Abstract
Virus infection of numerous cell types results in the transcriptional induction of a subset of virus- and interferon (IFN)-stimulated genes. The beta IFN (IFN-β) gene is one of these rapidly induced genes; it serves as a fundamental component of the cellular defense response in eliciting potent antiviral, immunomodulatory, and antiproliferative effects. One of the transcription factors involved in the stringent regulation of IFN-β production following virus infection is interferon regulatory factor (IRF) 3 (IRF-3). We have characterized an alternatively spliced isoform of IRF-3 that we have called IRF-3a. IRF-3a can selectively and potently inhibit virus-induced activation of the IFN-β promoter. IRF-3a lacks half of the DNA binding domain found in IRF-3 and is unable to bind to the classical IRF binding elements, IFN-stimulated response elements. These studies suggest that IRF-3a may act as a modulator of IRF-3.
Virus infection of mammalian cells results in the immediate and transient transcriptional induction of a number of cytokine and chemokine genes. Included among these are the type I interferons (IFNs), alpha IFN (IFN-α) and beta IFN (IFN-β). The IFNs are a large family of multifunctional cytokines involved in the antiviral response, regulation of cell growth, and activation of the immune system (reviewed in reference 26). IFN-β production is a complex process that is controlled at many levels, with the primary regulatory step occurring at the level of transcription. The virus-inducible enhancer of the IFN-β gene has been well defined and contains both positive regulatory domains (PRDs) and negative regulatory domains that are bound by specific transcription factors. Upon virus infection, repressor proteins bound to the negative regulatory domains appear to dissociate and novel transactivators bind to the PRDs of the promoter (19). In this way, the production of IFN-β is stringently regulated by the orchestrated association and dissociation of transcriptional regulators, allowing for the rapid response of the cell to a variety of environmental stimuli (19, 22). Two PRDs, PRD I and PRD III, of the IFN-β promoter are responsible for virus activation. They closely resemble the IFN-stimulated response elements (ISREs) found in the promoters of a large number of virus and IFN-stimulated genes. ISREs are known to bind the family of IFN regulatory factors (IRFs).
The IRFs are involved in a large number of cellular responses, including cellular growth control, resistance to bacterial infection, commitment to transformation by oncoproteins, T- and B-cell development, response to DNA damage, apoptosis, and the response to virus infection (reviewed in references 10, 18, and 24). There are presently nine members of the IRF family, all of which share significant structural homology in the amino-terminal DNA binding domain (DBD). The IRF DBD contains a characteristic tryptophan repeat that has been implicated in the interaction of IRF molecules with DNA. The crystal structures of the IRF-1 and IRF-2 DBDs bound to PRD I revealed that three of the five tryptophan residues contained within the helix-turn-helix motif are in direct contact with DNA (5, 7).
One IRF family member, IRF-3, has been implicated in the virus- and double-stranded RNA (dsRNA)-mediated induction of IFN-β, of the chemokine RANTES, and of a subset of interferon-stimulated genes (ISGs) (14, 15, 23, 27–29). Under normal conditions, IRF-3 exists in a latent form in the cytoplasm. Virus infection or the presence of dsRNA triggers the phosphorylation and translocation into the nucleus of IRF-3. IRF-3 then associates with the transcriptional coactivators p300 and CREB binding protein (CBP) to form virus-activated factor or dsRNA-activated factor 1 (27, 28). The site-specific DNA binding proteins which bind the PRDs of the IFN-β promoter, namely, IRF-3, IRF-7, NF-κB, ATF-2, and c-Jun, recruit the coactivators p300 and CBP to the IFN-β promoter after virus infection. These proteins, together with high-mobility-group protein I(Y), constitute a higher-order transcription-enhancing complex, the enhanceosome (19, 22). IRF-3 has also been identified in the cytomegalovirus-induced ISRE binding factor (31). The cytomegalovirus-induced ISRE binding factor is distinct from the virus-activated factor and dsRNA-activated factor 1 complexes, however, in that only CBP and not p300 has been identified as a binding partner in the complex.
The timing and duration of the IFN-β response to virus infection are likely controlled by the availability of transactivators, the regulation of which occurs at multiple levels. For example, two transactivators known to bind PRDs, NF-κB and IRF-3, reside mainly in the cell cytoplasm and are shuttled to the nucleus after virus infection (3, 15, 27–29). Furthermore, IRF-3 is subsequently but rapidly degraded, thereby providing an efficient mechanism for down-modulating IFN-β promoter activity (15, 25). However, given the complexity of the combinatorial control of IFN-β promoter activation, additional regulatory mechanisms likely will be revealed.
We have recently described alternative splicing for the IRF-3 gene (13, 17). Here we characterize the alternative splice isoform, which we have called IRF-3a, and show that its expression confers an additional level of regulation of IRF-3 activity. IRF-3a lacks a portion of the DBD at its amino terminus and in its place contains a unique amino acid sequence. IRF-3a is ubiquitously expressed in all tissues and cell lines tested, with the highest ratio of IRF-3a to IRF-3 being found in the brain. We demonstrate that IRF-3a can function in a dominant-negative manner and selectively impede IFN-β production in response to virus infection.
MATERIALS AND METHODS
Expression vectors.
The IRF-3a expression vector was obtained by subcloning the IRF-3a coding region into the KpnI and XbaI sites of plasmid pCMV4. The AU epitope tag was added at the amino terminus by PCR with the elimination of the initiator methionine. All clones were verified by sequence analysis.
Chloramphenicol acetyltransferase (CAT) reporters have been previously described (for Gal4-thymidine kinase [TK]-CAT) (9, 27). The simian virus 40 (SV-40) β-galactosidase plasmid has been described previously.
Cell lines, transfections, virus infections, and lysates.
HEC1B and 293 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. M059J cells were maintained in 1:1 Dulbecco modified Eagle medium-F12 medium supplemented with 10% fetal bovine serum (11). HEC1B cells were transfected by calcium phosphate precipitation (see below). 293 cells were transfected with Lipofectamine 2000 (GIBCO-BRL) according to the manufacturer's instructions. Sendai virus (SV) (Spafas) infections were carried out as described previously (25) for 6 h unless stated otherwise. For immunoprecipitations and Western blot analyses, cells were lysed in RIPA-300 buffer (50 mM Tris-HCl [pH 8.0], 0.1% sodium dodecyl sulfate [SDS], 0.5% deoxycholate, 1% Nonidet P-40, 300 mM NaCl). Human tissue lysates were obtained from GenoTechnology. The KCl extraction buffer used for the coimmunoprecipitation experiments has been described previously (25).
Electrophoretic mobility shift assays (EMSA).
For the ISG15 gene ISRE (5′-CTCGGGAAAGGGAAACCGAAACTGAAGCC-3′), a 32P-labeled probe was incubated with 5 μl of in vitro-translated proteins. The presence of similar amounts of different proteins was verified by comparison of [35S]Met-containing in vitro translation reactions performed in parallel. The binding mixture (20 μl) contained 10 mM HEPES (pH 7.9), 6% glycerol, 37.5 mM KCl, 1 mM dithiothreitol, 1.25 mM MgCl2, and 0.5 mM EDTA. Poly(dI-dC) (2.5 μg) was added to reduce nonspecific binding. After 20 min of incubation with the probe, extracts were loaded on an 8% polyacrylamide gel (75:1 acrylamide-bisacrylamide) prepared in Tris-glycine buffer. After running at 30 mA for 2 h, the gel was dried and exposed to Kodak film at −70°C for 4 h.
For the ISRE of the IFN-β promoter, the 32P-labeled PRD I/PRD III region (5′-GAAAACTGAAAGGGAGAAGTGAAAGTG-3′) was incubated with full-length glutathione S-transferase (GST)–IRF-3 or GST–IRF-3a. The binding mixture (20 μl) contained 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 50 mM NaCl, 2 mM dithiothreitol, 5% glycerol, 0.5% Nonidet P-40, and 10 mg of bovine serum albumin/ml. Poly(dI-dC) (1.5 μg) was added to reduce nonspecific binding. After 20 min of incubation with the probe, reactions were loaded on a 5% polyacrylamide gel (60:1 acrylamide-bisacrylamide) prepared in 0.5× Tris-borate-EDTA. After running at 30 mA for 2 h at 4°C, the gel was dried and exposed to Kodak film at −70°C for 4 h.
CAT assays.
HEC1B cells were cotransfected with 5 μg of β-galactosidase-encoding vector, 10 μg of the corresponding CAT reporter, and increasing amounts of a plasmid encoding IRF-3a. The total amount of DNA was adjusted to 25 μg with the empty vector in each case. The DNA mixture was removed after 10 h, and cells were washed twice with phosphate-buffered saline. At 48 h posttransfection, cells were infected with SV for 6 h or left uninfected. Cells were scraped in phosphate-buffered saline, spun down, and resuspended in 110 μl of 0.25 M Tris-HCl (pH 8.0). Lysis was performed by five cycles of freezing on dry ice for 15 min followed by thawing at 37°C for 1 min. Cellular debris was spun out at 4°C for 5 min at top speed in an Eppendorf tabletop centrifuge. Fifteen microliters of the supernatant was used for the liquid β-galactosidase assay, and 35 μl was used for the CAT assay. Products of the reaction were resolved by thin-layer chromatography, and the percent acetylation was determined using a Bio-Rad phosphorimager.
Immunoprecipitation and immunoblotting.
Immunoprecipitations and Western blot analyses with antibodies were performed as previously described (25), and the results were analyzed on a phosphorimager.
RESULTS
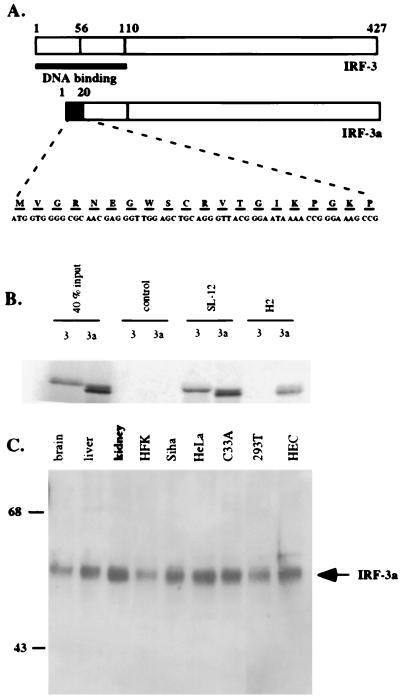
We have recently described a second mRNA that is generated from the IRF-3 gene by alternative splicing (13). Translation of this mRNA leads to the production of an alternative splice isoform of IRF-3, which we have called IRF-3a. The domain structure of IRF-3a is identical to that of IRF-3, except that a stretch of 20 unique amino acids replaces the N-terminal half of the DBD (Fig. 1A).
FIG. 1.
IRF-3a protein is ubiquitously expressed. (A) Schematic representation of the IRF-3 and IRF-3a proteins. The black box designates the novel 20 amino acids encoded by the IRF-3a-specific exon, which replace the first 56 out of 110 amino acids in the DBD of IRF-3. (B) H2 antibody is specific for IRF-3a. IRF-3 and IRF-3a were in vitro translated in the presence of [35S]Met and incubated with the indicated antibodies. Immunocomplexes were resolved by SDS-PAGE, and precipitated proteins were visualized by radiography. (C) Immunoprecipitation and Western blotting demonstrate ubiquitous expression of IRF-3a. Immunoprecipitation using IRF-3a-specific antibody H2 covalently cross-linked to protein A/G beads was carried out with total cell lysates. The immunoprecipitates were resolved by SDS-PAGE, and Western blotting was performed using antibody SL-12, which recognizes both IRF-3a and IRF-3. HFK, human foreskin keratinocytes; HEC, HEC1B cells.
The IRF-3a-specific mRNA is ubiquitously expressed (13). To determine whether the ubiquitous expression of the IRF-3a message is reflected in the expression of the protein, polyclonal antibodies were raised against a peptide corresponding to the unique region of IRF-3a. The antibodies recognized a band of the expected size of approximately 50 kDa in Western blotting and showed no cross-reactivity with IRF-3 (data not shown and Fig. 1B). To ascertain the identity of the recognized protein, immunoprecipitation with an IRF-3a-specific antibody, H2, was performed, followed by Western blotting with antibody SL-12, which recognizes both IRF-3 and IRF-3a. Figure 1B shows that the IRF-3a protein could be detected in all cell types examined.
IRF-3a does not bind to the ISG15 ISRE or the PRD I/III element.
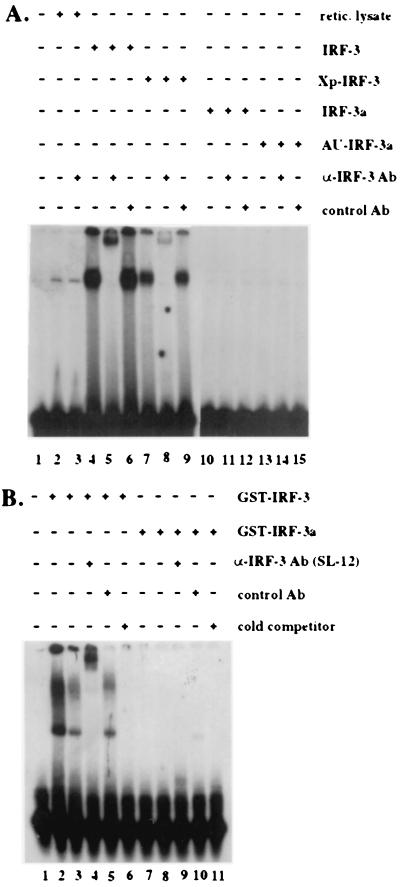
IRF family members bind highly conserved purine-rich elements, the most well-studied ones being the PRD I/III elements of the IFN-β promoter and the ISG15 ISRE. The amino-terminal DBD is a conserved functional domain within the IRF transcription factor family. As IRF-3a retains half of the DBD present in IRF-3 and possesses a stretch of 20 amino acids that is unique and unrelated to the N terminus of IRF-3, we sought to determine whether IRF-3a could bind to either of these two types of regulatory elements. Comparable amounts of either in vitro-translated proteins or bacterially produced GST fusion proteins were used in EMSA with probes representing either the ISRE of the ISG15 gene or the PRD I/III sequence of the IFN-β promoter (Fig. 2). Specific binding by IRF-3 was detected with both the ISG15 ISRE (Fig. 2A) and the PRD I/PRD III element (Fig. 2B). Interestingly, two different complexes were observed in the gel shift with PRD I/III; both of these could be supershifted by the anti-IRF-3 antibody SL-12. In contrast, no binding to either probe was detected for IRF-3a. Therefore, IRF-3 and IRF-3a differ in their respective abilities to bind specific ISREs. Additional experiments will need to be conducted to determine whether IRF-3a exhibits any sequence-specific DNA binding capacity and, if so, which other DNA elements IRF-3a may bind.
FIG. 2.
IRF-3a does not bind to the ISG15 ISRE or the PRD I/III element. (A) Comparable amounts (5 μl) of in vitro-translated IRF-3, Xpress-IRF-3 (Xp-IRF-3), IRF-3a, and AU1-IRF-3a were used for EMSA along with a 32P-labeled probe corresponding to the ISRE of the ISG15 gene (5′-CTCGGGAAAGGGAAACCGAAACTGAAGCC-3′). Ab, antibody. Xpress and AU1 are epitope tags. (B) Fifty nanograms (lanes 2 and 8 to 11) or 5 ng (lanes 3 to 7) of full-length GST–IRF-3 or GST–IRF-3a was used for EMSA with a 32P-labeled probe corresponding to the PRD I-PRD III region of the IFN-β promoter (5′-GAAAACTGAAAGGGAGAAGTGAAAGTG-3′). In all cases, an anti-IRF-3 antibody (SL-12) or a nonspecific control antibody was added to the binding reaction to demonstrate the specificity of the complexes. Cold probe was added at a 100-fold excess. Note that the difference in complex migration between the two gels reflects the difference in the type of gel used.
IRF-3a can inhibit the activity of IRF-3 in reporter assays.
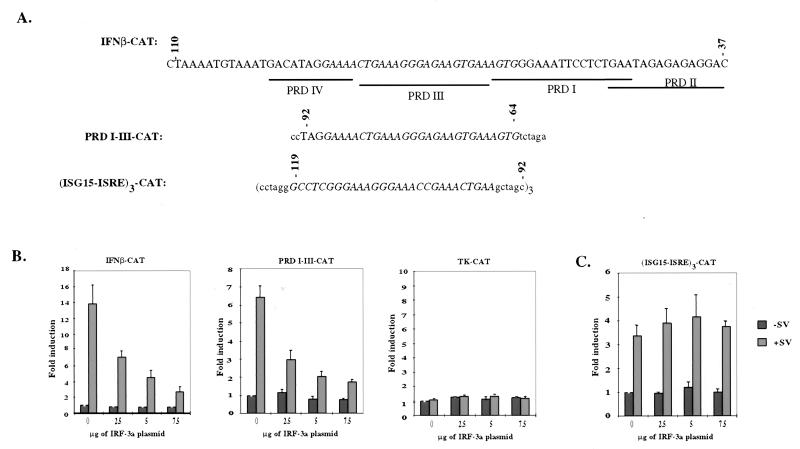
The inability of IRF-3a to bind the ISG15 ISRE and the PRD I/III element suggested that it may not function as a transcriptional activator but may instead function as an inhibitor of the virus-inducible pathway. Furthermore, Yoneyama et al. showed that truncation of the first 57 amino acids of IRF-3 generated a dominant-negative molecule capable of inhibiting the virus-induced expression of IFN-α and IFN-β (29). To address whether IRF-3a could function as a transactivator or as a dominant-negative regulator of the virus-inducible pathway, the ability of IRF-3a to affect the activity of a number of virus-inducible elements was studied. HEC1B cells were cotransfected with various CAT reporters, along with increasing amounts of IRF-3a and β-galactosidase expression vectors. CAT activity was then assayed in the presence or absence of virus infection. Three different ISRE-containing CAT reporters were used in this series of experiments (Fig. 3A). The first contained the entire virus-inducible promoter region of the IFN-β gene from positions −110 to −37. The second contained only the PRD I/III element (positions −90 to −65) of the IFN-β promoter, which have been implicated in the binding of IRF-3 and IRF-7 (15, 27, 29). The third contained three copies of the ISRE from the promoter of the virus and the IFN-inducible gene ISG15. As a control, a CAT reporter containing the promoter of herpes simplex virus TK (Gal4-TK-CAT) was used.
FIG. 3.
IRF-3a inhibits virus-induced activation of a subset of IRF-3-responsive promoters. (A) Schematic representation of the promoter elements of the reporter constructs. Lowercase letters represent sequence included in the construct that are not part of the enhancer element. (B) IRF-3a inhibits virus-induced activation of the IFN-β promoter through its ISRE, PRD I-PRD III. HEC1B cells were cotransfected with 10 μg of the reporter construct, 5 μg of β-galactosidase, and increasing amounts of IRF-3a. The total amount of DNA per transfection was adjusted to 25 μg with empty vector. At 48 h posttransfection, cells were infected with SV or left uninfected for 6 h, and CAT activity was determined. After normalization for β-galactosidase activity, results were plotted relative to the CAT activity of uninfected cells carrying no IRF-3a plasmid. (C) IRF-3a overexpression does not have an effect on virus-induced activation of the IRF-3-responsive ISG15 ISRE. Error bars in panels B and C indicate standard deviations.
Figure 3B shows that IRF-3a failed to enhance the activity of virus-inducible promoters, as has been demonstrated for IRF-3 (12, 15, 27, 29). Instead, IRF-3a inhibited virus-induced activation of the IFN-β promoter in a dose-dependent manner. At the highest concentration of IRF-3a tested, a fivefold reduction in CAT activity was observed. Similar results were obtained with the PRD I/III subdomain of the IFN-β promoter, with which up to a fourfold reduction in CAT activity was observed. This decrease in transcription is not the result of general toxicity from IRF-3a overexpression, since the data were normalized for β-galactosidase activity. Overexpression of IRF-3a had no effect on the activity of the herpes simplex virus TK promoter. In contrast to the inhibition shown for the first two reporters, IRF-3a had almost no effect on the virus-induced activity of the concatemerized ISG15 ISRE (Fig. 3C).
IRF-3a inhibits the activity of the endogenous IFN-β promoter
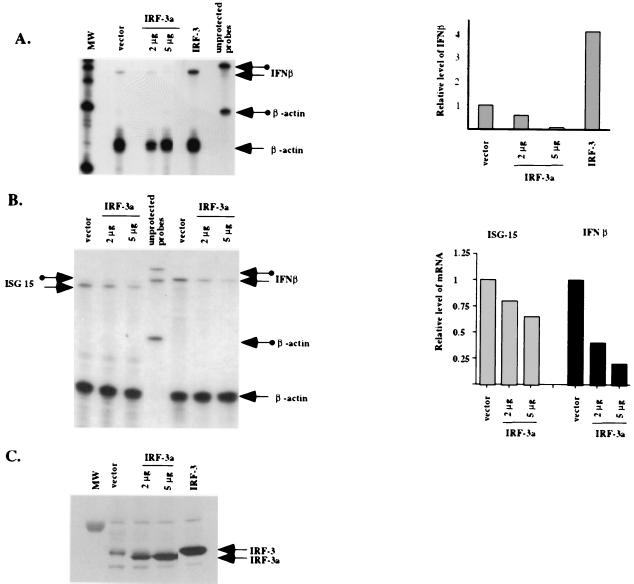
We next examined the ability of IRF-3a to inhibit expression from the endogenous IFN-β promoter. Numerous attempts to establish stable cell lines expressing IRF-3a were unsuccessful. Therefore, transient transfections with human embryonic kidney cell line 293 were used. Cells were transfected with a control plasmid or a plasmid expressing either IRF-3a or IRF-3. The expression of IRF-3 and IRF-3a was analyzed by Western blotting using whole-cell lysates (Fig. 4C). Cells were analyzed for IFN-β mRNA by RNase protection 24 h after transfection and 6 h after treatment with SV (Fig. 4A).
FIG. 4.
IRF-3a inhibits virus-induced activation of the endogenous IFN-β promoter. 293 cells were transfected with increasing amounts of IRF-3a, IRF-3, or empty vector plasmid. At 24 h after transfection, cells were split into two plates for subsequent protein and RNA analyses. At 36 h posttransfection, cells were infected with SV for 6 h; total RNA was isolated and used for an RNase protection assay. Protein lysates were made in parallel and analyzed for IRF-3 and IRF-3a expression. (A) Thirty micrograms of total cell lysates was resolved on a 4 to 12% Bis-Tris gel (Novex), and Western blotting was performed using antibody SL-12, which recognizes both IRF-3 and IRF-3a. MW, molecular weight standards. (B) Five micrograms of total RNA was subjected to an RNase protection assay with IFN-β and β-actin antisense riboprobes. Arrows with circles indicate unprotected probes; plain arrows indicate protected fragments. Quantitation of the relative IFN-β mRNA levels was performed by normalizing for the level of β-actin. (C) Comparison of the effect of IRF-3a overexpression on the induction of endogenous ISG15 and IFN-β transcripts. RNase protection and quantitation were carried out as described for panel B.
As expected, cells overexpressing exogenous IRF-3 showed fourfold stimulation of the IFN-β promoter relative to the controls. In contrast, the overexpression of IRF-3a resulted in up to 10-fold inhibition of IFN-β mRNA production (Fig. 4A). Western blotting using an antibody that recognizes both IRF-3 and IRF-3a showed that four- to fivefold overexpression of IRF-3a protein relative to the endogenous IRF-3 protein resulted in a strong inhibition of IFN-β transcription (Fig. 4C). Furthermore, as can be predicted from the results of the CAT assays, the overexpression of IRF-3a had essentially no effect on the virus-induced activation of the ISG15 gene promoter (Fig. 4B).
Physiologic levels of IRF-3a may be sufficient for effective modulation of IRF-3 activity.
The data from the previous experiment indicated that severalfold overexpression of IRF-3a in relation to IRF-3 was required for significant modulation of IRF-3 activity. This result raised the question of whether physiologic levels of IRF-3a observed in cells are sufficient for this regulatory mechanism to be important. Therefore, the levels of the two IRF-3 isoforms in normal human tissues and following virus infection were compared.
To compare the relative levels of IRF-3 and IRF-3a proteins in normal human tissues, both immunoprecipitation and Western blotting were performed using antibody SL-12, which recognizes both isoforms (Fig. 5A). The levels of IRF-3 and IRF-3a varied among the different tissues examined. In most cases, the level of IRF-3 was slightly higher than the level of IRF-3a, with the notable exception of the brain, in which IRF-3a was the predominant isoform.
FIG. 5.
Physiologic levels of IRF-3 and IRF-3a. (A) IRF-3 and IRF-3a are expressed in normal human tissues. Immunoprecipitation with antibody SL-12 and 150 μg of lysates from different tissues was followed by Western blotting with the same antibody. (B) Lack of degradation of IRF-3a results in a higher IRF-3a/IRF-3 ratio following virus infection. M059J cells were infected with virus for 9 h, and immunoprecipitations were performed with either antibody H2 or antibody SL-12. Immunoprecipitates were analyzed by Western blotting with antibody SL-12.
To determine if the relative ratio of the two isoforms changes following virus stimulation, the levels of each protein were compared after virus treatment. IRF-3 is known to be rapidly degraded following virus infection. If the kinetics of IRF-3a degradation are different from those of IRF-3, then the effect of IRF-3a on the pathway at later stages of the response will be stronger. In this experiment, M059J cells, in which IRF-3 is almost entirely degraded by 9 h postinfection, were infected with virus for various times. Immunoprecipitations were performed with lysates and either antibody H2 or antibody SL-12. Immunocomplexes were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and protein levels were visualized by Western blotting with antibody SL-12. Figure 5B shows that whereas almost all of IRF-3 had been degraded by 9 h postinfection, no significant change in IRF-3a levels was observed by that time point. Thus, an increase in the ratio of IRF-3a to IRF-3 following virus infection might contribute to the rapid down-regulation of the IFN response.
IRF-3a forms a heterodimer with IRF-3 after virus infection.
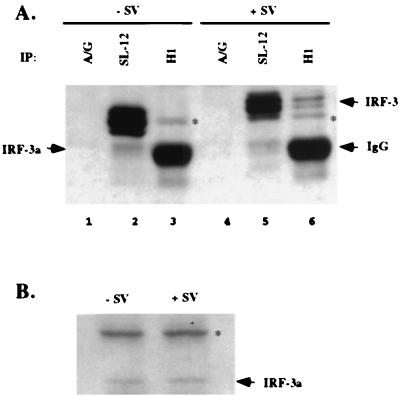
Given that IRF-3a is incapable of binding to ISRE sequences, a putative heterodimer between IRF-3 and IRF-3a is likely to be impaired in the transcriptional activation of promoters with multimeric IRF binding sites. IRF-3 has been shown to form homodimers after virus infection, and all of the regions implicated in the dimerization of IRF-3 are present in the alternative splice isoform (16). Therefore, we sought to determine whether the two proteins interact in vivo. Coimmunoprecipitation assays using an antibody specific for IRF-3a were performed with lysates from HEC1B cells that had been infected with virus or left uninfected. The immunocomplexes were resolved by SDS-PAGE, and bound IRF-3 and IRF-3a proteins were detected using an antibody which recognizes both isoforms. As expected, antibody SL-12 precipitated both IRF-3 and IRF-3a from lysates of both uninfected and infected cells (Fig. 6A). The IRF-3a-specific antibody, H1, however, failed to coprecipitate IRF-3 from uninfected cell lysates. This result is not surprising, as it has been shown that prior to virus infection, IRF-3 exists in a closed conformation through an intramolecular interaction and forms homo- and heterodimers only following stimulation (16). Significantly, IRF-3 could be coimmunoprecipitated with IRF-3a after virus infection (compare lanes 3 and 6 in Fig. 6A). The IRF-3 signal in Fig. 6A, lane 6, could not represent a phosphorylated form of IRF-3a, since there was no change in the migration of IRF-3a following virus infection, as assayed by Western blotting with whole-cell lysates (Fig. 6B). Thus, IRF-3a is capable of forming a heterodimer with IRF-3 following virus infection. An additional band in Fig. 6 cross-reacted with both antibodies and was variably observed in the immunoprecipitates. Given the specificity of the IRF-3a-specific antibody, this band likely represents a modified form of IRF-3a, although the presence of an additional isoform containing both the IRF-3a-specific region and the region common to IRF-3 and IRF-3a can be ruled out. Further studies are needed to characterize this protein.
FIG. 6.
IRF-3a and IRF-3 form a heterodimer after virus infection. (A) Immunoprecipitations with HEC1B cell lysates were carried out either with protein A/G beads (lanes 1 and 4), antibody SL-12 cross-linked to protein A/G beads (lanes 2 and 5), or antibody H1 (anti-IRF-3a) (lanes 3 and 6). Immunocomplexes were resolved by SDS-PAGE, and Western blotting was performed with antibody SL-12. IgG, immunoglobulin G. (B) Western blotting with HEC1B whole-cell lysates and antibody H1 in the absence or presence of virus infection demonstrates a lack of change in migration for the IRF-3a protein. Asterisks indicate a nonspecific band recognized by antibody H1.
DISCUSSION
IRF family members are multifunctional regulators of transcription capable of both transcriptional activation and repression, depending upon the context of the target promoter (reviewed in references 10, 18, and 24). IRF-3 has been functionally characterized as a transcriptional activator involved in the induction of the cellular cytokine response after virus infection. This study describes an alternatively spliced isoform of IRF-3 and suggests that alternative splicing of IRF-3 provides an additional level of regulation of virus-induced IFN-β gene expression.
Our results suggest that the splicing of the IRF-3 and IRF-3a transcripts may be regulated in a tissue-specific manner (13). We postulate that the relative levels of IRF-3a and IRF-3 in a given cell type may dictate the extent of IFN-β production after virus infection. Most cell types have been shown to produce IFN-α and IFN-β in response to virus infection; however, there is some variation in the extent of IFN production among various tissue and cell types. For instance, neuronal cell lines are impaired in the up-regulation of class I molecules relative to glial cell lines after virus infection, and this differential regulation correlates with the failure of virus infection to stimulate IFN-β (4). Indeed, chronic expression of IFN-α and IFN-β in the central nervous system has been found to elicit pathological effects, including encephalopathy, gliosis, and neurodegeneration (1, 4). Thus, IFN-β production is likely to be restricted in certain cell types due to the toxic effects of IFN-β exposure.
Our results demonstrate that the brain contains a much higher IRF-3a/IRF-3 ratio than other tissue types. A high IRF-3a/IRF-3 ratio would be expected to lead to significant inhibition of IRF-3-dependent genes, such as the IFN-β gene. The level of IRF-3a in tissues other than the brain was found to be 0.5 to 0.9 that of IRF-3. These levels of IRF-3a may be sufficient to affect IRF-3 activity. The differential expression of IRF-3 and IRF-3a in specific tissues could be an important determinant of the magnitude of the IFN-β response of certain cell types following virus infection. Indeed, since IRF-3 is targeted for ubiquitination and proteolysis rapidly after activation of the IFN-β promoter, stable pools of IRF-3a may set a threshold to inhibit further IFN-β expression. The precise physiologic role of IRF-3a, however, will become clear only in isoform-specific gene targeting studies.
The list of transcription factors whose functions are affected by splice variations is growing rapidly and includes other members of the IRF family (IRF-1 and IRF-7) and members of the STAT family (9, 20, 21, 30). It is particularly noteworthy that negative regulation by a splice isoform has been recently described for IRF-7, which had been implicated in the later stages of the IFN response. Furthermore, deletions within the N-terminal DBD of IRF-7 resulted in dominant-negative activity similar to what we describe here for IRF-3a (2, 20). These data provide a unifying picture of the regulation of IRF protein activity and underscore the importance of this regulatory mechanism for the tight control of the activity of this family of transcription factors.
The strongly conserved N-terminal DBD containing a tryptophan pentad is a characteristic feature of the IRF family of proteins. The crystal structures of the DBDs of two family members, IRF-1 and IRF-2, bound to ISRE oligonucleotides have been analyzed (5, 7). These two structures can easily be superimposed using the secondary structure elements, suggesting that the overall fold of this domain is conserved across the family. The global fold of the DBDs of IRF proteins is similar to that of helix-turn-helix proteins. All but one of the specific contacts with DNA are made by residues in the recognition helix of the fold. The crystal structure of IRF-2, but not that of IRF-1, has revealed that an additional specific contact with a base pair is made by a conserved histidine in the large loop preceding the first helix of the fold, His40, through a bridging water molecule. Furthermore, residues in the above-mentioned loop, as well as several others throughout the DBD, contribute to the stability of the structure by making nonspecific contacts with the phosphodiester backbone. Three out of five tryptophan residues (W11, W38, and W58) are involved in such nonspecific interactions.
From these structural studies, it is possible to infer which residues in IRF-3 have direct contact with DNA. The IRF-3a protein, on the other hand, can be predicted to have some of these structural features but is missing others. IRF-3a contains an intact recognition helix and, thus, is potentially capable of making most of the specific contacts with the ISRE sequences (Pro74-Arg86 in IRF-3). Although IRF-3a is missing two out of three tryptophans involved in the stabilizing interaction (W11 and W38 in IRF-3), its unique N-terminal region contains a single tryptophan residue (W11) which could contribute to the stabilization of DNA binding by IRF-3a. Furthermore, the unique region of IRF-3a contains a number of basic amino acids, which could play a role similar to that of the basic residues involved in the nonspecific binding of the large loop in IRF-3. However, the potentially critical His40 is not present in IRF-3a, and the first helix of the helix-turn-helix motif, which is known to be crucial for the positioning of the rest of the protein against bound DNA, is partially replaced by a novel sequence in IRF-3a. Therefore, the N-terminal domain of IRF-3a might preclude its binding to the ISRE sequences. Thus, it is not surprising that no specific DNA binding to the ISRE sequences was detected for this isoform.
The absence of intrinsic DNA binding ability suggests that IRF-3a is unlikely to interfere with IRF-3 function through direct competition for binding to DNA. Nevertheless, several possible mechanisms of inhibition can be envisioned. In one model, IRF-3a could sequester a binding partner(s) of IRF-3 required for synergistic transcriptional activation. Given the difference in the inhibitory activities of IRF-3a at the IFN-β promoter and the ISG15 promoter, this model would require IRF-3a to be able to sequester a protein(s) specific to the IFN-β promoter but not to the ISG15 promoter. Although the transcriptional enhancer complex at the ISG15 promoter has not been as extensively characterized, it has not been reported to contain the ATF-2–c-Jun transactivator that is part of the IFN-β promoter complex. Recent evidence suggests that an interaction between the DBDs of ATF-2 and IRF-3 is critical for fixing the ATF-2–c-Jun heterodimer in the correct orientation at the IFN-β promoter (6). It is possible that IRF-3a contains the domain responsible for the interaction with ATF-2 but is incapable of bringing it to the IFN-β promoter.
In a second model, IRF-3a and IRF-3 could nucleate a nonproductive enhanceosome complex at the IFN-β promoter. Coimmunoprecipitation experiments showed that IRF-3a can form a heterodimer with IRF-3 upon virus infection (Fig. 6). Given the inability of IRF-3a to bind the ISRE sequences and the structural distortion of the DNA induced by IRF protein binding (5, 7), an IRF-3–IRF-3a heterodimer is likely to be attenuated in its ability to activate transcription from promoters containing several IRF binding sites, such as the IFN-β promoter. In contrast, for promoters at which only one molecule of IRF-3 needs to bind in order to activate transcription, transcriptional activity might not be affected by IRF-3a, since the IRF-3a–IRF-3 heterodimer could then be transcriptionally competent. Analysis of the crystal structure of the IRF-2 DBD bound to the ISRE sequence revealed that the extended binding site for the IRF proteins contains the AAXXGAAA motif (7). While some IRF-responsive genes (including IFN-β) contain two such complete motifs, others (including ISG15) contain only one nonoverlapping site for IRF binding. This may provide one explanation for the different effects of IRF-3a on the IFN-β and ISG15 promoters. A possible different explanation for the selectivity of the inhibitory effect of the IRF-3a–IRF-3 heterodimer proposed in this second model may involve the ATF-2–c-Jun transactivator. As discussed above, IRF-3a may contain the domain responsible for the interaction with ATF-2, which is an IFN-β promoter-specific partner of IRF-3 in transcriptional activation. By virtue of its inability to bind DNA, IRF-3a may then prevent the establishment of the activation-competent orientation of the ATF-2–c-Jun heterodimer and selectively inhibit the induction of the IFN-β promoter. Either of the two explanations within the second model would account for the results we observed with the IFN-β and ISG15 reporter constructs (Fig. 3 and 4), since both predicted that the IRF-3–IRF-3a heterodimer would be inactive at the IFN-β promoter but not at the ISG15 promoter. Other models of the selective inhibitory action of IRF-3a undoubtedly exist, and further work is required to fully address the question of how IRF-3a functions.
Thus, alternative splicing of the IRF-3 gene-encoded transcript leads to the production of two isoforms with antagonistic functions. The expression of IRF-3a leads to potent and specific negative regulation of IRF-3 transcriptional activity, suggesting that the relative levels of IRF-3a and IRF-3 may provide a mechanism for the fine-tuning of the virus-induced activation of the IFN response. Our results demonstrate that the expression of IRF-3a is ubiquitous but that the levels of IRF-3a, compared to those of IRF-3, vary in a tissue-specific manner. Such regulated production of the IRF-3a protein would result in controlled inhibition of IRF-3 activity at the IFN-β promoter as well as at other promoters where dimerization of IRF proteins is required for transcriptional induction. Although the formation of an inactive IRF-3–IRF-3a heterodimer provides a possible explanation for the observed transcriptional inhibition, other models, including the ability of IRF-3a to disrupt an interaction between IRF-3 and an activator specific for the IFN-β promoter, cannot be excluded and await further investigation.
ACKNOWLEDGMENTS
We thank Charles Ro for expert technical assistance. We thank Maren Trost for helpful discussions and critical review of the manuscript.
L.V.R. was supported by fellowship grant 5 F32 AI09167-02 from the National Institute of Allergy and Infectious Diseases and a grant from Aid for Cancer Research. A.Y.K. is a Howard Hughes Medical Institute predoctoral fellow. This research was supported by National Institute of Health grant PO1 AI 42257 to P.M.H.
Alla Y. Karpova and Lucienne V. Ronco contributed equally to the work described in this article.
REFERENCES
- 1.Akwa Y, Hassett D E, Eloranta M L, Sandberg K, Masliah E, Powell H, Whitton J L, Bloom F E, Campbell I L. Transgenic expression of IFN-alpha in the central nervous system of mice protects against lethal neurotropic viral infection but induces inflammation and neurodegeneration. J Immunol. 1998;161:5016–5026. [PubMed] [Google Scholar]
- 2.Au W-C, Yeow W S, Pitha P M. Analysis of functional domains of interferon regulatory factor 7 and its association with IRF-3. Virology. 2001;280:273–282. doi: 10.1006/viro.2000.0782. [DOI] [PubMed] [Google Scholar]
- 3.Baeuerle P A, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 4.Campbell I L, Krucker T, Steffensen S, Akwa Y, Powell H C, Lane T, Carr D J, Gold L H, Henriksen S J, Siggins G R. Structural and functional neuropathology in transgenic mice with CNS expression of IFN-alpha. Brain Res. 1999;835:46–61. doi: 10.1016/s0006-8993(99)01328-1. [DOI] [PubMed] [Google Scholar]
- 5.Escalante C R, Yie J, Thanos D, Aggarwal A K. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 6.Falvo J V, Parekh B S, Lin C H, Fraenkel E, Maniatis T. Assembly of a functional beta interferon enhanceosome is dependent on ATF-2-c-jun heterodimer orientation. Mol Cell Biol. 2000;20:4814–4825. doi: 10.1128/mcb.20.13.4814-4825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galvin K M, Shi Y. Multiple mechanisms of transcriptional repression by YY1. Mol Cell Biol. 1997;17:3723–3732. doi: 10.1128/mcb.17.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harada H, Kondo T, Ogawa S, Tamura T, Kitagawa M, Tanaka N, Lamphier M S, Hirai H, Taniguchi T. Accelerated exon skipping of IRF-1 mRNA in human myelodysplasia/leukemia; a possible mechanism of tumor suppressor inactivation. Oncogene. 1994;9:3313–3320. [PubMed] [Google Scholar]
- 10.Harada H, Taniguchi T, Tanaka N. The role of interferon regulatory factors in the interferon system and cell growth control. Biochimie. 1998;80:641–650. doi: 10.1016/s0300-9084(99)80017-0. [DOI] [PubMed] [Google Scholar]
- 11.Hoppe B S, Jensen R B, Kirchgessner C U. Complementation of the radiosensitive MO59J cell line. Radiat Res. 2000;153:125–130. doi: 10.1667/0033-7587(2000)153[0125:cotrmc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 12.Juang Y, Lowther W, Kellum M, Au W C, Lin R, Hiscott J, Pitha P M. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karpova A Y, Howley P M, Ronco L V. Dual utilization of an acceptor/donor splice site governs the alternative splicing of the IRF-3 gene. Genes Dev. 2000;14:2813–2818. doi: 10.1101/gad.813800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin R, Heylbroeck C, Genin P, Pitha P M, Hiscott J. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol Cell Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin R, Mamane Y, Hiscott J. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol Cell Biol. 1999;19:2465–2474. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowther W J, Moore P A, Carter K C, Pitha P M. Cloning and functional analysis of the human IRF-3 promoter. DNA Cell Biol. 1999;18:685–692. doi: 10.1089/104454999314962. [DOI] [PubMed] [Google Scholar]
- 18.Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant M J, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: the next generation. Gene. 1999;237:1–14. doi: 10.1016/s0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis T, Falvo J V, Kim T H, Kim T K, Lin C H, Parekh B S, Wathelet M G. Structure and function of the interferon-beta enhanceosome. Cold Spring Harbor Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 20.Marie I, Smith E, Prakash A, Levy D E. Phosphorylation-induced dimerization of interferon regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol Cell Biol. 2000;20:8803–8814. doi: 10.1128/mcb.20.23.8803-8814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meinke A, Barahmand-Pour F, Wohrl S, Stoiber D, Decker T. Activation of different Stat5 isoforms contributes to cell-type-restricted signaling in response to interferons. Mol Cell Biol. 1996;16:6937–6944. doi: 10.1128/mcb.16.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 23.Navarro L, Mowen K, Rodems S, Weaver B, Reich N, Spector D, David M. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol Cell Biol. 1998;18:3796–3802. doi: 10.1128/mcb.18.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen H, Hiscott J, Pitha P M. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 25.Ronco L V, Karpova A Y, Vidal M, Howley P M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilcek J, Sen G S. Interferons and other cytokines. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press, Ltd.; 1996. pp. 375–400. [Google Scholar]
- 27.Wathelet M, Lin C H, Parekh B, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 28.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu H, Cong J P, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]