolated from a population of approximately 2,000 colonies to ensure representation of the repertoire of established oriP plasmids from the cell clone. This rescued p220 DNA was then reintroduced into 293 cells and the establishment efficiency was measured (Table 2). The rescued p220 plasmid population was established in only 5.5% of transfected, clonable cells, in a manner analogous to the establishment of the native p220 plasmid (P [two sided] = 0.44, Wilcoxon rank sum test). The plasmid p220ΔDS, in which the DS of oriP was deleted from p220, served as a control plasmid that cannot support efficient, EBNA-1-dependent DNA synthesis. pHEBo, the parental plasmid of p220 that lacks EBNA-1-encoding sequences, was used as a control oriP plasmid that cannot support long-term replication in the absence of EBNA-1. The establishment efficiency of these plasmids was significantly different from that of the native p220 plasmid (P [two sided] = 0.04 when each plasmid is compared to p220; Wilcoxon rank sum test). These experiments demonstrate that genetic alteration(s) of an oriP plasmid does not contribute to its establishment. Rather, the establishment of an oriP replicon is dependent upon an epigenetic event with respect to the replicon, which occurs infrequently and therefore is detected in only a minority of transfected cells.
TABLE 2.
Establishment of rescued oriP plasmid populations in 293 cells
| Plasmid | Avg no. of drug-resistant coloniesa | No. of transfected cells plated (103)b | Cloning efficiency (%)b | Establishment efficiency (%)b |
|---|---|---|---|---|
| p220 | 57.5 | 3.3 | 5.6 | |
| 49.0 | 2.7 | 31 | 5.9 | |
| 60.5 | 3.5 | 5.6 | ||
| rescued p220 | 36.0 | 2.4 | 31 | 4.8 |
| 58.0 | 2.7 | 6.9 | ||
| 67.5 | 3.4 | 6.4 | ||
| 46.0 | 2.8 | 5.2 | ||
| 39.0 | 3.0 | 4.2 | ||
| 51.0 | 3.1 | 5.3 | ||
| p220ΔDS | 3.0c | 3.2 | 31 | 0.3 |
| 3.0c | 3.3 | 0.3 | ||
| 1.0c | 3.4 | 0.09 | ||
| pHEBo | 1.0 | 3.2 | 31 | 0.1 |
| 1.0 | 3.3 | 0.1 | ||
| 3.0 | 2.9 | 0.3 |
Numbers reflect the average of 2 dilutions of cells plated.
Transfection, cloning, and establishment efficiencies are as defined in footnotes b, c, and d of Table 1.
Number reflects a single dilution of cells plated.
DISCUSSION
Epigenetic modification of an oriP replicon is required for its establishment.
We have monitored the fate of oriP plasmids from approximately 4 days posttransfection to more than 2 weeks posttransfection in the absence and presence of EBNA-1 in multiple cell lines. We have found that the level of replicated oriP plasmid present at 4 to 6 days posttransfection in the absence of EBNA-1 is approximately fivefold greater than the level of replicated prokaryotic backbone plasmid, consistent with the findings of Aiyar et al. (3). (These plasmids are isogenic except for the presence of oriP and a length polymorphism for distinction by PCR.) However, when EBNA-1 is provided in trans, the level of replicated oriP plasmid increased up to 64-fold. Unexpectedly, we found that these replicated oriP plasmids were lost at a rate of approximately 30% per cell generation during the time course in cell lines in which an oriP plasmid expressing EBNA-1 was cointroduced (H1299, 293, and 143) and in cell lines in which EBNA-1 was expressed stably (293/EBNA-1 and C33A/EBNA-1). This precipitous loss of oriP plasmids from recipient cells was not due to a growth disadvantage of the transfected cells, as equivalent numbers of transfected and untransfected cells accumulated after 17 days in culture. The rapid loss of oriP plasmids from the transiently transfected cell population contrasts with the loss of oriP plasmids from established cell clones, in which oriP plasmids are lost at a rate of 2 to 4% per cell generation after removal of selection (20, 47). These discrepant findings indicated that only a fraction of cells transfected with an oriP plasmid might support its efficient replication. Indeed, we found that only 1 to 10% of those cells transfected with an oriP plasmid expressing EBNA-1 and hygromycin phosphotransferase gave rise to drug-resistant clones in which an oriP replicon was established. When oriP plasmids were introduced into established oriP-positive cell clones, the newly introduced oriP plasmids supported replication but were lost precipitously while the resident oriP plasmids were stable. This experiment demonstrates that a hereditable alteration in the cell clones is not responsible for establishment of an oriP replicon. In addition, genetic alteration(s) of the plasmid does not promote the replicon's establishment. Rather, our experiments reveal that an infrequent, epigenetic event is required for the establishment of an oriP replicon. This epigenetic event must allow efficient DNA synthesis and/or partitioning, perhaps in a manner analogous to one of the epigenetic events required for origin function in S. cerevisiae and for faithful partitioning by CEN elements in S. pombe (11, 14, 41, 42). By defining this event, we will gain insight into the regulatory events involved in the stable replication of oriP plasmids, and perhaps into those events essential for replication fidelity in mammalian cells.
Why is an epigenetic event required for the establishment of a stable oriP replicon? The study of plasmids containing derivatives of oriP has provided some clues. oriP is composed of two cis-acting elements—the family of repeats (FR) and the DS. The FR, which contains 20 imperfect 30-bp repeats recognized by EBNA-1 dimers (5, 33), is thought to contribute to the partitioning of plasmids. For example, plasmids containing the FR and a putative chromosomal or viral origin support long-term replication in EBNA-1-expressing cells (21, 24). The DS contains four EBNA-1 binding sites of lower affinity (5, 33) and is the site at or near which DNA synthesis initiates (12). While both the FR and DS are required for efficient replication in some cell lines (35), we and others have found that plasmids containing only the DS of oriP support replication as efficiently as oriP plasmids at 2 to 4 days posttransfection in 293, HeLa, and C33A cells when EBNA-1 is provided in trans (15, 38, 50; Leight and Sugden, unpublished observations). These findings indicate that the DS is competent to support DNA synthetic and partitioning functions in some cell lines. We therefore monitored the replication of plasmids containing only the DS over 2 weeks posttransfection in 293/EBNA-1 cells that stably express EBNA-1. Surprisingly, we observed that while replicated oriP plasmids were lost precipitously, the replicated DS plasmids were stable during the time course and were present at greater than 100 copies per transfected cell, indicative of efficient synthesis and partitioning each cell generation (E. Leight and B. Sugden, unpublished data). That is, DS plasmids are established efficiently in some cell lines. Based on these findings, we propose that an epigenetic event is required to overcome an inhibitory activity of the FR of oriP. An epigenetic event may modulate the timing of origin firing to ensure completion of DNA synthesis despite the presence of a replication fork barrier imposed by EBNA-1 dimers bound to the FR (9, 12). Alternatively, an epigenetic event may promote efficient partitioning of oriP plasmids by favoring the formation of intermolecular interactions between oriP and mitotic chromosomes to the formation of intramolecular interactions (i.e., looping) between EBNA-1 dimers bound to the FR and DS (27, 28, 40, 44). Given that global methylation has no effect on the establishment of an oriP replicon (B. Sugden, unpublished observations), we favor a mechanism in which chromatin structure, as opposed to covalent modification, affects the replication efficiency of oriP plasmids.
Implications for studies of EBV.
We have found that oriP plasmids are not intrinsically stable in transfected cells but rather must be modified epigenetically in order to support efficient DNA synthesis and partitioning each cell cycle. But is such modification required for the establishment of EBV itself? Two lines of evidence demonstrate that EBV DNA is not intrinsically stable in newly infected cells, indicating that an epigenetic event is required for the establishment of EBV's genome. Firstly, Reisman and Sugden have shown that upon infection of the EBV-positive B-lymphoblast cell line TG8 with the B95-8 strain of EBV, 85% of the superinfecting EBV DNA is lost over 7 days, while the resident EBV DNA (P3HR1) remains stable (34). The B95-8 strain of EBV established a transient infection in these experiments in that 20 to 30% of the B95-8 genomes were circularized by 2 days postinfection. (This circularized DNA was undetectable by 7 days postinfection.) In addition, while EBNAs are undetectable in TG8 cells, 60% of the cells were EBNA positive at 2 days postinfection with the B95-8 strain of EBV. By 10 to 24 days postinfection, only 1% of cells remained EBNA positive. The findings of Reisman and Sugden (34) mirror the precipitous loss of oriP plasmids observed in our studies and indicate that inefficient establishment occurs upon viral infection as well as transfection of an oriP replicon.
A second line of evidence demonstrates that EBV is established inefficiently in cells, as are other oriP replicons. In these studies a recombinant Akata strain of EBV (carrying the neomycin resistance gene) was used to infect epithelial and B-cell lines. The infection efficiency was monitored by detection of EBNAs at 3 to 5 days postinfection, and the resulting drug-resistant colonies were enumerated. The establishment efficiency of 15 epithelial cell lines ranged from 0.06 to 14.4%, with an average of 4.0% (18). Likewise, when the recombinant Akata strain of EBV was used to infect an EBV-negative Burkitt's lymphoma cell line (Akata), 1% of infected cells gave rise to drug-resistant, EBV-positive cell clones (48). These observations with infecting EBV are consistent with our data demonstrating an inefficient establishment of transfected oriP plasmids and indicate that establishment of EBV DNA may be dependent upon its epigenetic modification.
ACKNOWLEDGMENTS
We are grateful to Paul Ahlquist, Paul Lambert, and our colleagues for commenting on the manuscript. We thank Jun Komano for sharing plasmids and Hirt-extracted DNA from the 293/p220#2 cell line. We thank Ping Hua for construction of plasmids 2275, 2276, and 2278.
This work was supported by Public Health Service grants CA-22443, CA-07175 and T32-CA-09135. Bill Sugden is an American Cancer Society Research Professor.
REFERENCES
- 1.Adams A. Replication of latent Epstein-Barr virus genomes in Raji cells. J Virol. 1987;61:1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar A, Sugden B. Fusions between Epstein-Barr viral nuclear antigen-1 of Epstein-Barr virus and the large T-antigen of simian virus 40 replicate their cognate origins. J Biol Chem. 1998;273:33073–33081. doi: 10.1074/jbc.273.49.33073. [DOI] [PubMed] [Google Scholar]
- 3.Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandru G, Zachariae W, Schleiffer A, Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambinder R F, Shah W A, Rawlins D R, Hayward G S, Hayward S D. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J Virol. 1990;64:2369–2379. doi: 10.1128/jvi.64.5.2369-2379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boddy M N, Russell P. DNA replication checkpoint control. Front Biosci. 1999;4:D841–D848. doi: 10.2741/boddy. [DOI] [PubMed] [Google Scholar]
- 7.Clarke D J, Gimenez-Abian J F. Checkpoints controlling mitosis. Bioessays. 2000;22:351–363. doi: 10.1002/(SICI)1521-1878(200004)22:4<351::AID-BIES5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 8.Dahmann C, Diffley J F, Nasmyth K A. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 9.Dhar V, Schildkraut C L. Role of EBNA-1 in arresting replication forks at the Epstein-Barr virus oriP family of tandem repeats. Mol Cell Biol. 1991;11:6268–6278. doi: 10.1128/mcb.11.12.6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimitrova D S, Gilbert D M. Regulation of mammalian replication origin usage in Xenopus egg extract. J Cell Sci. 1998;111:2989–2998. doi: 10.1242/jcs.111.19.2989. [DOI] [PubMed] [Google Scholar]
- 11.Ekwall K, Olsson T, Turner B M, Cranston G, Allshire R C. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 12.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert D M, Miyazawa H, DePamphilis M L. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol Cell Biol. 1995;15:2942–2954. doi: 10.1128/mcb.15.6.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grewal S I, Bonaduce M J, Klar A J. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics. 1998;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison S, Fisenne K, Hearing J. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1994;68:1913–1925. doi: 10.1128/jvi.68.3.1913-1925.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayles J, Fisher D, Woollard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 17.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 18.Imai S, Nishikawa J, Takada K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J Virol. 1998;72:4371–4378. doi: 10.1128/jvi.72.5.4371-4378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchmaier A L, Sugden B. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J Virol. 1997;71:1766–1775. doi: 10.1128/jvi.71.3.1766-1775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchmaier A L, Sugden B. Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J Virol. 1995;69:1280–1283. doi: 10.1128/jvi.69.2.1280-1283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchmaier A L, Sugden B. Rep∗: a viral element that can partially replace the origin of plasmid DNA synthesis of Epstein-Barr virus. J Virol. 1998;72:4657–4666. doi: 10.1128/jvi.72.6.4657-4666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knutson J C, Yee D. Electroporation: parameters affecting transfer of DNA into mammalian cells. Anal Biochem. 1987;164:44–52. doi: 10.1016/0003-2697(87)90365-4. [DOI] [PubMed] [Google Scholar]
- 23.Koshland D, Kent J C, Hartwell L H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985;40:393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- 24.Krysan P J, Haase S B, Calos M P. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989;9:1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leight E R, Sugden B. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev Med Virol. 2000;10:83–100. doi: 10.1002/(sici)1099-1654(200003/04)10:2<83::aid-rmv262>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 26.Lupton S, Levine A J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985;5:2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackey D, Sugden B. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol Cell Biol. 1999;19:3349–3359. doi: 10.1128/mcb.19.5.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marechal V, Dehee A, Chikhi B R, Piolot T, Coppey M M, Nicolas J C. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J Virol. 1999;73:4385–4392. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell T, Sugden B. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navas T A, Zhou Z, Elledge S J. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 32.Piatti S, Bohm T, Cocker J H, Diffley J F, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 33.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 34.Reisman D, Sugden B. An EBNA-negative, EBV-genome-positive human lymphoblast cell line in which superinfecting EBV DNA is not maintained. Virology. 1984;137:113–126. doi: 10.1016/0042-6822(84)90014-x. [DOI] [PubMed] [Google Scholar]
- 35.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowles A, Blow J J. Chromatin proteins involved in the initiation of DNA replication. Curr Opin Genet Dev. 1997;7:152–157. doi: 10.1016/s0959-437x(97)80123-2. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Shirakata M, Hirai K. Identification of minimal oriP of Epstein-Barr virus required for DNA. Biochemistry (Tokyo) 1998;123:175–181. doi: 10.1093/oxfordjournals.jbchem.a021907. [DOI] [PubMed] [Google Scholar]
- 39.Shirakata M, Imadome K I, Hirai K. Requirement of replication licensing for the dyad symmetry element-dependent replication of the Epstein-Barr virus oriP minichromosome. Virology. 1999;263:42–54. doi: 10.1006/viro.1999.9965. [DOI] [PubMed] [Google Scholar]
- 40.Shire K, Ceccarelli D F, Avolio H T, Frappier L. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J Virol. 1999;73:2587–2595. doi: 10.1128/jvi.73.4.2587-2595.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson R T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 42.Steiner N C, Clarke L. A novel epigenetic effect can alter centromere function in fission yeast. Cell. 1994;79:865–874. doi: 10.1016/0092-8674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 43.Stevenson J B, Gottschling D E. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su W, Middleton T, Sugden B, Echols H. DNA looping between the origin of replication of Epstein-Barr virus and its enhancer site: stabilization of an origin complex with Epstein-Barr nuclear antigen 1. Proc Natl Acad Sci USA. 1991;88:10870–10874. doi: 10.1073/pnas.88.23.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugden, B., and E. R. Leight. EBV's plasmid replicon: an enigma in cis and trans. Curr Top Microbiol Immunol., in press. [DOI] [PubMed]
- 46.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugden B, Warren N. Plasmid origin of replication of Epstein-Barr virus, oriP, does not limit replication in cis. Mol Biol Med. 1988;5:85–94. [PubMed] [Google Scholar]
- 48.Yang L, Maruo S, Takada K. CD21-mediated entry and stable infection by Epstein-Barr virus in canine and rat cells. J Virol. 2000;74:10745–10751. doi: 10.1128/jvi.74.22.10745-10751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yates J L, Camiolo S M, Bashaw J M. The minimal replicator of Epstein-Barr virus oriP. J Virol. 2000;74:4512–4522. doi: 10.1128/jvi.74.10.4512-4522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]