Abstract
Background: The role of bacterial co-infection and superinfection among critically ill COVID-19 patients remains unclear. The aim of this study was to assess the rates and characteristics of pulmonary infections, and associated outcomes of ventilated patients in our facility. Methods: This was a retrospective study of ventilated COVID-19 patients between March 2020 and March 2021 that underwent BioFire®, FilmArray® Pneumonia Panel, testing. Community-acquired pneumonia (CAP) was defined when identified during the first 72 h of hospitalization, and ventilator-associated pneumonia (VAP) when later. Results: 148 FilmArray tests were obtained from 93 patients. With FilmArray, 17% of patients had CAP (16/93) and 68% had VAP (64/93). Patients with VAP were older than those with CAP or those with no infection (68.5 vs. 57–59 years), had longer length of stay and higher mortality (51% vs. 10%). The most commonly identified FilmArray target organisms were H. influenzae, S. pneumoniae, M. catarrhalis and E. cloacae for CAP and P. aeruginosa and S. aureus for VAP. FilmArray tests had high negative predictive values (99.6%) and lower positive predictive values (~60%). Conclusions: We found high rates of both CAP and VAP among the critically ill, caused by the typical and expected organisms for both conditions. VAP diagnosis was associated with poor patient outcomes.
Keywords: COVID-19, co-infection, superinfection, community-acquired pneumonia, ventilator-associated pneumonia, FimArray, PCR, critically ill, intensive-care unit
1. Introduction
Patients hospitalized in intensive care units (ICUs) with critical novel coronavirus disease 2019 (COVID-19) have a high mortality rate, reaching ~50% (range 16–78%) [1,2]. The causes contributing to mortality are respiratory failure with severe hypoxemia and its immediate consequences, multiorgan failure, thromboembolism, hemorrhage, and healthcare-associated infections (HAIs) associated with bacteria and filamentous fungi. Bacterial co-infections, diagnosed around the time of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, appear to be uncommon, occurring in 0.6 to 3.2% of patients [3], although some studies reported higher rates, reaching 26–28% [4,5,6] or more [7]. The co-infection rates in COVID-19 seem to be lower than those reported for influenza pandemics [8,9]. The main pathogens associated with co-infections are S. aureus, S. pneumoniae and H. influenzae [4,6,10], while in some studies S. pneumoniae takes a minor role [7,8,10,11,12].
As the pandemic unfolds and more data come to light, the role of secondary infections, i.e., HAIs, among critical COVID-19 patients in the ICU, becomes more pronounced. Superinfection rates in ICUs were reported in 13.5–48% of patients [10,13,14,15], and most reported cases of HAIs include ventilator-associated pneumonia (VAP), with P. aeruginosa the main causative agent [10]. Superinfection may be becoming more of an issue now because of increased length of ICU stay secondary to improved overall COVID-19 survival rates, the now universal use of corticosteroid therapy and increased use of other immunomodulatory drugs such as tocilizumab and baricitinib, and improved reports from focused studies conducted to assess COVID-19-related infectious complications. Outbreaks of multidrug resistant bacteria are also commonly reported, including a variety of organisms such as carbapenem resistant Acinetobacter baumannii, carbapenem resistant Enterobacterales, multidrug resistant Pseudomonas aeruginosa, methicillin resistant Staphylococcus aureus (MRSA), Candida auris and others [2,16,17,18]. Since most COVID-19 patients in the ICUs are treated empirically with antimicrobials [4,19], antibiotic stewardship programs along with enhanced infection control protocols are essential in controlling the emergence of these dreaded infections.
Israel is currently (October 2021) in the midst of the fourth COVID-19 wave. The three previous waves occurred between March 2020 and March 2021. Very few clinical data have emerged from Israel during this period of time regarding critically ill COVID-19 patients, rates of co-infection and superinfection, or their outcomes. In this study, we primarily aimed to analyze these aspects retrospectively among COVID-19 patients who were admitted to Sanz Medical center and needed mechanical ventilation. We focused on data from a rapid multiplex polymerase chain reaction (mPCR) kit for the molecular identification of lower respiratory tract pathogens (Biofire®, FilmArray® Pneumonia Panel, bioMérieux, Marcy-l’Étoile, France), as well as, and in comparison with, classical microbiology methods. Our secondary aims were to assess the antimicrobial decisions related to the results of this rapid test in the context of an antimicrobial stewardship program.
2. Materials and Methods
2.1. Study Design
This was a retrospective observational study including patients hospitalized between March 2020 and March 2021 in the COVID-19 ICU of Sanz Medical Center, a university-affiliated, 400-bed regional hospital, located in the city of Netanya, Israel.
2.2. Patient Selection
COVID-19 patients who were mechanically ventilated at some point during their hospitalization course were included in the study if they underwent FilmArray® (FA) testing. FA tests were conducted at physicians’ discretion with infectious diseases expert approval. Tests were used to assist physicians in the diagnosis of a secondary community-acquired pneumonia (CAP, when suspected during the first 72 h from admission), or VAP (after 72 h from admission). PCR and cultures were obtained when patients showed signs of clinical and respiratory deterioration; when they developed sepsis or septic shock; when bacterial infection was suspected (recurring fever, the appearance of a new infiltrate on imaging or elevation of inflammatory indexes) and typically, immediately after intubation that facilitated the acquisition of respiratory samples.
2.3. Microbiology Samples
FA tests were performed on sputum samples, endotracheal aspirates (ETA, suctioned from intubated patients), non-bronchoscopic lavage (NBL) fluid and from bronchoalveolar lavage (BAL, when performed by a pulmonologist) in accordance with the manufacturer’s instructions. The results were directly reported to the treating physician and to the infectious diseases expert that requested the test. The same specimens that were used for FA testing were typically also used for conventional microbiology methods. In cases for which this was unavailable, new samples were requested. For comparisons between the FA results and microbiology cultures, we included only cases for which the same sample was used for both analyses or the time interval between FA and culture samples was ±2 days.
Samples were performed, analyzed and results reported in accordance with the Clinical Microbiology Procedures Handbook [20]. Samples were inoculated on blood (trypticase soy agar enriched with 5% sheep blood), chocolate blood, Columbia nalidixic acid, MacConkey, CHROMagar™ Orientation agars (Hy Laboratories, Rehovot, Israel). Suspected colonies were identified using a VITEK® 2 system (bioMérieux, Marcy-l’Étoile, France). Antibiotic susceptibility testing was performed using a VITEK® 2 system and confirmed by disk diffusion. Susceptibility was interpreted according to current CLSI guidelines [21]. Methicillin (oxacillin) resistance in S. aureus was determined by resistance to cefoxitin by disk diffusion and confirmed by the presence of the SCCmec/mecA genetic elements [21].
COVID-19 was detected using PCR platforms (Xpert® Xpress SARS-CoV-2, Cepheid, Sunnyvale, CA, USA; BD SARS-CoV-2, Beckton Dickinson, Franklin Lakes, NJ, USA; Allplex™ 2019-nCoV Assay, Seegene Inc, Seoul, Korea).
2.4. Evaluation of FA Results as Positive
Since FA test results are reported with a quantitative assessment of the DNA load, the source of the sample (sputum vs. ETA, NBL or BAL) is important to assess the relevance of the positive result. For this retrospective analysis we used two sets of rules to assign a positive FA result as such. A strict set was used with thresholds of: ≥104 genome copies/mL for BAL samples, ≥105 for ETA and NBL samples and 107 for sputum samples as positive. A second less strict set was used: ≥104 for BAL, ETA and NBL and ≥106 for sputum samples.
For comparison between cultures and FA tests we considered all the microorganisms identified in each culture, with the exclusion of Candida species and Enterococcii, which were regarded as contamination. Sensitivity and specificity were calculated using culture as gold standard.
2.5. Statistical Analysis
Categorial variables were expressed as frequencies and percentages and compared using Fishers’ exact test. Continuous variables were presented as means and standard deviations (SD) or medians and interquartile range (IQR) and compared using Student’s t-test. Calculations were performed using GraphPad Prism® v.7.0(GraphPad Software LLC, San Diego, CA, USA). Statistical significance was defined when p < 0.05.
The study was authorized by the Institutional Review Board (IRB) of Sanz medical center, 0014-21-LND.
3. Results
3.1. Patients
Between March 2020 and March 2021, 887 COVID-19 patients were hospitalized in our facility, of which 132 (15%) needed mechanical ventilation at some point during their hospital stay and, of these, 93 (70%) were tested for secondary bacterial infection using the FA pneumonia panel. Of these 93 patients, 49 had one test performed during their hospitalization, 35 had two tests, seven had three tests and two had four tests; altogether 148 FA tests were included in the analysis. In most cases, each FA represented a situation in which the patient was suspected of having a bacterial infection. There were few specific cases in which young critically ill patients (for example, peri-partum ventilated women) underwent more frequent testing, especially when they were not treated with empiric antimicrobials. The mean (SD) and median (IQR) interval between repeated FA tests were 9.9 (8.2) and 8 (5–12) days, respectively.
The mean (SD) and median (IQR) patients’ age were 64.6 (15.1) and 67 (56–76.5) with male predominance (70%). Long term care facility residency was rare (5/93, 5.3%). The mean (SD) duration from COVID-19 diagnosis to hospitalization was 3.8 days (3.7) with median (IQR) of 3 days (0–6), while six patients were diagnosed with COVID-19 during their hospitalization. Mean and median length of stay (LOS) were 28.9 (17.3) and 24 days (17–38).The mean was 32.9 days for males and 19.7 days for females, p = 0.0006. In-hospital mortality was 36/93 (38.7%) and mean (SD) and median (IQR) interval from admission to death were 30.6 days (19.9) and 25 (16–39.7).
3.2. CAP and VAP Diagnosis
Forty patients were tested during the first 72 h of hospital stay for CAP diagnosis. The mean (SD) and median (IQR) interval from admission to first FA test were 1.3 (0.6) and 1 (1–2) days. Of the 93 patients, 16 (17%) had CAP, when considering the less strict set of rules. Most of these patients (15/16, 94%) were treated with antimicrobials directed for CAP and in 6/16 (37.5%) there was a concomitant positive culture result. With the strict set of rules, there were 12/93 (13%) positive FA results, of which six (50%) also had a positive culture result.
74 patients were tested for VAP (including 21 patients also tested for CAP and 53 that were tested only for VAP). The mean (SD) and median (IQR) interval from admission to first FA directed for VAP diagnosis were 8 (6.7) and 6 (4–9) days. With the less strict rules, 64/93 patients (68%) had VAP (58 patients with VAP only and 6 also had CAP).
The 58 patients tested for VAP had altogether 108 FA tests. 75/108 (69%) were FA-positive, and 33 tests were FA-negative. In 62/75 cases (82%) there was also a concurrent positive culture and in 51/75 cases (68%) the results triggered antibiotic therapy change (commencing treatment or either upgrading or downgrading a given drug) to treat VAP.
With the strict set of rules, there were 62/108 (57%) positive FA tests and in 53/62 (85%) a positive culture was recorded. More data are shown in Table 1 and Table 2.
Table 1.
Ventilated COVID-19 patients and microbiology samples characteristics.
| Patients’ Characteristics | Number, Median (IQR) Total |
|---|---|
| Patients (n = 93) | |
| Age, years | 67 (56–76.5) |
| Male (%) | 65 (70) |
| LTCF residence, number (%) | 5 (5.3) |
| Length of stay, days | 24 (17–38) |
| COVID-19 diagnosis to hospitalization *, days (n = 87) | 3 (0–6) |
| Hospitalization to FA testing, days | 3 (1.5–6) |
| For diagnosis of CAP, days (n = 40) | 1 (1–2) |
| For diagnosis of VAP, days (n = 53) | 6 (4–9) |
| Patients diagnosed with ** (%): | |
| CAP | 16 (17) |
| VAP (without CAP) | 58 (62) |
| Neither | 19 (20) |
| C-reactive protein, mg/dL | 133 (76–220) |
| Mortality (%) | 36 (38.7) |
| Hospitalization to death, days (n = 36) | 25 (16–39.7) |
| VAP *** diagnosis to death, days (n = 33) | 17 (8.5–35.5) |
| Molecular and microbiology samples | |
| FA samples n = 148 (%) | |
| ETA | 102 (69) |
| Sputum | 22 (15) |
| BAL | 16 (11) |
| NBL | 8 (5) |
| Cultures samples n = 128 (%) | |
| ETA | 88 (69) |
| Sputum | 16 (12.5) |
| BAL | 16 (12.5) |
| NBL | 8 (6) |
* Six patients were diagnosed with COVID-19 while hospitalized; ** using the less strict set of rules; *** including 6 patients who were also previously diagnosed with CAP. Abbreviations: IQR—interquartile range; LTCF—long term care facility; FA—FilmArray; CAP—community acquired pneumonia; VAP—ventilator associated pneumonia; ETA—endotracheal aspirate, BAL—bronchoalveolar lavage; NBL—non-bronchoscopic lavage.
Table 2.
Comparisons between patients’ characteristics and outcomes according to diagnosis of pneumonia using the less strict set of rules.
| Patients with Only CAP n = 10 |
Patients with VAP * n = 64 |
Patients with Neither CAP nor VAP, n = 19 |
CAP vs. VAP * p (95% CI ** or OR ***) |
CAP vs. Neither p (95% CI or OR) |
VAP * vs. Neither p (95% CI or OR) |
|
|---|---|---|---|---|---|---|
| Age (years); median (IQR) | 57 (48.2–67.5) | 68.5 (63–77) | 59 (39–69) | 0.025 (1.1 to 17.5) | NS | 0.001 (−19.6 to −5) |
| Male (%) | 6 (60) | 48 (75) | 12 (63) | NS | NS | NS |
| LTCF residence (%) | 0 | 5 (7.8) | 0 | NS | NS | NS |
| Length of stay-days, median (IRQ) | 22.5 (16.2–29.5) | 29 (21–42.2) | 17 (12–29) | 0.08 (−1.3 to 23.3) | NS | 0.003 (−23.3 to −4.7) |
| Mortality ** (%) | 1 (10) | 33 (51.5) | 2 (10.5) | 0.001 (9.5) | NS | 0.0014 (9) |
* Including 6 patients who also had CAP. ** using student’s t-test. *** using Fisher’s exact test. Abbreviations: CAP—community acquired pneumonia; VAP—ventilator-associated pneumonia; OR—odds ratio; CI—confidence interval; IQR—interquartile range; LTCF—long term care facility; NS—nonsignificant.
3.3. FA Results
Most FA tests were performed on ETA samples (102/148, 69%), and the rest were: 22 sputa (15%), 16 BAL (11%) and 8 NBL (5%).
For the strict set of rules for evaluation of FA results, positive rates were higher for tests performed for VAP vs. CAP diagnosis (62/108 (57%) vs. 12/40 (30%), (odds ratio (OR) 0.31, 95% confidence interval (CI) 0.14–0.7, p = 0.005). The most common bacterial targets were P. aeruginosa (27 samples, 36.4%), S. aureus (24 samples, 32.4%)—12 methicillin-susceptible S. aureus (MSSA) and 12 methicillin-resistant S. aureus (MRSA), and H. influenzae (15 samples, 20.2%).
Twelve FA tests were considered positive for CAP diagnosis. Of these 12, the most common targets were H. influenzae (7/12, 58.3%), S. pneumoniae (6/12, 50%) and M. catarrhalis and E. cloacae (2/12, 16%, each). Of the 62 positive FA tests performed for VAP, the most common targets found were: P. aeruginosa (26/62, 42%), S. aureus (23/62, 37%, 12 MRSA and 11 MSSA), K. pneumoniae (14/62, 22.5%), and H. influenzae (8/62, 13%), Table 3.
Table 3.
Bacteria identified by FA tests using two sets of rules among patients with CAP and VAP.
| Strict Rules * | Less Strict Rules ** | |||||
|---|---|---|---|---|---|---|
| CAP (n = 12) |
VAP (n = 62) |
Total (n = 74) |
CAP (n = 16) |
VAP (n = 75) |
Total (n = 91) |
|
| Gram negative, number of targets detected (% ***) | ||||||
| E. cloacae | 2 (16.6) | 6 (9.6) | 8 (10.8) | 2 (12.5) | 10 (13.3) | 12 (13.1) |
| E. coli | 0 | 2 (3.2) | 2 (2.7) | 0 | 3 (4) | 3 (3.2) |
| A. baumannii | 1 (8.3) | 1 (1.6) | 2 (2.7) | 1 (6.2) | 2 (2.6) | 3 (3.2) |
| K. aerogenes | 0 | 5 (8) | 5 (6.7) | 0 | 7 (9.3) | 7 (7.6) |
| K. oxytoca | 0 | 0 | 0 | 0 | 2 (2.6) | 2 (2.1) |
| K. pneumoniae | 0 | 14 (22.5) | 14 (18.9) | 2 (12.5) | 15 (20) | 17 (18.6) |
| Proteus spp. | 0 | 2 (3.2) | 2 (2.7) | 0 | 3 (4) | 3 (3.2) |
| P. aeruginosa | 1 (8.3) | 26 (41.9) | 27 (36.4) | 3 (18.7) | 28 (37.3) | 31 (34) |
| M. catarrhalis | 2 (16.6) | 2 (3.2) | 4 (5.4) | 2 (12.5) | 2 (2.6) | 4 (4.3) |
| H. influenzae | 7 (58.3) | 8 (12.9) | 15 (20.2) | 9 (56.2) | 12 (16) | 21 (23) |
| S. marcescens | 0 | 4 (6.4) | 4 (5.4) | 0 | 6 (8) | 6 (6.5) |
| Total | 13 | 70 | 83 | 19 | 90 | 109 |
| Gram positive, number of targets detected (% ***) | ||||||
| S. agalactiae | 0 | 3 (4.8) | 3 (4) | 0 | 3 (4) | 3 (3.2) |
| S. pyogenes | 0 | 0 | 0 | 2 (12.5) | 0 | 2 (2.1) |
| S. aureus | 1 (8.3) | 23 (37) | 24 (32.4) | 4 (25) | 29 (38.6) | 33 (36.2) |
| MSSA | 1 | 11 | 12 | 3 | 15 | 18 |
| MRSA | 0 | 12 | 12 | 1 | 14 | 15 |
| S. pneumoniae | 6 (50) | 4 (6.4) | 10 (13.5) | 6 (37.5) | 7 (9.3) | 13 (14.2) |
| Total | 7 | 30 | 37 | 12 | 39 | 51 |
| Total | 20 | 100 | 120 | 31 | 129 | 160 |
* Positive results were ≥104 for BAL, ≥105 for ETA and NBL, 107 for sputum. ** Positive results were ≥104 for BAL, ETA and NBL and ≥106 for sputum. *** Totals exceed 100% since many patients had more than 1 target found. Abbreviations: CAP—community acquired pneumonia, VAP—ventilator-associated pneumonia, FA—FilmArray, MSSA—Methicillin-sensitive S. aureus, MRSA—Methicillin-resistant S. aureus.
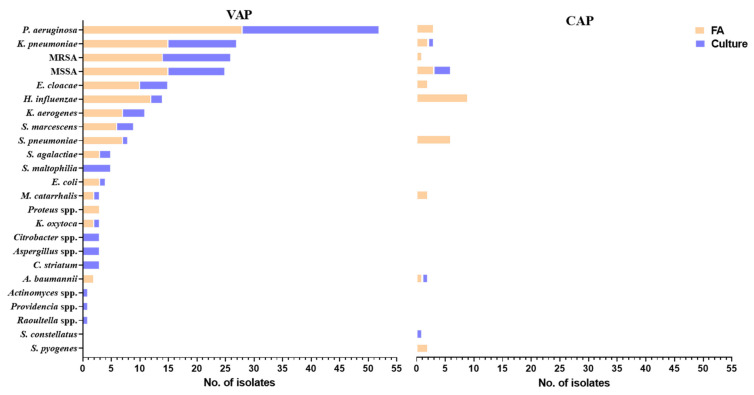
For the less stringent set of rules, positive results were more common: 91/148 (61%); 16/40 (40%) for CAP and 75/108 (69%), OR = 0.29, 95% CI = 0.13–0.64, p = 0.002. The same patterns of bacterial distribution as in the strict set were found and are depicted in Table 3 and Figure 1.
Figure 1.
Absolute number of isolates identified by FA and standard cultures, in patients with CAP and VAP.
Three FA tests were positive for viruses: two were positive to Rhinovirus/Enterovirus and one for Parainfluenza virus. CTX-M was positive in 17 and 20 FA results (including one case of sputum 104 and one of NBL 105) according respectively to the two set of rules. None of the resistance genes were detected among the tests performed for CAP diagnosis.
3.4. Associated Outcomes Related to FA Results
Patients with VAP diagnosis were significantly older than those with CAP or those with neither diagnosis (median age 68.5 vs. 57 and 59, p = 0.025 and p = 0.001, respectively). These patients also had longer hospitalization duration medians (29 vs. 22.5 and 17 days, p = 0.08 and p = 0.003). In-hospital mortality was 1/10 and 2/19 (10%) among patient with CAP or with neither diagnosis respectively, vs. 33/64 (51.5%) in patients with VAP (p = 0.001) (Table 2).
3.5. Culture Results
Out of 148 FA tests, for 128 (86%) there was a comparable microbiologic culture, of which a total of 105 bacteria/fungi were identified. In 54 cases the cultures were negative, one microorganism was cultured in 44 cases, two microorganisms in 30 cases and three bacteria in one case. ETA was the source of the culture in 88/128 cases (68%), and the rest were: 16 BAL, 16 sputa and eight NBL. The bacteria found were: 24 P. aeruginosa, 13 MSSA, 13 K. pneumoniae, 12 MRSA, 8 E. cloacae, five S. maltophilia, four K. aerogenes, three Citrobacter spp., three Aspergillus spp. and 20 other bacteria (Figure 1).
3.6. Comparison between FA and Culture Results
Of the 128 comparable FA-culture couples, 46 were FA-negative of which 38 (82%) were also culture-negative. Of the 82 FA-positive samples, 66 (80%) were also culture positive (the same pathogens appearing as in the FA panel results in 60/82, 73%). Of the 16 FA-positive/culture negative cases, eight were treated with antimicrobials at sampling time, a fact that could affect culture positivity. Interestingly, in four of these 16 cases, a subsequent culture was positive for the same microorganism that was previously detected in the FA panel (1 each: E. cloacae, K. pneumoniae, H. influenzae and S. pyogenes).
We evaluated the performance of FA as compared with the gold standard of classical microbiology cultures using the strict and less strict rules (Table 4). By applying the strict rules, the overall sensitivity and specificity of the FA test were 78.4% and 98.1% respectively, with positive and negative predictive values (PPV and NPV) of 66.3% and 98.9%. With less strict panel of rules, the sensitivity increased to 92.2% with a reciprocal reduction in the specificity to 96.6%. The lowest sensitivities were notable in cases of H. influenzae, A. baumannii, E. cloacae and MSSA
Table 4.
Analytical performance of FA compared to culture using a strict and less strict sets of rules (Strict */Less strict **).
| FA(+)/Culture(+) | FA(+)/Culture(−) | FA(−)/Culture(+) | FA(−)/Culture(−) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|---|---|
| Gram negative | ||||||||
| E. cloacae | 7/7 | 1/5 | 2/2 | 118/114 | 77.7/77.7 | 99.1/95.8 | 87.5/58.3 | 98.3/98.3 |
| E. coli | 1/1 | 0/1 | 0/0 | 127/126 | 100/100 | 100/99.2 | 100/50.0 | 100/100 |
| A. baumannii | 0/0 | 1/2 | 1/1 | 126/125 | 0/0 | 99.2/98.4 | 0/0 | 99.2/99.2 |
| K. aerogenes | 2/4 | 2/2 | 2/0 | 122/122 | 50/100 | 98.4/98.4 | 50/66.6 | 98.4/100 |
| K. oxytoca | 0/1 | 0/1 | 1/0 | 127/126 | 0/100 | 100/99.2 | -/50 | 99.2/100 |
| K. pneumoniae | 10/13 | 4/4 | 4/1 | 110/110 | 71.4/92.8 | 96.5/96.5 | 71.4/76.4 | 96.5/99.1 |
| Proteus spp. | 0/1 | 2/2 | 0/0 | 126/125 | -/100 | 98.4/98.4 | 0/33.3 | 100/100 |
| P. aeruginosa | 22/24 | 0/2 | 2/0 | 104/102 | 91.6/100 | 100/98 | 100/92.3 | 98.1/100 |
| M. catarrhalis | 1/1 | 1/1 | 0/0 | 126/126 | 100/100 | 99.2/99.2 | 50/50 | 100/100 |
| H. influenzae | 2/2 | 11/17 | 1/1 | 114/108 | 66.6/66.6 | 91.2/86.4 | 15.3/10.5 | 99.1/99.1 |
| S. marcescens | 1/2 | 2/3 | 0/0 | 125/123 | 100/100 | 98.4/97.6 | 33.3/40 | 100/100 |
| Total | 46/56 | 24/40 | 13/5 | 1325/1307 | 77.9/91.8 | 98.2/97 | 65.7/58.3 | 99.0/99.6 |
| Δ *** | +10 | +16 | −8 | −18 | +13.9 | −1.2 | −7.4 | +0.6 |
| Gram positive | ||||||||
| S. agalactiae | 2/2 | 1/1 | 0/0 | 125/125 | 100/100 | 99.2/99.2 | 66.6/66.6 | 100/100 |
| S. pyogenes | 0/1 | 0/1 | 1/0 | 127/126 | 0/100 | 100/99.2 | NA/50 | 99.2/100 |
| S. pneumoniae | 1/1 | 7/10 | 0/0 | 120/117 | 100/100 | 94.4/92.1 | 12.5/9.1 | 100/100 |
| S. aureus | 20/23 | 3/9 | 5/2 | 100/94 | 80/92 | 97.1/91.2 | 86.9/71.8 | 95.2/97.9 |
| MSSA | 9/11 | 2/6 | 4/2 | 113/109 | 69.2/84.6 | 98.3/94.8 | 81.8/64.7 | 96.6/98.2 |
| MRSA | 11/12 | 1/5 | 1/0 | 115/111 | 91.6/100 | 99.1/95.7 | 91.6/70.6 | 99.1/100 |
| Total | 23/27 | 11/21 | 6/2 | 472/462 | 79.3/93.1 | 97.7/95.6 | 67.6/56.2 | 98.7/99.5 |
| Δ | +4 | +10 | −4 | −10 | +13.8 | −2.1 | −11.4 | +0.8 |
| Total | 69/83 | 35/61 | 19/7 | 1797/1769 | 78.4/92.2 | 98.1/96.6 | 66.3/57.6 | 98.9/99.6 |
| Δ | +14 | +26 | −12 | −28 | +13.8 | −1.5 | −8.7 | +0.7 |
| Resistance genes | ||||||||
| CTX-M | 13/14 | 4/6 | 4/3 | 107/105 | 76.4/82.3 | 96.4/94.6 | 76.4/70 | 96.4/97.2 |
| NDM | 0/0 | 0/1 | 0/0 | 128/127 | - | 100/99.2 | NA/0 | 100/100 |
| IMP | 0/0 | 0/0 | 0/0 | 128/128 | - | 100/100 | - | 100/100 |
| KPC | 0/0 | 0/0 | 0/0 | 128/128 | - | 100/100 | - | 100/100 |
| OXA-48-like | 0/0 | 0/0 | 0/0 | 128/128 | - | 100/100 | - | 100/100 |
* Strict set of rules: ≥104 genome copies/mL for BAL, ≥105 for ETA/NBL and ≥107 for sputum. ** Less strict set of rules: ≥104 genome copies/mL for BAL/ETA/NBL and ≥106 for sputum. *** Difference between the strict and less strict sets of rules. Abbreviations: FA—FilmArray, PPV—positive predictive value, NPV—negative predictive value, NA—not applicable.
In 17 samples, the cultures were positive for bacteria that were not part of the FA panel, and these included five S. maltophilia, three Aspergillus spp., three C. striatum, two Citrobacter spp. and four others. One Providencia stuartii was cultured while the FA detected Proteus spp., probably a misidentification of the FA panel.
The sensitivity, specificity, PPV and NPV for CTX-M detection as compared with phenotypic culture results, using the less strict rules, were 82.3%, 94.6%, 70% and 97.2%, respectively (Table 4). One case of NDM was detected by the FA panel with no phenotypic counterpart in the respiratory or gastrointestinal tracts. No other resistant genes were detected.
3.7. Effect of FA Results on Antimicrobial Therapy
In 107/148 (72%) cases, FA results influenced antimicrobial decisions including avoidance of using antimicrobials, starting, stopping, upgrading or downgrading an already given treatment. In 41 cases (27%) it triggered the initiation of treatment—34 cases that were not previously treated and seven cases that were previously treated but were at the time on “antimicrobial treatment window”. 18/34 cases were treated for community-acquired organisms with ceftriaxone, four were given MSSA-directed therapy. All seven patients that were re-treated were given extended spectrum antimicrobials, that included coverage for MRSA, Extended-spectrum beta-lactamase (ESBL) producing Enterobacterales and P. aeruginosa. Antimicrobials were stopped in 13 cases, 11 of which had negative FA tests, and in two cases with positive FA results. Antimicrobials were avoided in 21 cases, all of which had negative FA results (Table 5).
Table 5.
Cases in which antimicrobials were avoided, started, or stopped as a result of FA testing.
| Action Triggered by FA Result n (%) |
Number of Patients | FA Results (n) | ABx Administered after FA Result | Corresponding Culture Results (n) | |||
|---|---|---|---|---|---|---|---|
| Avoiding ABx 21 (28.0) |
21 (100.0) | Negative FA | N/A | Normal flora (20) H. influenzae (1) |
|||
| Starting ABx 41 (54.7) |
Patients not treated previously 34 (82.9) |
18 (52.9) |
H. influenzae (11) S. pneumoniae (6) MSSA (5) M. catarrhalis (2) |
K. pneumoniae (2) E. coli (1) GBS (1) Negative FA (2) |
ABx for community pathogens CRO (18) |
MSSA (4) K. pnuemoniae (2) E. cloacae (2) S. pneumoniae (1) GBS (1) E. coli (1) |
A. baumannii (1) H. influenzae (1) M. catarrhalis (1) Citrobacter spp. (1) Aspergillus spp. (1) Normal flora (8) |
| 7 (20.6) |
P.aeruginosa (6) E. coli (1) E. cloacae (2) H. influenzae (1) |
K. pneumoniae (2) MSSA (1) A. baumannii (1) Serratia spp. (1) |
ABx for non-fermenters CAZ (5), TZP (2) |
P.aeruginosa (3) E. cloacae (2) MSSA (1) |
K. pnuemoniae (1) Serratia spp. (1) Normal flora (2) |
||
| 4 (11.8) | MSSA (3, one was sputum 104) H. influenzae (2) S. pneumoniae (1) GAS (1) Negative FA (1) |
ABx for MSSA CFZ (3), AMC (1) |
MSSA (2) Normal flora (1) |
||||
| 1 (2.9) | K. pneumoniae, CTX-M (1) | ABx for ESBL ETP (1) |
K. pneumoniae (CRO-Res) | ||||
| 4 (11.8) | MRSA (4, one was sputum 105) P. aeruginosa (1) GAS (1) |
ABx for MRSA VAN (4) |
MRSA (2) P. aeruginosa (1) Aspergillus spp. (1) MSSA (1) Normal flora (1) |
||||
| Patients in “ABx window” 7 (17.1) |
1 (14.3) | MRSA, P.aeruginosa | VAN + TZP | MRSA, P.aeruginosa | |||
| 1 (14.3) | MRSA, E. cloacae (ETA—104), CTX-M | VAN | MRSA, S. maltophilia | ||||
| 1 (14.3) | MSSA, K. pneumoniae | CIP + CFZ | MSSA, K. pneumoniae (CRO-Sus) | ||||
| 1 (14.3) | K. aerogenes | TZP | K. aerogenes (CRO-Sus) | ||||
| 1 (14.3) | MRSA, E. cloacae, CTX-M | VAN + CIP | MRSA, E. cloacae (CRO-Res) | ||||
| 1 (14.3) | MRSA, K. aerogenes | VAN + MEM |
K. aerogenes (CRO-Res) C. striatum |
||||
| 1 (14.3) | P. aeruginosa | CAZ | P. aeruginosa | ||||
| Stopping ABx 13 (17.3) |
11 (84.6) | Negative FA (11) | TZP (2) CXM + AZM (1) CRO (4) CRO + LVX (2) LVX (1) CHL (1) |
Normal flora/yeasts (10) N/A (1) |
|||
| 2 (15.4) |
K. aerogenes (1) MRSA (1) |
MEM (1) TZP (1) |
K. aerogenes (1) MRSA (1) |
||||
FA—FilmArray, ABx—Antibiotics, N/A—not available (no corresponding culture), ETA—endotracheal aspiration, CRO-Res—ceftriaxone resistance, CRO-Sus—ceftriaxone susceptible, MSSA—methicillin sensitive S. aureus, MRSA—methicillin resistant S. aureus, GBS—Streptococcus agalactiae, GAS—Streptococcus pyogenes, CRO—ceftriaxone, TZP—piperacillin-tazobactam, VAN—vancomycin, CFZ—cefazolin, ERT—ertapenem, CIP—ciprofloxacin, MEM—meropenem, CXM—cefuroxime, AZM—azithromycin, LVX—levofloxacin, CHL—chloramphenicol, CST—colistin, SXT—trimethoprim/sulfamethoxazole, VRC—voriconazole, LZD—linezolid, CAZ—ceftazidime, OXA—oxacillin, AMP—ampicillin, AMB—amphotericin B deoxycholate, AMK-amikacin.
Downgrading the coverage spectrum was noted in five cases, and upgrading in 27 cases, mostly for P. aeruginosa coverage (n = 13), as well as for MRSA and ESBLs (four each). In 41 cases, FA results did not affect the given treatment, 17/41 had negative FA, but antimicrobials were still considered appropriate, and in the rest the patients were already covered with the presumed antimicrobials for the identified FA targets found (Table 6).
Table 6.
Cases in which antimicrobials were downgraded, upgraded, or left unchanged as result of FA testing.
| Action Triggered by FA Result n (%) |
Number of Patients | FA Results (n) | ABx Given before FA Results | ABx Given after FA Results | Culture Results (n) | |
|---|---|---|---|---|---|---|
| Downgrading ABx 5 (6.8) |
1 | E. cloacae, P. aeruginosa | LVX | CIP | P. aeruginosa | |
| 1 | MSSA | TZP | OXA | Citrobacter spp. | ||
| 1 | Negative FA | VAN + TZP | TZP | Normal flora | ||
| 1 | MSSA | CRO | CFZ | MSSA | ||
| 1 | Negative FA | CRO + LVX | CRO | Normal flora | ||
| Upgrading ABx 27 (37.0) |
For P. aeruginosa coverage 13 (48.1) |
13 |
P. aeruginosa (13) and: K. pneumoniae (3) Serratia spp. (3) MSSA (2) MRSA (1) K. aerogenes (2) Proteus spp. (2) H. influenzae (2) E. cloacae (1) A. baumannii (1) CRO-M (2) |
CRO (5) VAN & LVX (1) VAN (1) CRO + AZM (1) LVX (1) CAZ (1) CHL (2) ERT + VAN (1) CXM (1) |
TZP (8) MEM (1) MEM + VAN (1) MEM + CST (1) CRO + LVX (1) CIP (1) |
P. aeruginosa (10), MSSA (2), MRSA (1), K. pneumoniae CRO-Res (1), Providencia spp. (1), Roultella spp. (1), K. aerogenes (1), Normal flora (1), N/A (1) |
| For MRSA coverage 4 (14.8) |
1 | MRSA, K. oxytoca, CTX-M | CRO + AZM | SXT | MRSA, K. oxytoca CRO-Res | |
| 1 | MRSA, K. oxytoca, CTX-M | CRO | VAN + TZP | MRSA, E. cloacae CRO-Res | ||
| 1 | MRSA | TZP | VAN + LVX | MRSA | ||
| 1 | MRSA, K. pneumoniae | ERT | SXT | K. pneumoniae Non-CP-CRE | ||
| For ESBL coverage 4 (14.8) |
1 | CTX-M, E. cloacae, E. coli, Serratia spp. | SXT | MEM | Serratia spp., S. maltophilia | |
| 1 | CTX-M, E. cloacae | AZM | TZP | E. cloacae CRO-Res | ||
| 1 | CTX-M, K. pneumoniae | ERT | MEM + LZD | Yeast | ||
| 1 | CTX-M, K. pneumoniae | TZP | MEM | K. pneumoniae CRO-Res | ||
| Others 6 (22.2) |
1 | Negative FA | CST | AMB + VAN | N/A | |
| 1 | Negative FA | CFZ | SXT | Normal flora | ||
| 1 | E. cloacae (Sputum 104) | AZM | CIP | NA | ||
| 1 | MSSA | TZP | TZP + CFZ | Normal flora | ||
| 1 | H. influenzae | AMP | CRO | Normal flora | ||
| 1 | H. influenzae, S. pneumoniae, MSSA | AMK (due to bacteremia) | CRO | MSSA | ||
| No change in ABx 41 (56.2) |
Negative FA (n = 17) |
17 | Negative FA | CRO (9) TZP (2) CRO + VAN (1) CRO + LVX (1) LVX (1) VAN (1) VAN + VRC (1) CHL (1) |
Same | Normal flora (9), MSSA (2), S. maltophilia (1), Actinomyces spp. (1), C. striatum (1), Aspergillus flavus (1), P. aeruginosa (1), S. constellatus (1) |
|
P. aeruginosa in FA (n = 8) |
8 |
P. aeruginosa and: CTX-M (3) MRSA (1) MSSA (1) Proteus spp. (1) K. pneumoniae (1) E. cloacae (1) K. aerogenes (1) |
TZP (2) CAZ (2) CAZ + CST (1) CST (1) VAN + TZP (1) MEM + VAN (1) |
Same | P. aeruginosa (8) and K. pneumoniae CRO-Res (1), MRSA (1), Citrobacter spp. (1) | |
| CTX-M in FA (n = 3) |
3 |
E. cloacae (1) K. pneumoniae (1) Serratia spp. (1) |
ERT (2) LVX (1) |
Same | S. maltophilia (1), Normal flora (1), K. pneumoniae CRO-Res (1) | |
| MRSA in FA (n = 2, one was sputum 104) | 2 | MRSA and S. pneumoniae (2) | MEM + VAN (2) | Same | Normal flora (2) | |
| MSSA in FA (n = 2) | 2 | MSSA (1) MSSA and GBS and S. pneumoniae (1) |
LVX (1) CRO (1) |
Same | S. maltophilia (1), Normal flora (1) | |
| Others (n = 9) | 9 |
H. influenzae (3) K. pneumoniae (3) M. catarrhalis (2) GBS (1) S. pneumoniae (1) A. baumannii (1) E. cloacae (1) K. aerogenes (1) |
CRO (8) ERT (1) |
Same | K. pneumoniae (2), E. cloacae CRO-Res & K. pneumoniae (1), GBS (1), K. aerogenes (1), C. striatum (1) | |
FA—FilmArray, ABx—Antibiotics, N/A—not available (no corresponding culture), ETA—endotracheal aspiration, CRO-Res—ceftriaxone resistance, CRO-Sus—ceftriaxone susceptible, Non-CP-CRE—non carbapenemase-producing carbapenem resistant Enterobacteriaceae, MSSA—methicillin sensitive S. aureus, MRSA—methicillin resistant S. aureus, GBS—Streptococcus agalactiae, GAS—Streptococcus pyogenes, CRO—ceftriaxone, TZP—piperacillin-tazobactam, VAN—vancomycin, CFZ—cefazolin, ERT—ertapenem, CIP—ciprofloxacin, MEM—meropenem, CXM—cefuroxime, AZM—azithromycin, LVX—levofloxacin, CHL—chloramphenicol, CST—colistin, SXT—trimethoprim/sulfamethoxazole, VRC—voriconazole, LZD—linezolid, CAZ—ceftazidime, OXA—oxacillin, AMP—ampicillin, AMB—amphotericin B deoxycholate, AMK-amikacin.
Of the 57 negative or insignificant FA results, in 34 (59%) these results were associated with stopping, downgrading or withholding antimicrobial therapy. In six cases, negative FA results triggered commencing antimicrobial therapy (two ceftriaxone and one cefazolin) or upgrading it (one from cefazolin to trimethoprim/sulfamethoxazole, one from azithromycin to ciprofloxacin and one from colistin to vancomycin and amphotericin B).
3.8. Effect of Post-FA Culture Result on Antimicrobial Therapy
Most cases of negative FA tests were confirmed by negative culture results (38/46, 82%). Among the eight discordances, there were four cases that were not covered until the result of the culture (one ceftriaxone-susceptible E. cloacae, one H. influenzae, one S. maltophilia and 1 Aspergillus flavus). The other four cases (one C. striatum, one MSSA, 1 Actinomyces spp. and 1 S. constellatus) had already been treated empirically. Two cases of ceftriaxone-susceptible Citrobacter spp. (an organism that does not appear in the FA panel), were seen in cultures, but both cases were adequately treated empirically.
4. Discussion
The main findings of this study are the relative high rates of both co-infections and superinfections among critically ill, ventilated COVID-19 patients and the association between the occurrence of positive FA results and poor patients’ outcomes. In concurrence with recent literature [4,5,6], we also encountered high rates of positive mPCR tests from COVID-19 patients’ secretions (mostly ETAs). Depending on the strictness of bacterial DNA loads evaluation, we had 13–17% positive FA results for patients with CAP and 68% positive FA results for VAP. It is important to note that a considerable part (50–65%) of these positive early FA test results for CAP were not met with positive cultures later, suggesting, at least in part, low bacterial loads probably representing bacterial colonization or non-viable bacteria.
High rates of COVID-19-bacterial co-infections identified with molecular methods among COVID-19 patients were reported recently by our group [7] and others [5,12,22,23]. The bacteria encountered by the tests performed on admission were typical for community-acquired infections, including mainly H. influenzae, S. pneumoniae and M. catarrhalis. VAP was significantly more prevalent than CAP, occurring in 62% of the cohort, and with better correlation to the microbiology results. As was previously reported [10,24], P. aeruginosa and S. aureus including MRSA, MSSA and Enterobacterales were the most common microorganisms seen in this study. A recent Italian study prospectively followed 248 patients in eight ICUs, and found 36% secondary bacterial infection, primarily VAP, with the same dominant bacteria as were reported in our study but without the use of molecular diagnostic methods [25].
Mortality among critically ill, ventilated COVID-19 patients is high. In our cohort, 38% of patients died, which is lower than typically reported, although the range varies considerably [26]. In our study, mortality was considerably lower (10%) among ventilated patients who were diagnosed with CAP (without subsequently also developing VAP) and among those who had only negative FA results. This was in clear contrast to the 51% mortality rate among patients with VAP diagnosis based on positive FA tests. Thus, VAP may be a surrogate for mortality, although causality should not be readily assumed. The patients with VAP in our cohort were older by nearly a decade as compared to those with CAP or neither diagnosis, and older age, typically associated with higher rates of comorbidities, is a very strong independent predictor of death from COVID-19 [26,27,28]. Hence, older, more fragile and pre-morbid patients, had longer duration of ICU stay and consequently more exposure to other infectious and non-infectious ICU-related complications. Aside from the severe pulmonary failure associated with SARS-CoV-2 infection, these patients suffered a myriad of complications due to prolonged ICU stay, including bleeding diathesis, arterial and venous thromboembolic events, spontaneous and iatrogenic pneumothorax and pneumomediastinum, and other HAIs including central line related bloodstream infections. Typically, patients succumbed to these complications (and primarily to infectious causes) and not to the acute respiratory distress syndrome (ARDS) associated with COVID-19. A prospective study on 45 critically ill patients in Switzerland in which 478 lower respiratory samples were assessed longitudinally, has reported 42% respiratory superinfections and association with poor outcomes, including longer ICU stay and lower ventilator-free survival rate. In this cohort, there were no differences between the patients with superinfection and those without, including their age and comorbidities, implying that the superinfection itself may be the surrogate for unfavorable consequences [29].
HAIs among COVID-19 patients are commonly caused by multidrug resistant organisms (MDROs) and filamentous fungi. In our patients, P. aeruginosa, S. aureus (MRSA, but also MSSA) and Enterobacterales were the main causes of VAP, identified by FA tests, but S. maltophilia and Aspergillus spp. were also encountered, posing a great treatment challenge. The short turn-around time of diagnosing a MDRO, using FA as opposed to classical methods, confers significant advantages in both providing an immediate, tailored antibiotic therapy to cover an unexpected organism (as MRSA or A. baumannii), avoiding the empiric use of wide-spectrum combination therapy for a deteriorating critically ill patients while awaiting Gram-stains results (which could be more difficult to obtain in laboratories without the means to inactivate infectious materials), and implementing contact precautions when a suspected organism or resistance mechanism appear in the FA panel result. On the other hand, care should be applied not to automatically withdraw antibiotic therapy in response to negative FA results, since important organisms may be missing from the panel (such as Citrobacter spp., S. maltophilia and others, including fungi). Discrepancies between the FA panel and cultures should also be considered.
The performance of the FA kit in our study was comparable to previous reports [30]. Using the less strict set of rules, we found that the overall NPV was 99.6% and was similar between Gram-negative and Gram-positive bacteria. Hence, a negative FA result in a critically ill patient could be relied on, and unnecessary antibiotic therapy withdrawn, as long as pathogens that do not appear in the FA panel are taken into consideration. The PPV using these rules was 57.6%, representing alleged relatively high rates of false-positive FA results, that could result in antimicrobial overuse. It should be noted that specifically for P. aeruginosa, which was the most common organism in our cohort, the PPV was the highest (92.3%). When stricter rules were applied, the sensitivity decreased and the PPV increased to 66.3%, with slight decrease in the NPV (to 99%). Specifically for P. aeruginosa the PPV in this setting was 100%. In real-life, many infectious diseases experts would probably choose not to ignore a positive ETA sample for MRSA, for example, because only 105 copies/mL and not 106 copies/mL were reported.
This study has limitations. The cohort of patients was small, and it was conducted in a single, medium-sized single medical center. Nevertheless, this cohort represents most of the ventilated COVID-19 patients seen in this facility during three pandemic phases. In this study, FA tests and cultures were not always performed on the same samples, which could have an impact on the comparative performance results. However, out of 128 comparative samples, in 85 (66%) the samples used were identical, and in 30 (23%) the culture was taken before FA testing—hence antimicrobials, if administered in the interval, should not have influenced the culture results, and may have had only minor effects on FA tests. In 13 cases (10%) the cultures were taken a day or two after FA testing and in all but three cases there was still a perfect correlation between the tests. In two of the three uncorrelated cases, the cultures grew MSSA and H. influenzae while the FA was negative, and in only one case K. pneumoniae detected by FA did not grow in culture. We did not collect data on the co-morbidities of the patients and other related data concerning other infectious and non-infectious complications during the ICU stay. Similarly, we did not conduct a multi-variate analysis for the mortality causes.
To conclude, we have shown high rates of both co-infection (CAP) and superinfection (VAP) among critically ill, ventilated COVID-19 patients caused by the typical and expected organisms for both conditions. The patients suffering from VAP were older and had longer ICU stays and mortality rates, as compared to those having only CAP or not having any positive FA test results. The correlation of FA tests with classical cultures was higher for VAP patients, with very high NPV and acceptable PPV, especially for P. aeruginosa. The timely accessible results of FA tests had an impact on antibiotic stewardship decisions, with negative results typically assisting in termination of therapy while positive results assisted in expanding or commencing antibiotic therapy. The performance of the FA tests was comparable to previous studies.
Author Contributions
Conceptualization, R.C.; methodology, R.C.; software, R.C.; validation, R.C., T.F., F.B., M.S., M.U., K.G., H.A., C.D. and J.L.; formal analysis, R.C. and J.L.; investigation, R.C.; resources, R.C.; data curation, R.C.; writing—original draft preparation, R.C.; writing—review and editing, R.C., F.B., T.F., K.G., H.A., C.D., M.U., M.S., S.P. and J.L.; visualization, R.C. and J.L.; supervision, R.C., J.L. and M.S.; project administration, R.C.; funding acquisition—not relevant. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board (IRB) of the Sanz Medical Center, 0014-21-LND.
Informed Consent Statement
Patient consent was waived since the study was a retrospective, observational, non-interventional study.
Data Availability Statement
Data may be provided per request from the authors.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Grasselli G., Greco M., Zanella A., Albano G., Antonelli M., Bellani G., Bonanomi E., Cabrini L., Carlesso E., Castelli G., et al. Risk Factors Associated With Mortality Among Patients With COVID-19 in Intensive Care Units in Lombardy, Italy. JAMA Intern. Med. 2020;180:1345–1355. doi: 10.1001/jamainternmed.2020.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clancy C.J., Nguyen M.H. Coronavirus Disease 2019, Superinfections, and Antimicrobial Development: What Can We Expect? Clin. Infect. Dis. 2020;71:2736–2743. doi: 10.1093/cid/ciaa524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudiol C., Dura-Miralles X., Aguilar-Company J., Hernandez-Jimenez P., Martinez-Cutillas M., Fernandez-Aviles F., Machado M., Vazquez L., Martin-Davila P., de Castro N., et al. Co-infections and superinfections complicating COVID-19 in cancer patients: A multicentre, international study. J. Infect. 2021;83:306–313. doi: 10.1016/j.jinf.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrasa H., Martin A., Maynar J., Rello J., Fernandez-Torres M., Aguirre-Quinonero A., Canut-Blasco A., Alava C.-S.I. High rate of infections during ICU admission of patients with severe SARS-CoV-2 pneumonia: A matter of time? J. Infect. 2021;82:186–230. doi: 10.1016/j.jinf.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contou D., Claudinon A., Pajot O., Micaelo M., Longuet Flandre P., Dubert M., Cally R., Logre E., Fraisse M., Mentec H., et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann. Intensive Care. 2020;10:119. doi: 10.1186/s13613-020-00736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreitmann L., Monard C., Dauwalder O., Simon M., Argaud L. Early bacterial co-infection in ARDS related to COVID-19. Intensive Care Med. 2020;46:1787–1789. doi: 10.1007/s00134-020-06165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen R., Finn T., Babushkin F., Geller K., Alexander H., Shapiro M., Uda M., Mostrchy A.R., Amash R., Shimoni Z., et al. High rate of bacterial respiratory tract co-infections upon admission amongst moderate to severe COVID-19 patients. Infect. Dis. 2021 doi: 10.1080/23744235.2021.1985732. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fattorini L., Creti R., Palma C., Pantosti A., Unit of Antibiotic R., Special P., Unit of Antibiotic R., Special Pathogens of the Department of Infectious Diseases Bacterial coinfections in COVID-19: An underestimated adversary. Ann Ist Super Sanita. 2020;56:359–364. doi: 10.4415/ANN_20_03_14. [DOI] [PubMed] [Google Scholar]
- 9.Morris D.E., Cleary D.W., Clarke S.C. Secondary Bacterial Infections Associated with Influenza Pandemics. Front Microbiol. 2017;8:1041. doi: 10.3389/fmicb.2017.01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elabbadi A., Turpin M., Gerotziafas G.T., Teulier M., Voiriot G., Fartoukh M. Bacterial coinfection in critically ill COVID-19 patients with severe pneumonia. Infection. 2021;49:559–562. doi: 10.1007/s15010-020-01553-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes S., Troise O., Donaldson H., Mughal N., Moore L.S.P. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020;26:1395–1399. doi: 10.1016/j.cmi.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langford B.J., So M., Raybardhan S., Leung V., Westwood D., MacFadden D.R., Soucy J.R., Daneman N. Bacterial co-infection and secondary infection in patients with COVID-19: A living rapid review and meta-analysis. Clin. Microbiol. Infect. 2020;26:1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B., et al. Detectable Serum Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load (RNAemia) Is Closely Correlated With Drastically Elevated Interleukin 6 Level in Critically Ill Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong X., Cao Y.Y., Lu X.X., Zhang J.J., Du H., Yan Y.Q., Akdis C.A., Gao Y.D. Eleven faces of coronavirus disease 2019. Allergy. 2020;75:1699–1709. doi: 10.1111/all.14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farfour E., Lecuru M., Dortet L., Le Guen M., Cerf C., Karnycheff F., Bonnin R.A., Vasse M., Lesprit P. Carbapenemase-producing Enterobacterales outbreak: Another dark side of COVID-19. Am. J. Infect. Control. 2020;48:1533–1536. doi: 10.1016/j.ajic.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Toole R.F. The interface between COVID-19 and bacterial healthcare-associated infections. Clin. Microbiol. Infect. 2021 doi: 10.1016/j.cmi.2021.06.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez J.Y., Le Pape P., Lopez O., Esquea K., Labiosa A.L., Alvarez-Moreno C. Candida auris: A latent threat to critically ill patients with COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai C.C., Wang C.Y., Hsueh P.R. Co-infections among patients with COVID-19: The need for combination therapy with non-anti-SARS-CoV-2 agents? J. Microbiol. Immunol. Infect. 2020;53:505–512. doi: 10.1016/j.jmii.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clinical Microbiology Procedures Handbook. 4th ed. ASM Press; Washington, DC, USA: 2016. pp. 297–316. [Google Scholar]
- 21.The Clinical and Laboratory Standards Institute (Annapolis Junction, MD, USA 20701) Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. CLSI; Wayne, PA, USA: 2021. [Google Scholar]
- 22.Lansbury L., Lim B., Baskaran V., Lim W.S. Co-infections in people with COVID-19: A systematic review and meta-analysis. J. Infect. 2020;81:266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rawson T.M., Moore L.S.P., Zhu N., Ranganathan N., Skolimowska K., Gilchrist M., Satta G., Cooke G., Holmes A. Bacterial and Fungal Coinfection in Individuals With Coronavirus: A Rapid Review To Support COVID-19 Antimicrobial Prescribing. Clin. Infect. Dis. 2020;71:2459–2468. doi: 10.1093/cid/ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldman C., Anderson R. The role of co-infections and secondary infections in patients with COVID-19. Pneumonia. 2021;13:5. doi: 10.1186/s41479-021-00083-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Santis V., Corona A., Vitale D., Nencini C., Potalivo A., Prete A., Zani G., Malfatto A., Tritapepe L., Taddei S., et al. Bacterial infections in critically ill patients with SARS-2-COVID-19 infection: Results of a prospective observational multicenter study. Infection. 2021 doi: 10.1007/s15010-021-01661-2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim Z.J., Subramaniam A., Ponnapa Reddy M., Blecher G., Kadam U., Afroz A., Billah B., Ashwin S., Kubicki M., Bilotta F., et al. Case Fatality Rates for Patients with COVID-19 Requiring Invasive Mechanical Ventilation. A Meta-analysis. Am. J. Respir. Crit. Care Med. 2021;203:54–66. doi: 10.1164/rccm.202006-2405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imam Z., Odish F., Gill I., O’Connor D., Armstrong J., Vanood A., Ibironke O., Hanna A., Ranski A., Halalau A. Older age and comorbidity are independent mortality predictors in a large cohort of 1305 COVID-19 patients in Michigan, United States. J. Intern Med. 2020;288:469–476. doi: 10.1111/joim.13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallo Marin B., Aghagoli G., Lavine K., Yang L., Siff E.J., Chiang S.S., Salazar-Mather T.P., Dumenco L., Savaria M.C., Aung S.N., et al. Predictors of COVID-19 severity: A literature review. Rev. Med. Virol. 2021;31:1–10. doi: 10.1002/rmv.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buehler P.K., Zinkernagel A.S., Hofmaenner D.A., Wendel Garcia P.D., Acevedo C.T., Gomez-Mejia A., Mairpady Shambat S., Andreoni F., Maibach M.A., Bartussek J., et al. Bacterial pulmonary superinfections are associated with longer duration of ventilation in critically ill COVID-19 patients. Cell Rep. Med. 2021;2:100229. doi: 10.1016/j.xcrm.2021.100229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maataoui N., Chemali L., Patrier J., Tran Dinh A., Le Fevre L., Lortat-Jacob B., Marzouk M., d’Humieres C., Rondinaud E., Ruppe E., et al. Impact of rapid multiplex PCR on management of antibiotic therapy in COVID-19-positive patients hospitalized in intensive care unit. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:2227–2234. doi: 10.1007/s10096-021-04213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data may be provided per request from the authors.